Abstract
In the absence of x-ray structures of calcium channels, their homology models are used to rationalize experimental data and design new experiments. The modeling relies on sequence alignments between calcium and potassium channels. Zhen et al. (2005. J. Gen. Physiol. doi:10.1085/jgp.200509292) used the substituted cysteine accessibility method (SCAM) to identify pore-lining residues in the Cav2.1 channel and concluded that their data are inconsistent with the symmetric architecture of the pore domain and published sequence alignments between calcium and potassium channels. Here, we have built Kv1.2-based models of the Cav2.1 channel with 2-(trimethylammonium)ethyl methanethiosulfonate (MTSET)-modified engineered cysteines and used Monte Carlo energy minimizations to predict their energetically optimal orientations. We found that depending on the position of an engineered cysteine in S6 and S5 helices, the ammonium group in the long flexible MTSET-modified side chain can orient into the inner pore, an interface between domains (repeats), or an interface between S5 and S6 helices. Different local environments of equivalent positions in the four repeats can lead to different SCAM results. The reported current inhibition by MTSET generally decreases with the predicted distances between the ammonium nitrogen and the pore axis. A possible explanation for outliers of this correlation is suggested. Our calculations rationalize the SCAM data, validate one of several published sequence alignments between calcium and potassium channels, and suggest similar spatial dispositions of S5 and S6 helices in voltage-gated potassium and calcium channels.
INTRODUCTION
Voltage-gated Ca2+ channels are involved in many physiological functions. The channels are targets for drugs used to treat arrhythmias, hypertension, myocardial ischemia, chronic pain, neuronal degeneration, and other disorders (Hockerman et al., 1997). The pore-forming α1 subunit of Ca2+ channels is formed by a single polypeptide chain that contains four homologous repeats (Hockerman et al., 1997). Each repeat includes six transmembrane helices: the voltage sensor (S1–S4), the outer helices S5, and the pore-lining helices S6. Ion selectivity is controlled by a ring of four glutamates (EEEE), which are located at the membrane-reentering P loops between S5s and S6s. Ca2+ and Na+ channels have evolved from K+ channels (Anderson and Greenberg, 2001), and the three families of channels are believed to have a similar folding of the pore-forming domains and transmembrane topology of S5s and S6s.
In the absence of x-ray structures of voltage-gated Ca2+ and Na+ channels, their homology models based on x-ray structures of K+ channels in the closed and open states (Doyle et al., 1998; Jiang et al., 2003; Long et al., 2005, 2007) are used to explain experimental data and suggest new experiments. Homology modeling relies on the sequence alignments of K+ channels with Na+ and Ca2+ channels. Various sequence alignments have been proposed (Huber et al., 2000; Zhorov et al., 2001; Lipkind and Fozzard, 2003; Stary et al., 2008). The models of the pore-forming domain (S5-P-S6) based on these alignments have different patterns of exposure of residues to the inner pore. The substituted cysteine accessibility method (SCAM) is used to define the architecture of ion channels (Karlin and Akabas, 1998). SCAM data are usually interpreted based on the cysteine orientation concept, according to which the application of a methanethiosulfonate (MTS) reagent to a channel with an engineered cysteine in a pore-facing position results in the chemical modification of the cysteine. If the ammonium group of the MTS-modified cysteine is exposed to the permeation pathway, it is expected to inhibit the current. Orientation of the engineered cysteine toward the lipid bilayer or the protein interior is believed to suppress ionization of the thiol group and its reaction with an MTS reagent. The current in such channels is expected to be similar to control channels with the native residue in the respective position.
The location of the activation gate in the Shaker channel was predicted using SCAM (Liu et al., 1997), and major conclusions from this study were later confirmed by the x-ray structures of K+ channels. More recently, SCAM was used to identify pore-lining residues in the Cav2.1 channel (Zhen et al., 2005). The authors of this meticulous study interpret their results as inconsistent with known sequence alignments between K+ and Ca2+ channels and suggest an asymmetric architecture of the inner pore of Cav2.1. This conclusion sheds doubts on published homology models of Ca2+ channels. The above interpretation of the SCAM experiments is apparently based on the cysteine orientation concept, which does not take into consideration the conformational flexibility of long side chains of MTS-modified cysteines. Due to this flexibility, the exposure of the MTS ammonium groups to the permeation pathway and hence the current-inhibiting effects of MTS may not correlate with the orientation of the Cα-Cβ vector to the pore axis. Such possibilities can be explored by homology modeling of the channel with MTS-modified cysteines.
Here, we have built 44 models of Cav2.1 with MTS-modified engineered cysteines and used Monte Carlo (MC) minimizations (MCMs) to predict energetically possible orientations of MTS-modified side chains in the channels. We found that the residual current observed upon 2-(trimethylammonium)ethyl MTS (MTSET) application (Zhen et al., 2005) correlates with the predicted distance between the ammonium nitrogen in the MTS-modified cysteine and the pore axis, but does not correlate with the orientation of the Cα-Cβ vector of the cysteine. Our results support the alignment between K+ and Ca2+ channels, which was used in our previous modeling studies, and suggest a similar disposition of transmembrane helices in the pore-forming domains of voltage-gated K+ and Ca2+ channels.
MATERIALS AND METHODS
Homology models of Cav2.1 (CAC1A_RABIT) with MTSET-modified engineered cysteines were built using the x-ray structure of Kv1.2 (Long et al., 2005) and sequence alignment shown in Table I. The models include the outer helices (S5s), P loops, and inner helices (S6s). The ascending limbs of P loops, including the selectivity filter residues, were built using the Nav1.4 model (Tikhonov and Zhorov, 2005) as a template. Those parts of the channel, which are far from the inner pore, were not modeled. Repeats I–IV were arranged clockwise when viewed extracellularly (Dudley et al., 2000). We use the terms “previous repeat” and “next repeat” to designate sequential neighbors of a mutated repeat (e.g., neighbors of repeat I are next repeat II and previous repeat IV).
Table I.
| Channel | Segment | No. | 1 | 11 | 21 | ||
| KcsA | M1 | o | 23 | ALHWRAAGAA | TVLLVIVLLA | GSYLAVLAER | |
| Kv1.2 | S5 | o | 322 | KASMRELGLL | IFFLFIGVIL | FSSAVYFAEA | |
| Cav2.1 | IS5 | 1o | 220 | MKAMIPLLQI | GLLLFFAILI | FAIIGLEFYM | |
| IIS5 | 2o | 608 | LNSMKSIISL | LFLLFLFIVV | FALLGMQLFG | ||
| IIIS5 | 3o | 1380 | VNSLKNVFNI | LIVYMLFMFI | FAVVAVQLFK | ||
| IVS5 | 4o | 1695 | VQSFKALPYV | CLLIAMLFFI | YAIIGMQVFG | ||
| Pore-facing position | * * | * * | |||||
| 1 | 11 | 21 | 31 | ||||
| KcsA | M2 | i | 86 | LWGRLVAVVV | MVAGITSFGL | VTAALATWFV | GREQ |
| Kv1.2 | S6 | i | 385 | IGGKIVGSLC | AIAGVLTIAL | PVPVIVSNFN | YFYH |
| Cav2.1 | IS6 | 1i | 336 | TWNWLYFIPL | IIIGSFFMLN | LVLGVLSGEF | AKER |
| IIS6 | 2i | 690 | MVFSIYFIVL | TLFGNYTLLN | VFLAIAVDNL | ANAQ | |
| IIIS6 | 3i | 1485 | MEMSIFYVVY | FVVFPFFFVN | IFVALIIITF | QEQG | |
| IVS6 | 4i | 1785 | EFAYFYFVSF | IFLCSFLMLN | LFVAVIMDNF | EYLT | |
| Nav1.4 | IVS6 | 4i | 1565 | SIGICFFCSY | IIISFLIVVN | MYIAIILENF | NVAT |
| Pore-facing position | * * ** | ** ** | |||||
| Cytoplasm-facing position | ** *** * | * | |||||
Data for Nav1.4 are from (Sunami et al., 2004).
Shown alignments of S5 (Huber et al., 2000) and S6 (Zhorov et al., 2001) segments were used to build the Cav2.1 models.
Bold-oblique and underlined characters indicate positions where MTSET inhibits channels with engineered cysteines ≥30 and <30%, respectively (Zhen et al., 2005).
All calculations were performed using the ZMM program (http://www.zmmsoft.com). Non-bonded energy was calculated using the AMBER force field (Weiner et al., 1984, 1986) with a cutoff distance of 8 Å. Hydration energy was calculated using the implicit solvent method (Lazaridis and Karplus, 1999). Electrostatic interactions were calculated using the distance-dependent dielectric function. Ionizable residues, including those in the selectivity filter, were modeled in their neutral forms (Lazaridis and Karplus, 1999), except for the acidic residues in the cytoplasmic side of KcsA-based closed-channel models. MTS-modified cysteines (designated mC) are incorporated in the ZMM program as nonstandard amino acids. The atomic charges of the mC residues have been calculated by the semi-empirical method AM1 (Dewar et al., 1985) using MOPAC. The charge of +1 proton charge unit is distributed among the ammonium nitrogen and surrounding methylene and methyl groups. ZMM program with MTS-modified cysteines was previously used in theoretical studies of glutamate-gated ion channels (Tikhonov, 2007). The MCM method (Li and Scheraga, 1987) was used to optimize the models. During energy minimizations, Cα atoms were constrained to corresponding positions of the template using pins. A pin is a flat-bottom energy function, which allows an atom to deviate penalty-free up to 1 Å from the template and imposes a penalty of 10 kcal mol−1 Å−1 for deviations >1 Å.
Each model was MC minimized until 2,000 consecutive minimizations did not update the apparent global minimum. Then, the multi-MCM protocol (Bruhova and Zhorov, 2007) was used to predict all low energy orientations of the MTS-modified cysteine. The side chain torsions of the mC residue were sampled from 60,000 random starting points. Each starting point was optimized in an MCM trajectory of 10 steps. The top 1,000 low energy conformations were further MC minimized for 1,000 steps. All conformations in which the interaction energy between the mC residue and the rest of the channel did not exceed 4 kcal/mol from the apparent global minimum were analyzed. No specific energy terms were used for cation–π interactions, which were accounted for due to partial negative charges at the aromatic carbons (Bruhova et al., 2008). Further details of methodology can be found elsewhere (Bruhova and Zhorov, 2007; Tikhonov and Zhorov, 2007; Bruhova et al., 2008).
To validate our methodology, we predicted orientations of mC223 in a cysteine transpeptidase, Sortase B. Comparison of the lowest energy orientation and the x-ray structure (Zong et al., 2004) shows that the ammonium nitrogen of mC223 in the model is just 0.66 Å away from the experimental position (Fig. S1). We use a labeling scheme that is universal for P loop channels (Zhorov and Tikhonov, 2004). A residue label includes the repeat number (1–4), which may be omitted when the label is pertinent to all four repeats, segment type (o, outer helix; p, P loop; i, inner helix), and the residue relative number in the segment (Table I).
Online supplemental material
Table S1 presents the experimental data on the current inhibition by MTSET (Zhen et al., 2005). The predicted mobility of ammonium groups of respective mC residues is shown in Fig. S1, which demonstrates the validity of our methodology of predicting conformations of mC residues using the x-ray structure of a cysteine transpeptidase. Figs. S2–S4 display conformations and local environments of residues mC1i23, mC1i24, mCi21, and mCi25 in respective open channels, and Fig. S5 shows energetically most favorable orientations of mCi24 residues in the KcsA-based model of the closed Cav2.1 channel. Table S1 and Figs. S1–S5 are available at http://www.jgp.org/cgi/content/full/jgp.200910288/DC1.
RESULTS
SCAM data and the cysteine-orientation concept
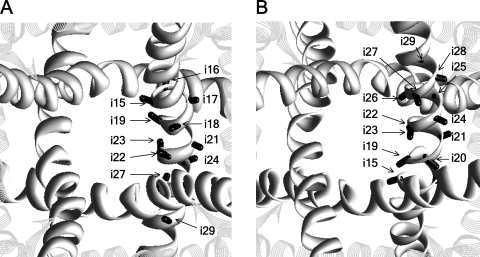
In the Kv1.2 template, vectors Cα-Cβ in positions i15, i18, i19, i22, and i23 direct to the pore axis, in positions i16, i17, i20, i21, i25, and i29 direct to a neighboring S6 or S5, and in positions i24, i26, i27, i28, i30, and i31 direct to the cytoplasm (Fig. 1). Assuming generally similar 3-D dispositions of S5s and S6s in Cav2.1 and Kv1.2, and correctness of the alignment in Table I, the SCAM data (Zhen et al., 2005), which are shown in Table S1, can be divided into two categories. In agreement with the cysteine orientation concept, the first-category data show that MTSET inhibits channels Ci15, Ci19, Ci23, C2i18, and C4i18, in which respective Cα-Cβ vectors direct to the pore axis. The second-category data, which disagree with the cysteine orientation concept, include three groups of observations: (1) MTSET does not inhibit channels C1i18, C3i18, and Ci22, despite the fact that respective vectors Cα-Cβ point to the pore axis; (2) MTSET inhibits channels C2o10 and C4o10, despite the fact that respective positions are far from the pore; and (3) MTSET inhibits channels Ci16, C2i20, C3i20, C3i21, Ci24, and C2i25, despite the fact that respective vectors Cα-Cβ direct away from the pore axis.
Figure 1.
The extracellular (A) and cytoplasmic (B) views of the Kv1.2 x-ray structure, with the Cα-Cβ bonds of positions i15–i29 shown as sticks. The S5 and S6 helices are shown as strands and ribbons, respectively. The P loops are not shown for clarity.
Below, we describe the Cav2.1 models and consider the SCAM data in view of orientations of Cα-Cβ vectors in mC residues relative to the pore axis. Unless otherwise mentioned, the described orientation of an mC residue corresponds to the lowest energy conformation.
MTS-modified cysteines in the pore-facing positions of S6s
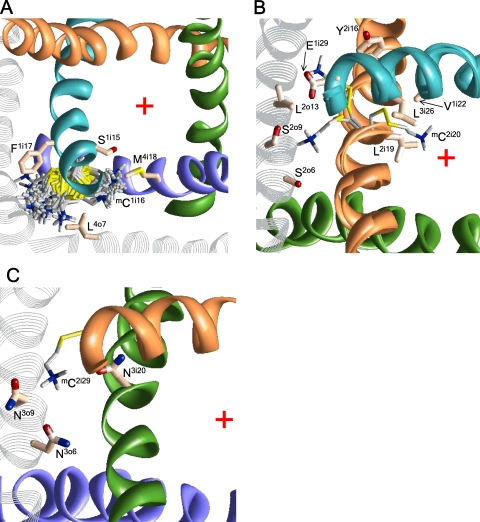
The current of mCi15 channels is inhibited by MTSET by 39.0–65.5%. According to our calculations, the ammonium groups of mCi15 occur in the pore, where they are stabilized by electrostatic interactions with the nucleophilic C termini of P helices (residues p47–p49) and with the EEEE locus and interact with residues at the same level (i15) and lower levels i18 and i19 (Fig. 2, A and B).
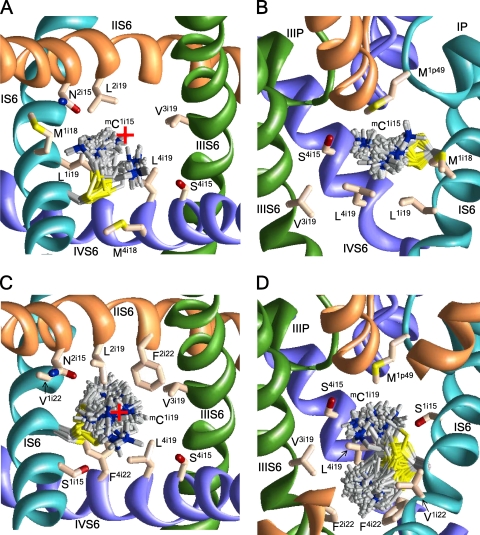
Figure 2.
The extracellular and side views of mC1i15 (A and B) and mC1i19 (C and D) in the open Cav2.1 channel. The side chains of the mC residues in different conformations within 4 kcal/mol from the apparent global minima are superimposed and shown as gray sticks with blue nitrogen and yellow sulfur atoms. Native residues are shown in the lowest energy conformation as pale orange sticks with red oxygens, blue nitrogens, and yellow sulfur atoms. The P loops and S6s in repeats I, II, III, and IV are cyan, orange, green, and violet, respectively. For clarity, P loops in A and C, IIS6 in B, and S5s in A–D are not shown. The ammonium group of mC1i15 is inside the pore (A) between levels i15 and i18 (B). The ammonium group of mC1i19 is close to the pore axis (C) approaching either the focus of P helices or level i22 (D). The red cross at A and C indicates the pore axis.
MTSET inhibits channels C2i18 and C4i18 more than C1i18 and C3i18. Calculations predict two orientations of mCi18 side chains (Fig. 3). In the pore-facing orientations, the ammonium groups are attracted to the C ends of the P helices, the EEEE locus, and residues i15 and i19. In the repeat interface orientation, the ammonium groups occur between S5 and next repeat S6. The energetically preferable orientations depend on the neighboring residues, which are distinct in different repeats (Table I). The pore orientation of mC2i18 is energetically most preferable. The side chain of mC4i18 is equally stable in the pore and interface IV/I, where it experiences cation–π interactions with F1i16. The side chain of mC1i18 is most stable in interfaces I/II, where it interacts with L1o10, L2i12, and Y2i16. The side chain of mC3i18 is most stable in interface III/IV, where it experiences cation–π interactions with F3i22 and F4i12 and hydrophobic interactions with I3o10.
Figure 3.
The extracellular views of various orientations of mCi18 residues in Cav2.1. For clarity, the P loops are not shown. The red cross indicates the pore axis. (A) Cation–π interactions with Y2i16 stabilize the repeat interface orientation mC1i18; the pore orientations have higher energy. (B) In the most preferable conformations, mC2i18 is oriented in the pore. (C) Cation–π interactions with F4i12 and F3i22 stabilize orientation of mC3i18 in the repeat interface; the pore orientations are less preferable. (D) Both pore and repeat interface orientations of mC4i18 are energetically favorable.
MTSET strongly inhibits Ci19 channels. The mCi19 side chains are in the pore and orient either upward to the focus of P helices (Fig. 2 C) or downward (Fig. 2 D). The latter orientation is more stable due to cation–π interactions with F2i22, F3i22, and F4i22. In both orientations, the mCi19 ammonium groups are close to the pore axis.
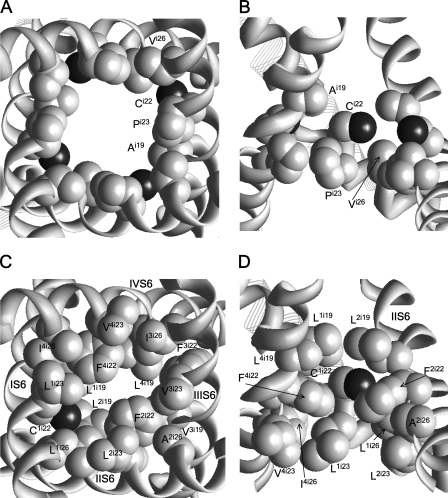
Channels Ci22 are only weakly sensitive to MTSET, despite the fact that their respective Cα-Cβ vectors direct to the pore axis. The mCi22 side chains can adopt two orientations: to the pore, where they interact with a neighboring Fi22, or to the repeat interfaces, where they interact with aromatic residues in positions i12, i16, or i18 (Fig. 4). The pore-facing orientations of mC2i22, mC3i22, and mC4i22 are energetically more preferable than the repeat interface orientations. The weak effect of MTSET on respective channels may be due to large hydrophobic residues i19, i22, and i26 (Fig. 5) that would prevent the ionization of Ci22 and thus the reaction with MTSET.
Figure 4.
The extracellular views of mC1i22 (A), mC2i22 (B), mC3i22 (C), and mC3i22 (D). The red cross indicates the pore axis. P loops are not shown for clarity. Orientations of the mCi22 side chains in the pore are energetically more preferable than repeat interface orientations.
Figure 5.
The cytoplasmic (A and C) and side (B and D) views of the environment for C1i22 in the Shaker (A and B) and Cav2.1 (C and D) channels. Side chains in positions i19, i22, i23, and i26 are space-filled with gray carbon and black sulfur atoms.
MTSET inhibits the Ci23 channels by 43.6–87.7%. The ammonium groups of mCi23s are oriented into the pore. For example, the pore orientation of mC1i23 is stabilized by interactions with L1i26, F2i22, F4i30, F4i22, and I4i26 (Fig. S2).
S6 positions that do not face the pore
The mCi16 side chains have two orientations (Fig. 6 A), with the ammonium group either approaching the pore or being away from the pore. In either orientation, the ammonium groups are rather far from the pore axis.
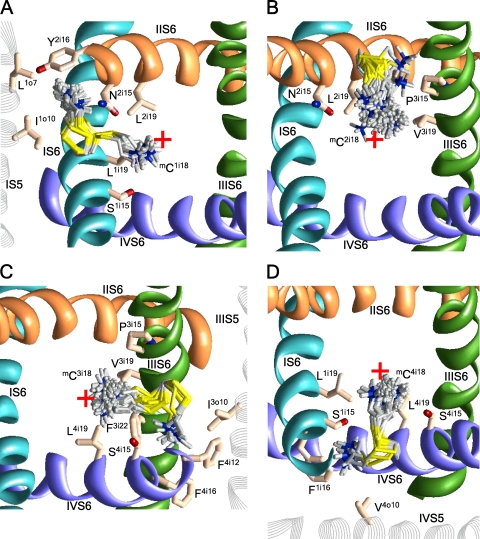
Figure 6.
(A) The extracellular view of mC1i16 that orients either along IVS6 or toward IS5 and IVS5. (B) The cytoplasmic view of three possible orientations of mC2i20. (C) The cytoplasmic view of mC2i29 interacting with N3o6, N3o9, and N3i20. The red cross indicates the pore axis.
Cysteines at positions i20 substitute native asparagines that are highly conserved in Ca2+ and Na+ channels. Mutants C1i20 and C4i20 are nonfunctional. MTSET inhibits channels C2i20 and C3i20 by 56.2 and 42.0%, respectively. The side chains of mC2i20 and mC3i20 can adopt three orientations. In the energetically most preferable orientations, the ammonium groups face S5s (Fig. 6 B), whereas orientation to the pore is less preferable.
The Cα-Cβ vectors of Ci21s direct to S5s. The C4i21 mutant is not functional. MTSET does not inhibit channels C1i21 and C2i21, but it inhibits the C3i21 channel by ∼40%. The mC1i21 and mC2i21 side chains fit between the mutated repeat S5 and the next repeat S6 (Fig. S3). Cation–π interactions with F4i30 and electrostatic interactions with Q1o9 stabilize the ammonium group of mC1i21 in interface IV/I. Similar interactions with F1i30 and E1i29 stabilize the ammonium group of mC2i21 in interface I/II. The side chain of mC3i21 orients into the pore, where it experiences cation–π interactions with F3i18 and F3i22.
MTSET inhibits channels Ci24 by 73.6–100%, despite the fact that vectors Cα-Cβ direct toward cytoplasm rather than to the pore axis, (Fig. 1 B). In the most preferable conformations, cation–π interactions with F1i30, F3i30, and F4i30 attract the ammonium groups of mC2i24, mC4i24, and mC1i24, respectively, toward the pore. However, in these orientations, the ammonium nitrogen is as far as 6–8 Å from the pore axis. In the alternative orientations, toward S5, the ammonium group is even farther from the pore axis (Fig. S2 B). The ammonium group of mC3i24 lacks an aromatic partner (note L2i30 vs. F1i30, F3i30, and F4i30; Table I) and faces away from the pore in the most preferable orientation. Thus, our open Cav2.1 models are inconsistent with the strong current inhibition by mCi24s. Possible explanations of this fact are provided in the Discussion.
Vectors Cα-Cβ in positions i25 direct away from the pore (Fig. 1). MTSET weakly inhibits channels C1i25, C3i25, and C4i25, but it strongly inhibits channel C2i25. Aromatic residues i16 stabilize the orientations of mC1i25, mC3i25, and mC4i25 away from the pore (Fig. S4), whereas large residues L1i26, L3i26, and V4i26 preclude orientations into the pore. In contrast, cation–π interactions with F2i22 stabilize the orientation of mC2i25 to the pore, which is not precluded by small A2i26.
C2i29 is the only channel in which MTSET increases the current. In our model, the mC2i29 ammonium group is oriented into the II/III interface and binds between the side chains of N3o9, N3o6, and N3i20. Position i29 is four helical turns closer to the cytoplasm than position i14, where the gating-hinge glycine is located in K+ channels. Superposition of the x-ray structures of K+ channels in the open and closed conformations shows that position i29 shifts significantly between these structures. We suggest that the electrostatic attraction of the mC2i29 ammonium group to N3o9, N3o6, and N3i20 stabilizes the open-gate conformation of the pore domain. This can explain the unique characteristics of the mC2i29 channel.
MTS-modified cysteines in S5s
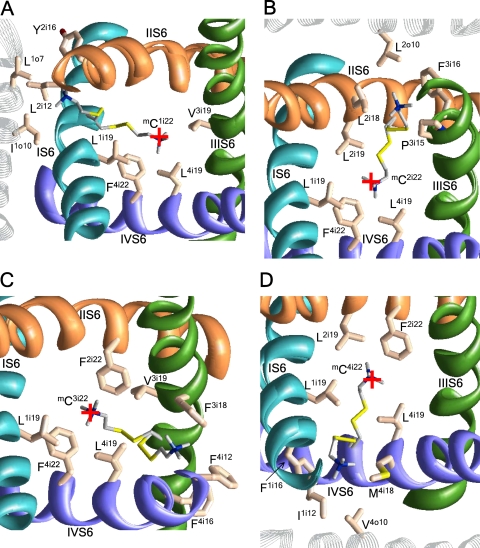
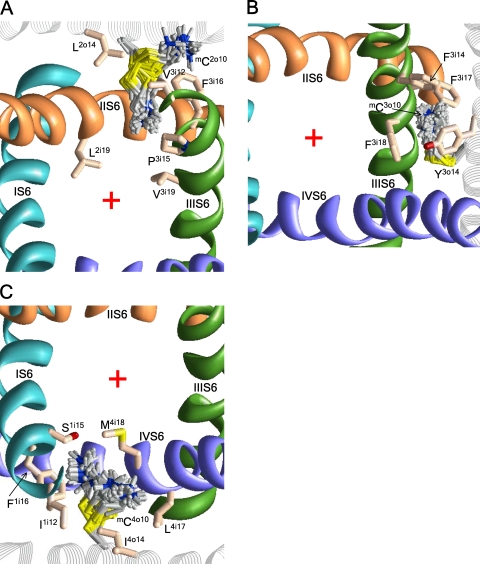
Despite the fact that S5 helices do not line the pore, MTSET inhibits channels C2o10 and C4o10 (but not C3o10). Experimental data for the C1o10 channel are unavailable. In the energetically preferable conformations, the ammonium groups of mC2o10 and mC4o10 extend between S6s and approach the pore (Fig. 7, A and C), whereas the orientation of mC3o10 to the repeat interface is stabilized by cation–π interactions with F3i14, F3i17, F3i18, and Y3o14 (Fig. 7 B). MTSET inhibits C3o15, C4o12, and C4o17 channels by ∼30–40%. In our models, the side chain of mC4o17 extends toward the pore, whereas the side chains of mC3o15 and mC4o12 face away from the pore.
Figure 7.
The extracellular view of possible orientations of mC residues in S5s. mC2o10 (A) and mC4o10 (C) can extend their ammonium groups toward the pore. (B) mC3o10 is stabilized inside the repeat interface by cation–π interaction with F3i18 and Y3o14. The red cross indicates the pore axis.
Currents correlate with the distance of mC_N+ from the pore axis
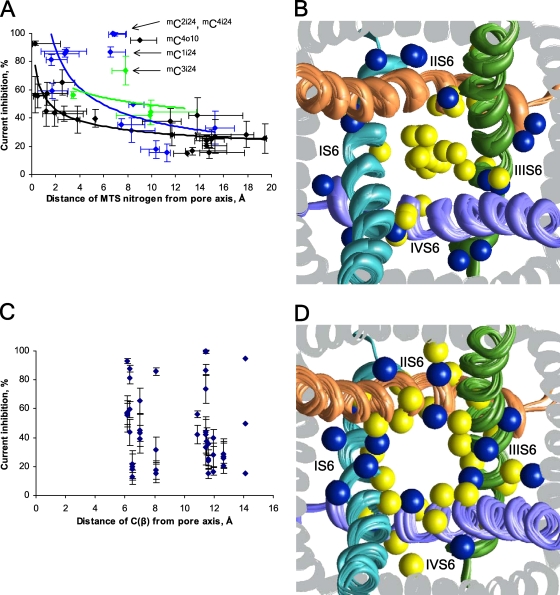
The above results provide multiple examples showing that the mC side chains adopt essentially different orientations relative to the pore. Electrostatic and cation–π interactions between the mC ammonium group and its surrounding residues stabilize these particular orientations. Importantly, the MTSET potency correlates with the distance between the pore axis and the N+ atom of the respective mC residue (Fig. 8 A). The current inhibition decreases with the distance, approaching the level of ∼20% at distances >16 Å. The inhibition of the channels at large distances cannot be explained by electrostatic repulsion between the MTSET ammonium group and permeating cations; rather, it reflects the fact that MTSET inhibits by 19 ± 5.9% the “control channel” in which the α1 subunit lacks both native and engineered cysteines (Zhen et al., 2005). In Fig. 8 B, the energetically preferable orientations of mC residues in the pore are shown by the N+ atoms, which are colored according to the MTSET effect on the respective channels. Substantial current inhibition is usually observed when the N+ atom (colored yellow) is inside the pore, and weak inhibition is usually observed when the N+ atom (colored blue) is outside the pore. In contrast, the Cβ atom position of the engineered cysteine does not correlate with the current inhibition by MTSET (Fig. 8, C and D).
Figure 8.
The residual current upon MTSET application correlates with the distance of the MTS atom N+_mC (A and B), but not atom Cβ_mC (C and D) from the pore axis. (A and C) The experimental values of the current inhibition with standard deviations (Zhen et al., 2005) are plotted against the predicted distances of atoms N+_mC (A) or Cβ (C) from the pore axis. Data are shown for channels with engineered cysteines in positions i15–i21, i23–i25, and o10. Black dots represent the apparent global minima of channels in which all minimum energy conformations of mC side chains are unambiguously oriented in respect to the pore (e.g., inside the pore for channels mCi15 or outside the pore for channels mCi17). Blue dots represent the apparent global minima of the channels in which the mC side chain adopts low energy conformations with distinct orientation in respect to the pore (e.g., channels mCi18). A green dot represents a local minimum (within 2 kcal/mol from the apparent global minimum) of a channel in which the mC side chain adopts conformations with distinct orientation in respect to the pore (e.g., channels mCi16). Horizontal lines show the N+_mC atom mobility in conformations within 2 kcal/mol from the apparent global minimum (Table S1). Note a smooth decrease of the current inhibition with increase of the distance between the MTS nitrogen and the pore axis. The current inhibition of ∼20% at distances >16 Å corresponds to MTSET block of the “control channel,” in which eight native cysteines in the α1 subunit have been replaced with alanines and no engineered cysteines have been introduced (Zhen et al., 2005). (B and D) The extracellular view of Cav2.1, with atoms mC_N+ (B) and mC_Cβ (D) shown as spheres. P loops are omitted for clarity. Yellow and blue spheres represent the respective atoms in the channels, which are inhibited by MTSET by >30 and ≤30%, respectively. (B) In most of the channels, which are strongly inhibited by MTSET, the yellow-colored ammonium nitrogen (mC_N+) is located either close to the pore axis or at the inner surface of the pore, whereas in the channels, which are weakly inhibited by MTSET, the blue-colored ammonium nitrogen is not inside the pore. (D) Location of β carbons does not correlate with the level of current inhibition by MTSET. Both yellow and blue spheres are randomly distributed at different sides of the inner helices.
DISCUSSION
In the absence of x-ray structures of Na+ and Ca2+ channels, their homology models based on x-ray structures of K+ channels are used to interpret data from mutational, electrophysiological, and ligand-binding experiments. The most crucial step in homology modeling is the sequence alignment. Because the sequence similarity between Ca2+ and Na+ channels is rather high, alignment between these channels is unambiguous (Zhorov and Tikhonov, 2004). In contrast, due to poor sequence similarity between K+ channels on one hand and Ca2+ and Na+ channels on the other hand, there is no consensus alignment of S5 and S6 segments between these channels. In particular, the proposed alignments for S6s (Huber et al., 2000; Zhorov et al., 2001; Lipkind and Fozzard, 2003; Shafrir et al., 2008; Stary et al., 2008) differ in positions of asparagines that are highly conserved in every repeat of eukaryotic Ca2+ and Na+ channels and are present in the homotetrameric bacterial channel NaChBac (Ren et al., 2001). In the alignment proposed by Lipkind and Fozzard (2003), these asparagines do not appear in matching positions of the four repeats. Kv1.2-based models of the Cav1.2 (Stary et al., 2008) and NaChBac (Shafrir et al., 2008) have been built with the alignment in which an insertion is introduced at the conserved asparagines.
Intensive studies identified residues that, when mutated, affect the action of ligands targeting the pore of voltage-gated Ca2+ (Hockerman et al., 1997) and Na+ channels (Catterall et al., 2005). Homology models of these channels were used to visualize the binding sites and propose atomic mechanisms of various drugs, including benzothiazepines (Tikhonov and Zhorov, 2008), dihydropyridines (Zhorov et al., 2001; Lipkind and Fozzard, 2003; Cosconati et al., 2007; Tikhonov and Zhorov, 2009), phenylalkylamines (Lipkind and Fozzard, 2003; Cheng et al., 2009), local anesthetics (Lipkind and Fozzard, 2005; Tikhonov and Zhorov, 2007; Bruhova et al., 2008), steroidal sodium channel activators (Wang et al., 2006), and pyrethroid insecticides (O’Reilly et al., 2006; Du et al., 2009).
The above models have been built using different templates and different alignments. How sensitive are these results to the choice of the template and alignment? In a recent study (Cheng et al., 2009), several Cav1.2 models based on different open-channel templates (KvAP, MthK, and Kv1.2) and different alignments were compared in terms of interactions with a flexible phenylalkylamine ligand devapamil. The alignment, which we use here (Table I), and all three templates were found to be consistent with the ligand–channel contacts known from experiments. The reason for such promiscuity of the modeled ligand–channel interactions to the choice of the open-channel template is the flexibility of both devapamil and side chains of devapamil-interacting tyrosines. This flexibility compensated rather small differences in the templates, all of which have similar patterns of the pore-facing residues. However, a shift of the S6 alignment between K+ and Ca2+ channels by just one position resulted in such dramatic reorientation of devapamil-interacting residues in the models that it was not possible to establish critical devapamil-Cav1.2 contacts known from experiments.
Because mC residues can be considered as long, flexible tethered ligands, predicted contacts of the mC ammonium groups should also be highly sensitive to the sequence alignment, but rather insensitive to the choice of the open-channel x-ray template. A single-position shift in the alignment would turn a pore-directing vector Cα-Cβ away from the pore and vice versa. For example, Ci15 channels are sensitive to MTSET, whereas Ci17 channels are not (Zhen et al., 2005). In agreement with these data and the cysteine orientation concept, our models built using alignment shown in Table I (Zhorov et al., 2001) have vectors Cα-Cβ in positions i15 and i17, which direct to the pore axis and away from it, respectively. Models built using other alignments (Huber et al., 2000; Lipkind and Fozzard, 2003) are unlikely to provide a correlation with experimental SCAM data (Zhen et al., 2005). Guy and coworkers (Durell and Guy, 2001; Stary et al., 2008) proposed an S6 alignment, which is similar to that shown in Table I, but it has an insertion at position i20 of each repeat of the Ca2+ channel. A model built with this alignment does not resolve the problem of the pore-away orientation of mCi24 residues in the outlying channels (Fig. 8), whereas Cα-Cβ vectors in residues downstream of positions i20 orient differently than in our models, which are based on our alignment (Table I) and which explain the SCAM data.
Despite different alignments underlying these models, and different details of predicted ligand–channel interactions, the above studies agree that the x-ray structures of K+ channels provide reasonable templates for the homology modeling of Na+ and Ca2+ channels. This opinion was undermined by the interpretation of SCAM experiments with Cav2.1, which suggests that Ca2+ and K+ channels have different patterns of pore-lining residues and questions the symmetric arrangement of the Ca2+ channel repeats around the pore axis (Zhen et al., 2005).
We do not doubt the experimental observations of the SCAM study, but we show here that interpretation of these observations requires analysis of some factors, which were apparently not considered in the original study (Zhen et al., 2005). These factors are conformational flexibility of long side chains of mC residues and their interaction with neighboring residues. In this study, we used the Kv1.2-based models of Cav2.1, which are based on the alignment shown in Table I. We reasoned that if our results explain the SCAM data, this supports the alignment as well as the generally similar spatial disposition of S5s and S6s in K+ and Ca2+ channels.
Interpreting SCAM experiments is not straightforward. Several factors should be taken into consideration (Karlin and Akabas, 1998). MTS reagents, such as MTSET, react with water-accessible ionized cysteines to form a covalent bond. If a cysteine is exposed to the lipid bilayer or buried inside the protein, the ionization of the thiol group is suppressed. It is assumed that the MTS reagent covalently bound to the engineered cysteine and exposed to the pore decreases the current. The current may be unaffected because of two causes. First, the reaction does not proceed due to hydrophobic environment, steric constraints, lack of ionized Cys residues, or other grounds. Second, the reaction proceeds, but the MTS-modified cysteine does not affect ion permeation.
A brief application of an MTS reagent may result in incomplete chemical modification of cysteines (Liu et al., 1997), but prolonged exposure increases the probability of disulfide formation, even with partially buried cysteines. The prolonged Cav2.1 exposure to MTSET suggests that cysteines in different sides of S6s and S5s were modified (Zhen et al., 2005), despite the fact that some positions do not face the ion permeation pathway.
Besides the kinetic effects, other factors should be considered to interpret SCAM data. An MTS-modified cysteine has a long flexible side chain: in the all-trans conformation, the distance between atoms Cα and N+ is 8.4 Å. Prediction of energetically optimal conformations of an mC residue can be considered as docking of a tethered ligand to the channel. The energetically optimal position of the mC ammonium group depends on interactions with neighboring residues, among which electrostatic attractions (including those with the nucleophilic C ends of P helices) and cation–π interactions play the major role. In some positions where vector Cα-Cβ directs to the pore axis, the mC ammonium group does not bind in the pore. And in some positions where vector Cα-Cβ directs away from the axis, the ammonium group can reach the pore through the repeat interface or by wrapping around the mutated helix. Thus, the exposure of the mC ammonium group to the pore may not correlate with the angle between the Cα-Cβ vector and the vector drawn from the Cα atom to the pore axis.
Our calculations predict that the reported current inhibition by MTSET generally decreases with the predicted distances between the ammonium nitrogen and the pore axis (Fig. 8, A and B). This trend is important. First, it shows that interpretation of the SCAM data is possible in gradual rather than discrete terms. Second, it supports the underlying sequence alignment between Ca2+ and K+ channels (Table I). Third, it implies the fourfold symmetry of transmembrane helices in the pore-forming domain of Ca2+ channels and a similar disposition of S5s and S6s in K+ and Ca2+ channels. Fourth, it shows that significant block is observed only when the ammonium group occurs in the pore, but partial inhibition is possible when the ammonium group is rather far from the pore axis. This is in agreement with the single-channel recordings, which demonstrate that MTSET decreases the current amplitude (Lu et al., 1999).
In homotetrameric K+ channels, a single mutation to Cys yields four identical potential targets for the reaction with an MTS reagent. Linking the subunits in a single polypeptide chain allowed the expression of channels with one, two, three, or four cysteines at a given S6 position (Lu et al., 1999). MTSET application to the channels with one, two, and three cysteines in position i18 inhibited the current by 24, 55, and 80%, respectively. MTSET application to channels with one, two, and three engineered cysteines in position i22 resulted in the current inhibition by 51, 80, and 89%, respectively (Lu et al., 1999). Thus, the introduction of one positive charge into the open pore of the K+ channel does not fully inhibit current. Furthermore, protonation at the selectivity filter of Ca2+ channels reduces single-channel conductance, but it does not produce a complete channel block (Prod’hom et al., 1987; Pietrobon et al., 1989). The above observations are consistent with the fact that the mC residues at pore-facing positions, such as i15 and i19, reduce the current but do not completely inhibit it. On the other hand, incomplete inhibition could indicate that the MTS application did not modify 100% of the respective channels.
Fig. 8 A was obtained using data for 40 channels. In 36 cases, the data point represents the energetically most preferable conformation of the respective mC residue, and in four channels, green points represent local minima with the energies up to 2 kcal/mol above the apparent global minima. Thus, only 10% of the data points represent the less populated conformations. The fact that our method yields a poorer correlation when only the apparent global minima were considered is understandable in view of the limited precision of the homology modeling. Because of these limitations, quantitative analysis is difficult, particularly when an mC side chain has two distinct groups of conformers, one group inside the pore (the ammonium nitrogen within 4 Å from the pore axis) and another away from the pore. Two distinct groups of conformers were observed, e.g., for mCi16 and mCi18 (Figs. 6 A and 3).
In Fig. 8, we did not include the Ci22 channels, which are not inhibited by MTSET. Respective Cα-Cβ vectors face the pore, and in the most preferable conformations of mCi22 residues, the N+ atoms occur inside the pore. There are local minima with the N+ atoms beyond the pore, and the Ci22 channels could be represented by green points in Fig. 8 A. However, we believe that Ci22 channels are insensitive to MTSET because large hydrophobic residues in the pore-facing positions i19, i22, and i26 (Table I) create a highly hydrophobic environment that precludes the reaction with MTSET. In Na+ channels, Y4i22 was proposed to face the pore and interact with local anesthetics (Tikhonov and Zhorov, 2007; Bruhova et al., 2008). However, the Y4i22C mutant is insensitive to MTS ethylammonium or MTSET (Sunami et al., 2004). A possible cause is the hydrophobic environment of C4i22 created by hydrophobic residues in the pore-facing positions i19, i22, and i26. In contrast to Ca2+ and Na+ channels, MTSET inhibits the Ci22 mutants of Shaker (Liu et al., 1997), Kir2.1 (Lu et al., 1999), Kir6.2 (Phillips et al., 2003), and KCa3.1 (Klein et al., 2007). Position i22 of the Shaker is surrounded by Ai19, Ci22, and Vi26, which provide a favorable environment for reaction with MTSET (Fig. 5). The ring i22 of Cav2.1 is unique in terms of the completely hydrophobic environment at its own level and the levels of the pore-facing residues i19 and i26 above and below the ring, respectively. The correlation in Fig. 8 A suggests a common mechanism of current inhibition by MTSET, but we cannot rule out that incomplete block of some channels also results from slow reaction with MTSET.
The correlation in Fig. 8 A has five prominent outliers, which correspond to channels C4o10, C1i24, C2i24, C3i24, and C4i24. These channels are strongly blocked by MTSET, despite the fact that corresponding mC_ N+ atoms are 6.5–9 Å from the pore axis. Vectors Cα-Cβ in positions i24 are close to the cytoplasm in the x-ray structures of Kv1.2, KcsA, and the closed-Kv1.2 model (Pathak et al., 2007), suggesting that MTSET could attack Ci24s not from the open pore, but from the cytoplasm. The strong current inhibition in channels mCi24 may arise from stabilization of the closed-channel conformation. To explore this possibility, we have built KcsA-based models of the mCi24 mutants of Cav2.1 and sought for possible contacts of the mCi24 ammonium group with nearby acidic residues, which were modeled in the ionized forms (Fig. S5). MC minimizations with distance constraints biasing inter-repeat salt bridges yielded low energy structures with the following salt bridges: mC1i24–D4i28, mC2i24–E1i29, mC3i24–D2i28, and mC4i24–E3i32. These remained stable in subsequent MC minimizations without the constraints. These salt bridges could be formed upon MTSET application to the hyperpolarized membrane and preclude channel opening upon membrane depolarization. However, in the absence of an x-ray structure of a closed voltage-gated channel, we cannot rule out other possible mechanisms of closed-channel stabilization, e.g., interaction of mCi24 residues with the β subunit or cytoplasmic segments of the α1 subunit.
Experiments with fluorinated aromatic residues (Santarelli et al., 2007; Ahern et al., 2008; Xiu et al., 2009) proved the long-proposed role of cation–π interactions in ligand receptor recognition. Despite the fact that the AMBER force field lacks a specific energy term for cation–π interactions, these interactions can be detected in structures where the ammonium group is attracted to partial negative charges of aromatic carbons (Bruhova et al., 2008). Such structures were earlier predicted for complexes of Na+ channels with local anesthetics (Fozzard et al., 2005; Lipkind and Fozzard, 2005; Tikhonov and Zhorov, 2007). Here, we found many structures in which the mC ammonium groups are attracted to aromatic residues via cation–π interactions. These interactions were particularly important in stabilizing the ammonium groups of mC3o10, mC1i18, and mC3i18 in the repeat interfaces, as well as mCi19 in the pore.
According to our models, the repeat interfaces would provide the access paths for MTSET to engineered cysteines in those positions of S5s and S6s that do not face the pore. Furthermore, the ammonium group of an mC residue can extend through a repeat interface into the pore and decrease the current. Interesting examples are mC2o10 and mC4o10, whose ammonium groups can approach the pore only through the repeat interface. This prediction is consistent with our studies, which suggest that the III/IV repeat interface provides the extracellular access route for local anesthetics into Na+ channels (Bruhova et al., 2008) as well as benzothiazepines (Tikhonov and Zhorov, 2008) and dihydropyridines (Tikhonov and Zhorov, 2009) in Ca2+ channels.
In conclusion, here we used molecular modeling to reinterpret the results of the SCAM study of Cav2.1 (Zhen et al., 2005). We found that the residual current upon MTSET application does not correlate with the orientation of the Cα-Cβ vector in the modified residue to the pore, but generally decreases with the distance between the pore axis and the N+ atom of the respective mC residue. Our models suggest that different local environments of equivalent positions in the four repeats lead to different SCAM results reported for such positions. Our study supports the sequence alignments between K+ and Ca2+ channels earlier proposed for S5s (Huber et al., 2000) and S6s (Zhorov et al., 2001), and suggests that the x-ray structure of Kv1.2 is a suitable template to model Ca2+ channels in the open-state conformation.
Acknowledgments
We thank Denis Tikhonov for helpful discussions.
This study was supported by a grant (MOP-53229) from the Canadian Institutes of Health Research (CIHR) to B.S. Zhorov. I. Bruhova is a recipient of the CIHR Canada Graduate Scholarships. Computations were made possible by the facilities of the Shared Hierarchical Academic Research Computing Network (SHARCNET; http://www.sharcnet.ca).
Lawrence G. Palmer served as editor.
Footnotes
Abbreviations used in this paper:
- MC
- Monte Carlo
- MCM
- MC minimization
- MTS
- methanethiosulfonate
- MTSET
- 2-(trimethylammonium)ethyl MTS
- SCAM
- substituted cysteine accessibility method
References
- Ahern C.A., Eastwood A.L., Dougherty D.A., Horn R. 2008. Electrostatic contributions of aromatic residues in the local anesthetic receptor of voltage-gated sodium channels. Circ. Res. 102:86–94 10.1161/CIRCRESAHA.107.160663 [DOI] [PubMed] [Google Scholar]
- Anderson P.A., Greenberg R.M. 2001. Phylogeny of ion channels: clues to structure and function. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 129:17–28 10.1016/S1096-4959(01)00376-1 [DOI] [PubMed] [Google Scholar]
- Bruhova I., Zhorov B.S. 2007. Monte Carlo-energy minimization of correolide in the Kv1.3 channel: possible role of potassium ion in ligand-receptor interactions. BMC Struct. Biol. 7:5 10.1186/1472-6807-7-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruhova I., Tikhonov D.B., Zhorov B.S. 2008. Access and binding of local anesthetics in the closed sodium channel. Mol. Pharmacol. 74:1033–1045 10.1124/mol.108.049759 [DOI] [PubMed] [Google Scholar]
- Catterall W.A., Goldin A.L., Waxman S.G. 2005. International Union of Pharmacology. XLVII. Nomenclature and structure-function relationships of voltage-gated sodium channels. Pharmacol. Rev. 57:397–409 10.1124/pr.57.4.4 [DOI] [PubMed] [Google Scholar]
- Cheng R.C., Tikhonov D.B., Zhorov B.S. 2009. Structural model for phenylalkylamine binding to L-type calcium channels. J. Biol. Chem. 284:28332–28342 10.1074/jbc.M109.027326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosconati S., Marinelli L., Lavecchia A., Novellino E. 2007. Characterizing the 1,4-dihydropyridines binding interactions in the L-type Ca2+ channel: model construction and docking calculations. J. Med. Chem. 50:1504–1513 10.1021/jm061245a [DOI] [PubMed] [Google Scholar]
- Dewar M.J., Zoebisch E.G., Healy E.F., Stewart J.J. 1985. Development and use of quantum mechanical molecular models. 76. AM1: a new general purpose quantum mechanical molecular model. J. Am. Chem. Soc. 107:3902–3909 10.1021/ja00299a024 [DOI] [Google Scholar]
- Doyle D.A., Morais Cabral J., Pfuetzner R.A., Kuo A., Gulbis J.M., Cohen S.L., Chait B.T., MacKinnon R. 1998. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 280:69–77 10.1126/science.280.5360.69 [DOI] [PubMed] [Google Scholar]
- Du Y., Lee J.E., Nomura Y., Zhang T., Zhorov B.S., Dong K. 2009. Identification of a cluster of residues in transmembrane segment 6 of domain III of the cockroach sodium channel essential for the action of pyrethroid insecticides. Biochem. J. 419:377–385 10.1042/BJ20082082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley S.C., Jr., Chang N., Hall J., Lipkind G., Fozzard H.A., French R.J. 2000. µ-Conotoxin GIIIA interactions with the voltage-gated Na+ channel predict a clockwise arrangement of the domains. J. Gen. Physiol. 116:679–690 10.1085/jgp.116.5.679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durell S.R., Guy H.R. 2001. A putative prokaryote voltage-gated Ca(2+) channel with only one 6TM motif per subunit. Biochem. Biophys. Res. Commun. 281:741–746 10.1006/bbrc.2001.4408 [DOI] [PubMed] [Google Scholar]
- Fozzard H.A., Lee P.J., Lipkind G.M. 2005. Mechanism of local anesthetic drug action on voltage-gated sodium channels. Curr. Pharm. Des. 11:2671–2686 10.2174/1381612054546833 [DOI] [PubMed] [Google Scholar]
- Hockerman G.H., Peterson B.Z., Johnson B.D., Catterall W.A. 1997. Molecular determinants of drug binding and action on L-type calcium channels. Annu. Rev. Pharmacol. Toxicol. 37:361–396 10.1146/annurev.pharmtox.37.1.361 [DOI] [PubMed] [Google Scholar]
- Huber I., Wappl E., Herzog A., Mitterdorfer J., Glossmann H., Langer T., Striessnig J. 2000. Conserved Ca2+-antagonist-binding properties and putative folding structure of a recombinant high-affinity dihydropyridine-binding domain. Biochem. J. 347:829–836 10.1042/0264-6021:3470829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., Lee A., Chen J., Ruta V., Cadene M., Chait B.T., MacKinnon R. 2003. X-ray structure of a voltage-dependent K+ channel. Nature. 423:33–41 10.1038/nature01580 [DOI] [PubMed] [Google Scholar]
- Karlin A., Akabas M.H. 1998. Substituted-cysteine accessibility method. Methods Enzymol. 293:123–145 10.1016/S0076-6879(98)93011-7 [DOI] [PubMed] [Google Scholar]
- Klein H., Garneau L., Banderali U., Simoes M., Parent L., Sauvé R. 2007. Structural determinants of the closed KCa3.1 channel pore in relation to channel gating: results from a substituted cysteine accessibility analysis. J. Gen. Physiol. 129:299–315 10.1085/jgp.200609726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazaridis T., Karplus M. 1999. Effective energy function for proteins in solution. Proteins. 35:133–152 [DOI] [PubMed] [Google Scholar]
- Li Z., Scheraga H.A. 1987. Monte Carlo-minimization approach to the multiple-minima problem in protein folding. Proc. Natl. Acad. Sci. USA. 84:6611–6615 10.1073/pnas.84.19.6611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipkind G.M., Fozzard H.A. 2003. Molecular modeling of interactions of dihydropyridines and phenylalkylamines with the inner pore of the L-type Ca2+ channel. Mol. Pharmacol. 63:499–511 10.1124/mol.63.3.499 [DOI] [PubMed] [Google Scholar]
- Lipkind G.M., Fozzard H.A. 2005. Molecular modeling of local anesthetic drug binding by voltage-gated sodium channels. Mol. Pharmacol. 68:1611–1622 [DOI] [PubMed] [Google Scholar]
- Liu Y., Holmgren M., Jurman M.E., Yellen G. 1997. Gated access to the pore of a voltage-dependent K+ channel. Neuron. 19:175–184 10.1016/S0896-6273(00)80357-8 [DOI] [PubMed] [Google Scholar]
- Long S.B., Campbell E.B., Mackinnon R. 2005. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science. 309:897–903 10.1126/science.1116269 [DOI] [PubMed] [Google Scholar]
- Long S.B., Tao X., Campbell E.B., MacKinnon R. 2007. Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature. 450:376–382 10.1038/nature06265 [DOI] [PubMed] [Google Scholar]
- Lu T., Nguyen B., Zhang X., Yang J. 1999. Architecture of a K+ channel inner pore revealed by stoichiometric covalent modification. Neuron. 22:571–580 10.1016/S0896-6273(00)80711-4 [DOI] [PubMed] [Google Scholar]
- O’Reilly A.O., Khambay B.P., Williamson M.S., Field L.M., Wallace B.A., Davies T.G. 2006. Modelling insecticide-binding sites in the voltage-gated sodium channel. Biochem. J. 396:255–263 10.1042/BJ20051925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak M.M., Yarov-Yarovoy V., Agarwal G., Roux B., Barth P., Kohout S., Tombola F., Isacoff E.Y. 2007. Closing in on the resting state of the Shaker K(+) channel. Neuron. 56:124–140 10.1016/j.neuron.2007.09.023 [DOI] [PubMed] [Google Scholar]
- Phillips L.R., Enkvetchakul D., Nichols C.G. 2003. Gating dependence of inner pore access in inward rectifier K(+) channels. Neuron. 37:953–962 10.1016/S0896-6273(03)00155-7 [DOI] [PubMed] [Google Scholar]
- Pietrobon D., Prod’hom B., Hess P. 1989. Interactions of protons with single open L-type calcium channels. pH dependence of proton-induced current fluctuations with Cs+, K+, and Na+ as permeant ions. J. Gen. Physiol. 94:1–21 10.1085/jgp.94.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prod’hom B., Pietrobon D., Hess P. 1987. Direct measurement of proton transfer rates to a group controlling the dihydropyridine-sensitive Ca2+ channel. Nature. 329:243–246 10.1038/329243a0 [DOI] [PubMed] [Google Scholar]
- Ren D., Navarro B., Xu H., Yue L., Shi Q., Clapham D.E. 2001. A prokaryotic voltage-gated sodium channel. Science. 294:2372–2375 10.1126/science.1065635 [DOI] [PubMed] [Google Scholar]
- Santarelli V.P., Eastwood A.L., Dougherty D.A., Horn R., Ahern C.A. 2007. A cation-pi interaction discriminates among sodium channels that are either sensitive or resistant to tetrodotoxin block. J. Biol. Chem. 282:8044–8051 10.1074/jbc.M611334200 [DOI] [PubMed] [Google Scholar]
- Shafrir Y., Durell S.R., Guy H.R. 2008. Models of the structure and gating mechanisms of the pore domain of the NaChBac ion channel. Biophys. J. 95:3650–3662 10.1529/biophysj.108.135327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stary A., Shafrir Y., Hering S., Wolschann P., Guy H.R. 2008. Structural model of the Ca(V)1.2 pore. Channels (Austin). 2:210–215 [DOI] [PubMed] [Google Scholar]
- Sunami A., Tracey A., Glaaser I.W., Lipkind G.M., Hanck D.A., Fozzard H.A. 2004. Accessibility of mid-segment domain IV S6 residues of the voltage-gated Na+ channel to methanethiosulfonate reagents. J. Physiol. 561:403–413 10.1113/jphysiol.2004.067579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikhonov D.B. 2007. Ion channels of glutamate receptors: structural modeling. Mol. Membr. Biol. 24:135–147 10.1080/09687860601008806 [DOI] [PubMed] [Google Scholar]
- Tikhonov D.B., Zhorov B.S. 2005. Modeling P-loops domain of sodium channel: homology with potassium channels and interaction with ligands. Biophys. J. 88:184–197 10.1529/biophysj.104.048173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikhonov D.B., Zhorov B.S. 2007. Sodium channels: ionic model of slow inactivation and state-dependent drug binding. Biophys. J. 93:1557–1570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikhonov D.B., Zhorov B.S. 2008. Molecular modeling of benzothiazepine binding in the L-type calcium channel. J. Biol. Chem. 283:17594–17604 10.1074/jbc.M800141200 [DOI] [PubMed] [Google Scholar]
- Tikhonov D.B., Zhorov B.S. 2009. Structural model for dihydropyridine binding to L-type calcium channels. J. Biol. Chem. 284:19006–19017 10.1074/jbc.M109.011296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikhonov D.B., Bruhova I., Zhorov B.S. 2006. Atomic determinants of state-dependent block of sodium channels by charged local anesthetics and benzocaine. FEBS Lett. 580:6027–6032 10.1016/j.febslet.2006.10.035 [DOI] [PubMed] [Google Scholar]
- Wang S.Y., Mitchell J., Tikhonov D.B., Zhorov B.S., Wang G.K. 2006. How batrachotoxin modifies the sodium channel permeation pathway: computer modeling and site-directed mutagenesis. Mol. Pharmacol. 69:788–795 10.1124/mol.106.022368 [DOI] [PubMed] [Google Scholar]
- Weiner S.J., Kollman P.A., Case D.A., Singh U.C., Ghio C., Alagona G., Profeta S., Weiner P. 1984. A new force-field for molecular mechanical simulation of nucleic-acids and proteins. J. Am. Chem. Soc. 106:765–784 10.1021/ja00315a051 [DOI] [Google Scholar]
- Weiner S.J., Kollman P.A., Nguyen D.T., Case D.A. 1986. An all atom force field for simulations of proteins and nucleic acids. J. Comput. Chem. 7:230–252 10.1002/jcc.540070216 [DOI] [PubMed] [Google Scholar]
- Xiu X., Puskar N.L., Shanata J.A., Lester H.A., Dougherty D.A. 2009. Nicotine binding to brain receptors requires a strong cation-pi interaction. Nature. 458:534–537 10.1038/nature07768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhen X.G., Xie C., Fitzmaurice A., Schoonover C.E., Orenstein E.T., Yang J. 2005. Functional architecture of the inner pore of a voltage-gated Ca2+ channel. J. Gen. Physiol. 126:193–204 10.1085/jgp.200509292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhorov B.S., Tikhonov D.B. 2004. Potassium, sodium, calcium and glutamate-gated channels: pore architecture and ligand action. J. Neurochem. 88:782–799 [DOI] [PubMed] [Google Scholar]
- Zhorov B.S., Folkman E.V., Ananthanarayanan V.S. 2001. Homology model of dihydropyridine receptor: implications for L-type Ca(2+) channel modulation by agonists and antagonists. Arch. Biochem. Biophys. 393:22–41 10.1006/abbi.2001.2484 [DOI] [PubMed] [Google Scholar]
- Zong Y., Mazmanian S.K., Schneewind O., Narayana S.V. 2004. The structure of sortase B, a cysteine transpeptidase that tethers surface protein to the Staphylococcus aureus cell wall. Structure. 12:105–112 10.1016/j.str.2003.11.021 [DOI] [PubMed] [Google Scholar]