Loss of the condensin complex components Smc2 and -4 disrupts epigenetic modifications required for embryonic stem cell survival.
Abstract
In an RNA interference screen interrogating regulators of mouse embryonic stem (ES) cell chromatin structure, we previously identified 62 genes required for ES cell viability. Among these 62 genes were Smc2 and -4, which are core components of the two mammalian condensin complexes. In this study, we show that for Smc2 and -4, as well as an additional 49 of the 62 genes, knockdown (KD) in somatic cells had minimal effects on proliferation or viability. Upon KD, Smc2 and -4 exhibited two phenotypes that were unique to ES cells and unique among the ES cell–lethal targets: metaphase arrest and greatly enlarged interphase nuclei. Nuclear enlargement in condensin KD ES cells was caused by a defect in chromatin compaction rather than changes in DNA content. The altered compaction coincided with alterations in the abundance of several epigenetic modifications. These data reveal a unique role for condensin complexes in interphase chromatin compaction in ES cells.
Introduction
Embryonic stem (ES) cells are derived from the inner cell mass of the mammalian blastocyst, a cell population which exists transiently from approximately embryonic day (E) 3.5 to E4.5 in mice (Dard et al., 2008). In vitro, ES cells can proliferate indefinitely in an undifferentiated state and maintain the potential to differentiate into all tissues of the animal (Evans and Kaufman, 1981; Martin, 1981).
The ability to form every embryonic cell type is not the only feature that distinguishes ES cells from somatic cells. The cell cycle of ES cells is very rapid, with altering phases of DNA replication and mitosis separated by minimal gap phases (Liu et al., 2007), a feature which likely promotes rapid expansion of the pluripotent cell population in the early embryo. Another difference relates to cellular checkpoints. Somatic cells often respond to cellular stresses such as DNA damage by the activation of checkpoint pathways, which lead to cell cycle arrest until the damage is repaired (Niida and Nakanishi, 2006). In contrast, ES cells lack DNA damage checkpoints and largely respond to treatment with DNA-damaging agents by undergoing apoptosis (Aladjem et al., 1998). This feature may have evolved to remove cells more likely to carry mutations from the pool of progenitor cells that will populate the organism (Hong et al., 2007). Finally, ES cells have unusually dynamic chromatin, in which many architectural proteins are loosely bound, a feature which is proposed to contribute to the maintenance of developmental plasticity (Meshorer and Misteli, 2006; Meshorer et al., 2006).
In an RNAi screen examining the role of chromatin structural and regulatory factors and DNA/RNA helicases in mouse ES cells, 62 of 1,008 targets exhibited cell death upon knockdown (KD; Fazzio et al., 2008). The majority of genes required for ES cell viability were known or predicted to function in processes crucial to cell growth and division, including DNA replication, chromosome cohesion and condensation, RNA processing, and ribosome biogenesis. In this study, we show that, for the majority of the genes required for ES cell viability, KD in somatic cells is tolerated with little or no phenotype. Further characterization of condensin proteins (Smc2 and -4), the replicative helicases (Mcm proteins), and Integrator (Ints) proteins that play a role in the 3′ end processing of small nuclear RNAs (snRNAs) revealed that perturbations of different processes in ES cells all result in DNA damage and apoptosis. These data support a model in which ES cells undergo apoptosis in response to a wide variety of cellular stresses that are tolerated in somatic cells.
Smc2 and -4 form a heterodimer that is the catalytic subunit of both the condensin I and II complexes, which play roles in mitotic and meiotic chromosome condensation and rigidity, interphase ribosomal DNA compaction, and removal of cohesin during mitosis and meiosis (Losada and Hirano, 2005; Belmont, 2006; Ivanovska and Orr-Weaver, 2006; Tsang et al., 2007). Although mitotic chromosome condensation absolutely requires condensin I in Xenopus laevis egg extracts, this is not the case in mammalian somatic cells. In mammalian somatic cells, depletion of condensins I and II results in an increased frequency of defects during mitotic anaphase, including lagging chromosomes and anaphase bridges (Hirota et al., 2004; Ono et al., 2004). However, despite these mitotic defects, condensin-depleted mammalian somatic cells continue to proliferate, indicating that the low levels of condensin complexes remaining after RNAi-mediated depletion are sufficient to sustain cellular viability. In spite of the fact that subunits of condensin II are localized to the nucleus throughout interphase, condensin KD has no widespread effect on interphase chromatin compaction in mammalian somatic cells.
In this study, we show that condensin I and II complexes are redundantly required for ES cell proliferation. ES cells depleted of both condensin complexes exhibit two unique phenotypes: severely delayed initiation of anaphase and accumulation of enlarged and misshapen interphase nuclei. Using FISH to assess the organization of individual loci, we show that the nuclear enlargement in condensin KD ES cells correlates with an alteration in higher-order chromatin folding. Furthermore, condensin KD also results in changes in the amounts or distribution of interphase histone H3 (H3) phosphorylated on serine 10 (H3S10P), H3 trimethylated on lysine 9 (H3K9me3), and DNA methylation. Together, these data point to a novel role for condensins in the maintenance of ES cell–specific interphase chromatin structure.
Results
Many chromatin factors that are essential for viability in ES cells can be depleted in somatic cells
In an RNAi screen assaying the effects of depletion of 1,008 chromatin and RNA-binding proteins in mouse ES cells, the most common phenotype was cell death. Of 68 genes with KD phenotypes, depletion of 62 reduced ES cell viability (Fazzio et al., 2008). The vast majority of these 62 genes encode proteins that are broadly expressed, with known or predicted functions in essential processes such as RNA processing, DNA replication, ribosome biogenesis, and chromosome condensation (Table I). However, no viability defects were reported for KD of several of these genes in somatic mammalian cells (Ono et al., 2003; Cortez et al., 2004; Baillat et al., 2005; Farhana et al., 2007; Suzuki et al., 2008), suggesting that ES cells may be unusually sensitive to perturbations of some essential processes.
Table I.
Targets with RNAi phenotypes
| Gene name | ESC death | MEF death | Known or predicted function |
| Rnf2a | 3 | 0 | Histone ubiquitylation |
| Mettl5 | 2 | 0 | DNA, RNA, or ribosomal protein methylase |
| Setd8a | 4 | 3 | Histone H4K20 methyltransferase |
| Brca1 | 1 | 0 | E3 ubiquitin ligase |
| Ighmbp2 | 4 | 0 | Transcription regulation |
| Sin3aa | 4 | 0 | Scaffold for HDACs |
| Arid4a | 2 | 0 | Recruitment of HDACs |
| Cbx3a | 3 | 2 | Chromatin binding |
| Brd4 | 3 | 3 | Transcription initiation |
| Chaf1b | 3 | 0 | Chromatin assembly |
| Tpr | 3 | 0 | Nuclear envelope |
| Tcp1 | 4 | 0 | Protein chaperone, chromosome binding |
| Rif1 | 3 | 0 | Telomere length regulation |
| Stat3 | 1 | 0 | Transcription factor |
| Ints4 | 3 | 0 | RNA 3′ end processing |
| Trim28a | 3 | 0 | Transcriptional corepressor |
| Cdc5l | 3 | 0 | Transcription factor |
| Ssrp1b | 4 | 0/2 | Transcriptional elongation |
| Supt16h | 4 | 4 | Transcriptional elongation |
| Ruvbl1a | 3 | 0 | Chromatin remodeling |
| Ruvbl2a | 2 | 0 | Chromatin remodeling |
| Dmap1a | 1 | 0 | Chromatin remodeling |
| Ep400a | 2 | 0 | Chromatin remodeling |
| Htatipa | 1 | 0 | Chromatin remodeling |
| Trrapa | 1 | 0 | Chromatin remodeling |
| Yeats4a | 2 | 0 | Chromatin remodeling |
| H2afz | 1 | 0 | Chromatin structure |
| Smc2 | 4 | 0 | Mitotic chromosome condensation |
| Smc4a | 4 | 0 | Mitotic chromosome condensation |
| Smc1a | 1 | 0 | Chromosome cohesion |
| Gmnn | 4 | 0 | DNA replication |
| Mcm7 | 3 | 0 | DNA replication |
| Mcm2a | 3 | 0 | DNA replication |
| Mcm6 | 3 | 0 | DNA replication |
| Mcm4 | 3 | 0 | DNA replication |
| Eif4a3a | 4 | 4 | NMD, ribosome biogenesis |
| Ddx54 | 4 | 3 | Ribosome biogenesis |
| Ddx56 | 2 | 0 | Ribosome biogenesis |
| Ddx18a | 4 | 0 | Ribosome biogenesis |
| Ddx47a | 3 | 0 | Ribosome biogenesis |
| Utp3 | 2 | 0 | Processome, ribosome biogenesis |
| Snrpd1 | 4 | 0 | RNA splicing |
| Snrpd2 | 4 | 3 | RNA splicing |
| Snrpd3 | 4 | 0 | RNA splicing |
| Snrpe | 4 | 2 | RNA splicing |
| Snrpfa | 4 | 0 | RNA splicing |
| Snrpg | 4 | 0 | RNA splicing |
| Snrpn | 4 | 0 | RNA splicing |
| Lsm2a | 3 | 0 | RNA splicing |
| Lsm6a | 4 | 0 | RNA splicing |
| Lsm7 | 3 | 0 | RNA splicing |
| Dhx15 | 3 | 0 | RNA splicing |
| Ascc3l1 | 4 | 0 | RNA splicing |
| Skiv2l2 | 3 | 0 | RNA splicing |
| Ddx41 | 3 | 0 | RNA splicing |
| Dhx37 | 3 | 0 | RNA splicing |
| Dhx8 | 4 | 3 | RNA splicing |
| Ddx39 | 4 | 4 | RNA splicing |
| Aqr | 4 | 3 | RNA splicing |
| Sf3b1 | 4 | 3 | RNA splicing |
| Phf5a | 4 | 0 | RNA splicing |
| Ddx19a | 4 | 0 | mRNA export |
ESC, ES cell; HDAC, histone deacetylase complex; NMD, nonsense-mediated mRNA decay. For ES cell and MEF death, the numerical scale of cell division/viability defects is as follows: 0, no effect; 1, minor; 2, moderate; 3, moderate to severe; and 4, severe. Genes indicated in bold exhibit viability defects upon KD in both ES cells and iMEFs.
Tested with second esiRNA.
Initial esiRNA did not produce a phenotype in iMEFs, whereas a second esiRNA produced moderate cell death.
To test this possibility, we assayed the effect of KD of each of these 62 genes on the viability of a somatic cell type, immortal mouse embryonic fibroblasts (MEFs [iMEFs]). For 49 of the 62, KD had no apparent effect on viability (Table I, nonbold rows). We next tested 20 of the 62 genes using a second, nonoverlapping siRNA pool. The phenotypes observed in the initial set of KDs were highly reproducible: 19 of 20 had the same iMEF KD phenotype (or lack of phenotype) as the corresponding initial siRNA pool (Table I). For several candidates with ES cell–specific KD phenotypes, the lack of any proliferation defect upon KD was confirmed in two additional somatic cell types, primary MEFs and immortal mouse mammary epithelial cells (Fig. S1). These data suggest that a significant number of proteins that are essential for ES cell viability are either dispensable or are required at very low levels in somatic cells.
We focused further analyses on subunits of the Mcm, condensin, and Ints complexes, which function in three distinct but important cellular processes. KD of multiple subunits of each complex increased levels of apoptosis in ES cells (Fig. 1, A–C; and not depicted) but had no apparent effect on the viability of iMEFs (Fig. 1, D and E; and Table I). The Mcm2-7 helicase complex functions in DNA replication (Maiorano et al., 2006). All four Mcm genes in our siRNA library (Mcm2, -4, -6, and -7) showed ES cell–specific lethality (Fig. S1 and Table I). The condensin complexes function in chromosome compaction during mitosis and meiosis as well as in the regulation of gene expression (Losada and Hirano, 2005; Belmont, 2006). KD of Smc2 and -4, core subunits of both condensin complexes, also exhibited ES cell–specific lethality (Table I). The Ints complex is necessary for the 3′ end processing of U1 and U2 snRNAs. Although Ints4 was the sole member of the Ints complex included in the RNAi screen, we found that depletion of two additional Ints subunits, Ints1 and -11, also caused cell death in ES cells but not somatic cells (Fig. 1, A–E; and Fig. S1). In addition, we found no significant increase in apoptotic (TUNEL+) cells in Smc2, Mcm2, or Ints1 KD iMEFs (Fig. 1 D). Consistent with these results, KD of MCM7 (Cortez et al., 2004), MCM4 (Sakwe et al., 2007), SMC2 (Ono et al., 2003), INTS1, or INTS11 (Baillat et al., 2005) was not reported to affect viability of somatic human cells.
Figure 1.
RNAi phenotypes of ES cells and iMEFs. (A) Proportion of TUNEL+ ES cells in KDs indicated. Means and standard deviations of triplicate samples are shown. (B) Colony morphology of ES cells depleted of the indicated proteins on day 4 of KD. Smc2, Mcm2, and Ints1 KDs exhibit a large number of rounded up and detached dead or dying cells. (C) Western blots for Smc2, Mcm, Ints1, and actin in ES cells depleted of the indicated proteins for 4 d. The arrow indicates Mcm2 in the pan-Mcm blot. (D) Morphology of iMEFs depleted of the indicated proteins. Cells were transfected on days 0, 2, 5, 8, and 11 and split back 1:8 on days 4, 7, and 10. Images were taken on day 11. Numbers in the bottom right corner represent the percentage of TUNEL+ cells in separate KD experiments. (E) Western blots for Smc2, Mcm, Ints1, and actin in iMEFs depleted of the indicated proteins at various points during the time course. “d” indicates length of KD in days. (C and E) The positions of molecular mass standards (in kilodaltons) are shown. (F) Proliferation of iMEFs depleted of the indicated proteins during the time course described in D. KD cells were split and counted at 3-d intervals. One of two independent experiments with comparable results is shown. Bars, 120 µm.
We considered two trivial explanations for the ES cell–specific lethality. First, KD may be significantly more effective in ES cells than somatic cells. However, when we compared the efficiency of KD of Smc2, Mcm2, and Ints1 in ES cells and iMEFs, we consistently observed comparable KD in both cell types for all three genes (Fig. 1, C and E; Fig. S1, D and E; and not depicted). Therefore, the ES cell–specific lethality is not caused by differences in KD efficiency between ES cells and somatic cells. Second, because ES cells proliferate more rapidly than most somatic cells (Liu et al., 2007), RNAi phenotypes that require several cell divisions to emerge may be evident in ES cells but not in iMEFs in the 4-d period of our initial screen. In this case, KD of some of these genes might be lethal in iMEFs if the KD is performed longer. To test this possibility, proliferation was assayed in Smc2, Mcm2, and Ints1 KD iMEFs over a period of 13 d (Fig. 1, D–F). During this time, iMEFs completed ∼17 divisions, significantly more than the approximately four divisions completed by ES cells in the 3 d required for the KD phenotype to become evident in our screen. Although Smc2 KD did cause a slight decrease in the proliferation rate (Fig. 1 F), we observed no viability defect in iMEFs during long-term Smc2, Mcm2, or Ints1 KD. This demonstrates that an increase in the number of cell divisions does not reveal a significant iMEF KD phenotype for these genes. These data suggest that although ES cells are dependent on proper expression of many proteins that control key cellular functions, somatic cells are not. However, 13 of 62 genes did exhibit moderate to severe viability defects upon KD in iMEFs (Table I), indicating that although some proteins are required at higher levels in ES cells relative to somatic cells, this requirement is not a general feature.
ES cells depleted of essential chromatin regulators accumulate DNA damage and undergo apoptosis
We next characterized the apoptotic cell death pathway for ES cells depleted of Smc2, Mcm2, or Ints1. In ES cells, the tumor suppressor p53 is required to activate apoptosis in response to several cellular stresses (Aladjem et al., 1998). Apoptotic triggers have been shown to stabilize p53 protein, which promotes expression of some proapoptotic genes (Roos and Kaina, 2006). We found that p53 levels were increased in response to KD of Smc2, Mcm2, or Ints1 in ES cells but not in primary MEFs (Fig. 2 A), suggesting that the different cellular stresses induced by depletion of each of these factors in ES cells may converge on a common pathway that functions through p53. The transcriptional regulatory activity of p53 is regulated by several posttranslational modifications, including phosphorylation of residues within its amino-terminal transcriptional activation domain (Lakin and Jackson, 1999). We observed increased levels of p53 phosphorylated on serines 15 and 20 upon KD of Smc2 or -4 in ES cells but not MEFs, indicating that p53 was activated upon condensin KD in ES cells alone (Fig. 2 B). Activation of p53 did not result in increased expression of p21 protein (Fig. 2 B), which is consistent with the lack of a G1 checkpoint in ES cells (Hong and Stambrook, 2004).
Figure 2.
ES cells depleted of regulators of essential processes accumulate DNA damage but exhibit distinct cell cycle phenotypes. (A, left) Western blots for p53, γ-H2AX, and actin in ES cells (ESCs) and primary MEFs depleted of the indicated proteins. (right) p53 protein levels with (+) or without (−) UV exposure. For all blots, actin levels are shown as a loading control. Asterisks denote background bands of unknown identity. (B) Western blots for p53, p53 phosphorylated on serine 15, p53 phosphorylated on serine 20, p21, and actin in ES cells and primary MEFs depleted of the indicated proteins. (A and B) The positions of molecular mass standards (in kilodaltons) are shown. (C) Cell cycle profiles of ES cells knocked down for the proteins indicated. Cell number is plotted on the vertical axis, and DNA content is plotted on the horizontal axis. The arrowhead denotes a small 8N population observed in Smc2 KD ES cells, and the dashed lines center on the 2N and 4N peaks in control cells. The percentages of cells in each phase of the cell cycle or exhibiting tetraploidy (8N) are shown to the right.
One common trigger for activation of the intrinsic apoptotic pathway by p53 is DNA damage. In response to DNA damage, histone H2AX is phosphorylated on serine 139 near the lesion (a mark referred to as γ-H2AX), which leads to recruitment of proteins that function in the DNA damage response and activation of p53 (Paull et al., 2000; Schultz et al., 2000; Rappold et al., 2001). In ES cells, small foci of γ-H2AX staining are commonly observed during normal ES cell proliferation, whereas large, brightly staining γ-H2AX foci accumulate upon DNA damage (Banáth et al., 2009). ES cells depleted of Smc2, Mcm2, or Ints1 had elevated levels of γ-H2AX (Fig. 2 A) and accumulated large foci of γ-H2AX staining (Fig. S2), suggesting that defects in chromosome condensation, DNA replication, and snRNA processing all result in DNA damage in ES cells, which triggers apoptosis. Levels of γ-H2AX were not significantly increased in primary MEFs or iMEFs depleted of these factors (Fig. 2 A and Fig. S2 D), which is consistent with the minimal effects of condensin, Mcm, or Ints KD on MEF proliferation.
Activation of the DNA damage response in somatic cells can lead to an arrest in the cell cycle until the damage is repaired. In ES cells depleted of Ints subunits, there was no notable alteration in the cell cycle (Fig. 2 C and Fig. S3). KD of Mcm subunits caused a slight shift from the S-phase to the G1-phase population (Fig. 2 C and Fig. S3), suggesting that they might have a slight defect in entry into S phase. Smc2 KD ES cells showed the most dramatic alteration in cell cycle profile: there was a notable increase in the fraction of G2/M cells (Fig. 2 C). Therefore, depletion of the condensin, Mcm, or Ints complexes each has a distinct effect on the ES cell cycle, which is consistent with each KD causing a different cellular stress, all of which converge on the DNA damage/apoptotic response. In all three KDs, apoptotic cells were equally distributed in all stages of the cell cycle (unpublished data). This is consistent with the lack of DNA damage checkpoints in ES cells (Aladjem et al., 1998).
Condensins I and II are redundantly required for ES cell viability
The two condensin complexes, condensin I and II, function by temporally and spatially distinct mechanisms during mitotic chromosome condensation (Ono et al., 2003; Hirota et al., 2004; Gerlich et al., 2006). In our screen, we identified Smc2 and -4, subunits common to both complexes, as factors essential for ES cell viability (Table I). Although siRNAs targeting subunits specific to condensin I (NcapD2) or II (NcapD3) were present in our siRNA library, we failed to identify those subunits in the screen, suggesting that the functions of condensin I and II may be redundant for ES cell viability. To determine whether the Smc2/Smc4 KD phenotype was a consequence of disruption of either condensin I or II or both of the condensin complexes, we determined the effect of depletion of NcapD2 and -D3 separately and in combination. Although depletion of either condensin complex alone had little effect on ES cell viability or cell cycle profile, simultaneous KD of both complexes had strong cell cycle and cell death phenotypes, similar to those of the common condensin subunit Smc2 (Fig. 3 and Fig. S3). These data suggest that the individual functions of the two condensin complexes are redundant for ES cell proliferation.
Figure 3.
Condensins I and II are redundantly required for ES cell proliferation. (A) Morphology of ES cell colonies depleted of NcapD2, NcapD3, both NcapD2 and -D3, or Smc2. (B) Proportion of apoptotic (TUNEL+) ES cells depleted of condensin complexes as in A. (C) Proportion of apoptotic (TUNEL+) E14 ES cells or iMEFs upon mitotic arrest by nocodazole (noc) treatment. (B and C) Means and standard deviations of triplicate samples are shown. Bars, 120 µm.
Condensins are required for the metaphase to anaphase transition in ES cells
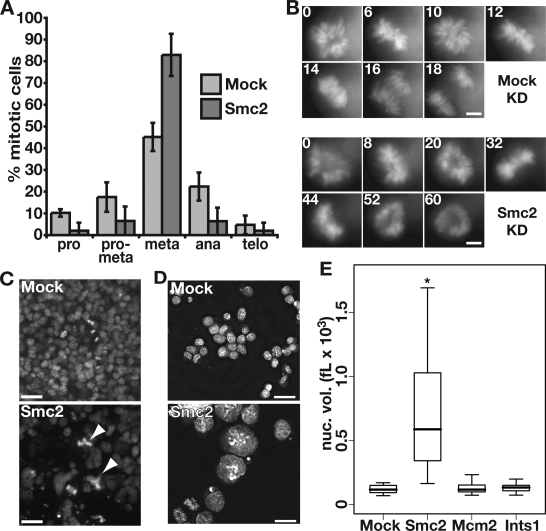
ES cells depleted of Smc2 or both NcapD2 and -D3 exhibited a large increase in G2/M (4N) cells (Fig. 2 C and Fig. S3), suggesting a defect in mitotic progression. The mitotic index of Smc2-depleted ES cells (13.2 ± 2%) was higher than that of mock-depleted ES cells (3.8 ± 1%), which is consistent with a block in mitosis. We next determined whether depletion of condensins blocked cells at any specific stage of mitosis. In control KD experiments, metaphase cells were most abundant (45% of mitotic cells), with moderate fractions of prophase, prometaphase, anaphase, and telophase cells (Fig. 4 A). In contrast, the vast majority of mitotic cells depleted of Smc2 were in metaphase (83%), with very few cells in each of the other mitotic phases. These data suggest that depletion of condensin I and II leads to a block or severe delay at metaphase in ES cells.
Figure 4.
Condensin-depleted ES cells arrest at metaphase and accumulate large, misshapen interphase nuclei. (A) Fractions of mitotic cells in each stage of mitosis in mock- or Smc2-depleted ES cells. Live cells expressing H2A-GFP were counted and scored for each phase of mitosis. Means and standard deviations of triplicate samples are shown. (B) Representative time-lapse live cell imaging of mock- or Smc2-depleted H2A-GFP ES cells starting in metaphase and imaged at the times (in minutes) indicated in each box. ES cells grow in clusters and rotate within the cluster, resulting in different views of the metaphase plate during the time lapse. (C) Fields of H2A-GFP–expressing ES cells either mock or Smc2 depleted for 4 d. White arrowheads denote multipolar mitoses. (D) ES cells either mock or Smc2 depleted for 4 d were fixed, cytospun onto slides, and DAPI stained, and 1-µm z sections were captured. Deconvolved z sections at the midpoint of each stack are shown. (E) Box plot of the nuclear volumes (nuc. vol.) in thousands of femtoliters (fL × 103) of 10–20 ES cell nuclei for each KD indicated. The top and bottom edges of each box represent the 75th and 25th percentiles, respectively, whereas the bold line represents the median of each group. The asterisk indicates a statistically significant difference between the nuclear volumes of mock and Smc2 KD cells (P < 2 × 10−5; Student’s t test). Bars: (B) 2.5 µm; (C) 30 µm; (D) 10 µm.
To further analyze this mitotic block, we performed time-lapse imaging on live mock- or Smc2-depleted ES cells expressing GFP-tagged histone H2A (H2A-GFP). We identified metaphase cells and imaged them for 30 min. Six of eight mock-depleted ES cells initiated or completed anaphase within this time frame (Fig. 4 B and Video 1), which is consistent with previous measurements of mitotic timing in ES cells (Hadjantonakis and Papaioannou, 2004). In contrast, only 1 of 16 Smc2-depleted ES cells proceeded to anaphase during the 30-min interval (Fig. 4 B and Video 2). Smc2-depleted cells imaged starting from prophase exhibited rapid chromosome condensation and formation of a metaphase plate on a similar time scale to that of normal ES cells (Video 2 and not depicted), after which cells remained in metaphase for the duration of imaging. Furthermore, during 3-h time-lapse experiments, Smc2-depleted ES cells exhibited mitotic arrest for the duration of the experiments (unpublished data). These data indicate that although condensin-depleted ES cells are competent for chromosome condensation and formation of a metaphase plate, they are defective in the initiation of anaphase. We did not observe any effect on formation of the mitotic spindle in condensin-depleted ES cells (unpublished data), suggesting that the metaphase block occurs downstream of spindle formation. A small fraction of Smc2 KD cells appeared to escape mitotic arrest without completing mitosis and likely rereplicated their genome, as suggested by the presence of a minor 8N population (Fig. 2 C). Consistent with this interpretation, Smc2 KD ES cells exhibited aberrant mitoses with multipolar mitotic figures at low frequency (Fig. 4 C). These phenotypes are distinct from the catastrophic anaphase defect in Smc2-null chicken DT40 cells (Hudson et al., 2003) and the brief mitotic delay and perturbed anaphase phenotype of somatic mammalian cells depleted of SMC2 (Ono et al., 2003; Hirota et al., 2004). Thus, the condensins appear to play a unique role in promoting the metaphase to anaphase transition in ES cells.
Condensins are required for normal interphase chromatin organization in ES cells
In addition to their defect in mitotic progression, Smc2 KD ES cells exhibited a high proportion of enlarged and misshapen interphase nuclei (Fig. 4, C–E). We considered the possibility that this phenotype could reflect tetraploid cells that arise when KD ES cells reenter the cell cycle without completing mitosis. However, the 4.9% of 8N ES cells (corresponding to tetraploid cells in G2/M) that accumulated upon Smc2 KD (Fig. 2 C) was too low for tetraploidy to explain the large proportion of cells with this phenotype (52% of Smc2 KD ES cells had nuclear volumes at or above the 95th percentile of control KD ES cells; Fig. 4, C–E).
An alternative explanation is that the enlarged and misshapen nuclei indicate a role for condensin complexes in interphase chromatin compaction. In somatic cells, condensin I, which constitutes the majority of the condensin complexes in the cell, is excluded from the nucleus until nuclear envelope breakdown (Hirota et al., 2004; Ono et al., 2004). Although condensin II–specific subunits are localized to the nucleus throughout interphase in somatic cells (Hirota et al., 2004; Ono et al., 2004), most of the Smc2/Smc4 catalytic ATPase is excluded from the nucleus until mitotic prophase (Schmiesing et al., 2000; Hirota et al., 2004), and the Smc2/Smc4 ATPase activity is not strongly activated until mitosis (Kimura et al., 2001; Takemoto et al., 2004). We speculated that if condensin complexes function during interphase in ES cells to maintain chromatin compaction, condensin proteins may be more highly expressed in ES cells than differentiated cells and that a portion of condensin complexes may be nuclear localized during interphase in ES cells. Indeed, we observed high levels of Smc4 in both the cytoplasm and nucleus in interphase ES cells (Fig. S4 A). Furthermore, we found that Smc2 and -4 were expressed at higher levels overall in ES cells than MEFs (Fig. S1 D). These data are consistent with a role for condensin in interphase chromatin organization in ES cells.
We next examined whether the enlarged nuclei were caused by an alteration in chromatin organization in condensin KD ES cells. We reasoned that if higher-order chromatin folding was perturbed in condensin KD cells, the spatial relationships between adjacent loci on individual chromosomes might be altered in condensin KD ES cells relative to control cells. To test this possibility, we performed two color DNA-FISH to detect two loci, Pgk1 and Mecp2, which lie ∼30 Mb apart on the X chromosome, in control or condensin KD male ES cells. Both loci gave a single pinpoint or two closely spaced pinpoint (doublet) FISH signals in control KD ES cells, corresponding to cells before or after replication of these loci, respectively. In contrast, the Pgk1 and Mecp2 signals in condensin KD cells were fragmented and spread out over a larger portion of the nucleus (Fig. 5 A). This increase in the area occupied by FISH signals was not unique to X-linked genes and was also observed for two autosomal genes, Fn1 and OR2-1 (Fig. 5 B). We did not observe enlarged or misshapen nuclei in iMEFs depleted of Smc2 (Fig. S5), suggesting that the interphase function of condensin complexes in chromatin compaction is ES cell specific. Together, these data reveal a role for condensin complexes in higher-order chromatin organization during interphase in ES cells.
Figure 5.
Condensin KD results in interphase chromatin decondensation. (A) DNA-FISH for Mecp2 and Pgk1 in control (mock) or condensin KD (Smc2 and Smc4 KD) E14 ES cells. The merged panel (right) consists of Mecp2 (red), Pgk1 (green), and DAPI (blue). (B) Area occupied by Mecp2, Pgk1, Fn1, or OR2-1 loci in control and condensin KD ES cells, as measured by FISH. The maximum area occupied by each of the four loci was plotted against nuclear diameter for 10–20 cells per locus for control (mock) and condensin KD E14 ES cells. Bars, 5 µm.
ES cells contain condensin-dependent interphase chromatin domains marked by H3S10P
We next examined whether condensin KD affected other aspects of chromatin organization by assessing the distribution of modified histones and DNA in control and condensin KD ES cells. We used immunostaining to assess markers of heterochromatin (H3K9me3, macroH2A, H4K20me1, H3K27me3, and 5-methylcytosine [5meC]), euchromatin (H3K4me3 and H4ac), and mitotic chromatin (H3S10P and H3S28P).
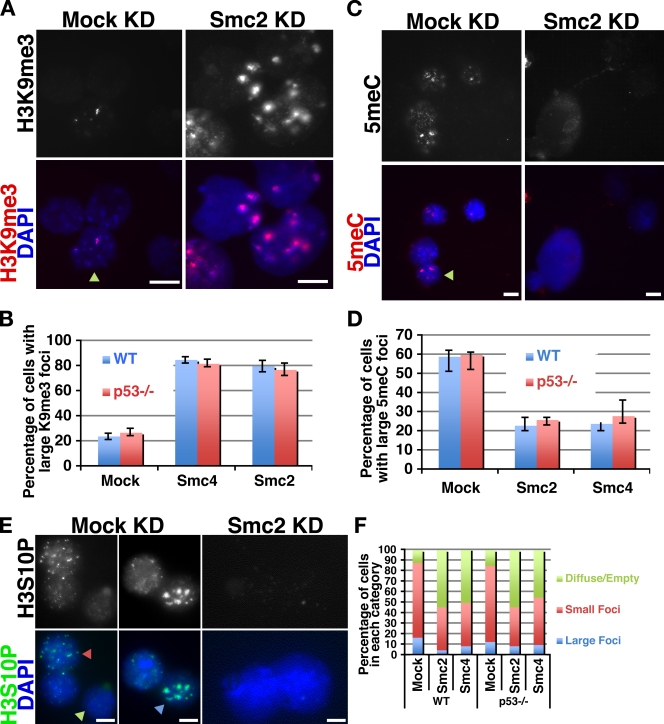
The majority of the epigenetic modifications assayed showed no change in distribution between control and condensin KD cells (Fig. S4 and not depicted). However, the staining patterns of H3S10P, H3K9me3, and 5meC were altered upon condensin KD. H3K9me3 is highly enriched on pericentric heterochromatin in somatic cells but does not exhibit this marked enrichment in many ES cell lines (Meshorer et al., 2006; Gaspar-Maia et al., 2009). Upon Smc2 KD, there was a significant increase in the proportion of cells with large foci of H3K9me3 that overlapped with pericentric heterochromatin (Fig. 6, A and B). This increase in H3K9me3 staining suggested the possibility that condensin KD may cause ES cells to differentiate. However, the only notable change in the gene expression profile in condensin KD ES cells was increased expression of some apoptotic genes (unpublished data), indicating that KD was not triggering differentiation.
Figure 6.
Alterations in chromatin modifications in condensin KD ES cells. (A) H3K9me3 localization in control (mock KD) and condensin KD (Smc2 KD) E14 ES cells, as shown by immunofluorescence. The arrowhead indicates a cell with large pericentric heterochromatin H3K9me3 foci. (B) Quantification of control and condensin KD E14 (WT) and p53−/− ES cells with large foci of H3K9me3 staining. Means and standard deviations of three replicate experiments are shown. (C) 5meC in control (mock KD) and condensin KD (Smc2 KD) E14 ES cells. The arrowhead indicates a cell with large 5meC foci. (D) Quantification of 5meC staining, as in B. (E) H3S10P localization in control and condensin KD E14 ES cells. The green arrowhead indicates a cell with diffuse or weak H3S10P interphase staining. The red arrowhead indicates an interphase cell with small H3S10P foci (a pattern similar to that is transiently seen when somatic cells are serum stimulated). The blue arrowhead indicates an interphase cell with large H3S10P foci at pericentric heterochromatin. (F) Percentage of E14 (WT) and p53−/− ES cells in each category indicated on the right. Bars, 5 µm.
5meC also marked large foci in ES cells; however, these foci did not overlap with pericentric heterochromatin and tended to be localized to the nuclear periphery. In contrast to H3K9me3, there was an overall decrease in 5meC staining in condensin KD ES cells (Fig. 6, C and D). In control ES cells, H3S10P was present in both interphase and mitotic nuclei (Fig. 6 E and not depicted). The interphase staining patterns we observed were similar to H3S10P enrichment on pericentric heterochromatin during late S and G2 phases of the somatic cell cycle (Hendzel et al., 1997) as well as focal enrichment transiently seen in somatic cells upon stimulation by growth factors or cytokines (Dormann et al., 2006). In contrast, condensin KD ES cells were significantly depleted of H3S10P staining within interphase nuclei, whereas the stronger H3S10P staining of mitotic chromosomes was maintained (Fig. 6 E and not depicted). We observed relatively normal distributions of all other epigenetic marks assayed (Fig. S4 and not depicted), suggesting that the loss of H3S10P and 5meC staining was not simply caused by dilution of signal as the chromatin volume expanded.
Phosphorylation of H3S10 by the Aurora B kinase is necessary for binding of condensin I to mitotic chromosomes (Hagstrom et al., 2002; Kaitna et al., 2002; Lipp et al., 2007; Takemoto et al., 2007), suggesting a link between interphase chromatin domains marked by H3S10P and condensin complexes. However, the observation that mitotic H3S10P staining, which is deposited by Aurora B, was unaffected by condensin KD suggested that interphase H3S10P-marked chromatin domains were Aurora B independent. When we examined the staining of H3S28P, which is also catalyzed by Aurora B, in control and condensin KD ES cells, we observed H3S28P staining only in mitotic cells, and this staining was unaffected by condensin KD (Fig. S4). This finding is consistent with the hypothesis that interphase H3S10P domains in ES cells are established independently of Aurora B. Together, these data suggest that, in interphase ES cells, both higher-order chromatin folding and the maintenance of specific chromatin domains depend on the function of the condensin complexes.
p53 is required for induction of apoptosis but not disruption of chromatin structure in condensin KD ES cells
Because p53 is induced upon condensin KD in wild-type ES cells, we tested whether p53 was necessary for the several ES cell–specific phenotypes we observed in condensin KD cells. To test this possibility, we knocked down Smc2 or -4 in p53−/− ES cells (Aladjem et al., 1998) and determined mitotic index, interphase nuclear size, the presence of epigenetic marks H3S10P, H3K9me3, and 5meC, and the proportion of apoptotic cells. Similar to the phenotypes we observed in wild-type ES cells, condensin KD in p53−/− ES cells resulted in a metaphase block, enlargement of interphase nuclei, disruption of H3S10P and 5meC, and an increase in the proportion of cells with pericentric foci of H3K9me3 (Fig. 6, B, D, and F; Fig. S4, E–G; and not depicted). In contrast, we observed no increase in apoptotic cells upon condensin KD in p53−/− ES cells compared with controls (Fig. S4 H). Therefore, p53 is required for induction of apoptosis upon condensin KD but is dispensable for alteration of chromatin structure and chromatin decompaction.
Discussion
In this study, we have described a group of proteins required for ES cell viability that are largely dispensable for proliferation of somatic cells. Most of these proteins are widely expressed and function in essential processes such as DNA replication, RNA processing, or chromosome condensation. This raises the question of how somatic cells can survive upon depletion of these factors, in most cases with no obvious phenotype. One possibility is that somatic cells require only a small fraction of their normal steady-state levels of these proteins. In contrast, ES cells depleted of proteins involved in several different essential processes accumulate DNA damage, up-regulate or stabilize p53 protein, and undergo apoptosis. There may be an evolutionary advantage to such sensitivity, as damaged cells would be eliminated before they have a chance to contribute to the developing embryo.
In addition, we describe two unusual phenotypes resulting from condensin KD that appear to be unique to ES cells. A portion of condensin KD ES cells exhibit prolonged metaphase arrest. This phenotype may be unique to ES cells because somatic cells depleted of condensin typically exhibit a slight delay in the initiation and completion of anaphase but, nonetheless, are able to complete mitosis and proceed through subsequent rounds of cell division (Hirota et al., 2004). Therefore, the metaphase arrest in condensin KD ES cells may simply be a more severe manifestation of the same phenotype. Second, many ES cells depleted of condensins exhibit enlarged and misshapen interphase nuclei. The vast majority of these cells have normal G1 (2N) or G2/M (4N) DNA content, indicating that the nuclear enlargement phenotype is not simply a consequence of increased ploidy. Close inspection of individual loci using DNA-FISH analysis revealed that chromatin is less compact in condensin KD ES cells than controls. Furthermore, we found that three epigenetic marks, H3K9me3, H3S10P, and 5meC, were altered in interphase nuclei of condensin-depleted ES cells.
The mechanism by which condensin KD leads to p53-dependent apoptosis is unclear. Although there were significant numbers of ES cells exhibiting either mitotic arrest or chromatin decompaction during interphase, it is not clear whether these phenotypes directly lead to DNA damage and induction of apoptosis. Alternatively, condensin KD may indirectly lead to apoptotic death by sensitizing ES cells to naturally occurring cellular stresses such as DNA replication or oxidative damage.
How might condensins function to regulate interphase chromatin folding? Interphase decompaction may be a cause or consequence of the epigenetic alterations in condensin KD ES cells or may be unrelated to these alterations. The decrease in DNA methylation in condensin KD cells is unlikely to be the sole cause of the increased nuclear volume, as DNA methyltransferase 1 mutant ES cells are normal in size (Panning and Jaenisch, 1996). It may be that interphase decompaction alters the accessibility of chromatin for some modifying enzymes. In addition, several chromatin-modifying activities are regulated in a cell cycle–dependent fashion (Probst et al., 2009), raising the possibility that the altered cell cycle in condensin KD ES cells contributes to the changes in epigenetic marks. H3S10P and H3K9me3 are antagonistic marks (Nowak and Corces, 2004; Dormann et al., 2006), which may explain why H3S10P decreases while H3K9me3 increases upon condensin KD. In mitotic cells, H3S10P is necessary for the condensin complex to bind chromatin. Therefore, our observation that H3S10P is lost in interphase nuclei upon condensin KD raises the possibility that condensin binding and phosphorylation of H3S10 are mutually reinforcing events. Future work examining the roles of H3S10P and H3K9me3 in the maintenance of interphase chromatin compaction, as well as examination of the effects of condensin depletion on various levels of chromatin higher-order folding during interphase should help to answer these questions.
Why are condensin complexes necessary to maintain interphase chromatin folding only in ES cells? Major architectural chromatin proteins bind more loosely to chromatin in ES cells (Meshorer et al., 2006). Furthermore, in ES cells, a much smaller fraction of the genome is packaged into heterochromatin. Therefore, it is possible that ES cells lack some of the mechanisms of regulating interphase chromatin folding that are used by somatic cells and have co-opted condensin complexes for this purpose. In this model, condensin complexes might function similarly during interphase and mitosis, but their localization or activity may be regulated differently in ES cells compared with somatic cells.
In sum, our data suggest that ES cells and somatic cells may exhibit significant differences in the regulation of fundamental cellular processes. Understanding the basis of these differences will be useful for the potential therapeutic applications of ES cells. Furthermore, it will be useful to determine whether additional stem cell types have similar sensitivities to perturbations of core cellular processes. For example, if cancer stem cells were found to be similar to ES cells in this regard, some of these proteins might make useful targets for novel drug therapies. In addition, the unique role for condensin complexes in interphase chromatin folding adds to a growing list of differences in chromatin structure regulation between ES cells and somatic cells. The unique features of chromatin regulation may be necessary both for their ability to self-renew and remain pluripotent as well as their rapid proliferation and unique cell cycle.
Materials and methods
Cells and antibodies
Mouse ES cells used in this study were E14 (Hooper et al., 1987) and E14 H2A-GFP (Fazzio et al., 2008). Spontaneously immortalized fibroblasts were obtained by serial passaging, following a 3T3 protocol. Primary MEFs were isolated from d 14.5 mouse embryos using standard methods. Immortal mouse mammary epithelial cells (X3) were described previously (Silver et al., 2007). Antibodies used in this study were Smc2 (Abcam), Smc4 (LifeSpan BioSciences), pan-MCM (BD), Ints1 (Abgent), actin (Sigma-Aldrich), p53 (Vector Laboratories), γ-H2AX (LP Bio), H3S10P (Abcam and Millipore), H3K9me3 (Abcam), H3K27me3 (Abcam), H4ac (Active Motif), H4K20me1 (Abcam), 5meC (EMD), and H3S28P (Millipore).
RNAi
RNAi was performed using RNase III–digested double-stranded RNAs (esiRNAs; Yang et al., 2002) transfected using Lipofectamine 2000 for ES cells or Lipofectamine RNAiMAX (Invitrogen) for somatic cells as described previously (Fazzio et al., 2008). D. Yang and J.M. Bishop (University of California, San Francisco, San Francisco, CA) provided the RNase III expression vector.
Cell death, cell cycle, and mitotic index analysis
TUNEL staining was performed as described previously (Darzynkiewicz and Juan, 1997) and measured using FACS. Cell cycle profiling of propidium iodide–stained cells was performed as described previously (Benanti et al., 2002). For mitotic index calculations, cells were fixed with 4% paraformaldehyde and stained with the mitotic marker H3S28P.
DNA-FISH and immunofluorescence
FISH was performed as described previously (Mlynarczyk-Evans et al., 2006) using directly labeled bacterial artificial chromosomes containing Mecp2 (CHORI RP23-77L16), Pgk1 (CHORI R24-90H17), Fn1 (CHORI RP23-27O4), or OR2-1 (an array of odorant receptors on chromosome 2; CHORI RP23-52P17) sequences as probes. Immunostaining was performed as described previously (Nusinow et al., 2007). Before 5meC immunostaining, DNA was denatured by a 30-min incubation in 2 N HCl. For both FISH and immunostaining, nuclei were stained with DAPI. For immunofluorescence experiments, we used Cy3- and/or FITC-labeled secondary antibodies. FISH probes were directly labeled with Cy3-dCTP or FITC-dUTP.
Imaging
Brightfield and H2A-GFP still images of live cells in growth media were captured on an inverted microscope (DM 1RB; Leica) at room temperature using a 10× NA 0.25 air lens. Images were captured with a charge-coupled device (CCD) camera (ORCA AG; Hamamatsu Photonics) using Volocity software (PerkinElmer). Time-lapse imaging on live H2A-GFP ES cells, enclosed within a 37°C incubation chamber supplied with 5% CO2, was performed on an inverted microscope (TE2000E; Nikon) with Perfect Focus using a 20× NA 0.75 air lens. Images were captured on a CCD camera (CoolSNAP HQ; Photometrics) using NIC Elements software (Nikon). All immunofluorescence, FISH, and nuclear volume measurements were performed at room temperature on paraformaldehyde-fixed cells mounted on slides with Vectashield (Vector Laboratories). For nuclear volume calculations, z series (1-µm sections) of DAPI-stained nuclei were gathered using a microscope (IX70; Olympus) equipped with a 20× NA 0.75 air lens. Images were captured with a CoolSNAP HQ CCD camera. The stage and filters were controlled by the program SoftWoRx (DeltaVision). Image processing was performed using this program’s deconvolution feature. Nuclear volumes were obtained from polygon summation calculated from z series using the EditPolygon function in SoftWoRx. FISH and immunostaining images were captured using a fluorescent microscope (BX60; Olympus) with a 100× NA 1.30 oil immersion objective equipped with a digital camera (ORCA-ER; Hamamatsu Photonics) and Openlab 4.0.1 software (PerkinElmer). FISH signal areas and nuclear diameters were determined from images captured using both of the aforementioned systems and calculated using Openlab 4.0.1 or SoftWoRx. Photoshop (Adobe) was used for linear adjustments (identical for all images) of brightfield images, pseudocoloring of fluorescent images, and cropping of all images. The Photoshop Levels tool was used to adjust the upper and lower input levels for each channel to match the upper and lower boundaries of the image histogram. No gamma adjustments were made.
Online supplemental material
Fig. S1 shows the KD phenotypes of several genes in primary MEFs and a mouse epithelial cell line, X3, along with expression levels of condensins in control and KD cells. Fig. S2 shows the pattern of γ-H2AX staining in ES cells and the frequency of γ-H2AX staining in ES cells and MEFs. Fig. S3 shows that similar phenotypes are exhibited upon KD of different subunits of the same essential chromatin regulatory complexes in ES cells. Fig. S4 illustrates histone modifications that remain unchanged by condensin depletion in ES cells as well as histone modifications that change upon condensin KD in p53−/− ES cells. Fig. S5 shows that histone modifications that are altered in ES cells upon condensin KD are unchanged when condensin is knocked down in iMEFs. Videos 1 and 2 show live cell imaging of control and Smc2 KD ES cells, respectively, in interphase and mitosis. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200908026/DC1.
Acknowledgments
We thank J. Benanti, M. McCleland, P. O’Farrell, J. Huff, M. Royce-Tolland, K. Worringer, and F. Chu for comments on the manuscript and helpful discussions, B. Habermann, J. Benanti, M. McCleland, and D. Nusinow for technical advice, A. Shermoen, M. McCleland, J. Fung, and K. Thorn for help with imaging, and D. Yang and J.M. Bishop for the RNase III expression vector. We also thank J.M. Bishop, in whose laboratory some of these experiments were performed. Live cell imaging data were acquired at the Nikon Imaging Center at the University of California, San Francisco (UCSF).
This work was supported by grants from the National Institutes of Health (NIH; RO1 GM63671 and R01 GM085186). T.G. Fazzio was supported by the Jane Coffin Childs Memorial Fund for Medical Research, a National Cancer Institute training grant (T32CA09043) to UCSF, an NIH grant (1K99CA140854-01), and the G.W. Hooper Research Foundation.
Footnotes
Abbreviations used in this paper:
- 5meC
- 5-methylcytosine
- CCD
- charge-coupled device
- ES
- embryonic stem
- iMEF
- immortalized MEF
- KD
- knockdown
- MEF
- mouse embryonic fibroblast
- snRNA
- small nuclear RNA
References
- Aladjem M.I., Spike B.T., Rodewald L.W., Hope T.J., Klemm M., Jaenisch R., Wahl G.M. 1998. ES cells do not activate p53-dependent stress responses and undergo p53-independent apoptosis in response to DNA damage. Curr. Biol. 8:145–155 10.1016/S0960-9822(98)70061-2 [DOI] [PubMed] [Google Scholar]
- Baillat D., Hakimi M.A., Näär A.M., Shilatifard A., Cooch N., Shiekhattar R. 2005. Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repeat of RNA polymerase II. Cell. 123:265–276 10.1016/j.cell.2005.08.019 [DOI] [PubMed] [Google Scholar]
- Banáth J.P., Bañuelos C.A., Klokov D., MacPhail S.M., Lansdorp P.M., Olive P.L. 2009. Explanation for excessive DNA single-strand breaks and endogenous repair foci in pluripotent mouse embryonic stem cells. Exp. Cell Res. 315:1505–1520 10.1016/j.yexcr.2008.12.007 [DOI] [PubMed] [Google Scholar]
- Belmont A.S. 2006. Mitotic chromosome structure and condensation. Curr. Opin. Cell Biol. 18:632–638 10.1016/j.ceb.2006.09.007 [DOI] [PubMed] [Google Scholar]
- Benanti J.A., Williams D.K., Robinson K.L., Ozer H.L., Galloway D.A. 2002. Induction of extracellular matrix-remodeling genes by the senescence-associated protein APA-1. Mol. Cell. Biol. 22:7385–7397 10.1128/MCB.22.21.7385-7397.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez D., Glick G., Elledge S.J. 2004. Minichromosome maintenance proteins are direct targets of the ATM and ATR checkpoint kinases. Proc. Natl. Acad. Sci. USA. 101:10078–10083 10.1073/pnas.0403410101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dard N., Breuer M., Maro B., Louvet-Vallée S. 2008. Morphogenesis of the mammalian blastocyst. Mol. Cell. Endocrinol. 282:70–77 10.1016/j.mce.2007.11.004 [DOI] [PubMed] [Google Scholar]
- Darzynkiewicz Z., Juan G. 1997. Analysis of DNA Content and DNA Strand Breaks for Detection of Apoptotic Cells. In Current Protocols in Cytometry Robinson J.P., Darzynkiewicz Z., Hoffman R., Nolan J., Orfao A., Rabinovitch P.S., Watkins S., John Wiley and Sons, Inc., Hoboken, NJ: 7.4.1–7.4.8 [Google Scholar]
- Dormann H.L., Tseng B.S., Allis C.D., Funabiki H., Fischle W. 2006. Dynamic regulation of effector protein binding to histone modifications: the biology of HP1 switching. Cell Cycle. 5:2842–2851 [DOI] [PubMed] [Google Scholar]
- Evans M.J., Kaufman M.H. 1981. Establishment in culture of pluripotential cells from mouse embryos. Nature. 292:154–156 10.1038/292154a0 [DOI] [PubMed] [Google Scholar]
- Farhana L., Dawson M.I., Leid M., Wang L., Moore D.D., Liu G., Xia Z., Fontana J.A. 2007. Adamantyl-substituted retinoid-related molecules bind small heterodimer partner and modulate the Sin3A repressor. Cancer Res. 67:318–325 10.1158/0008-5472.CAN-06-2164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazzio T.G., Huff J.T., Panning B. 2008. An RNAi screen of chromatin proteins identifies Tip60-p400 as a regulator of embryonic stem cell identity. Cell. 134:162–174 10.1016/j.cell.2008.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar-Maia A., Alajem A., Polesso F., Sridharan R., Mason M.J., Heidersbach A., Ramalho-Santos J., McManus M.T., Plath K., Meshorer E., Ramalho-Santos M. 2009. Chd1 regulates open chromatin and pluripotency of embryonic stem cells. Nature. 460:863–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlich D., Hirota T., Koch B., Peters J.M., Ellenberg J. 2006. Condensin I stabilizes chromosomes mechanically through a dynamic interaction in live cells. Curr. Biol. 16:333–344 10.1016/j.cub.2005.12.040 [DOI] [PubMed] [Google Scholar]
- Hadjantonakis A.K., Papaioannou V.E. 2004. Dynamic in vivo imaging and cell tracking using a histone fluorescent protein fusion in mice. BMC Biotechnol. 4:33 10.1186/1472-6750-4-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagstrom K.A., Holmes V.F., Cozzarelli N.R., Meyer B.J. 2002. C. elegans condensin promotes mitotic chromosome architecture, centromere organization, and sister chromatid segregation during mitosis and meiosis. Genes Dev. 16:729–742 10.1101/gad.968302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendzel M.J., Wei Y., Mancini M.A., Van Hooser A., Ranalli T., Brinkley B.R., Bazett-Jones D.P., Allis C.D. 1997. Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma. 106:348–360 10.1007/s004120050256 [DOI] [PubMed] [Google Scholar]
- Hirota T., Gerlich D., Koch B., Ellenberg J., Peters J.M. 2004. Distinct functions of condensin I and II in mitotic chromosome assembly. J. Cell Sci. 117:6435–6445 10.1242/jcs.01604 [DOI] [PubMed] [Google Scholar]
- Hong Y., Stambrook P.J. 2004. Restoration of an absent G1 arrest and protection from apoptosis in embryonic stem cells after ionizing radiation. Proc. Natl. Acad. Sci. USA. 101:14443–14448 10.1073/pnas.0401346101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Y., Cervantes R.B., Tichy E., Tischfield J.A., Stambrook P.J. 2007. Protecting genomic integrity in somatic cells and embryonic stem cells. Mutat. Res. 614:48–55 [DOI] [PubMed] [Google Scholar]
- Hooper M., Hardy K., Handyside A., Hunter S., Monk M. 1987. HPRT-deficient (Lesch-Nyhan) mouse embryos derived from germline colonization by cultured cells. Nature. 326:292–295 10.1038/326292a0 [DOI] [PubMed] [Google Scholar]
- Hudson D.F., Vagnarelli P., Gassmann R., Earnshaw W.C. 2003. Condensin is required for nonhistone protein assembly and structural integrity of vertebrate mitotic chromosomes. Dev. Cell. 5:323–336 10.1016/S1534-5807(03)00199-0 [DOI] [PubMed] [Google Scholar]
- Ivanovska I., Orr-Weaver T.L. 2006. Histone modifications and the chromatin scaffold for meiotic chromosome architecture. Cell Cycle. 5:2064–2071 [DOI] [PubMed] [Google Scholar]
- Kaitna S., Pasierbek P., Jantsch M., Loidl J., Glotzer M. 2002. The aurora B kinase AIR-2 regulates kinetochores during mitosis and is required for separation of homologous chromosomes during meiosis. Curr. Biol. 12:798–812 10.1016/S0960-9822(02)00820-5 [DOI] [PubMed] [Google Scholar]
- Kimura K., Cuvier O., Hirano T. 2001. Chromosome condensation by a human condensin complex in Xenopus egg extracts. J. Biol. Chem. 276:5417–5420 10.1074/jbc.C000873200 [DOI] [PubMed] [Google Scholar]
- Lakin N.D., Jackson S.P. 1999. Regulation of p53 in response to DNA damage. Oncogene. 18:7644–7655 10.1038/sj.onc.1203015 [DOI] [PubMed] [Google Scholar]
- Lipp J.J., Hirota T., Poser I., Peters J.M. 2007. Aurora B controls the association of condensin I but not condensin II with mitotic chromosomes. J. Cell Sci. 120:1245–1255 10.1242/jcs.03425 [DOI] [PubMed] [Google Scholar]
- Liu N., Lu M., Tian X., Han Z. 2007. Molecular mechanisms involved in self-renewal and pluripotency of embryonic stem cells. J. Cell. Physiol. 211:279–286 10.1002/jcp.20978 [DOI] [PubMed] [Google Scholar]
- Losada A., Hirano T. 2005. Dynamic molecular linkers of the genome: the first decade of SMC proteins. Genes Dev. 19:1269–1287 10.1101/gad.1320505 [DOI] [PubMed] [Google Scholar]
- Maiorano D., Lutzmann M., Méchali M. 2006. MCM proteins and DNA replication. Curr. Opin. Cell Biol. 18:130–136 10.1016/j.ceb.2006.02.006 [DOI] [PubMed] [Google Scholar]
- Martin G.R. 1981. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl. Acad. Sci. USA. 78:7634–7638 10.1073/pnas.78.12.7634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meshorer E., Misteli T. 2006. Chromatin in pluripotent embryonic stem cells and differentiation. Nat. Rev. Mol. Cell Biol. 7:540–546 10.1038/nrm1938 [DOI] [PubMed] [Google Scholar]
- Meshorer E., Yellajoshula D., George E., Scambler P.J., Brown D.T., Misteli T. 2006. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev. Cell. 10:105–116 10.1016/j.devcel.2005.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlynarczyk-Evans S., Royce-Tolland M., Alexander M.K., Andersen A.A., Kalantry S., Gribnau J., Panning B. 2006. X chromosomes alternate between two states prior to random X-inactivation. PLoS Biol. 4:e159 10.1371/journal.pbio.0040159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niida H., Nakanishi M. 2006. DNA damage checkpoints in mammals. Mutagenesis. 21:3–9 10.1093/mutage/gei063 [DOI] [PubMed] [Google Scholar]
- Nowak S.J., Corces V.G. 2004. Phosphorylation of histone H3: a balancing act between chromosome condensation and transcriptional activation. Trends Genet. 20:214–220 10.1016/j.tig.2004.02.007 [DOI] [PubMed] [Google Scholar]
- Nusinow D.A., Sharp J.A., Morris A., Salas S., Plath K., Panning B. 2007. The histone domain of macroH2A1 contains several dispersed elements that are each sufficient to direct enrichment on the inactive X chromosome. J. Mol. Biol. 371:11–18 10.1016/j.jmb.2007.05.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono T., Losada A., Hirano M., Myers M.P., Neuwald A.F., Hirano T. 2003. Differential contributions of condensin I and condensin II to mitotic chromosome architecture in vertebrate cells. Cell. 115:109–121 10.1016/S0092-8674(03)00724-4 [DOI] [PubMed] [Google Scholar]
- Ono T., Fang Y., Spector D.L., Hirano T. 2004. Spatial and temporal regulation of Condensins I and II in mitotic chromosome assembly in human cells. Mol. Biol. Cell. 15:3296–3308 10.1091/mbc.E04-03-0242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panning B., Jaenisch R. 1996. DNA hypomethylation can activate Xist expression and silence X-linked genes. Genes Dev. 10:1991–2002 10.1101/gad.10.16.1991 [DOI] [PubMed] [Google Scholar]
- Paull T.T., Rogakou E.P., Yamazaki V., Kirchgessner C.U., Gellert M., Bonner W.M. 2000. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr. Biol. 10:886–895 10.1016/S0960-9822(00)00610-2 [DOI] [PubMed] [Google Scholar]
- Probst A.V., Dunleavy E., Almouzni G. 2009. Epigenetic inheritance during the cell cycle. Nat. Rev. Mol. Cell Biol. 10:192–206 10.1038/nrm2640 [DOI] [PubMed] [Google Scholar]
- Rappold I., Iwabuchi K., Date T., Chen J. 2001. Tumor suppressor p53 binding protein 1 (53BP1) is involved in DNA damage–signaling pathways. J. Cell Biol. 153:613–620 10.1083/jcb.153.3.613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos W.P., Kaina B. 2006. DNA damage-induced cell death by apoptosis. Trends Mol. Med. 12:440–450 10.1016/j.molmed.2006.07.007 [DOI] [PubMed] [Google Scholar]
- Sakwe A.M., Nguyen T., Athanasopoulos V., Shire K., Frappier L. 2007. Identification and characterization of a novel component of the human minichromosome maintenance complex. Mol. Cell. Biol. 27:3044–3055 10.1128/MCB.02384-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmiesing J.A., Gregson H.C., Zhou S., Yokomori K. 2000. A human condensin complex containing hCAP-C-hCAP-E and CNAP1, a homolog of Xenopus XCAP-D2, colocalizes with phosphorylated histone H3 during the early stage of mitotic chromosome condensation. Mol. Cell. Biol. 20:6996–7006 10.1128/MCB.20.18.6996-7006.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz L.B., Chehab N.H., Malikzay A., Halazonetis T.D. 2000. p53 binding protein 1 (53BP1) is an early participant in the cellular response to DNA double-strand breaks. J. Cell Biol. 151:1381–1390 10.1083/jcb.151.7.1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver D.P., Dimitrov S.D., Feunteun J., Gelman R., Drapkin R., Lu S.D., Shestakova E., Velmurugan S., Denunzio N., Dragomir S., et al. 2007. Further evidence for BRCA1 communication with the inactive X chromosome. Cell. 128:991–1002 10.1016/j.cell.2007.02.025 [DOI] [PubMed] [Google Scholar]
- Suzuki C., Murakumo Y., Kawase Y., Sato T., Morinaga T., Fukuda N., Enomoto A., Ichihara M., Takahashi M. 2008. A novel GDNF-inducible gene, BMZF3, encodes a transcriptional repressor associated with KAP-1. Biochem. Biophys. Res. Commun. 366:226–232 10.1016/j.bbrc.2007.11.118 [DOI] [PubMed] [Google Scholar]
- Takemoto A., Kimura K., Yokoyama S., Hanaoka F. 2004. Cell cycle-dependent phosphorylation, nuclear localization, and activation of human condensin. J. Biol. Chem. 279:4551–4559 10.1074/jbc.M310925200 [DOI] [PubMed] [Google Scholar]
- Takemoto A., Murayama A., Katano M., Urano T., Furukawa K., Yokoyama S., Yanagisawa J., Hanaoka F., Kimura K. 2007. Analysis of the role of Aurora B on the chromosomal targeting of condensin I. Nucleic Acids Res. 35:2403–2412 10.1093/nar/gkm157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang C.K., Wei Y., Zheng X.F. 2007. Compacting DNA during the interphase: condensin maintains rDNA integrity. Cell Cycle. 6:2213–2218 [DOI] [PubMed] [Google Scholar]
- Yang D., Buchholz F., Huang Z., Goga A., Chen C.Y., Brodsky F.M., Bishop J.M. 2002. Short RNA duplexes produced by hydrolysis with Escherichia coli RNase III mediate effective RNA interference in mammalian cells. Proc. Natl. Acad. Sci. USA. 99:9942–9947 10.1073/pnas.152327299 [DOI] [PMC free article] [PubMed] [Google Scholar]