Evidence is presented for an unconventional protein secretion pathway that is conserved from yeast to Dictyostelium discoideum in which Acb1 may be sequestered into autophagosomal vesicles, which then fuse (either directly or indirectly) with the plasma membrane (see also the companion paper from Duran et al. in this issue).
Abstract
In contrast to the enormous advances made regarding mechanisms of conventional protein secretion, mechanistic insights into the unconventional secretion of proteins are lacking. Acyl coenzyme A (CoA)–binding protein (ACBP; AcbA in Dictyostelium discoideum), an unconventionally secreted protein, is dependent on Golgi reassembly and stacking protein (GRASP) for its secretion. We discovered, surprisingly, that the secretion, processing, and function of an AcbA-derived peptide, SDF-2, are conserved between the yeast Pichia pastoris and D. discoideum. We show that in yeast, the secretion of SDF-2–like activity is GRASP dependent, triggered by nitrogen starvation, and requires autophagy proteins as well as medium-chain fatty acyl CoA generated by peroxisomes. Additionally, a phospholipase D implicated in soluble N-ethyl-maleimide sensitive fusion protein attachment protein receptor–mediated vesicle fusion at the plasma membrane is necessary, but neither peroxisome turnover nor fusion between autophagosomes and the vacuole is essential. Moreover, yeast Acb1 and several proteins required for its secretion are necessary for sporulation in P. pastoris. Our findings implicate currently unknown, evolutionarily conserved pathways in unconventional secretion.
Introduction
Unconventional protein secretion was discovered more than 20 years ago, and we know more than 20 examples of such proteins (Nickel and Rabouille, 2009). These include insulin-degrading enzymes (Zhao et al., 2009), FGF2, β-galactoside–specific lectins (galectin 1; Seelenmeyer et al., 2008), certain interleukins such as IL-1β, IL-18, and IL-33 (Keller et al., 2008), nuclear proteins such as HMGB1 (high mobility group protein B1; Gardella et al., 2002) and the homeoprotein engrailed (Maizel et al., 2002), and Dictyostelium discoideum AcbA (Kinseth et al., 2007). In yeast, the only protein known to be secreted in an ER/Golgi-independent manner is MAT a-factor, whose secretion is mediated by the ATP-binding cassette transporter, Ste6 (McGrath and Varshavsky, 1989).
Proteins that are secreted in an unconventional manner typically lack an N-terminal secretion signal, fail to traffic thorough the ER and Golgi, and do not possess protein modifications indicative of transit through the secretory pathway (Nickel and Seedorf, 2008). Many of these proteins are evolutionarily conserved, reinforcing their physiological relevance, and their secretion is often regulated. Many mechanisms for unconventional secretion, including vesicular and nonvesicular modes, have been debated and can be classified into four groups (Nickel and Seedorf, 2008): (1) direct translocation from the cytoplasm across the plasma membrane by transporters, (2) exosome secretion by fusion of multivesicular bodies (MVBs) with the plasma membrane, (3) plasma membrane blebbing followed by the shedding of extracellular vesicles, and (4) uptake of proteins into endosomes or lysosomes followed by their fusion with the plasma membrane. However, these models remain speculative in the absence of information regarding the proteins and pathways involved. In this study, we define the novel pathways and proteins necessary for AcbA-like protein (Acb1 in Pichia pastoris) secretion in yeast.
Results
A bioassay for yeast Acb1 using D. discoideum cells as the reporter
Encapsulation of prespore cells of D. discoideum is controlled by several intercellular signals to ensure appropriate timing during fruiting body formation. Acyl coenzyme A (CoA)–binding protein (ACBP), AcbA, is secreted by prespore cells and processed by a trypsin-like prestalk protease, TagC, to form the 34–amino acid peptide, SDF-2, which triggers rapid spore encapsulation (Anjard et al., 1998; Anjard and Loomis, 2005). The cell surface exposure of TagC protease and subsequent production of SDF-2 by D. discoideum requires activation by γ-amino butyric acid (GABA), which is produced by prespore cells in response to a steroid signal (Anjard et al., 2009). Previous experiments have shown that addition of recombinant AcbA alone does not induce sporulation in D. discoideum when TagC is not exposed on the surface; however, the addition of the SDF-2 peptide to these cells promotes sporulation (Anjard and Loomis, 2005).
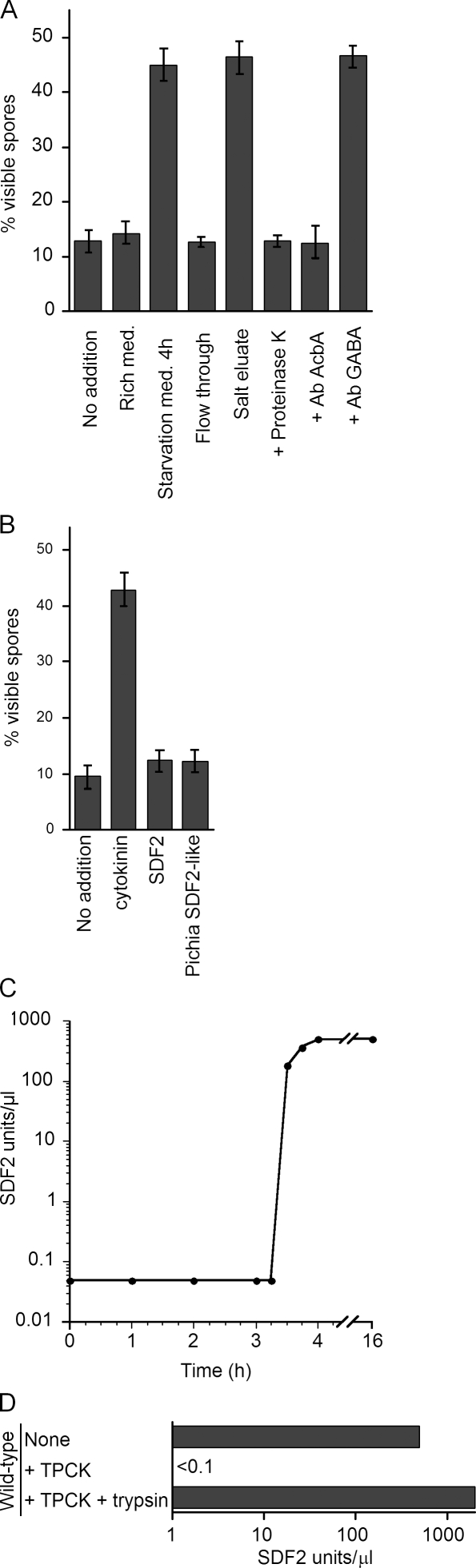
We tested whether P. pastoris culture supernatants display an SDF-2–like activity that could be revealed by a bioassay in D. discoideum. For this to work, the P. pastoris cells would have to secrete Acb1 and process it proteolytically into an SDF-2–like peptide, which would then bind to the D. discoideum SDF-2 receptor, DhkA, to promote sporulation. No activity was detected in the culture supernatant from wild-type P. pastoris cells grown in glucose medium supplemented with a nitrogen source (Fig. 1 A). However, after 4 h incubation in nitrogen starvation medium, SDF-2–like activity was detected in the supernatant. Like SDF-2, this activity was enriched upon purification by anion exchange chromatography. Moreover, the proteinaceous nature of the SDF-2–like activity was confirmed by its inactivation by proteinase K treatment. Preincubating the P. pastoris culture supernatant with antibodies to D. discoideum AcbA completely blocked the SDF-2–like activity. Moreover, purified recombinant P. pastoris Acb1 (PpAcb1) itself did not demonstrate this activity but did so upon trypsin digestion, suggesting the generation of SDF-2–like peptide activity.
Figure 1.
P. pastoris secretes Acb1 upon nitrogen starvation and processes it to generate SDF-2–like activity. (A) Cell-free supernatants were collected from either wild-type P. pastoris cells growing in rich medium or after 4 h in nitrogen starvation medium and tested for SDF-2–like activity using the bioassay as explained in Materials and methods. Samples were passed through an anion exchange chromatography, and the activity was eluted in 400 mM NaCl (salt eluate). An aliquot of this sample was protease treated (+ proteinase K; 20 µg) or incubated with either antibody to D. discoideum AcbA (1:500) or with antibody against GABA (1:5,000) and tested for SDF-2–like activity (Anjard and Loomis, 2005). Because KP cells can sporulate when presented with GABA, the experiment with antibody against GABA is a control to test whether the P. pastoris culture supernatant activated SDF-2 production in the KP cells by generating GABA. 10 nM recombinant P. pastoris HIS-Acb1 (PpAcb1) either untreated or digested with trypsin and purified (10 pM) were tested for SDF-2–like activity. (B) D. discoideum dhkA−/K (SDF-2 receptor null) cell bioassay. Induction was performed using 10 pM synthetic D. discoideum cytokinin and 1 pM SDF-2 peptides as controls. 1 µl supernatant from wild-type P. pastoris cells starved for 4 h was also tested. (C) Time course of the SDF-2–like activity secreted by P. pastoris cells. Samples were collected at the indicated times, purified, and quantified by the bioassay. (D) Acb1 is secreted and processed to generate SDF-2–like activity. Wild-type cells were nitrogen starved for 3 h before 1 µM TPCK was added to an aliquot of cells. 1 h later, the supernatant was collected from the untreated (none) and treated (+ TPCK) cells. TPCK was removed from part of the TPCK-treated sample and processed with trypsin as described in Materials and methods. All the samples were tested for SDF-2–like activity. Error bars indicate mean ± SD.
The specificity of the SDF-2–like activity was shown using D. discoideum cells lacking the histidine kinase SDF-2 receptor, DhkA (Wang et al., 1999). As expected, these dhkA-null mutant cells produced spores when treated with the unrelated cytokinin sporulation signal but not when incubated in the presence of either SDF-2 peptides or P. pastoris culture supernatants that displayed SFD-2–like activity in the bioassay (Fig. 1 B). These results and the fact that P. pastoris acb1Δ also showed no activity in the bioassay (see Fig. 2 B) provide strong evidence that upon nitrogen starvation, P. pastoris cells secrete Acb1 that is processed to produce SDF-2–like activity similar to that seen in D. discoideum.
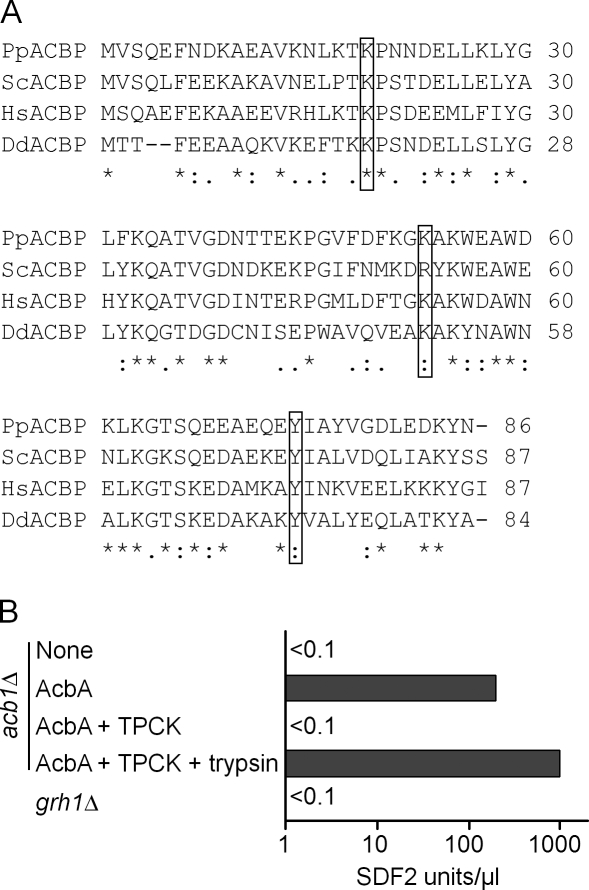
Figure 2.
Primary sequence of P. pastoris Acb1 and activity of a P. pastoris protease that processes AcbA to SDF-2. (A) The primary sequences of Acbps from P. pastoris (NCBI Protein database accession no. XP_002490495), S. cerevisiae (NCBI Protein database accession no. NP_011551), Homo sapiens (NCBI Protein database accession no. NP_001073331), and D. discoideum (NCBI Protein database accession no. XP_646321) were aligned in CLUSTALW using the default alignment parameters (*, conserved residues). Putative, conserved trypsin-like cleavage site residues (K) are highlighted in boxed regions. The MatGAT program was used to calculate percent identity (I) and similarity (S) of the P. pastoris Acb1 protein with that of S. cerevisiae (I = 63.2%; S = 77%), H. sapiens (I = 55.2%; S = 77%), and D. discoideum (I = 45.3%; S = 62.8%). (B) Trypsin-like protease activity of acb1Δ cells. The acb1Δ cells were incubated in nitrogen starvation medium for 4 h without any addition (none) or in the presence of 100 pM recombinant AcbA, and extracellular media were collected, purified, and assayed for SDF-2–like activity (AcbA). 1 µM TPCK inhibitor was added 3 h after transfer to starvation medium and assayed (AcbA + TPCK). Part of the sample was digested with trypsin (see Materials and methods), purified, and assayed using the bioassay (AcbA + TPCK + trypsin). The GRASP-null mutant (grh1Δ) was also processed and tested for SDF-2–like activity like the other samples. Experiments were performed three times.
Kinetics of Acb1 release and dependence on nitrogen starvation
Investigation of the kinetics of Acb1 release revealed that there was no activity in the supernatant during the first 3 h after nitrogen starvation, but there was a sudden rapid increase in Acb1 activity thereafter (Fig. 1 C). To ascertain whether the secretion was sustained after ∼4 h of starvation, the cells were subsequently resuspended in fresh starvation medium and incubated overnight. The overnight culture supernatant did not demonstrate any activity in the bioassay, indicating that P. pastoris cells most likely release Acb1 in a single burst upon nitrogen starvation. Addition of a nitrogen source at any time between 0–3 h after the switch to starvation medium blocked Acb1 production, suggesting that nitrogen starvation induces in a precise temporal manner the extracellular production of the SDF-2–like activity in yeast (unpublished data).
The processing of AcbA to SDF-2 in D. discoideum requires the activity of the TagC protease (Anjard and Loomis, 2005). To test whether a similar processing step was involved in the generation of SDF-2–like activity in P. pastoris, wild-type cells were incubated with the trypsin inhibitor TPCK (L-[tosylamido-2-phenyl] ethyl chloromethyl ketone) in starvation medium. Under these conditions, no SDF-2–like activity was detected (Fig. 1 D). Interestingly, when the Acb1 in this TPCK-treated culture supernatant was purified free of TPCK and activated by trypsin, the SDF-2–like activity was restored. Moreover, the SDF-2–like activity recovered after trypsin treatment was almost fourfold higher than when processed by the putative P. pastoris trypsin-like activity, an observation similar to one made for D. discoideum TagC. However, P. pastoris and other yeasts have no obvious TagC homologue.
The P. pastoris ACB1 gene is responsible for the secretion of SDF-2–like activity
Using the D. discoideum AcbA protein sequence, the BLAST program revealed a single ORF, Acb1, in the P. pastoris genome (De Schutter et al., 2009) with NCBI Protein database accession no. CAY68214.1 (Fig. 2 A). The primary sequences of ACBPs from human, D. discoideum, S. cerevisiae, and P. pastoris are highly conserved. The putative lysines (K19, K33, and K53 in P. pastoris) and the following amino acids (P20, Q34, and A54) that mark the cleavage sites for generating SDF-2 peptides are identical between the proteins.
A knockout strain of ACB1 (acb1Δ) in P. pastoris, like its S. cerevisiae counterpart, grew slowly even in rich media. This strain failed to generate SDF-2–like activity upon nitrogen starvation, suggesting that this ORF was responsible for the activity (Fig. 2 B). Furthermore, when nitrogen-starved acb1Δ cells were incubated with recombinant AcbA from D. discoideum, SDF-2–like activity was recovered. This activity was not seen if the extracellular medium from acb1Δ cells was incubated with TPCK 3 h after starvation followed 1 h later by a test for SDF-2–like activity. These results indicate that nitrogen-starved acb1Δ cells are unable to produce SDF-2–like activity but are still able to process recombinant D. discoideum AcbA to generate SDF-2.
Involvement of GRASP and autophagy proteins in Acb1 secretion
Previously, the Golgi reassembly and stacking protein (GRASP) was shown to be necessary for unconventional secretion of D. discoideum AcbA (Kinseth et al., 2007). Therefore, we asked whether Acb1 secretion in P. pastoris also depended on its GRASP counterpart, GRH1. Unlike the wild-type cells, the grh1Δ cells did not produce any SDF-2–like activity, confirming that the secretion of Acb1 in P. pastoris also requires the GRASP protein (Fig. 2 B).
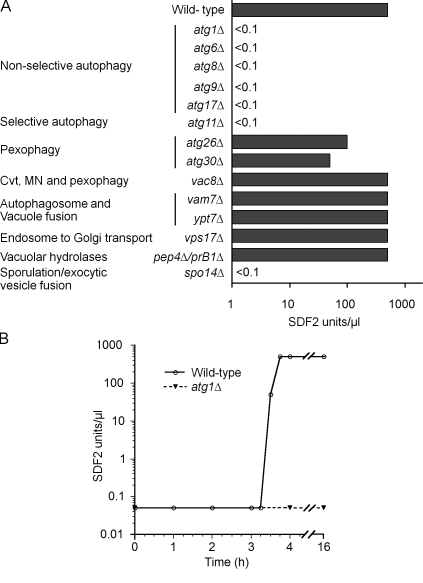
Based on our unpublished observations about the possible involvement of the autophagy machinery in secretion of AcbA in D. discoideum, we tested this hypothesis with P. pastoris autophagy mutants. P. pastoris atg5Δ and atg7Δ mutants failed to show any SDF-2–like activity (unpublished data). During autophagy, these proteins play critical roles in the formation of double-membrane vesicles, the autophagosomes, whose formation involves initiation, nucleation, expansion, and completion steps that are orchestrated by a cohort of proteins (Xie and Klionsky, 2007). We tested autophagy mutants atg1Δ (impaired in initiation, nucleation, and expansion steps of autophagosome formation), atg6Δ (required for all stages), atg8Δ (required for autophagosome completion), atg9Δ (provides membrane for autophagosomes), and atg17Δ (scaffold protein for organization of the autophagy-specific phagophore assembly site). None of these core autophagy mutants involved in the generation of autophagosomes released any SDF-2–like activity (Fig. 3 A).
Figure 3.
Autophagy and protein trafficking mutants affected in production of SDF-2–like activity. (A) Wild-type cells and various atg mutants were incubated in nitrogen starvation medium for 4 h, and cell supernatants were processed and tested for SDF-2–like activity. Cvt, cytosol to vacuole transport; MN, micronucleophagy. (B) Rapamycin induces Acb1 release. Wild-type (open circles) and atg1Δ cells (closed circles) were grown in SD+N medium up to 1 A600/ml, rapamycin (200 ng/ml final concentration) was added to the medium, and cell supernatants were collected at the indicated times, purified, and quantified by the bioassay. Experiments were performed three times.
Role of selective autophagy and autophagy-related pathways in Acb1 secretion
Atg11, which is necessary for selective, receptor-dependent autophagy-related pathways but not for autophagy of nonselective cargoes (Xie and Klionsky, 2007), was required for SDF-2–like activity. Next, we examined autophagy-related mutants that are not impaired in autophagosome formation. Mutants such as atg26Δ and atg30Δ are involved in the selective autophagic degradation of peroxisomes (pexophagy) but not in general autophagy (Nazarko et al., 2009). These mutants displayed SDF-2–like activity, albeit lower than that of wild-type cells, as did cells deficient in Vac8, a protein necessary for several selective autophagy-related pathways (cytosol to vacuole transport, pexophagy, and micronucleophagy; Fig. 3 A; Farré et al., 2009). These data suggest that although the requirement of Atg11 for activity in the bioassay points to some selectivity in the capture, transport, and secretion of Acb1 via autophagosomes, proteins implicated in known selective autophagy-related pathways do not play any direct role in Acb1 secretion.
Late steps of autophagy are not required for Acb1 secretion
During autophagy, autophagosomes eventually fuse with the vacuole, wherein their contents are degraded by resident vacuolar hydrolases. We explored whether autophagosome fusion with the vacuole and its subsequent degradation contribute to SDF-2–like activity. Deletion of the VAM7 gene, encoding a vacuolar membrane protein involved in the fusion of autophagosomes with the vacuole (Sato et al., 1998), or the YPT7 gene, encoding a small GTPase required for homotypic fusion during vacuole inheritance as well as for endosome–vacuole fusion events (Sato et al., 1998; Fratti and Wickner, 2007), did not impair SDF-2–like activity (Fig. 3 A). A mutant lacking Vps17, a component of the retromer complex required for endosome to Golgi retrograde protein transport (Seaman and Williams, 2002), also generated activity, as did the pep4Δ/prB1Δ double mutant, compromised in hydrolases required for the vacuolar breakdown of autophagic bodies (Fig. 3 A). Therefore, the vacuolar delivery of autophagosomes and the vacuolar turnover of autophagic bodies are not a prerequisite for Acb1 secretion.
Plasma membrane fusion events in Acb1 secretion
If Acb1 in vesicles is to be secreted, we would expect Acb1 vesicles to be involved directly or indirectly in membrane fusion events at the plasma membrane, a process that would likely involve SNARE proteins (Rothman and Warren, 1994). In S. cerevisiae, the plasma membrane t-SNARE is comprised of redundant subunits, allowing the formation of alternative t-SNARE complexes that could act at different sites or under varying conditions (Neiman, 2005). The t-SNARE is comprised of the either Sso1 or its partner Sso2 and the mammalian SNAP-25 homologues Sec9 or Spo20. Likewise, the v-SNARE contains either Snc1 or its homologue Snc2 (Neiman, 2005). Interestingly, P. pastoris has only single genes encoding Sso1/Sso2, Sec9/Spo20, and Snc1/Snc2 (De Schutter et al., 2009), predicting a t- and v-SNARE comprised of unique subunits for vesicle fusion at the plasma membrane. This lack of subunit redundancy complicated our analysis of the requirement of these subunits for Acb1 secretion because a deletion of any of these subunits is likely to render P. pastoris cells inviable. Indeed, we were unsuccessful in generating a haploid P. pastoris sso1Δ strain. To circumvent this problem, we deleted the SPO14 gene, which encodes a PLD that generates phosphatidic acid required for localization of the Spo20-containing S. cerevisiae t-SNARE at specific membranes (Neiman, 2005). When assayed for SDF-2–like activity, spo14 cells were negative (Fig. 3 A), indicating that it is required for secretion of SDF-2–like activity.
Acb1 secretion is triggered by rapamycin, an inducer of autophagy
We wondered if triggering autophagy during vegetative conditions would result in secretion of SDF-2–like activity. Exponentially growing wild-type cells were treated with rapamycin to pharmacologically induce autophagy. Culture supernatants were collected for 16 h after rapamycin addition and assayed. Interestingly, a burst of SDF-2 activity was detected at the same time after addition of rapamycin as observed after nitrogen starvation (3.5 h). The atg1Δ cells did not produce this activity under similar conditions, indicating that induction of autophagy is required for the activity (Fig. 3 B).
Peroxisomal membrane and matrix protein import is necessary for Acb1 secretion
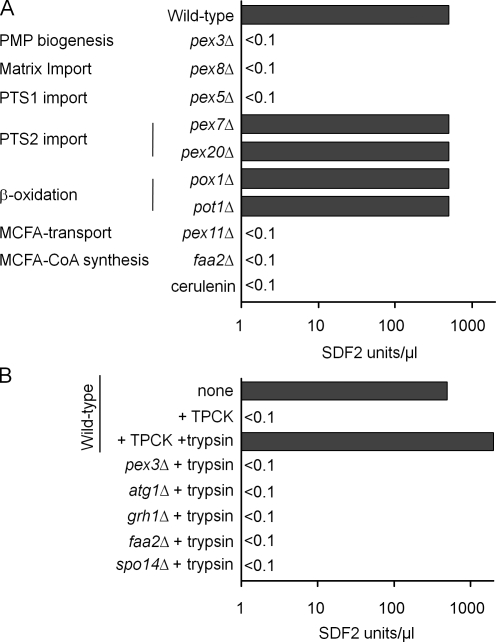
Because the D. discoideum and mammalian homologues of Acb1 bind medium- and long-chain fatty acyl-CoA (Mikkelsen and Knudsen, 1987; Anjard and Loomis, 2005), we asked whether peroxisomes, which generate and metabolize fatty acyl CoAs, influence Acb1 secretion. Pex3 and Pex19 are critical peroxins responsible for peroxisomal membrane protein assembly (Ma and Subramani, 2009). Protein import into the peroxisome matrix requires the presence of a multisubunit complex known as the importomer (comprised of Pex13, Pex14, Pex17, Pex3, Pex8, Pex2, Pex10, and Pex12) and another complex known as the receptor-recycling machinery (consisting of Pex1, Pex6, Pex4, and Pex22; Ma and Subramani, 2009). Both pex3Δ and pex8Δ cells were impaired in production of SDF-2–like activity (Fig. 4 A). Mutants lacking either Pex19 or each of the tested components of the importomer and receptor-recycling machineries also failed to yield activity in the bioassay (unpublished data).
Figure 4.
Acb1 secretion in mutants affecting peroxisome biogenesis and metabolism. (A) Wild-type cells and various mutants were incubated in nitrogen starvation medium at 1 A600/ml for 4 h, and cell supernatants were tested for SDF-2–like activity. An aliquot of the wild-type cells was treated with 100 µg/ml cerulenin upon shift to starvation medium and assayed for SDF-2–like activity after 4 h. (B) Cell supernatants from pex3Δ, atg1Δ, grh1Δ, faa2Δ, and spo14Δ mutants starved for 4 h in nitrogen-deficient medium were treated with trypsin to see whether any secreted Acb1 could be processed to generate SDF-2–like activity. The wild-type controls that were analyzed in this experiment are from Fig. 1 D but are shown again for clarity. Experiments were performed three times.
Peroxisomal matrix protein import is mediated by two peroxisome targeting signals (PTS1 and PTS2; Ma and Subramani, 2009). Pex5 is the PTS1 receptor, whereas Pex7 and Pex20 are the receptor/coreceptor for PTS2 cargo. Although pex5Δ cells did not show SDF-2–like activity, both pex7Δ and pex20Δ cells produced normal levels (Fig. 4 A). Thus, the PTS1 but not the PTS2 import pathway is required for secretion of Acb1.
Medium-chain fatty acyl CoA production, but not fatty acid β-oxidation, is necessary for Acb1 secretion
Because peroxisomes catalyze the β-oxidation of fatty acids (van Roermund et al., 2003), we investigated whether this pathway is essential for SDF-2 production. Acyl CoA oxidase (Pox1) and 3-ketoacyl-CoA thiolase (Pot1) are required for β-oxidation of long- and medium-chain fatty acids (MCFAs). The pot1Δ and pox1Δ mutants produced SDF-2–like activity (Fig. 4 A). Therefore, the peroxisomal β-oxidation of fatty acids is not required for SDF-2–like activity.
However, one mutant, pex11Δ, involved in both peroxisome division and the transport of MCFAs into peroxisomes, was deficient in the production of SDF-2–like activity (Fig. 4 A; van Roermund et al., 2000). We hypothesized that the fatty acid transport function of Pex11 and subsequent coupling of CoA to fatty acids might be necessary for Acb1 secretion. We tested this hypothesis by mutating the FAA2 gene encoding a PTS1-containing, peroxisomal matrix–localized, fatty acyl CoA synthetase that couples CoA to MCFAs transported into peroxisomes by Pex11 (Hettema et al., 1996). Interestingly, faa2Δ cells also had no SDF-2–like activity. These observations were corroborated when wild-type cells incubated in nitrogen starvation medium with cerulenin, an inhibitor of fatty acid synthesis (Marchesini and Poirier, 2003), yielded no SDF-2–like activity (Fig. 4 A). These results validate our hypothesis that the production of medium-chain fatty acyl CoA is necessary for SDF-2 production in P. pastoris.
Mutants that fail to generate SDF-2–like activity are deficient in Acb1 secretion and not its processing
To distinguish whether the pex3Δ, atg1Δ, and other mutants (grh1Δ, spo14Δ, and faa2Δ) that displayed no SDF-2–like activity are deficient in Acb1 secretion or its processing, we trypsinized the extracellular medium from representative mutants to artificially activate any Acb1 that might have been secreted from these cells and tested these samples. No activity was seen in the bioassay (Fig. 4 B). In contrast, Acb1 recovered from wild-type cells treated with TPCK can be activated by trypsin treatment (after removal of TPCK; Fig. 1 D). These results show that these P. pastoris mutants are deficient in Acb1 secretion not in its processing.
Physiological role of Acb1 in yeast
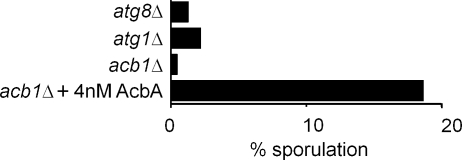
Because nitrogen starvation induces spore formation in yeast (Neiman, 2005), Acb1 might be required for sporulation. Using a P. pastoris sporulation assay, we found that atg1Δ and atg8Δ cells were severely compromised in their ability to form spores during nitrogen starvation (Fig. 5). Importantly, acb1Δ cells were drastically impaired in spore formation, and this phenotype was comparable with that of the atg1Δ and atg8Δ mutants. The sporulation defect of acb1Δ cells was complemented, albeit less efficiently, by the addition of recombinant AcbA, showing that extracellular Acb1 is necessary for sporulation (Fig. 5). Independent tests of the extracellular media from cultures confirmed that although wild-type cells did produce SDF-2–like activity, the sporulation-defective mutants (atg1Δ and atg8Δ) did not.
Figure 5.
Acb1 is necessary for sporulation. Wild-type, atg1Δ, atg8Δ, and acb1Δ cells were incubated in 1% potassium acetate solution at 30°C with rotation at 250 rpm. An aliquot of acb1Δ cells was incubated with 4 nM recombinant AcbA during the sporulation assay and analyzed for production of spores. 1 ml of the culture was processed as explained in Materials and methods to destroy viable cells. The spore preparation was appropriately diluted and plated on YPD plates. Plates were incubated for 2–3 d, and colonies were counted. Spore production for individual strains was expressed as the percentage of the wild-type spore count. One of two experiments is shown. Wild-type and acb1Δ cells were subjected to the sporulation assay as described in Materials and methods.
Discussion
A bioassay for yeast Acb1 secretion, processing, and function
In this study, we show that, upon nitrogen starvation, P. pastoris cells secrete an activity resembling that of SDF-2 produced by D. discoideum prespore cells (Fig. 1 A). In P. pastoris cells, this activity is derived from secreted Acb1, which is homologous to D. discoideum AcbA (Fig. 2 A). The Acb1 protein secreted by P. pastoris behaves like D. discoideum AcbA in that it is inactive in stimulating sporulation of D. discoideum KP cells unless it is activated either artificially by trypsin or by a trypsin-like protease of P. pastoris cells. Additionally, Acb1 is induced in a single burst in P. pastoris, just as AcbA is in D. discoideum, and induces sporulation in both organisms. This is consistent with the findings that proteins we show to be essential for Acb1 secretion, including several Atg proteins and Spo14, are also necessary for sporulation (Neiman, 2005). Similarly, cerulenin treatment of starved yeast cells impairs sporulation (Ohno et al., 1976). These facts establish beyond any doubt that P. pastoris cells secrete and produce SDF-2–like activity essentially like D. discoideum cells. Our bioassay for Acb1 using D. discoideum cells points to the remarkable evolutionary conservation of Acb1, its extracellular secretion, and its processing, allowing us to identify over a dozen components of this unconventional secretion pathway.
Acb1 is a model for an unconventional secretion pathway in yeast
Although AcbA is secreted by an unconventional pathway in D. discoideum (Kinseth et al., 2007), the underlying mechanisms have remained enigmatic. GRASP is required for the unconventional secretion of AcbA in D. discoideum (Kinseth et al., 2007) for Acb1 in yeasts as shown in this study (Fig. 2 B) and for the unconventional secretion of α-integrins in Drosophila melanogaster (Schotman et al., 2008). Although GRASP is a Golgi-associated protein, it is proposed to act at the plasma membrane as a tether for vesicle fusion events or in the trafficking of some other protein required at the plasma membrane for this function (Nickel and Rabouille, 2009). Consistent with this concept, GRASP is localized at the plasma membrane during epithelial cell remodeling in D. melanogaster (Schotman et al., 2008). Additionally, in D. discoideum cells lacking this protein, Acb1 is in punctate structures adjacent to the plasma membrane (Cabral et al., 2010), suggesting that GRASP is likely to act at the final fusion of Acb1 vesicles with the plasma membrane.
P. pastoris and S. cerevisiae have a single gene, GRH1, encoding GRASP. In contrast, mammals have two GRASP genes, whose roles in unconventional secretion remain unexplored, but these proteins are involved in cellular entry into mitosis and in the stacking of Golgi cisternae (Shorter et al., 1999; Feinstein and Linstedt, 2007).
Pathways involved in the unconventional secretion of Acb1
Peroxisomally generated medium-chain fatty acyl CoA but not fatty acid β-oxidation is required for Acb1 secretion. Mammalian ACBP (a homologue of AcbA) binds medium- and long-chain fatty acyl CoAs but not free fatty acids (Mikkelsen and Knudsen, 1987). Three lines of evidence prove that medium chain fatty acyl CoA generated in peroxisomes is necessary for Acb1 secretion. First, cells lacking Pex11, a protein required for peroxisome division and for the transport of MCFAs into peroxisomes (van Roermund et al., 2000), did not have SDF-2 activity (Fig. 4 A). Of the two functions of Pex11, it is the MCFA transporter function that is the key to Acb1 secretion, as indicated next. Second, Faa2, the PTS1-containing, peroxisomal fatty acyl CoA synthetase that couples CoA to MCFAs (Hettema et al., 1996) is necessary for Acb1 secretion (Fig. 4 A). Finally, the drug cerulenin, an inhibitor of fatty acid synthesis (Ohno et al., 1976), blocked Acb1 secretion (Fig. 4 A). Two possibilities could explain the MCFA CoA requirement for Acb1 secretion. The first is that although S. cerevisiae Acb1 binds fatty acyl CoAs of different chain lengths (Mikkelsen and Knudsen, 1987), perhaps only the small pool of Acb1 bound to MCFA CoA is secreted. Indeed, only ∼5% of the intracellular Acb1 is secreted in P. pastoris (unpublished data), which is similar to that observed in D. discoideum (Anjard and Loomis, 2005; Kinseth et al., 2007). This possibility is made likely by the observation that D. discoideum mutated AcbA affected in fatty acid binding is not secreted but can be processed to SDF-2 if exposed in vitro to the TagC protease (Cabral et al., 2010). Alternatively, there could be a role for MCFA CoA in the functions of compartments that are required for unconventional secretion (Faergeman et al., 2004).
The MCFA CoA requirement explains neatly why Acb1 secretion depends on peroxisomal membrane and matrix protein biogenesis and peroxisomal matrix protein import via the PTS1 but not the PTS2 import pathway (Fig. 4 A). Furthermore, as long as Pex11 can transport MCFA into peroxisomes for its activation, fatty acid β-oxidation is unnecessary for Acb1 secretion (Fig. 4 A). The dependence of unconventional secretion of Acb1 on lipid binding is not unprecedented in that FGF-2, another unconventionally secreted protein, must bind PIP2 for secretion (Temmerman et al., 2008).
Involvement of autophagosome formation proteins in Acb1 secretion
Atg proteins forming the core autophagic machinery (Atg1, Atg6, Atg8, Atg9, and Atg17) required for the initiation, nucleation, membrane expansion, and autophagosome formation steps of autophagy are necessary for Acb1 secretion in P. pastoris (Fig. 3 A; Xie and Klionsky, 2007). These results suggest that the Acb1 fatty acyl CoA conjugate must be captured into specialized autophagosomes (Acb1 vesicles) before Acb1 is secreted in yeast. An alternative possibility we have considered is that it is some inhibitor of Acb1 secretion that must be removed by autophagy, but we favor the model in which Acb1 is captured in autophagosomes based on data in D. discoideum that Acb1 is in punctate vesicular structures that accumulate near the plasma membrane in cells lacking GRASP (Cabral et al., 2010).
Acb1 secretion from Acb1 vesicles bypasses autophagosome–vacuole fusion and vacuolar hydrolytic events
The secretion of Acb1 implies that the normal fate of Acb1 vesicles must be different from that of general autophagosomes in that the fusion of Acb1 vesicles with the vacuole must be bypassed. Indeed, we demonstrate that the final stages of autophagy, involving fusion of autophagosomes with the vacuole (vam7 and ypt7 mutants; Fig. 3 A) and the subsequent degradation of the autophagosomal contents by vacuolar hydrolases (pep4 and prB1 mutants; Fig. 3 A) are unnecessary for SDF-2 production in P. pastoris. Additionally, a mutant that indirectly affects vacuolar protein sorting via a block in retrograde endosome to Golgi trafficking, vps17, does not affect Acb1 secretion (Fig. 3 A).
Fusion events at the plasma membrane are necessary for Acb1 secretion
Acb1 vesicles would need to fuse directly or indirectly with the plasma membrane if Acb1 is to be secreted. Support for a fusion event at the plasma membrane comes from the requirement of Spo14 for Acb1 secretion (Fig. 3 A).
The involvement of Spo14 in secretion of Acb1 may result from a requirement for phosphatidic acid produced by this PLD to direct the Sso1-containing t-SNARE to the plasma membrane (Neiman, 2005) of P. pastoris for the fusion of Acb1-containing vesicles. Consistent with this idea, Spo14-encoded PLD, along with Sso1, is required for exocytic vesicle fusion of prospore membrane precursor vesicles during meiosis-induced sporulation in yeast (Sreenivas et al., 1998; Nakanishi et al., 2006), and mammalian PLDs are involved in regulated exocytosis (Zeniou-Meyer et al., 2007; Temmerman et al., 2008; Disse et al., 2009). The capture of a small pool of an unconventionally secreted protein into intracellular vesicles independent of the normal secretory pathway followed by its exocytosis may also extend to mammalian cells as shown for α-synuclein, but it remains to be seen whether these vesicles are related to autophagosomes (Lee et al., 2005).
Do Acb1 vesicles fuse directly or indirectly at the plasma membrane?
Our model predicts that under certain circumstances, Acb1 vesicles, but not ordinary autophagosomes, are diverted for fusion with the plasma membrane. There is accumulating evidence that autophagosomes do not always fuse with vacuoles or lysosomes but instead can fuse with MVBs (an endosomal compartment) to form a novel compartment that can fuse with the plasma membrane. For example, in human-immortalized myelogenous leukemia line K562, which can differentiate in vitro to the reticulocyte lineage, fusion of autophagosomes with MVBs to create amphisomes is induced by starvation (Fader et al., 2008). These K562 cells capture proteins into MVBs and secrete exosomes generated by the fusion of MVBs with the plasma membrane (Fader et al., 2009). This is an unconventional secretion process whereby cytosolic and organelle-associated proteins are secreted under special conditions. In fact, exosome secretion is a property of many mammalian cell types (van Niel et al., 2006). The fusion of autophagosomes with MVBs and of MVBs with the plasma membrane provides a framework for the model for unconventional secretion of Acb1 in yeast (Fig. 6), as presented in this study and in the accompanying paper (see Duran et al. in this issue). Precedent for this idea comes from the recent demonstration that in MG6 microglial cells or primary microglia, the activation of purinergic P2X7 receptor by ATP results in the accumulation of autophagosome-like structures and the contents of the autophagolysosome or phagolysosome are released into the extracellular spaces (Takenouchi et al., 2009).
Figure 6.
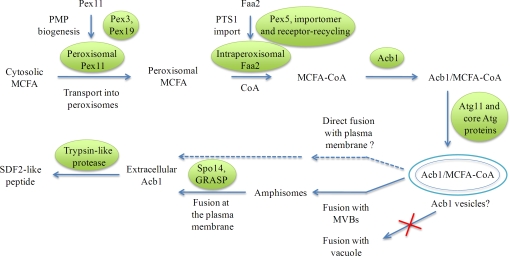
Working model for the unconventional secretion of Acb1 in P. pastoris. Pex11, a peroxisomal membrane protein associated with the peroxisome membrane, is transported to peroxisomes via the action of Pex3 and Pex19, whereas Faa2 is transported into peroxisomes via the PTS1 import pathway that relies on components of the peroxisomal importomer and the receptor-recycling machinery (see Results). Acb1 binds MCFA-CoA, which is derived by the Pex11-mediated transport of cytosolic MCFA into the peroxisome matrix, where the PTS1-containing protein Faa2 (fatty acyl CoA synthetase) couples it to CoA. The Acb1 bound to MCFA-CoA is captured in specialized autophagosomes (Acb1 vesicles) via the action of Atg11, involved in selective autophagy, and other core Atg proteins required for autophagosome formation. The normal fusion of Acb1 vesicles with vacuoles, but not that of general autophagic vesicles involved in macroautophagy, is bypassed during unconventional secretion. Acb1 vesicles might be delivered directly or indirectly for fusion with the plasma membrane, most likely via amphisomes. In addition to the proteins shown to be involved in Acb1 secretion, GRASP is also required. The fusion of a presently unknown vesicular compartment with the plasma membrane would release extracellular Acb1, which is then proteolytically cleaved by the P. pastoris trypsin-like protease, to generate extracellular SDF-2–like activity. We call this process the unconventional secretion via autophagosomes pathway.
Although this study does not distinguish whether Acb1 vesicles or some other compartment that has received their contents fuse with the plasma membrane, the literature cited in the previous paragraph and the accompanying paper (Duran et al., 2010) make us favor an indirect mechanism in which autophagosomes fuse with MVBs, which in turn fuse with the plasma membrane. To distinguish the secretion of Acb1 via an autophagy-related process distinct from general macroautophagy and other forms of selective autophagy, we have named this mode of Acb1 transport as the unconventional secretion via autophagosomes pathway (Fig. 6).
Unconventional secretion is regulated
Conventional secretion pathways are subject to many modes of regulation that are linked to dynamic physiological responses, and which, when dysfunctional, cause disease. Many unconventional secretion pathways are also regulated (Nickel and Seedorf, 2008). Our results show that Acb1 secretion is inactive in cells growing in rich medium but triggered by nitrogen starvation (Fig. 1, A and C). We also found stimulation of autophagy by rapamycin-induced Acb1 secretion in P. pastoris even in rich medium (Fig. 3 B). Interestingly, the secretion of the human homologue of Acb1 is also regulated (Swinnen et al., 1996; Qian et al., 2008). In K562 cells, the fusion of autophagosomes with MVBs and of amphisomes with the plasma membrane is induced by starvation (Fader et al., 2008).
Evolutionary implications of the unconventional secretion pathway
The AcbA-like proteins are remarkably conserved in S. cerevisiae, P. pastoris, D. discoideum, and mammals (Fig. 2 A), and as shown in this study, their modes of secretion and processing are also likely to be conserved evolutionarily (Knudsen et al., 1993). Interestingly, the human homologue of AcbA is the precursor of secreted neuropeptides that modulate GABAA receptor function in central nervous system neurons (Costa and Guidotti, 1991) and is involved in the regulation of multiple biological processes such as acyl CoA metabolism, steroidogenesis, and insulin secretion (Swinnen et al., 1996). Given the interest in the mammalian ACBP, the experiments presented in this study open several avenues for the exploration of the secretion and function of ACBP.
A second aspect of the conservation of Acb1 relates to its physiological function. It is interesting that a protein secreted by unicellular organisms such as P. pastoris and S. cerevisiae elicits a complex developmental response involving enhanced spore encapsulation in the multicellular D. discoideum. Because sporulation is induced by starvation (Crandall and Lawrence, 1980) and by rapamycin (Zheng and Schreiber, 1997), we wondered whether Acb1 might play a role in yeast sporulation, and indeed it did (Fig. 5).
This work elucidates the broad outlines of a complex pathway for unconventional secretion, shedding light on an unsolved problem that is relevant to many physiologically important proteins. The process of unconventional secretion could, in fact, be as complex, interesting, and physiologically relevant as conventional secretion and its regulation. What is fascinating is that Acb1 and its homologues, as well as its secretion pathway, are conserved between organisms that diverged over one billion years ago, making it likely that we have uncovered details of an evolutionarily ancient fundamental process.
Materials and methods
Yeast strains and growth conditions
Yeast strains used are summarized in Table S1. P. pastoris (PPY12; wild type) cells were grown in rich YPD medium (1% yeast extract, 2% peptone, and 2% glucose) at 30°C on a shaker set at 250 rpm. For starvation, the cells were incubated in nitrogen starvation medium (SD-N; 0.67% yeast nitrogen base without ammonium sulfate and 2% glucose).
Sample preparation for bioassay
Exponentially growing P. pastoris strains were transferred to nitrogen starvation medium at 1 A600/ml and incubated at 30°C on a shaker set at 250 rpm. At specific time points, 1 ml culture was collected and centrifuged at 14,000 g at room temperature. 500 µl supernatant was collected and centrifuged again to remove any yeast cells. 200 µl supernatant was collected and stored at −20°C until tested.
Activation of Acb1 by trypsin
50 µl of sample was incubated overnight in a final volume of 100 µl with 10 µg trypsin at 37°C in 50 mM Tris buffer, pH 8.0, and 1 mM CaCl2 to process Acb1. SDF-2–like activity was purified using an anion exchange resin, washed with starvation buffer containing 200 mM NaCl, and eluted with 50 µl with starvation buffer containing 400 mM NaCl. TPCK was removed before trypsin treatment using an anion exchange resin that binds the P. pastoris Acb1 protein at low salt concentration. After washing in starvation buffer, the uncleaved Acb1 was eluted with starvation buffer containing 200 mM NaCl before being digested with trypsin. Samples were quantified using the bioassay.
Purification of SDF-2 activity and bioassay
The cell-free extracellular medium from yeast was adsorbed on exchange resin (A-25; Anion), washed with 200 mM NaCl, and eluted with 400 mM NaCl. The D. discoideum KP strain is a previously described derivative of strain Ax2-overexpressing protein kinase A under the control of its endogenous promoter (Anjard et al., 1998). Vegetative KP cells were harvested during exponential growth and resuspended at 4.5 × 104 cells in 12.5 ml starvation buffer (20 mM MES, pH 6.2, 20 mM NaCl, 20 mM KCl, 1 mM MgSO4, and 1 mM CaCl2) containing 5 mM cAMP. 500 µl aliquots were distributed in each well of a 24-well dish to a density of 2 × 103 cells/cm2. The cells were incubated overnight at 23°C before addition of test samples. Spore formation was scored by counting spores and undifferentiated cells 1 h after addition of the samples. SDF-2 activity was determined by serial dilution of the sample before addition to KP cells. 1 U corresponds to the lowest dilution giving full induction of spore formation.
The spore differentiation assay using dhkA−/K (SDF-2 receptor null) cells was performed as described previously with slight modifications of the assay used for KP cells (Wang et al., 1999). The dhkA−/K cells were collected at a density of 1–4 × 106/ml, pelleted, resuspended in starvation buffer, and plated at a density of 2 × 103 cells/cm2 in starvation buffer containing 5 mM cAMP. After overnight incubation at 23°C, the indicated inducers were added. Spore formation was scored 1 h later. Each experiment was repeated at least three times, and the error bars represent SD. For the other figures, because the bioassay relies on twofold serial dilutions, a statistical SD cannot be calculated to determine the error bars. However, based on the properties of serial dilution–based assays, we estimate that the values present a reliability range of ±50%. The assay has also been represented in a similar manner in our previous study (Anjard and Loomis, 2005).
Strain and plasmid construction
For gene deletions, the 5′ flanking regions (∼1 kb) of the respective ORFs (ACB1: NCBI Protein database accession no. CAY68214.1; GRH1: NCBI Protein database accession no. CAY72138.1; YPT7: NCBI Protein database accession no. CAY72169.1; and FAA2: NCBI Protein database accession no. CAY71596.1) were amplified from PPY12 genomic DNA and cloned into the 5′ multiple cloning site of pSEB44 (Léon et al., 2006). Next, the 3′ flanking regions (∼1 kb) of the ORFs were amplified and further cloned into the 3′ multiple cloning site of the pSEB44 construct containing the 5′ flanking regions. The disruption cassette was amplified from the resultant construct and transformed into the PPY12 strain. G418-resistant clones were screened by PCR and product size analysis. A similar strategy was used for the SPO14 (CAY70694.1) deletion using a zeocin disruption plasmid, pAP1.
Yeast sporulation assay
P. pastoris wild-type and mutant cells were grown to late exponential phase in rich medium and transferred to 1% potassium acetate solution at 1 A600/ml. After 3 d of incubation at 30°C in a rotator shaker set at 250 rpm, cells from 1 ml culture were washed and resuspended in 1 ml of sterile water. 50 µl of this suspension was treated with 1 µl zymolyase solution (1 KU/ml stock solution) at 30°C for 1 h to destroy any vegetative cells. The reaction was stopped by addition of 500 µl of 0.5 M ice-cold sorbitol solution. Appropriate serial dilutions were performed in sterile water and plated onto YPD agar plates. The plates were incubated at 30°C for 2–3 d, and the colonies were counted.
Online supplemental material
Table S1 lists the strains used in this study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200911149/DC1.
Supplementary Material
Acknowledgments
We thank Dr. Vivek Malhotra for discussions and the exchange of data, Adam Kuspa for sharing results and insights, Richard Mathewson and Jean-Claude Farré for strains, and Ms. Aparna Hebbar for technical help.
This work was supported by grants from the National Institutes of Health (DK41737 and GM069373 to S. Subramani and GM78175 to W.F. Loomis).
Footnotes
Abbreviations used in this paper:
- ACBP
- acyl CoA–binding protein
- CoA
- coenzyme A
- GABA
- γ-amino butyric acid
- GRASP
- Golgi reassembly and stacking protein
- MCFA
- medium-chain fatty acid
- MVB
- multivesicular body
References
- Anjard C., Loomis W.F.. 2005. Peptide signaling during terminal differentiation of Dictyostelium. Proc. Natl. Acad. Sci. USA. 102:7607–7611. 10.1073/pnas.0501820102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anjard C., Zeng C., Loomis W.F., Nellen W.. 1998. Signal transduction pathways leading to spore differentiation in Dictyostelium discoideum. Dev. Biol. 193:146–155. 10.1006/dbio.1997.8804 [DOI] [PubMed] [Google Scholar]
- Anjard C., Su Y., Loomis W.F.. 2009. Steroids initiate a signaling cascade that triggers rapid sporulation in Dictyostelium. Development. 136:803–812. 10.1242/dev.032607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabral M., Anjard C., Malhotra V., Fuller D., Loomis W.F., Kuspa A.. 2010. GRASP dependent protein secretion is mediated by vesicles. Eukaryot. Cell. In press. [Google Scholar]
- Costa E., Guidotti A.. 1991. Diazepam binding inhibitor (DBI): a peptide with multiple biological actions. Life Sci. 49:325–344. 10.1016/0024-3205(91)90440-M [DOI] [PubMed] [Google Scholar]
- Crandall M., Lawrence L.J.. 1980. Sporulation in Hansenula wingei is induced by nitrogen starvation in maltose-containing media. J. Bacteriol. 142:276–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Schutter K., Lin Y.C., Tiels P., Van Hecke A., Glinka S., Weber-Lehmann J., Rouzé P., Van de Peer Y., Callewaert N.. 2009. Genome sequence of the recombinant protein production host Pichia pastoris. Nat. Biotechnol. 27:561–566. 10.1038/nbt.1544 [DOI] [PubMed] [Google Scholar]
- Disse J., Vitale N., Bader M.F., Gerke V.. 2009. Phospholipase D1 is specifically required for regulated secretion of von Willebrand factor from endothelial cells. Blood. 113:973–980. 10.1182/blood-2008-06-165282 [DOI] [PubMed] [Google Scholar]
- Duran J.M., Anjard C., Stefan C., Loomis W.F., Malhotra V.. 2010. Unconventional secretion of Acb1 is mediated by autophagosomes. J. Cell Biol. 188:527–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fader C.M., Sánchez D., Furlán M., Colombo M.I.. 2008. Induction of autophagy promotes fusion of multivesicular bodies with autophagic vacuoles in k562 cells. Traffic. 9:230–250. [DOI] [PubMed] [Google Scholar]
- Fader C.M., Sánchez D.G., Mestre M.B., Colombo M.I.. 2009. TI-VAMP/VAMP7 and VAMP3/cellubrevin: two v-SNARE proteins involved in specific steps of the autophagy/multivesicular body pathways. Biochim. Biophys. Acta. 1793:1901–1916. 10.1016/j.bbamcr.2009.09.011 [DOI] [PubMed] [Google Scholar]
- Faergeman N.J., Feddersen S., Christiansen J.K., Larsen M.K., Schneiter R., Ungermann C., Mutenda K., Roepstorff P., Knudsen J.. 2004. Acyl-CoA-binding protein, Acb1p, is required for normal vacuole function and ceramide synthesis in Saccharomyces cerevisiae. Biochem. J. 380:907–918. 10.1042/BJ20031949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farré J.C., Krick R., Subramani S., Thumm M.. 2009. Turnover of organelles by autophagy in yeast. Curr. Opin. Cell Biol. 21:522–530. 10.1016/j.ceb.2009.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinstein T.N., Linstedt A.D.. 2007. Mitogen-activated protein kinase kinase 1-dependent Golgi unlinking occurs in G2 phase and promotes the G2/M cell cycle transition. Mol. Biol. Cell. 18:594–604. 10.1091/mbc.E06-06-0530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fratti R.A., Wickner W.. 2007. Distinct targeting and fusion functions of the PX and SNARE domains of yeast vacuolar Vam7p. J. Biol. Chem. 282:13133–13138. 10.1074/jbc.M700584200 [DOI] [PubMed] [Google Scholar]
- Gardella S., Andrei C., Ferrera D., Lotti L.V., Torrisi M.R., Bianchi M.E., Rubartelli A.. 2002. The nuclear protein HMGB1 is secreted by monocytes via a non-classical, vesicle-mediated secretory pathway. EMBO Rep. 3:995–1001. 10.1093/embo-reports/kvf198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hettema E.H., van Roermund C.W., Distel B., van den Berg M., Vilela C., Rodrigues-Pousada C., Wanders R.J., Tabak H.F.. 1996. The ABC transporter proteins Pat1 and Pat2 are required for import of long-chain fatty acids into peroxisomes of Saccharomyces cerevisiae. EMBO J. 15:3813–3822. [PMC free article] [PubMed] [Google Scholar]
- Keller M., Rüegg A., Werner S., Beer H.D.. 2008. Active caspase-1 is a regulator of unconventional protein secretion. Cell. 132:818–831. 10.1016/j.cell.2007.12.040 [DOI] [PubMed] [Google Scholar]
- Kinseth M.A., Anjard C., Fuller D., Guizzunti G., Loomis W.F., Malhotra V.. 2007. The Golgi-associated protein GRASP is required for unconventional protein secretion during development. Cell. 130:524–534. 10.1016/j.cell.2007.06.029 [DOI] [PubMed] [Google Scholar]
- Knudsen J., Mandrup S., Rasmussen J.T., Andreasen P.H., Poulsen F., Kristiansen K.. 1993. The function of acyl-CoA-binding protein (ACBP)/diazepam binding inhibitor (DBI). Mol. Cell. Biochem. 123:129–138. 10.1007/BF01076484 [DOI] [PubMed] [Google Scholar]
- Lee H.J., Patel S., Lee S.J.. 2005. Intravesicular localization and exocytosis of alpha-synuclein and its aggregates. J. Neurosci. 25:6016–6024. 10.1523/JNEUROSCI.0692-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Léon S., Zhang L., McDonald W.H., Yates J., III, Cregg J.M., Subramani S.. 2006. Dynamics of the peroxisomal import cycle of PpPex20p: ubiquitin-dependent localization and regulation. J. Cell Biol. 172:67–78. 10.1083/jcb.200508096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C., Subramani S.. 2009. Peroxisome matrix and membrane protein biogenesis. IUBMB Life. 61:713–722. 10.1002/iub.196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maizel A., Tassetto M., Filhol O., Cochet C., Prochiantz A., Joliot A.. 2002. Engrailed homeoprotein secretion is a regulated process. Development. 129:3545–3553. [DOI] [PubMed] [Google Scholar]
- Marchesini S., Poirier Y.. 2003. Futile cycling of intermediates of fatty acid biosynthesis toward peroxisomal beta-oxidation in Saccharomyces cerevisiae. J. Biol. Chem. 278:32596–32601. 10.1074/jbc.M305574200 [DOI] [PubMed] [Google Scholar]
- McGrath J.P., Varshavsky A.. 1989. The yeast STE6 gene encodes a homologue of the mammalian multidrug resistance P-glycoprotein. Nature. 340:400–404. 10.1038/340400a0 [DOI] [PubMed] [Google Scholar]
- Mikkelsen J., Knudsen J.. 1987. Acyl-CoA-binding protein from cow. Binding characteristics and cellular and tissue distribution. Biochem. J. 248:709–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi H., Morishita M., Schwartz C.L., Coluccio A., Engebrecht J., Neiman A.M.. 2006. Phospholipase D and the SNARE Sso1p are necessary for vesicle fusion during sporulation in yeast. J. Cell Sci. 119:1406–1415. 10.1242/jcs.02841 [DOI] [PubMed] [Google Scholar]
- Nazarko T.Y., Farré J.C., Subramani S.. 2009. Peroxisome size provides insights into the function of autophagy-related proteins. Mol. Biol. Cell. 20:3828–3839. 10.1091/mbc.E09-03-0221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiman A.M. 2005. Ascospore formation in the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 69:565–584. 10.1128/MMBR.69.4.565-584.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickel W., Rabouille C.. 2009. Mechanisms of regulated unconventional protein secretion. Nat. Rev. Mol. Cell Biol. 10:148–155. 10.1038/nrm2617 [DOI] [PubMed] [Google Scholar]
- Nickel W., Seedorf M.. 2008. Unconventional mechanisms of protein transport to the cell surface of eukaryotic cells. Annu. Rev. Cell Dev. Biol. 24:287–308. 10.1146/annurev.cellbio.24.110707.175320 [DOI] [PubMed] [Google Scholar]
- Ohno T., Awaya J., Omura S.. 1976. Inhibition of sporulation by cerulenin and its reversion by exogenous fatty acids in Saccharomyces cerevisiae. Antimicrob. Agents Chemother. 9:42–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Z., Bilderback T.R., Barmack N.H.. 2008. Acyl coenzyme A-binding protein (ACBP) is phosphorylated and secreted by retinal Müller astrocytes following protein kinase C activation. J. Neurochem. 105:1287–1299. 10.1111/j.1471-4159.2008.05229.x [DOI] [PubMed] [Google Scholar]
- Rothman J.E., Warren G.. 1994. Implications of the SNARE hypothesis for intracellular membrane topology and dynamics. Curr. Biol. 4:220–233. 10.1016/S0960-9822(00)00051-8 [DOI] [PubMed] [Google Scholar]
- Sato T.K., Darsow T., Emr S.D.. 1998. Vam7p, a SNAP-25-like molecule, and Vam3p, a syntaxin homolog, function together in yeast vacuolar protein trafficking. Mol. Cell. Biol. 18:5308–5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schotman H., Karhinen L., Rabouille C.. 2008. dGRASP-mediated noncanonical integrin secretion is required for Drosophila epithelial remodeling. Dev. Cell. 14:171–182. 10.1016/j.devcel.2007.12.006 [DOI] [PubMed] [Google Scholar]
- Seaman M.N., Williams H.P.. 2002. Identification of the functional domains of yeast sorting nexins Vps5p and Vps17p. Mol. Biol. Cell. 13:2826–2840. 10.1091/mbc.02-05-0064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seelenmeyer C., Stegmayer C., Nickel W.. 2008. Unconventional secretion of fibroblast growth factor 2 and galectin-1 does not require shedding of plasma membrane-derived vesicles. FEBS Lett. 582:1362–1368. 10.1016/j.febslet.2008.03.024 [DOI] [PubMed] [Google Scholar]
- Shorter J., Watson R., Giannakou M.E., Clarke M., Warren G., Barr F.A.. 1999. GRASP55, a second mammalian GRASP protein involved in the stacking of Golgi cisternae in a cell-free system. EMBO J. 18:4949–4960. 10.1093/emboj/18.18.4949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreenivas A., Patton-Vogt J.L., Bruno V., Griac P., Henry S.A.. 1998. A role for phospholipase D (Pld1p) in growth, secretion, and regulation of membrane lipid synthesis in yeast. J. Biol. Chem. 273:16635–16638. 10.1074/jbc.273.27.16635 [DOI] [PubMed] [Google Scholar]
- Swinnen J.V., Esquenet M., Rosseels J., Claessens F., Rombauts W., Heyns W., Verhoeven G.. 1996. A human gene encoding diazepam-binding inhibitor/acy1-CoA-binding protein: transcription and hormonal regulation in the androgen-sensitive human prostatic adenocarcinoma cell line LNCaP. DNA Cell Biol. 15:197–208. 10.1089/dna.1996.15.197 [DOI] [PubMed] [Google Scholar]
- Takenouchi T., Nakai M., Iwamaru Y., Sugama S., Tsukimoto M., Fujita M., Wei J., Sekigawa A., Sato M., Kojima S., et al. 2009. The activation of P2X7 receptor impairs lysosomal functions and stimulates the release of autophagolysosomes in microglial cells. J. Immunol. 182:2051–2062. 10.4049/jimmunol.0802577 [DOI] [PubMed] [Google Scholar]
- Temmerman K., Ebert A.D., Müller H.M., Sinning I., Tews I., Nickel W.. 2008. A direct role for phosphatidylinositol-4,5-bisphosphate in unconventional secretion of fibroblast growth factor 2. Traffic. 9:1204–1217. 10.1111/j.1600-0854.2008.00749.x [DOI] [PubMed] [Google Scholar]
- van Niel G., Porto-Carreiro I., Simoes S., Raposo G.. 2006. Exosomes: a common pathway for a specialized function. J. Biochem. 140:13–21. 10.1093/jb/mvj128 [DOI] [PubMed] [Google Scholar]
- van Roermund C.W., Tabak H.F., van Den Berg M., Wanders R.J., Hettema E.H.. 2000. Pex11p plays a primary role in medium-chain fatty acid oxidation, a process that affects peroxisome number and size in Saccharomyces cerevisiae. J. Cell Biol. 150:489–498. 10.1083/jcb.150.3.489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Roermund C.W., Waterham H.R., Ijlst L., Wanders R.J.. 2003. Fatty acid metabolism in Saccharomyces cerevisiae. Cell. Mol. Life Sci. 60:1838–1851. 10.1007/s00018-003-3076-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N., Söderbom F., Anjard C., Shaulsky G., Loomis W.F.. 1999. SDF-2 induction of terminal differentiation in Dictyostelium discoideum is mediated by the membrane-spanning sensor kinase DhkA. Mol. Cell. Biol. 19:4750–4756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Klionsky D.J.. 2007. Autophagosome formation: core machinery and adaptations. Nat. Cell Biol. 9:1102–1109. 10.1038/ncb1007-1102 [DOI] [PubMed] [Google Scholar]
- Zeniou-Meyer M., Zabari N., Ashery U., Chasserot-Golaz S., Haeberlé A.M., Demais V., Bailly Y., Gottfried I., Nakanishi H., Neiman A.M., et al. 2007. Phospholipase D1 production of phosphatidic acid at the plasma membrane promotes exocytosis of large dense-core granules at a late stage. J. Biol. Chem. 282:21746–21757. 10.1074/jbc.M702968200 [DOI] [PubMed] [Google Scholar]
- Zhao J., Li L., Leissring M.A.. 2009. Insulin-degrading enzyme is exported via an unconventional protein secretion pathway. Mol. Neurodegener. 4:4. 10.1186/1750-1326-4-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X.F., Schreiber S.L.. 1997. Target of rapamycin proteins and their kinase activities are required for meiosis. Proc. Natl. Acad. Sci. USA. 94:3070–3075. 10.1073/pnas.94.7.3070 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.