The L166P mutation in DJ-1 is associated with Parkinson’s disease. DJ-1–interacting protein BAG1 chaperones mutant DJ-1 and reverses its mutant phenotype.
Abstract
Mutations in the gene coding for DJ-1 protein lead to early-onset recessive forms of Parkinson’s disease. It is believed that loss of DJ-1 function is causative for disease, although the function of DJ-1 still remains a matter of controversy. We show that DJ-1 is localized in the cytosol and is associated with membranes and organelles in the form of homodimers. The disease-related mutation L166P shifts its subcellular distribution to the nucleus and decreases its ability to dimerize, impairing cell survival. Using an intracellular foldase biosensor, we found that wild-type DJ-1 possesses chaperone activity, which is abolished by the L166P mutation. We observed that this aberrant phenotype can be reversed by the expression of the cochaperone BAG1 (Bcl-2–associated athanogene 1), restoring DJ-1 subcellular distribution, dimer formation, and chaperone activity and ameliorating cell survival.
Introduction
Parkinson’s disease (PD) is the second most common neurodegenerative disease, affecting 1–2% of the population over the age of 65 (de Rijk et al., 2000). The majority of PD cases are of a sporadic nature with unknown etiology. However, mutations in several genes that result in familial forms of PD have been identified. Among them, the PARK7 locus corresponds to the gene coding for the DJ-1 protein (Bonifati et al., 2003a). Diverse mutations, including deletions and point mutations in the DJ-1 gene, have been described and were linked to autosomal recessive early-onset parkinsonism (Bonifati et al., 2003b). DJ-1 is a protein of 20 kD whose loss of function is hypothesized to constitute the pathological mechanism underlying the disease. DJ-1 is widely expressed in neurons and glia as well as peripheral tissues (Bandopadhyay et al., 2004; Olzmann et al., 2007). It was found to be localized in the cytosol and the nucleus, with a small fraction being associated with mitochondria (Zhang et al., 2005; Junn et al., 2009). Different functions have been suggested for DJ-1, including characterization as a redox sensor (Kinumi et al., 2004; Taira et al., 2004; Andres-Mateos et al., 2007; Batelli et al., 2008), a molecular chaperone (Xia et al., 2003; Olzmann et al., 2004), or a transcriptional cofactor (Bae et al., 2005). The association of DJ-1 with HSP70 (heat shock protein 70) and other molecular chaperones was described previously (Li et al., 2005). The crystal structure of DJ-1 indicated that the protein forms a homodimer in vitro (Honbou et al., 2003; Wilson et al., 2003). Interestingly, the most commonly studied DJ-1 L166P mutation appears to be impaired in dimer formation (Moore et al., 2003; Blackinton et al., 2005), suggesting that dimer formation is required for its physiological function. Some groups have shown that L166P is unstable in mammalian cells, implicating a faster turnover of the mutated protein by degradation via the proteasome system and nonproteasomal pathways (Miller et al., 2003; Moore et al., 2003; Görner et al., 2004; Olzmann et al., 2004).
BAG1 (Bcl-2–associated athanogene 1) is a cochaperone of HSP70 that stimulates its ATPase- and protein-folding activity (Takayama et al., 1997; Sondermann et al., 2001; Liman et al., 2005). BAG1 also binds to the proteasome, where it is believed to act as a shuttle protein, delivering chaperone-recognized misfolded substrates to the proteasome for degradation (Lüders et al., 2000; Demand et al., 2001). BAG1 is of particular significance for the nervous system. It was described to stimulate neuronal differentiation and to promote neuronal survival (Kermer et al., 2002; Liman et al., 2008). Mice lacking BAG1 die embryonically as a result of apoptosis in the nervous system and liver (Götz et al., 2005). Conversely, overexpression of BAG1 provides protection against various apoptotic insults (Takayama et al., 1995; Kermer et al., 2003; Townsend et al., 2003). Moreover, BAG1 proved to enhance axonal regeneration in mice (Planchamp et al., 2008). In this study, we describe BAG1 as a DJ-1–interacting protein that has the ability to restore DJ-1 L166P function.
Results
BAG1 interacts with DJ-1 and affects its dimer formation
Wild-type DJ-1 and DJ-1 L166P mutant were expressed in SH-SY5Y neuroblastoma cells in the presence or absence of Flag-BAG1. Coimmunoprecipitation showed that wild-type and mutant forms of DJ-1 as well as the endogenous protein interact with BAG1. To evaluate the specificity and to examine binding affinity, we included conditions in which the recombinant DJ-1 was absent or in which the endogenous DJ-1 protein was knocked down using siRNA. As indicated in Fig. 1, we were not able to detect a preferential association of BAG1 with either endogenous or recombinant DJ-1. At the same time, pull-down signals could be extinguished by DJ-1 siRNA.
Figure 1.
BAG1 interacts with recombinantly expressed and endogenous DJ-1. Wild-type DJ-1 (DJ-1 WT) and DJ-1 L166P mutant (DJ-1 Mut) with a V5/HIS tag were transiently transfected in SH-SY5Y cells in the presence or absence of Flag-BAG1 and Flag-BAGΔC. Knockdown of endogenous DJ-1 was performed with DJ-1 siRNA. Coimmunoprecipitation (CO-IP) shows the interaction between BAG1 and both forms of the DJ-1 protein. In addition, endogenous DJ-1 and HSP70 could also be detected via BAG1 pull-down. The BAGΔC deletion variant failed to interact either with DJ-1 or with HSP70. Both siRNA samples were grouped to the figure from a different gel. White lines indicate that intervening lanes have been spliced out. WB, Western blot.
As expected, we were able to detect endogenous HSP70 as a second interacting protein of Flag-BAG1. In line with a previous study (Kermer et al., 2003), expression levels of HSP70 were slightly increased in the case of BAG1 overexpression. The deletion mutant Flag-BAGΔC did not show an interaction with either HSP70 or DJ-1 (Fig. 1). Next, we performed the experiment with coimmunoprecipitation of BAG1 via Flag–DJ-1 constructs. Again, a strong interaction between DJ-1 and BAG1 could be observed, and HSP70 was also detected as a second interacting protein for the L166P mutant protein (Fig. 2).
Figure 2.
Immunoprecipitation of BAG1 through DJ-1 wild type and L166P DJ-1. BAG1 and BAGΔC were transiently transfected in SH-SY5Y cells in the presence or absence of Flag–DJ-1 wild type (DJ-1 WT) and Flag–DJ-1 mutant (DJ-1 Mut). Again, coimmunoprecipitation (CO-IP) shows the interaction between BAG1 and both forms of the DJ-1 protein. Endogenous HSP70 was only detected via pull-down of Flag–DJ-1 mutant. WB, Western blot.
We then performed subcellular fractionation into cytosolic, membranous/organellar, nuclear, and cytoskeletal fractions. Both DJ-1 wild type and L166P were found equally abundant in the cytosolic and membrane/organelle fractions (Fig. 3 A). However, the nuclear fraction contained only mutant DJ-1, whereas DJ-1 wild type was absent. Coexpression of BAG1 together with L166P DJ-1 decreased its amount in the nucleus, as shown by subcellular fractionation. Results were confirmed in situ using EGFP-tagged DJ-1. The nuclear accumulation of L166P mutant contrasted with the absence of nuclear staining of DJ-1 wild type (Fig. 3 B).
Figure 3.
BAG1 influences DJ-1 dimerization and subcellular localization. (A) Wild-type DJ-1 (DJ-1 WT) and DJ-1 L166P mutant (DJ-1 Mut) with a V5/HIS tag were transiently expressed in SH-SY5Y cells in the presence or absence of Flag-BAG1 and subjected to subcellular fractionation and immunoblotting. Both DJ-1 wild type and DJ-1 L166P mutant were found equally abundant in the cytosolic and membrane/organelle fractions. However, the nuclear fraction contained only mutant DJ-1, whereas DJ-1 wild type was absent. Compartment-specific proteins served as loading controls (bottom). In the membrane/organelle fraction, we detected an extra band at 50 kD, which likely corresponds to DJ-1 homodimers. (B) Nuclear accumulation of DJ-1 mutant in situ was confirmed by microscopy using EGFP-tagged DJ-1. (C) Coexpression of BAG1 together with DJ-1 increases the ratio of dimer/monomer staining intensity both for wild-type and mutant protein. Densitometry of immuno signal quantitates levels of DJ-1 protein. Error bars indicate SEM out of triplicates. Bars, 10 µm.
On immunoblots, in addition to a band running at 25 kD that corresponds to monomeric DJ-1, we could detect a band at 50 kD, which presumably corresponds to DJ-1 homodimers. This band was particularly strong in the membrane/organelle fraction. Coexpression of BAG1 together with DJ-1 significantly increased the ratio of dimer/monomer staining intensity both for wild-type and mutant protein (Fig. 3 C). All results were reproduced in the nonneuronal HEK293T cell line (unpublished data).
To further investigate the possibility that BAG1 influences the dimerization of DJ-1, we performed Förster resonance energy transfer (FRET)/fluorescence lifetime imaging (FLIM) experiments with fluorescently (CFP or YFP) labeled DJ-1. The occurrence of FRET between CFP- and YFP-labeled DJ-1, as judged quantitatively by a reduction in the CFP fluorescence lifetime, is indicative of its homodimerization. In agreement with the fractionation experiments, we observed that the FRET efficiency was significantly greater for DJ-1 wild type (25%) than DJ-1 L166P (10%). Coexpression of BAG1 together with DJ-1 L166P substantially increased FRET efficiency of the mutant proteins almost to the levels of DJ-1 wild type (Fig. 4, A–D; P < 0.001). Interestingly, BAG1 was also able to augment the dimerization of DJ-1 wild type.
Figure 4.
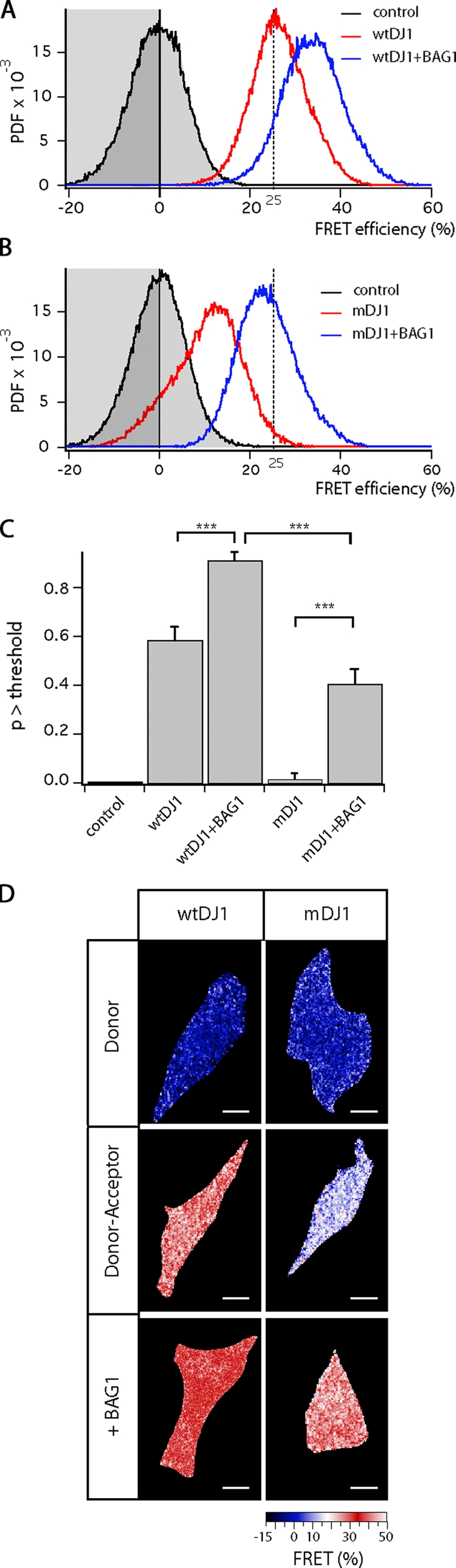
BAG1 influences DJ-1 dimerization (FRET/FLIM experiments). (A and B) Dimerization of DJ-1 isoforms was examined in SH-SY5Y neuroblastoma cells. Control lifetime distributions were obtained from cells expressing only the CFP-labeled DJ-1 (A and B, black trace, grayed area). Lifetimes were converted to FRET efficiencies using the mean lifetime of this condition. Coexpression of CFP- and YFP-labeled wild-type DJ-1 (wtDJ1) reduces the CFP lifetime, i.e., increases FRET, which is indicative of homodimerization. Coexpression of BAG1 further increases the level of dimerization (A, blue trace). The FRET efficiency between CFP- and YFP-labeled mutant DJ-1 (mDJ1) is lower (B, red trace) but restores to wild-type DJ-1 basal levels upon coexpression of DJ-1. The dashed line at 25% FRET indicates the adopted threshold value. (C) These relationships were confirmed by statistical analysis. FRET efficiency of different conditions (n = 15) by t test (***, P < 0.001); p > threshold. Error bars indicate SEM out of triplicates. (D) Images of the FRET efficiency, i.e., level of dimerization, in representative cells were color-coded as indicated in the activity scale. Bars, 10 µm.
DJ-1 possesses chaperone activity
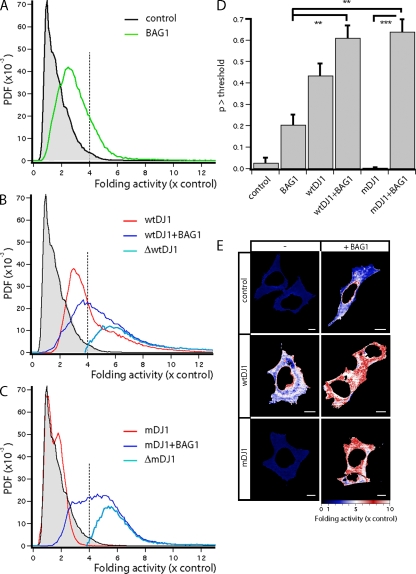
Several studies have suggested that DJ-1 might function as a molecular chaperone (Shendelman et al., 2004; Li et al., 2005; Zhou et al., 2006), but conclusive in situ evidence is still lacking. For this reason, we took advantage of a unique fluorescent biosensor for the activity of the cellular chaperone machinery (Liman et al., 2005). The sensor consists of a chaperone-dependent YFP (cdYFP) with low fluorescence intensity caused by a point mutation that impairs its folding, preventing the formation of the chromophore. The protein is tuned such that an increase in chaperone activity corrects its folding defect, giving rise to an increase in its fluorescence intensity. We expressed cdYFP together with DJ-1 wild type or L166P in neuronal CSM 14.1 cells and performed quantitative measurements of the fluorescence intensity of cdYFP relative to the Cy5 immunofluorescence signal, which constitutes an inert concentration reference to calculate folding activity (expressed as fold increase relative to the activity in cells expressing only the cdYFP sensor) with high spatiotemporal resolution inside intact cells. Under control conditions in which cells expressed cdYFP together with an empty vector, we observed a narrow distribution of low YFP fluorescence (Fig. 5 A, black trace with grayed area). Expression of DJ-1 resulted in a substantial and highly significant increase in cellular folding activity, with a median increase of threefold, reaching levels up to 8–10-fold (Fig. 5 B, red trace). Cells expressing DJ-1 L166P mutant showed a folding activity distribution that was essentially identical to control cells, indicating that the mutation completely abolishes foldase activity of DJ-1 (Fig. 5 C, red trace). As shown before, expression of BAG1 increased the cellular foldase activity up to sixfold as a result of the activation of chaperones (Fig. 5 A, green trace; Liman et al., 2005). Coexpression of BAG1 together with DJ-1 L166P resulted in a dramatic shift toward high folding activities, thus restoring DJ-1 chaperone function (Fig. 5 B, dark blue trace). These increased folding activity levels exceeded those reached by the expression of foldase-stimulating BAG1 alone, showing the additional separate activity of DJ-1. The light blue traces in Fig. 5 B represent the isolated DJ-1 folding activities, which were corrected for the distribution of folding activities generated by the direct action of BAG1 for the coexpression conditions. As observed for the dimerization behavior of DJ-1 in the presence of BAG1, the folding activity of DJ-1 wild type was also increased by BAG1 (Fig. 5 B, dark blue trace). Importantly, folding activity levels reached by BAG1 coexpression with mutant DJ-1 were similar to those in which DJ-1 wild type was coexpressed together with BAG1 (Fig. 5 C, dark blue trace).
Figure 5.
DJ-1 possesses chaperone activity. (A–C) Folding activity was quantified in SH-SY5Y cells expressing different constructs in addition to the cdYFP foldase sensor. Cells expressing only the biosensor serve as control and reference situation, with a foldase activity of unity (A, black trace with grayed area). The normalized cumulative distribution (PDF) is shown. Expression of BAG1 increases cellular foldase activity (A, green trace). Expression of wild-type DJ-1 (wtDJ1; B, red trace) increases foldase activity, and coexpression of BAG1 slightly increases foldase activity even further (B, dark blue trace). Correction of this distribution for the induction by BAG1 alone shows DJ-1 to be a high activity chaperone (B, light blue trace). Expression of mutant DJ-1 (mDJ1) does not induce foldase activity (C, red trace), but coexpression of BAG1 restores foldase activity to equal the BAG1-enhanced wild-type DJ-1 levels (C, dark blue trace). Consequently, the isolated DJ-1 activity (C, light blue trace) is similar to that of the wild-type protein. The dashed line at fourfold increased folding indicates the adopted threshold value. (D) Statistical analysis confirms these relationships. Induced foldase activity of different conditions (n = 10) by t test (**, P < 0.01; ***, P < 0.001); p > threshold. Error bars indicate SEM out of triplicates. (E) Images of the folding activity in representative cells, color-coded as indicated in the activity scale, are shown. Bars, 10 µm.
BAG1 prevents DJ-1 L166P–induced cell death
To assess the functional relevance of these findings, we cotransfected SH-SY5Y cells with fluorescently tagged DJ-1 (either wild type or L166P) and BAG1 or empty vector. Cells expressing DJ-1 wild type showed a 5% basal cell death rate. Transfection of DJ-1 L166P mutant led to an increase in the number of dead cells up to ∼20%. The toxicity of the DJ-1 mutant was ameliorated by cotransfection with BAG1, with only 8% of dead cells found under this condition. We confirmed these findings by the application of an independent bioluminescent cytotoxicity assay that quantifies the integrity of the plasma membrane by measurement of the release of the intracellular enzyme adenylate kinase into the medium. Here, we included expression of the BAGΔC deletion mutant as a condition. The inability of the BAGΔC deletion mutant to ameliorate DJ-1 L166P toxicity (Fig. 6) demonstrated the specificity of the cell death–preventing action of the BAG1 cochaperone.
Figure 6.
BAG1 prevents DJ-1 L166P–induced cell death. (A) Transient cotransfection of SH-SY5Y cells with fluorescently tagged DJ-1 (either wild type [WT] or L166P [Mut]) and BAG1 or empty vector. Cell count was based on DAPI staining, and cells presenting fragmented and pyknotic nuclei were considered apoptotic. Comparison of cell count of different conditions (n = 2,000) by t test (**, P < 0.0001) is shown. (B) We confirmed these findings in SH-SY5Y and HEK293T cells by measuring the release of adenylate kinase from damaged cells with the help of a bioluminescent cytotoxicity assay. Expression of the BAGΔC deletion mutant does not ameliorate DJ-1 mutant toxicity. The amount of cell death within conditions compared by t test (**, P < 0.0001) is shown. (A and B) Error bars indicate SEM out of triplicates. Bar, 10 µm.
Discussion
DJ-1 protein mutations have been linked to recessive familial forms of PD. Although these cases represent only a minor portion of all PD cases, the majority of which are sporadic, an understanding of the disease mechanisms underlying familial PD may shed light on the pathophysiological events that operate in sporadic PD as well. However, so far, no consensus on the exact function of DJ-1 and its role in PD could be achieved. In this study, we compared the known disease mutation DJ-1 L166P with the wild-type protein in terms of subcellular localization, dimer formation, and biological activity.
Using biochemical fractionation methods, we show that DJ-1 wild type is present in the cytosol and mitochondrial fraction. DJ-1 L166P is also present in these compartments but is found additionally in the nucleus. This is interesting because other proteins that are implicated in neurodegenerative disorders (e.g., huntingtin in Huntington’s disease [Saudou et al., 1998], ataxin-1 in SCA1 [Klement et al., 1998], and ataxin-3 in SCA3 [Schmidt et al., 1998]) are thought to cause disease at least in part through their abnormal accumulation in the nucleus. However, the relevance of the nuclear localization of mutant DJ-1 demands further investigation.
Structural investigations of DJ-1 revealed that it forms homodimers and that the L166P mutation impairs this dimer formation. Our results obtained by protein biochemistry and FRET microscopy in single intact cells confirm that DJ-1 wild type but not DJ-1 L166P has the capacity to form dimers under physiological conditions.
With respect to DJ-1 function, there have been several studies linking the protein to redox and chaperone systems, albeit without producing a unifying concept or conclusive results (Kinumi et al., 2004; Taira et al., 2004; Andres-Mateos et al., 2007; Batelli et al., 2008). We aimed at resolving this controversy in neuronal cells by the application of our fluorescent chaperone sensor. Indeed, we are able to show for the first time that DJ-1 wild type exerts a strong chaperone-like activity in intact single cells, which is lost in DJ-1 bearing the L166P mutation. From the combination of FRET/FLIM measurements quantifying dimerization of fluorescently labeled DJ-1, we can conclude that homodimerization is a prerequisite for chaperone function and that the L166P mutation severely impairs dimerization capacity, suggesting that this is the molecular defect that gives rise to disease pathophysiology. Additionally, we present BAG1, a cochaperone protein with previously described neuroprotective functions (Takayama et al., 1995; Kermer et al., 2003; Townsend et al., 2003), as new binding partner for DJ-1, with important consequences for DJ-1 function.
Strikingly, we found that BAG1 influences several parameters affected by the L166P mutation. First, it decreases the abundance of L166P mutant in the nuclear fraction; second, it stimulates dimer formation of both wild-type and L166P DJ-1; third, it enhances chaperone function as seen in the folding sensor assay. Interestingly, even though dimerization of DJ-1 was reestablished by BAG1 to levels of DJ-1 wild type and BAG1 increased the level of DJ-1 wild type even further, no additional increase in foldase activity was observed for DJ-1 wild type when coexpressed with BAG1. This seems to suggest that the targets of DJ-1 limit maximal stimulation under overexpression conditions.
Finally, we observed that overexpression of DJ-1 L166P in SH-SY5Y cells leads to an increase in cell death. Because it is assumed that loss of DJ-1 function causes toxicity, one can speculate that L166P-induced cell death is caused by the inability of the protein to form a functional homodimer, leading to anomalous nuclear targeting. Coexpression of BAG1 corrects dimer formation and proper subcellular distribution, thereby ameliorating the toxic action of the DJ-1 mutant.
In summary, we linked DJ-1 subcellular localization and levels of dimerization to its physiological function in neuronal cells. Our results provide evidence in support of the view that DJ-1 acts, in dimeric form, as a chaperone with very strong foldase activity. In fact, the folding activities achieved by DJ-1 are similar in magnitude to those observed upon overexpression of HSP70 (Liman et al., 2005; Ganesan et al., 2008). Because it is believed that it is the loss of DJ-1 function that underlies this genetic form of PD, stimulating the cellular chaperone machinery could prove to be of therapeutic potential for DJ-1–dependent PD. Accordingly, we show that BAG1, an antiapoptotic foldase-stimulating cochaperone, restores DJ-1 L166P dimerization and chaperone function. Our data add to the current understanding of PD pathophysiology and point toward novel putative therapeutic avenues.
Materials and methods
Constructs and transfection conditions
Constructs used were full-length cDNA constructs for DJ-1 wild type and L166P mutant carrying a C-terminal V5/HIS tag or a C-terminal Flag tag, full-length cDNA constructs for DJ-1 wild type and L166P mutant as a C-terminal CFP or as a N-terminal YFP fusion protein, full-length cDNA constructs for BAG1 and BAGΔC with or without a C-terminal Flag tag, and cdYFP with an N-terminal HA tag. All constructs were verified by sequencing at Seqlab, Sequence Laboratories Göttingen GmbH.
siGenome SMARTpool DJ-1 siRNA and siGenome nontargeting siRNA pool siRNA were purchased from Thermo Fisher Scientific. Cells were transiently transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions.
Preparation of protein extracts and Western blot analysis
Cells were washed, scraped in ice-cold PBS, and pelleted. Cell lysates were prepared using lysis buffer containing 50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% Triton-X 100, and complete protease inhibitor cocktail (Roche) for 15 min at 4°C, followed by 10-min centrifugation at 13,000 rpm at 4°C. Subcellular fractionation was performed using ProteoExtract Subcellular Proteome Extraction kit (EMD).
Lysates were mixed with 6× Laemmli loading buffer and boiled for 5 min at 95°C before performing the SDS-PAGE Western blot. Membranes were blocked for 1 h in 5% dried milk powder in TBST (TBS/Tween 20) at room temperature.
Quantitative densitometric analysis (Fig. 3 C) was performed on digitized images of immunoblots using the Fluor-S MultiImager (Bio-Rad Laboratories) with Quantity-One-4.2.2 software (Bio-Rad Laboratories). SEM refers to triplicate preparations of protein extracts. Densitometric levels of monomer and dimer fractions of DJ-1 protein were normalized to levels of cadherin loading control. The experiments were repeated at least three times using SH-SY5Y and HEK293T cells.
Coimmunoprecipitation
Cells were cultured and transfected in 6-well plates, and immunoprecipitation was performed with red anti-Flag M2 affinity gel (Sigma-Aldrich). After bead equilibration and cell lysate preparation, samples were incubated for 4 h at 4°C. The bead pellet was washed with TBS and centrifuged in a microcentrifuge (Biofuge Fresco; Heraeus) for 30 s at 8,200 g. Washing steps were repeated three times, and bound protein was eluted using SDS-PAGE sample buffer. The experiments were repeated at least three times using SH-SY5Y and HEK293T cells.
Immunocytochemistry
Cells cultured on coverslips and transfected with the biosensor HA-cdYFP for the chaperone assay were washed in PBS and fixed in PBS containing 4% PFA for 10 min at room temperature, followed by several washing steps in PBS. Permeabilization was performed with 0.1% Triton X-100 in PBS for 10 min with subsequent preblocking in PBS containing 3% bovine serum albumin. Cells were incubated in blocking solution containing anti-HA antibody (1:1,000 dilution; Covance) for 1 h at room temperature. After washing three times in PBST (PBS Tween 20) and incubation with Cy5-conjugated secondary antibody (1:400 dilution; Jackson ImmunoResearch Laboratories, Inc.) for 1 h at room temperature, cells were again washed three times in PBST and mounted on microscope slides using Roti-Histokitt 2 (Roth) imaging medium.
Cells cultured on coverslips and transfected with CFP–DJ-1 wild-type or mutant protein were washed in PBS and fixed in PBS containing 4% PFA for 10 min at room temperature, followed by several washing steps in PBS. Permeabilization was performed with 0.1% Triton X-100 in PBS for 10 min with subsequent preblocking in PBS containing 3% bovine serum albumin. Cells were incubated in blocking solution containing anti-BAG1 antibody (rabbit polyclonal BAG 1680; 1:1,000 dilution) for 1 h at room temperature. After washing three times in PBST and incubation with Cy5-conjugated secondary antibody (1:400 dilution; Jackson ImmunoResearch Laboratories, Inc.) for 1 h at room temperature, cells were again washed three times in PBST. Slides were stained with DAPI to detect nuclei. Roti-Histokitt 2 imaging medium was used.
Wide-field fluorescent images were obtained at room temperature using a microscope (Axioplan2; Carl Zeiss, Inc.) equipped with a 63× NA 1.4 oil objective. The camera used was an AxioCam (Carl Zeiss, Inc.), and Axiovision (Carl Zeiss, Inc.) was used as acquisition software. The experiments were repeated at least three times using SH-SY5Y and CSM 14.1 cells.
Fluorescence lifetime–based FRET imaging
For FLIM, CSM 14.1 or SH-SY5Y cells growing on coverslips were transfected using Lipofectamine and fixed after 24 h using 4% PFA. The mounted coverslips were imaged on a frequency domain FLIM setup (Esposito et al., 2005) built around a microscope (Axioplan; Carl Zeiss, Inc.) equipped with a 63× NA 1.4 oil objective. An Ar laser (Innova 304C; Coherent) tuned to 457 nm at 250 mW output power and intensity sine-modulated at 80 MHz by passing the beam through an acoustooptical modulator was used as excitation source. Images were demodulated by a directly gain-modulated multichannel-based image intensifier (PicoStar; LaVision GmbH). Cells mounted in Mowiol (Hoechst) were imaged at room temperature. Image acquisition and lifetime calculation were performed by custom-written routines in the DaVis software suite (Lavision GmbH). The filter cube used for CFP lifetime imaging contained a LP470 dichroic and a BP 490/20 emission filter from AHF analysentechnik AG. Eight phase step images were taken and used to construct an intensity image and a lifetime image (Esposito et al., 2005). Cells were segmented automatically in the intensity image. All further operations were performed with custom-written routines (A. Esposito [Hutchinson/Medical Research Council Research Centre, Cambridge, England, UK] and M. Gralle) in MatLab (MathWorks). The lifetime values of all pixels in the threshold masks from images taken in the same experimental condition are shown in pooled distributions, with distributions for individuals normalized to unity for equal weighing before renormalization of the cumulative distribution to allow comparison between conditions with different sample sizes (probability density function [PDF]). Lifetime values were converted to FRET efficiency values using the mean lifetime of the appropriate control. For further statistical analysis, a threshold of three times the standard deviation from the mean of the PDF of the control situation (i.e., where cells only express CFP–DJ-1 was defined at 25% FRET) and the area of the normalized distributions for the individual cells exceeding this threshold (considered statistically relevant events) were determined. These values were subjected to analysis of variance with Tukey postprocessing to test statistical significance between conditions. These calculations were performed using the Igor Analysis Package (WaveMetrics). Resulting histograms were obtained imaging 15 cells per condition. Experiments were repeated at least three times.
Fluorescent folding assay
CSM 14.1 or SH-SY5Y cells were imaged using a confocal laser-scanning microscope (SP2; Leica) equipped with an AOBS acoustooptical beamsplitter and using a 63× NA 1.4 oil objective. Cells mounted in Mowiol (Hoechst) were imaged at room temperature. For the detection of YFP fluorescence, cells were excited using the 514-nm Ar laser line, and emission was recorded between 525 and 600 nm. For Cy5 fluorescence, excitation was performed with the 633-nm HeNe laser line, with an emission range of 640–750 nm. Photomultiplier tube gains were kept constant for each emission range throughout the acquisitions. The Cy5 fluorescence served as an inert concentration reference for the total concentration of cdYFP. The folding efficiency was retrieved by simple image arithmetics, by dividing YFP fluorescence by reference fluorescence, using ImageJ software (National Institutes of Health). The image intensity values were corrected for different laser power settings needed to obtain proper photon statistics under the different conditions. The ratio images were masked based on thresholded fluorescence intensities in the CFP or Cy5 channel to remove noise in the noncellular background. The folding ratios were always normalized to the first, partial folding, peak in wild-type cells (see Results) so that the effects on the folding responses were expressed in units of native folding activity. Folding ratio frequency histograms were obtained from the masked images of 10 cells for each condition, unless indicated otherwise. The nucleus was also masked on the basis of its lower Cy5 signals that represent incomplete penetration of the labeled antibody, which would upset the folding statistics. Folding ratio histograms were analyzed and used to construct cumulative PDF distributions of folding activities. These calculations and the further statistical analysis of the different conditions were performed as described for the lifetime-based FRET imaging. Because of the non-Gaussian distribution of the control condition, we here adopted a threshold value of fourfold increased folding activity.
Cell death assays
Cell death was analyzed with the ToxiLight BioAssay kit (Lonza). It is a nondestructive bioluminescent cytotoxicity assay designed to measure the release of adenylate kinase from damaged cells. Cells were cultured in 6-well plates, and 36 h after transfection, cell supernatant was transferred to a luminescence-compatible 96-well plate. Analysis was performed with a microplate luminometer (Trilux Microbeta 1450; Wallac).
Transient cotransfection of SH-SY5Y cells was performed with fluorescently tagged DJ-1 (either wild type or L166P) and BAG1 or empty vector. Cells cultured on coverslips and transfected with CFP–DJ-1 wild type or mutant protein were washed in PBS and fixed in PBS containing 4% PFA for 10 min at room temperature, followed by several washing steps in PBS. Permeabilization was performed with 0.1% Triton X-100 in PBS for 10 min with subsequent preblocking in PBS containing 3% bovine serum albumin. Cells were incubated in blocking solution containing anti-BAG1 antibody (rabbit polyclonal BAG 1680; 1:1,000 dilution) for 1 h at room temperature. After washing three times in PBST and incubation with Cy5-conjugated secondary antibody (1:400 dilution; Jackson ImmunoResearch Laboratories, Inc.) for 1 h at room temperature, cells were again washed three times in PBST. Slides were stained with DAPI to detect nuclei. Roti-Histokitt 2 imaging medium was used.
Wide-field fluorescent images were obtained at room temperature using an Axioplan2 microscope equipped with a 63× NA 1.4 oil objective. The camera used was an AxioCam, and Axiovision was used as acquisition software. Cell counts were based on DAPI staining, and cells presenting fragmented and pyknotic nuclei were considered apoptotic. Up to 2,000 cells per condition were counted, and experiments were repeated at least three times using SH-SY5Y and CSM 14.1 cells.
Antibodies
The following antibodies were used at corresponding dilutions: 1:1,000 rabbit polyclonal BAG 1680, 1:1,000 HA (Covance), 1:1,000 goat polyclonal DJ-1 (Imgenex), 1:1,000 mouse monoclonal β-tubulin (Sigma-Aldrich), 1:2,000 mouse histone 2B (Abcam), 1:5,000 mouse actin (Millipore), 1:1,000 rabbit HSP70 (Sigma-Aldrich), 1:2,000 mouse cadherin (Sigma-Aldrich), and 1:2,000 mouse vimentin (Sigma-Aldrich).
Cell culture
CSM 14.1 wild-type cells were grown in DME supplemented with 10% FCS and penicillin/streptomycin at 32°C. SH-SY5Y and HEK293T cells were grown in DME supplemented with 10% FCS and penicillin/streptomycin at 37°C. For transient transfections, Lipofectamine 2000 was used.
Acknowledgments
This study was supported by Deutsche Forschungsgemeinschaft (DFG) Research Center for Molecular Physiology of the Brain and its Excellence cluster “Microscopy on the Nanometer Scale (EXC171).”
Footnotes
Abbreviations used in this paper:
- cdYFP
- chaperone-dependent YFP
- FLIM
- fluorescence lifetime imaging
- FRET
- Förster resonance energy transfer
- PD
- Parkinson’s disease
- probability density function
References
- Andres-Mateos E., Perier C., Zhang L., Blanchard-Fillion B., Greco T.M., Thomas B., Ko H.S., Sasaki M., Ischiropoulos H., Przedborski S., et al. 2007. DJ-1 gene deletion reveals that DJ-1 is an atypical peroxiredoxin-like peroxidase. Proc. Natl. Acad. Sci. USA. 104:14807–14812 10.1073/pnas.0703219104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae B.I., Xu H., Igarashi S., Fujimuro M., Agrawal N., Taya Y., Hayward S.D., Moran T.H., Montell C., Ross C.A., et al. 2005. p53 mediates cellular dysfunction and behavioral abnormalities in Huntington’s disease. Neuron. 47:29–41 10.1016/j.neuron.2005.06.005 [DOI] [PubMed] [Google Scholar]
- Bandopadhyay R., Kingsbury A.E., Cookson M.R., Reid A.R., Evans I.M., Hope A.D., Pittman A.M., Lashley T., Canet-Aviles R., Miller D.W., et al. 2004. The expression of DJ-1 (PARK7) in normal human CNS and idiopathic Parkinson’s disease. Brain. 127:420–430 10.1093/brain/awh054 [DOI] [PubMed] [Google Scholar]
- Batelli S., Albani D., Rametta R., Polito L., Prato F., Pesaresi M., Negro A., Forloni G. 2008. DJ-1 modulates alpha-synuclein aggregation state in a cellular model of oxidative stress: relevance for Parkinson’s disease and involvement of HSP70. PLoS One. 3:e1884 10.1371/journal.pone.0001884 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Blackinton J., Ahmad R., Miller D.W., van der Brug M.P., Canet-Avilés R.M., Hague S.M., Kaleem M., Cookson M.R. 2005. Effects of DJ-1 mutations and polymorphisms on protein stability and subcellular localization. Brain Res. Mol. Brain Res. 134:76–83 10.1016/j.molbrainres.2004.09.004 [DOI] [PubMed] [Google Scholar]
- Bonifati V., Rizzu P., Squitieri F., Krieger E., Vanacore N., van Swieten J.C., Brice A., van Duijn C.M., Oostra B., Meco G., Heutink P. 2003a. DJ-1 (PARK7), a novel gene for autosomal recessive, early onset parkinsonism. Neurol. Sci. 24:159–160 10.1007/s10072-003-0108-0 [DOI] [PubMed] [Google Scholar]
- Bonifati V., Rizzu P., van Baren M.J., Schaap O., Breedveld G.J., Krieger E., Dekker M.C., Squitieri F., Ibanez P., Joosse M., et al. 2003b. Mutations in the DJ-1 gene associated with autosomal recessive early-onset parkinsonism. Science. 299:256–259 10.1126/science.1077209 [DOI] [PubMed] [Google Scholar]
- de Rijk M.C., Launer L.J., Berger K., Breteler M.M., Dartigues J.F., Baldereschi M., Fratiglioni L., Lobo A., Martinez-Lage J., Trenkwalder C., Hofman A. 2000. Prevalence of Parkinson’s disease in Europe: A collaborative study of population-based cohorts. Neurology. 54:S21–S23 [PubMed] [Google Scholar]
- Demand J., Alberti S., Patterson C., Höhfeld J. 2001. Cooperation of a ubiquitin domain protein and an E3 ubiquitin ligase during chaperone/proteasome coupling. Curr. Biol. 11:1569–1577 10.1016/S0960-9822(01)00487-0 [DOI] [PubMed] [Google Scholar]
- Esposito A., Gerritsen H.C., Wouters F.S. 2005. Fluorescence lifetime heterogeneity resolution in the frequency domain by lifetime moments analysis. Biophys. J. 89:4286–4299 10.1529/biophysj.104.053397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganesan S., Rohde G., Eckermann K., Sroka K., Schaefer M.K.E., Dohm C.P., Kermer P., Haase G., Wouters F., Bähr M., Weishaupt J.H. 2008. Mutant SOD1 detoxification mechanisms in intact single cells. Cell Death Differ. 15:312–321 10.1038/sj.cdd.4402262 [DOI] [PubMed] [Google Scholar]
- Görner K., Holtorf E., Odoy S., Nuscher B., Yamamoto A., Regula J.T., Beyer K., Haass C., Kahle P.J. 2004. Differential effects of Parkinson’s disease-associated mutations on stability and folding of DJ-1. J. Biol. Chem. 279:6943–6951 10.1074/jbc.M309204200 [DOI] [PubMed] [Google Scholar]
- Götz R., Wiese S., Takayama S., Camarero G.C., Rossoll W., Schweizer U., Troppmair J., Jablonka S., Holtmann B., Reed J.C., et al. 2005. Bag1 is essential for differentiation and survival of hematopoietic and neuronal cells. Nat. Neurosci. 8:1169–1178 10.1038/nn1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honbou K., Suzuki N.N., Horiuchi M., Niki T., Taira T., Ariga H., Inagaki F. 2003. The crystal structure of DJ-1, a protein related to male fertility and Parkinson’s disease. J. Biol. Chem. 278:31380–31384 10.1074/jbc.M305878200 [DOI] [PubMed] [Google Scholar]
- Junn E., Jang W.H., Zhao X., Jeong B.S., Mouradian M.M. 2009. Mitochondrial localization of DJ-1 leads to enhanced neuroprotection. J. Neurosci. Res. 87:123–129 10.1002/jnr.21831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kermer P., Krajewska M., Zapata J.M., Takayama S., Mai J., Krajewski S., Reed J.C. 2002. Bag1 is a regulator and marker of neuronal differentiation. Cell Death Differ. 9:405–413 10.1038/sj.cdd.4400972 [DOI] [PubMed] [Google Scholar]
- Kermer P., Digicaylioglu M.H., Kaul M., Zapata J.M., Krajewska M., Stenner-Liewen F., Takayama S., Krajewski S., Lipton S.A., Reed J.C. 2003. BAG1 over-expression in brain protects against stroke. Brain Pathol. 13:495–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinumi T., Kimata J., Taira T., Ariga H., Niki E. 2004. Cysteine-106 of DJ-1 is the most sensitive cysteine residue to hydrogen peroxide-mediated oxidation in vivo in human umbilical vein endothelial cells. Biochem. Biophys. Res. Commun. 317:722–728 10.1016/j.bbrc.2004.03.110 [DOI] [PubMed] [Google Scholar]
- Klement I.A., Skinner P.J., Kaytor M.D., Yi H., Hersch S.M., Clark H.B., Zoghbi H.Y., Orr H.T. 1998. Ataxin-1 nuclear localization and aggregation: role in polyglutamine-induced disease in SCA1 transgenic mice. Cell. 95:41–53 10.1016/S0092-8674(00)81781-X [DOI] [PubMed] [Google Scholar]
- Li H.M., Niki T., Taira T., Iguchi-Ariga S.M., Ariga H. 2005. Association of DJ-1 with chaperones and enhanced association and colocalization with mitochondrial Hsp70 by oxidative stress. Free Radic. Res. 39:1091–1099 10.1080/10715760500260348 [DOI] [PubMed] [Google Scholar]
- Liman J., Ganesan S., Dohm C.P., Krajewski S., Reed J.C., Bähr M., Wouters F.S., Kermer P. 2005. Interaction of BAG1 and Hsp70 mediates neuroprotectivity and increases chaperone activity. Mol. Cell. Biol. 25:3715–3725 10.1128/MCB.25.9.3715-3725.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liman J., Faida L., Dohm C.P., Reed J.C., Bähr M., Kermer P. 2008. Subcellular distribution affects BAG1 function. Brain Res. 1198:21–26 10.1016/j.brainres.2008.01.010 [DOI] [PubMed] [Google Scholar]
- Lüders J., Demand J., Höhfeld J. 2000. The ubiquitin-related BAG-1 provides a link between the molecular chaperones Hsc70/Hsp70 and the proteasome. J. Biol. Chem. 275:4613–4617 10.1074/jbc.275.7.4613 [DOI] [PubMed] [Google Scholar]
- Miller D.W., Ahmad R., Hague S., Baptista M.J., Canet-Aviles R., McLendon C., Carter D.M., Zhu P.P., Stadler J., Chandran J., et al. 2003. L166P mutant DJ-1, causative for recessive Parkinson’s disease, is degraded through the ubiquitin-proteasome system. J. Biol. Chem. 278:36588–36595 10.1074/jbc.M304272200 [DOI] [PubMed] [Google Scholar]
- Moore D.J., Zhang L., Dawson T.M., Dawson V.L. 2003. A missense mutation (L166P) in DJ-1, linked to familial Parkinson’s disease, confers reduced protein stability and impairs homo-oligomerization. J. Neurochem. 87:1558–1567 [DOI] [PubMed] [Google Scholar]
- Olzmann J.A., Brown K., Wilkinson K.D., Rees H.D., Huai Q., Ke H., Levey A.I., Li L., Chin L.S. 2004. Familial Parkinson’s disease-associated L166P mutation disrupts DJ-1 protein folding and function. J. Biol. Chem. 279:8506–8515 10.1074/jbc.M311017200 [DOI] [PubMed] [Google Scholar]
- Olzmann J.A., Bordelon J.R., Muly E.C., Rees H.D., Levey A.I., Li L., Chin L.S. 2007. Selective enrichment of DJ-1 protein in primate striatal neuronal processes: implications for Parkinson’s disease. J. Comp. Neurol. 500:585–599 10.1002/cne.21191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planchamp V., Bermel C., Tönges L., Ostendorf T., Kügler S., Reed J.C., Kermer P., Bähr M., Lingor P. 2008. BAG1 promotes axonal outgrowth and regeneration in vivo via Raf-1 and reduction of ROCK activity. Brain. 131:2606–2619 10.1093/brain/awn196 [DOI] [PubMed] [Google Scholar]
- Saudou F., Finkbeiner S., Devys D., Greenberg M.E. 1998. Huntingtin acts in the nucleus to induce apoptosis but death does not correlate with the formation of intranuclear inclusions. Cell. 95:55–66 10.1016/S0092-8674(00)81782-1 [DOI] [PubMed] [Google Scholar]
- Schmidt T., Landwehrmeyer G.B., Schmitt I., Trottier Y., Auburger G., Laccone F., Klockgether T., Völpel M., Epplen J.T., Schöls L., Riess O. 1998. An isoform of ataxin-3 accumulates in the nucleus of neuronal cells in affected brain regions of SCA3 patients. Brain Pathol. 8:669–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shendelman S., Jonason A., Martinat C., Leete T., Abeliovich A. 2004. DJ-1 is a redox-dependent molecular chaperone that inhibits alpha-synuclein aggregate formation. PLoS Biol. 2:e362 10.1371/journal.pbio.0020362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sondermann H., Scheufler C., Schneider C., Hohfeld J., Hartl F.U., Moarefi I. 2001. Structure of a Bag/Hsc70 complex: convergent functional evolution of Hsp70 nucleotide exchange factors. Science. 291:1553–1557 10.1126/science.1057268 [DOI] [PubMed] [Google Scholar]
- Taira T., Saito Y., Niki T., Iguchi-Ariga S.M., Takahashi K., Ariga H. 2004. DJ-1 has a role in antioxidative stress to prevent cell death. EMBO Rep. 5:213–218 10.1038/sj.embor.7400074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama S., Sato T., Krajewski S., Kochel K., Irie S., Millan J.A., Reed J.C. 1995. Cloning and functional analysis of BAG-1: a novel Bcl-2-binding protein with anti-cell death activity. Cell. 80:279–284 10.1016/0092-8674(95)90410-7 [DOI] [PubMed] [Google Scholar]
- Takayama S., Bimston D.N., Matsuzawa S., Freeman B.C., Aime-Sempe C., Xie Z., Morimoto R.I., Reed J.C. 1997. BAG-1 modulates the chaperone activity of Hsp70/Hsc70. EMBO J. 16:4887–4896 10.1093/emboj/16.16.4887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend P.A., Cutress R.I., Sharp A., Brimmell M., Packham G. 2003. BAG-1: a multifunctional regulator of cell growth and survival. Biochim. Biophys. Acta. 1603:83–98 [DOI] [PubMed] [Google Scholar]
- Wilson M.A., Collins J.L., Hod Y., Ringe D., Petsko G.A. 2003. The 1.1-A resolution crystal structure of DJ-1, the protein mutated in autosomal recessive early onset Parkinson’s disease. Proc. Natl. Acad. Sci. USA. 100:9256–9261 10.1073/pnas.1133288100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J., Lee D.H., Taylor J., Vandelft M., Truant R. 2003. Huntingtin contains a highly conserved nuclear export signal. Hum. Mol. Genet. 12:1393–1403 10.1093/hmg/ddg156 [DOI] [PubMed] [Google Scholar]
- Zhang L., Shimoji M., Thomas B., Moore D.J., Yu S.W., Marupudi N.I., Torp R., Torgner I.A., Ottersen O.P., Dawson T.M., Dawson V.L. 2005. Mitochondrial localization of the Parkinson’s disease related protein DJ-1: implications for pathogenesis. Hum. Mol. Genet. 14:2063–2073 10.1093/hmg/ddi211 [DOI] [PubMed] [Google Scholar]
- Zhou W., Zhu M., Wilson M.A., Petsko G.A., Fink A.L. 2006. The oxidation state of DJ-1 regulates its chaperone activity toward alpha-synuclein. J. Mol. Biol. 356:1036–1048 10.1016/j.jmb.2005.12.030 [DOI] [PubMed] [Google Scholar]