Abstract
Swine Influenza Virus (H1N1) is a known causative agent of swine flu. Transmission of Swine Influenza Virus form pig to human is not a common event and may not always cause human influenza. The 2009 outbreak by subtype H1N1 in humans is due to transfer of Swine Influenza Virus from pig to human. Thus to analyze the origin of this novel virus we compared two surface proteins (HA and NA) with influenza viruses of swine, avian and humans isolates recovered from 1918 to 2008 outbreaks. Phylogenetic analyses of hemagglutinin gene from 2009 pandemic found to be clustered with swine influenza virus (H1N2) circulated in U.S.A during the 1999-2004 outbreaks. Whereas, neuraminidase gene was clustered with H1N1 strains isolated from Europe and Asia during 1992-2007 outbreaks. This study concludes that the new H1N1 strain appeared in 2009 outbreak with high pathogenicity to human was originated as result of re-assortment (exchange of gene). Moreover, our data also suggest that the virus will remain sensitive to the pre-existing therapeutic strategies.
Keywords: H1N1, pandemic, sequence, analysis, mutations
Background
A new H1N1 re-assortant “swine” influenza virus was recently detected in individuals from the United States who presented respiratory symptoms, and the same virus was subsequently confirmed in patients from several countries around the world [1]. The World Health Organization (WHO) reported 94,512 cases of influenza A (H1N1) infection with a pandemic potential, including 429 deaths in July 06, 2009 [2]. The circumstances surrounding the emergence of this pathogen, and the factors that facilitated the initial cross-species transmission, are still not fully understood. It became apparent in the early days of the outbreak that the virus can be directly transmitted between humans. Among the various efforts made to evaluate, diagnose and implement measures against the spread of virus, is the timely release of the genomic sequences from different viral isolates.
The notorious 1918 influenza pandemic‐deservedly known as the “mother of all pandemics”‐infected more than one quarter of the world's population and claimed an estimated 20 to 50 million human lives [3]. Two features of the influenza virus, compounded by current globalization trends, explain its ability to become a particularly worrisome zoonotic threat. One feature is the high error rate during genomic replication, which is typical of RNA viruses. The other is the segmented influenza virus genome, which facilitates re-assortment between different viral strains that infect the same cell. At a time when air travel makes it possible to reach the most remote corners of the planet in a matter of hours, any emerging or re-emerging infectious disease stops being a local issue and instead becomes a global medical and public health priority. Details of how the current influenza virus emerged, although still scarce, will enhance our knowledge about factors that enable viruses to cross the species barrier and cause disease in humans [4]. Hence, keeping above things in our mind our lab has started analyses of this new (2009) human H1N1 virus and compared it with others H1N1 and H1N2 viruses from human, swine and avian during the outbreak occurred in 1918-2008.
Methodology
Nucleotide and protein sequences of haemagglutinin (HA) and neuraminidase (NA) were downloaded form NCBI database (ftp://ftp.ncbi.nih.gov/genomes/INFLUENZA). Blast server at NCBI was used to identify the sequences identity (Nucleotide and Protein). Alignments of the selected sequences of HA and NA genes were performed by ClustalW program with default parameters [5]. Phylogenetic tree was built by Neighbour-joining method with Kimura-2 parameter, performed in Phylip3.65 package (http://evolution.genetics.washington.edu/phylip.html). Phylogenies were determined by bootstrap analyses of 1000 replicates using seqboot program available in Phylip3.65 package. Phylogenetic tree was drawn by TreeView1.6 program (http://taxonomy.zoology.gla.ac.uk/rod/treeview). Glycosylation sites were identified by Scanprosite (http://expasy.org/tools/scanprosite) at Expasy server.
Discussion
Pigs have been known as “mixing vessel” [6] and thus play a vital role in interspecies transmission of influenza viruses. A novel 2009 human H1N1 virus, circulating in USA, Europe and Asia is known to be a reassorted virus. Thus to trace out the origin of this novel human H1N1 virus, we performed the phylogenetic analyses of two surface proteins (HA and NA).
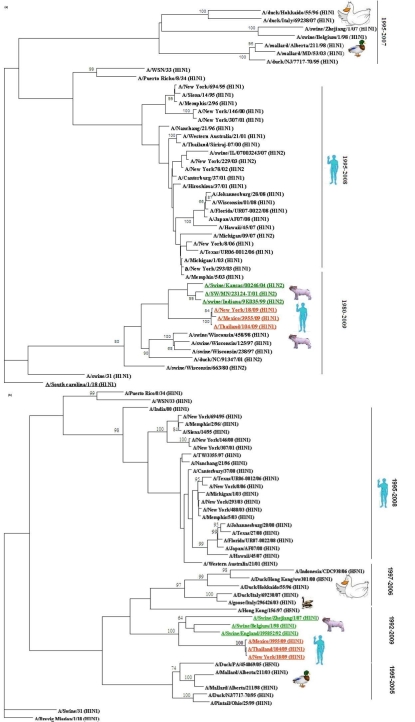
The haemagglutinin (HA) sequences of three strains form 2009 outbreak (two from U.S.A and one from Asia) clustered with the swine influenza sequences from H1N2 subtype recovered from 1999-2004 outbreaks in U.S.A (Figure 1A). The HA gene sequences of all the three isolates shared highest homology over 95% and 93% at nucleotide and protein level was found with H1N2 strain from Indiana and H1N1 strain from Kansas, respectively (Table 1 in supplementary material). In contrast, the segment coding for neuraminidase gene (NA) of new human virus clustered with the swine influenza viruses from Asia and Europe from 1992-2007 outbreaks (Figure 1B). However the NA genes of three isolates from 2009 outbreak showed highest homology over 94% and 92% to H1N1 strain from England and H1N1 strain from Belgium, respectively (Table 1 in supplementary material). Whereas H1N1 strain from Zhejiang shared at least 91% homology both at nucleotide and protein level. Thus our result suggests that the neuraminidase gene segment lies with in the Eurasian swine genetic lineage, which also corroborates with a recent report by Garten et al [7]. However all the humans isolates from H1N1 subtype recovered from 1995 to 2008 fail to clustered with these novel strains and able to make separate clustered . The HA and NA, genes from 2009 outbreak showed homology to 77% to 80% strains clustered in this group. Comparing the HA and NA genes from virus with 1918 outbreak we found that this novel virus shared at least 85% and 83% homology respectively.
Figure 1.
A Neighbour‐Joining phylogenetic tree from nucleotide sequences of HA (a) and NA(b) of selected influenza A viruses. The tree was rooted to A/South Carolina/1/18(H1N1) for HA and A/Brevig Mission/1/18(H1N1) for NA. Robustness of individual nodes of the tree was determined by using boot strap analyses at 1000 replicates.
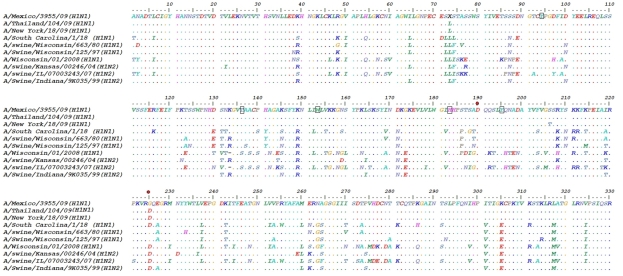
Alignment of HA1 amino acids sequences from 1918-2009 are shown in Figure 2. Five conserved amino acids Tyr95, Tyr136, Trp153, His183, Tyr195 are highlighted at receptor-binding site of H1 which have been described by Skehel and Wiley [8]. With H1, SA binding was specific for the NeuAcα2,6 Gal linkage due to amino acids Asp- 190 and Asp-225, Although Mexican (2009) H1 isolate had Gly225 which is similar to isolates from Swine Influenza Virus during 1980 and 1997 outbreaks in USA. Mutation at position 225 may effect the binding to NeuAcα2, 6 Gal SA but further needs to be validated by experimental approach. At least six potential glycosylation sites have been identified at position 27-30, 28-31, 40-43, 104-107, 293- 296, 304-307 at H1 in new human 2009 strains. All the glycosylation sites were related to Swine Influenza Virus H1 viruses recovered from 1999-2001 outbreaks and that include subtype H1N2 and H1N1 in U.S.A (data not shown). These data suggest novel H1N1 virus was genetically related to viruses form swine H1N2 and H1N1 subtypes which give strong indication of re‐assortment. With the novel 2009 virus, the amino acid sequence at receptor‐binding site was similar to H1 of Swine influenza virus and human isolates from 1918-2008 outbreaks, which further implicate the conserved pattern of receptor binding in present H1N1 virus.
Figure 2.
Amino acids comparison of the HA1 domain. Dots represent consensus similar amino acids. Conserved amino acids at receptor binding sites are shown as small rectangles. Red dots represent amino acids specific for NeuAcα2, 6 Gal SA.
Previous reports suggested that mutations at amino acid H274Y and N294S made strain resistant to available drug oseltamivir [9]. Thus to investigate whether available drugs (oseltamivir, zanamivir) may be effective against the recent circulating virus, we performed the amino acids sequence alignment of NA gene with previous strains. The result revealed that no mutation was found in the conserved region of active site (data not shown), which is the potential target for the existing drugs. Thus, conserved pattern at active site of NA suggests that the virus will remain sensitive to the pre-existing therapeutic strategies.
Conclusion
This study precisely intrigue to investigate the origin of novel virus (2009) subtype H1N1 (based on two surface proteins HA & NA) prevalent in humans. Our Insilco analyses revealed that this is a reassorted virus, where swine may constitute a crucial intermediate host for transmission. Furthermore, the study also predicts the presence of conserved domains in both the surface proteins, which could be of therapeutic relevance.
Supplementary material
Acknowledgments
We are thankful to Prof. M. Saleemuddin, coordinator, IBU for providing facility for the work. AUK acknowledges DBT grant BT/PR7507/BID/07/201/2006.
Footnotes
Citation:Danishuddin et al, Bioinformation 4(3): 94-97 (2009)
References
- 1.Shinde V, et al. N Engl J Med. 2009;360:2616. doi: 10.1056/NEJMoa0903812. [DOI] [PubMed] [Google Scholar]
- 2. http://www.who.int.
- 3.Xu X, et al. J Virol. 2008;82:10493. doi: 10.1128/JVI.00959-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anwar T, et al. Bioinformation. 2006;1:253. doi: 10.6026/97320630001253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thompson JD, et al. Nucleic Acids Res. 1994;22:4673. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Webster RG, et al. Microbiol Rev. 1992;56:152. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garten RJ, et al. Science. 2009 In press. [Google Scholar]
- 8.Skehel JJ, Wiley DC. Annu Rev Biochem. 2000;69:531. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- 9.de Jong MD, et al. N Engl J Med. 2005;25:2667. doi: 10.1056/NEJMoa054512. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.