Abstract
Fluorescence microscopy is one of the most powerful tools to investigate complex cellular processes such as cell division, cell motility, or intracellular trafficking. The availability of RNA interference (RNAi) technology and automated microscopy has opened the possibility to perform cellular imaging in functional genomics and other large-scale applications. Although imaging often dramatically increases the content of a screening assay, it poses new challenges to achieve accurate quantitative annotation and therefore needs to be carefully adjusted to the specific needs of individual screening applications. In this review, we discuss principles of assay design, large-scale RNAi, microscope automation, and computational data analysis. We highlight strategies for imaging-based RNAi screening adapted to different library and assay designs.
Microscopy for functional genomics and systems biology
High-throughput methodologies such as proteomics, expression profiling, or protein interaction mapping have established an era of systems biology, which aims at understanding biological processes through comprehensive identification of network components and their interplay (Hartwell et al., 1999). Such large-scale approaches can generate valuable parts lists and provide basic insight into their modular organization, but they do not resolve spatial and temporal aspects of protein function and regulation (Megason and Fraser, 2007). Most biological processes occur spatially confined at distinct subcellular sites and vary between different cells, thus calling for methods capable of sampling spatial and temporal patterns at the single cell level.
Fluorescence microscopy provides an ideal tool to study complex biological processes with high spatiotemporal resolution. Fluorescent proteins allow one to label virtually any cellular structure or signaling component under physiological conditions in live cells (Giepmans et al., 2006). A wide range of fluorescent biosensors and imaging modalities provides the possibility to detect steady-state protein dynamics, posttranslational modifications, protein–protein interactions, and small molecules (Lippincott-Schwartz et al., 2003; Giepmans et al., 2006). Microscopy has long been tedious and difficult to perform in a systematic and quantitative way. Therefore, imaging-based assays have in most cases been restricted to manual low-throughput experiments, for example, detailed mechanistic studies of few selected candidate genes. Recent developments in robotics for sample preparation and automation of microscope control now enable one to perform imaging at a large scale (Pepperkok and Ellenberg, 2006). The key challenge often remains the annotation of complex phenotypic patterns in huge image datasets. Many studies still rely on visual scoring and manual annotation, which is slow, error prone, and potentially biased by the user. Significant progress has been made through the implementation of computer vision methods for multidimensional data analysis (Gerlich et al., 2001; Gerlich and Ellenberg, 2003) and supervised machine learning approaches for automated classification of cellular and subcellular phenotypes (Conrad et al., 2004; Neumann et al., 2006; Glory and Murphy, 2007; Jones et al., 2009; Walter et al., 2009).
In this review, we provide an overview of imaging-based screening strategies. We focus on biological assay design, automated image acquisition, and computational analysis. We further discuss advanced imaging options and how throughput and content of screening assays can be balanced. Finally, we present a perspective on how integration of experimental robotics, image analysis tools, and large-scale data resources may be used to further automate the discovery process.
Biological assays: content versus throughput
The most basic readout for imaging-based assays is total cellular fluorescence intensity of immunodetected antigens or overexpressed fluorescent reporters (Fig. 1 A). For example, this can be used to score the expression of marker genes (Müller et al., 2005; Loo et al., 2007), DNA content for cell cycle progression (Kittler et al., 2007), lipoprotein uptake (Bartz et al., 2009), mitochondrial Ca2+ transport (Jiang et al., 2009), or virus entry into cells (Pelkmans et al., 2005; Brass et al., 2008; Krishnan et al., 2008; Plouffe et al., 2008).
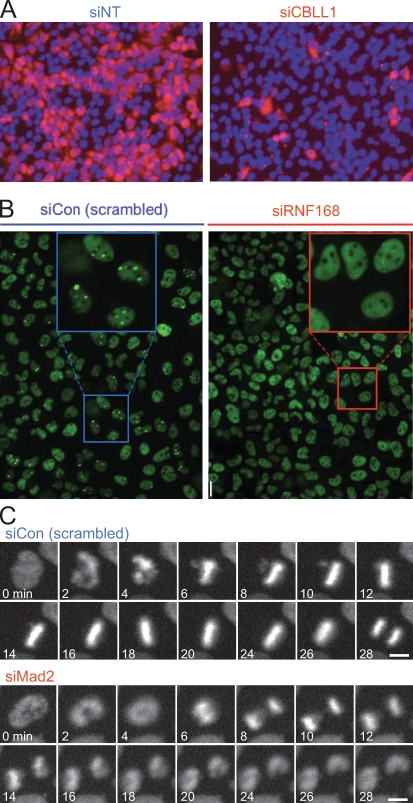
Figure 1.
Examples for imaging-based assays. (A) Intensity-based assay. In this screen for human genes associated with West Nile virus infection, cell nuclei were labeled with DAPI (blue) and stained by immunofluorescence against a viral epitope (red). Genes that reduced the intensity of viral epitope staining were scored as hits. (left) Negative control (NT-siRNA). (right) Hit (CBLL1-siRNA) identified in the screen. This panel is reprinted with permission from Nature Publishing Group (Krishnan et al., 2008). (B) Morphology-based assay for genes functioning in the assembly of DNA double-strand repair foci. Cells were stained by immunofluorescence for 53BP1, a protein which localizes to spontaneously occurring DNA damage foci, visible as bright green spots (see negative control; left). In a hit obtained in this screen (siRNF168), 53BP1 failed to target to spots and was instead dispersed throughout the nucleoplasm. This panel is reprinted with permission from Elsevier (Doil et al., 2009). (C) Live imaging-based assay for mitotic timing. HeLa cells stably expressing chromatin marker H2B-mCherry (Steigemann et al., 2009) were imaged for 24 h on an incubated screening microscope (Schmitz and Gerlich, 2009). Mitotic phenotypes were detected based on the timing from nuclear breakdown (2 min in siCon) until anaphase (28 min in siCon). Cells depleted for the spindle checkpoint protein Mad2 prematurely enter anaphase (8 min). Images provided by D.W. Gerlich. Bars, 10 µm.
Another class of assays scores cellular morphology features (Fig. 1 B). For example, the pattern of cytoskeletal or chromatin markers can serve to probe cellular morphologies (Bakal et al., 2007; Liu et al., 2009), cell division phenotypes (Gönczy et al., 2000; Echard et al., 2004; Sönnichsen et al., 2005; Neumann et al., 2006; Draviam et al., 2007; Goshima et al., 2007), cell cycle progression (Boutros et al., 2004; Kittler et al., 2007), or DNA double-strand break repair (Doil et al., 2009). Although manual annotation of such assays is possible, this way of analyzing the images is very tedious and may be user biased. Fortunately, computational machine learning methods allow efficient annotation even of subtle morphological features (see Computational image analysis for quantitative phenotyping).
Fluorescent proteins can also be used to assay biochemical events in live cells. GFP-based biosensors have been engineered for visualization of protein–protein interactions (Ciruela, 2008) and posttranslational modifications (Aye-Han et al., 2009) as well as enzyme activity and small molecules (VanEngelenburg and Palmer, 2008). Imaging modalities such as fluorescence correlation spectroscopy (Haustein and Schwille, 2007), photobleaching and photoactivation (Lippincott-Schwartz et al., 2003), and chemical labeling of engineered target proteins (Johnsson, 2009) further enable the study of steady-state protein dynamics in living cells. All of these methods can, in principle, be applied to high-throughput imaging assays, opening new possibilities to screen for factors in very specific aspects of cellular signaling.
Time-resolved live imaging (Fig. 1 C) provides the highest content for assays on complex dynamic processes such as cell division (Sönnichsen et al., 2005; Neumann et al., 2006). Live cell imaging can be automated to a level that enables genome-scale RNAi screens (MitoCheck project; Neumann et al., 2010). Several commercial microscope platforms support automated time-lapse imaging of live cells. The key to live imaging-based screening is a stable incubation and careful optimization of the light dose to avoid photodamage (Schmitz and Gerlich, 2009). The most severe limitation of live imaging in screening is the complexity of data annotation.
Screening of hypothesis-derived candidate gene sets
Many initial functional genomics screens relied on simple intensity-based readouts, which can be scaled relatively easily to the full genome level. However, this approach provides little information on the underlying phenotype and its variability within the cell population. The application of higher-content imaging assays that incorporate spatial or temporal phenotypic patterns is much more labor intensive and therefore can require preselection of candidate genes to perform low- to medium-throughput screens (Fig. 2 A).
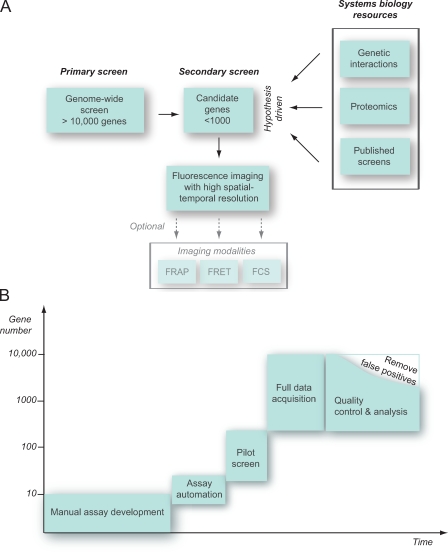
Figure 2.
Screening strategies. (A) Secondary screening on candidate gene sets either derived from genome-wide primary screens (left) or derived based on a hypothesis and systems biology resources such as genetic interaction, proteomics, or bioinformatics data (right). The smaller size of perturbation conditions allows high-resolution spatial and temporal imaging or assays sampling protein mobility (for example, FRAP) or protein–protein interaction (for example, by fluorescence resonance energy transfer [FRET] or fluorescence correlation spectroscopy [FCS]). (B) Typical time line for implementation of an RNAi screen. Screening assays typically derive from a manual visual assay development, which aims at maximizing the discrimination between selected positive and negative controls. In the next step, the automation of sample preparation, data acquisition, storage, and analysis parameters has to be implemented and tested in a pilot screen. After the acquisition of the entire genome, quality control procedures reduce the number of validated hits.
Candidate gene sets can be derived from a low-level functional assay in a genome-wide screen. Alternatively, published information (e.g., from other large-scale experiments or public databases) can serve to compile target gene lists. At this level, a basic hypothesis can be incorporated into the screening strategy. For example, it may be reasonable to screen only proteins localizing to a certain subcellular structure (Skop et al., 2004) or defined classes of enzymes like kinases (Pelkmans et al., 2005). The level of detail needed in a functional genomics assay as well as the number of experimental perturbation conditions will vary considerably depending on the biological question. In some applications, it may be advantageous to sample only very few experimental conditions with ultra high–content assays such as time-lapse imaging, whereas others may not require sophisticated assays and/or lack a rationale to define reasonable sets of candidate genes.
In any screening scenario, assay optimization affects both sample preparation as well as data analysis strategy. The assay optimization often requires more effort and time than that to perform the actual screen (Fig. 2 B). Once an assay has been established, it needs to be scaled and tested for realistic screening conditions. For this, automated microscopy and computational image analysis parameters have to be carefully adjusted, which still remains the most difficult part for cell biologists. Next, negative controls and selected perturbation conditions representative for the subsequent large-scale screens should be used to perform pilot screens. It is advisable to establish standardized quality control procedures already at this stage to ensure subsequent consistent recording of the full dataset.
RNAi in screening applications
The discovery of RNAi as a conserved gene-silencing mechanism (Fire et al., 1998; Meister and Tuschl, 2004) has been the starting point for functional genomics studies in organisms previously inaccessible to systematic genetic perturbations. Several types of reagents have been developed to induce RNAi, which have also been used in screening applications. In mammalian cells, the most common are chemically synthesized short double-stranded RNA (dsRNA) oligomers (siRNAs; Elbashir et al., 2001), which can be obtained commercially as libraries targeting the entire genome or subsets targeting specific processes or gene families. As an alternative to chemical synthesis of siRNAs, long dsRNAs can be enzymatically digested in vitro by Dicer or RNase III into heterogeneous pools of endo-RNase–prepared siRNAs (esiRNAs; Yang et al., 2002; Buchholz et al., 2006). A third method to induce RNAi uses the expression of short hairpin RNAs from plasmid vectors, which are cleaved into siRNAs by the endogenous Dicer enzyme (Root et al., 2006; Snøve and Rossi, 2006; Wiznerowicz et al., 2006). In contrast to the transient knockdown mediated by siRNA or esiRNA, the latter method enables stable RNAi in long-term gene-silencing studies. However, it requires transfection methods that can be less efficient and more toxic than transfection of siRNAs. Furthermore, the levels of vector-expressed short hairpin RNA are often difficult to control, which can be critical for potential concentration-dependent off-target effects (Jackson et al., 2006).
In some cell types like Drosophila melanogaster S2 cells, RNAi can be induced by simply adding dsRNA directly to the culture medium. However, most other cell types require transfection methods to deliver RNAi reagents. siRNAs or esiRNAs can be efficiently transfected by chemical transfection reagents using liquid-handling robotics and multiwell plates. As an alternative method, cell transfection arrays have been developed that contain dried spots of siRNA oligonucleotides mixed with transfection and matrix reagents. A uniform layer of cells seeded onto transfection arrays is then transfected locally at each spot by solid-phase transfection (Ziauddin and Sabatini, 2001; Erfle et al., 2004, 2007; Erfle and Pepperkok, 2007). Thereby, a large number of replica transfection plates can be generated in a single step. These can be distributed and stored for subsequent use in various screening applications without the need of further robotics (Neumann et al., 2006). Although transfection arrays are cheap and convenient to use, establishing the challenging production workflow and quality control procedures might be solved best in large central facilities. The main limitation of transfection arrays is the risk of cross-contamination from neighboring spots. However, solid-phase transfection can also be applied in multiwell plates, thereby avoiding cross-contamination between different siRNA oligonucleotides (Erfle et al., 2008).
Microscope automation
Initial imaging-based screening was often implemented on standard motorized epifluorescence microscopes, to which the automation was added by academic researchers. This requires motorized control of stage positioning, fluorescence filters, and camera acquisition, which is now available as preassembled systems from all major microscope providers. One of the most challenging tasks in microscope automation is the focus control (Shen et al., 2006). Initial systems implemented image-based autofocus methods that first record entire z stacks of images to determine the focal plane with maximal information content. This method has the advantage to correctly position the focus even when cellular geometry changes, for example, in morphological phenotypes. However, image-based autofocus is slow and it exposes the specimen to excessive light-causing photodamage. An alternative autofocusing method measures the reflection of a laser at the surface of the tissue culture dish or multiwell plate. This method is appropriate to image adherent cells at a defined z offset, and it is very robust, fast, and minimizes perturbation of the specimen by light exposure. Reflection-based autofocusing requires specific hardware that has recently become available from most major microscope companies.
Another difficulty in automating microscopy for screening applications is the supply of objective immersion media. Therefore, most screens use dry objectives. With short working distance, the numerical aperture available for dry objectives will suit the requirements for most image-based screening assays. However, systems for automated water supply on immersion objectives with higher numerical aperture have been developed by some microscope companies.
With the increased demand for automated microscopes, several companies developed dedicated screening microscopes. These were optimized for throughput and robustness, but they are often less flexible for specific assay requirements. For example, they use proprietary software and image formats, which limit their integration into specialized image analysis pipelines. As a more flexible alternative, powerful open source software has been developed for microscope control, database integration, and image processing (Text box 1).
Text box 1.
Open source software projects for image-based screening
μManager
μManager (http://www.micro-manager.org) is software used to control automated image acquisition on motorized microscopes. Supported hardware devices include motorized microscope stands from various companies as well as a large number of illumination sources, shutters, filter wheels, scanning stages, and digital cameras. μManager provides a graphical user interface through integration into ImageJ software (National Institutes of Health). μManager can be easily extended to support new hardware through standardized application programming interfaces. μManager can, in principle, control epifluorescence and spinning-disc microscopes but does not support laser-scanning microscopes.
Open Microscopy Environment (OME)
OME (http://www.openmicroscopy.org/site) was designed to establish standards in multidimensional microscopy (Swedlow et al., 2003). A suite of software tools supports standardized annotation and storage of images, microscope settings, and analysis results (Goldberg et al., 2005). A recent extension established OME as a server platform linked to different web-based application tools, for example, for visualization and basic image analysis (Moore et al., 2008). This client-server application architecture (OME Remote Objects called OMERO) enables implementation of visualization control in webpages to browse data on remote OME servers. OME also provides a common image format for microscopy (OME-TIFF) and, through Bio-Formats (http://www.loci.wisc.edu/ome/formats.html), provides software libraries that can parse almost all current microscope image data structures.
CellProfiler
CellProfiler (http://www.cellprofiler.org) is software for data exploration and image analysis, designed for the needs of high-throughput imaging. It provides a graphical user interface to manage complex image analysis workflows and offers some functionality in feature extraction and machine learning (Sigal et al., 2006; Jones et al., 2008, 2009; Rämö et al., 2009). The software CellClassifier provides support vector machine classification to CellProfiler (Rämö et al., 2009).
CellHTS
The software CellHTS is part of a software project Bioconductor (http://www.bioconductor.org), which adds functionality to the statistical software R for the analysis of high-content screens. CellHTS can be used to compute the Z' scores on the experiment-wide level of screens, offering various normalization graphical visualization options (Boutros et al., 2006). CellHTS also links to Gene Ontology and BioMart data warehouses to serve as a data mining tool.
Computational image analysis for quantitative phenotyping
Automated image processing is the key for reliable quantitative measurements in high-throughput microscopy. After image preprocessing by denoising and background correction, the starting point for any cell-based assay is the identification of objects or cells by segmentation algorithms (Fig. 3 A). Segmentation algorithms detect objects based on a priori knowledge of their properties (like brightness, size, homogeneity, or edge information). Which object detection algorithm performs best highly depends on the specific fluorescent markers and cellular morphologies of a particular assay. Image segmentation is still one of the most challenging aspects when implementing an image analysis workflow for screening. To facilitate cell detection, fluorescent DNA or chromatin markers are often used because labeled cell nuclei have well-defined contours and they are well separated from neighboring cells (Megason and Fraser, 2007). Dilation of cell nuclei contours can serve to define cytoplasmic regions.
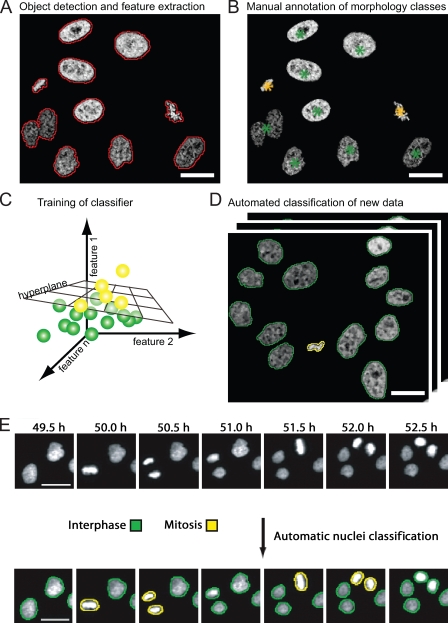
Figure 3.
Supervised machine learning for classification of cellular morphologies. (A) Detection of cells based on fluorescent chromatin label (core histone 2B fused to GFP; Kanda et al., 1998), as indicated by red contours. A set of quantitative texture and shape features is then extracted for each segmented object. (B) Manual annotation of morphology classes. In this example, interphase cell nuclei are annotated by green asterisks, and metaphase chromosome plates are annotated by yellow asterisks. (C) The hyperplane that optimally separates the two different morphology classes is automatically determined by the classification algorithm. The hyperplane can be defined by linear functions (as shown) or by radial base functions, depending on the classification algorithm. (D) The trained classifier can be applied to new data for automated classification of trained morphology classes. The yellow and green contours label interphase and metaphase morphology classes as shown in B. (E) Time-resolved assay for mitotic progression. Live HeLa cells expressing fluorescent core histone 2B (H2B-GFP) were imaged by automated time-lapse microscopy for 24 h using a 10× dry objective. Morphological classes were annotated by machine learning and pattern classification. This panel is reprinted with permission from Nature Publishing Group (Neumann et al., 2006). Bars, 20 µm.
Because of the high experimental variability of most staining procedures, absolute fluorescence intensities for quantification of marker levels usually do not provide reproducible measures within large-scale experiments. Therefore, intensity-based assays mostly rely on fluorescence ratios related to a counterstained reference marker in individual cells. With this, imaging-based ratio metric measurements can be as sensitive as flow cytometry devices (Gordon et al., 2007).
To annotate distinct morphological phenotypes, pattern recognition methods have been developed, among which supervised machine learning classifiers are most common. These methods first require detailed quantitative description of shape and texture of each segmented cell. Several collections of algorithms to calculate such statistical features have been published (Haralick et al., 1973; Prokop and Reeves, 1992; Walker and Jackway, 1996), which can be combined to define cellular morphologies by numerical vectors. In the next step of supervised machine learning, a classification algorithm needs to be trained on user-defined examples for cellular morphology classes (Fig. 3 B). A widely used classification algorithm, support vector machine, automatically determines a boundary (termed hyperplane) within the multidimensional feature space, which optimally discriminates the user-annotated cellular morphologies (Fig. 3 C). This classifier can be subsequently applied to annotate cell morphologies in large-scale image data (Fig. 3 D). In practice, supervised machine learning succeeded to accurately classify a variety of phenotypes typically in the range of 70–90%, including localization to subcellular structures, mitotic phenotypes, and virus infection (Boland and Murphy, 2001; Conrad et al., 2004; Neumann et al., 2006; Rämö et al., 2009). In fact, classifiers for subcellular objects have been shown to outperform human annotation for localization patterns of Golgi apparatus, endosomes, or lysosomes (Glory and Murphy, 2007). The orientation of the classification hyperplane and its distance to cellular objects in the feature space can also be directly used for detailed phenotypic profiling (Loo et al., 2007). In any case, a good representation of phenotypical variability in the training data is essential for accurate annotation of the full screening data.
Supervised machine learning can, in principle, also be used to annotate cellular dynamics in live imaging data. This requires a workflow combining morphology classification with the tracking of cells over time. However, because the maximal classification accuracy is rarely higher than 95% per object, cellular trajectories typically contain multiple annotation errors. Some efforts have been made to improve the classification accuracy in time series by suppressing biologically illegitimate transitions of morphological states. Although this has been shown to improve the annotation accuracy in an assay based on mitotic chromosome morphologies (Harder et al., 2009; Zhou et al., 2009), its application to other assays will require implementation of specific biological a priori models. To overcome this limitation, automatically extracted class transition probabilities can serve to correct classification errors without user supervision, providing a generic tool for time-resolved phenotype annotation (will be released as open source software CellCognition in 2010; unpublished data).
The most severe limitation of current machine learning approaches to RNAi screening is the dependence on a supervised training step. The definition of morphology classes is often based on a relatively small training dataset, which may be unrepresentative or lack rare phenotypic aberrations. Iterative supervised machine learning can be performed to raise the chance of discovering rare unknown phenotypes. Thereby, automated preselection of aberrant morphologies enables annotation of a more comprehensive classifier (Jones et al., 2009). Alternatively, iterative clustering of morphology classes can build a model that includes rare and unexpected phenotypes (Yin et al., 2008). Finally, unsupervised machine learning methods are available for clustering cellular morphologies into phenotype classes without any user interaction. Although these methods bear big promise, their performance in real-life biological screening applications still needs to be demonstrated.
Quality control and validation of hits
Optimizing an assay for screening requires a quantitative parameter for sensitivity and robustness of signal detection. A widely used measure for assay performance is the Z′ score, which defines the discriminative power of an assay between unperturbed negative controls and cells perturbed with a positive control that induces the respective phenotype (Zhang et al., 1999). Based on this quality score, the best experimental parameters can be determined, for example, the cell seeding density, transfection conditions, or image analysis parameters. Calculating Z′ scores is straightforward, but it has been found to be less robust in more complex screens, for example, when the siRNA transfection efficiency is variable (Birmingham et al., 2009). Improved quality scores for RNAi screening assays can be calculated by taking into account pools of siRNA targeting one gene (König et al., 2007) or multiple experimental replicas of siRNA conditions combined with plate to plate variation (Zhang et al., 2008).
Two recent RNAi screens on cell cycle regulation in mammalian cells demonstrated surprisingly little overlap (∼10%) of the hit lists (Mukherji et al., 2006; Kittler et al., 2007). This may in part reflect cell line–specific differences or the use of different RNAi reagents but raises concerns about a generally high rate of false hits in RNAi screens (Echeverri et al., 2006). A study on screening reproducibility indicated a complex pattern of how siRNA design, cell type, and the cellular context impact phenotypic readouts (DasGupta et al., 2007). Moreover, several cell population context–dependent parameters showed an impact on virus infection and endocytosis assays, which can serve to improve the phenotype annotation accuracy (Snijder et al., 2009).
A potential source of false positive hits in RNAi screens are off-target effects of the RNAi reagents. The main reason for potential off-target effects in mammalian RNAi screens is the relatively high tolerance of mismatches on the siRNA (Birmingham et al., 2006). Several validation strategies are available. RNAi phenotypes should be reproduced by at least two distinct siRNAs. A correlation of the phenotype penetrance caused by different siRNAs and the depletion level provides support for the specificity of an RNAi phenotype. A powerful validation strategy tests for RNAi phenotype complementation by overexpression of an RNAi-resistant version of the target gene (Echeverri et al., 2006). A resource for such RNAi phenotype rescue experiments is mouse transgenes cloned from bacterial artificial chromosomes for stable expression in cultured mammalian cells (Poser et al., 2008). Together, these studies highlight that hit lists resulting from primary RNAi screens bear limited value and that without detailed validation and follow up analysis, each “hit” should be considered not more than a candidate.
Linking screening data to systems biology resources
The ever-increasing content in image-based screening assays poses new challenges on the statistical analysis and data mining. Multiparametric phenotype descriptors, phenoprints, can be used to cluster genes in functional groups of similar RNAi phenotypes, which then can be related to potential common cellular processes, signaling pathways, or protein complex formation (Perlman et al., 2004; Skop et al., 2004; Pelkmans et al., 2005; Sönnichsen et al., 2005; Neumann et al., 2006; Loo et al., 2007). This requires sophisticated normalization and multivariate analysis methods to compensate potential experimental variations on the level of plates, wells, or cellular subpopulations (Text box 1; Boutros et al., 2004).
Comparison with multiple large-scale datasets, for example, subcellular protein localization, protein–protein interaction, and automated mining of published literature (Jensen et al., 2009), can be used to increase the relevance of a hit list obtained by RNAi screening. Furthermore, the comparison of different screens can be used for validation. This has been recently applied in a cell-based screen for antimalaria compounds, which were validated by comparison with >100 published unrelated cellular and enzymatic screens (Plouffe et al., 2008). Public access to original image data and their phenotypic annotations is necessary for comparing multiple large-scale datasets. This has been provided by RNAi screening databases for Caenorhabditis elegans (Gunsalus et al., 2004; Sönnichsen et al., 2005) and Drosophila (Flockhart et al., 2006), and similar resources for mammalian cells will be available in the near future (MitoCheck database; Neumann et al., 2010). However, the lack of standards for cell types, RNAi reagents, assay design, data analysis, and phenotype ontology still severely limits the integrated analysis of multiple large-scale datasets. Future efforts should address such standards, as it has been achieved, for example, in expression array profiling (Brazma et al., 2001).
Conclusions
The past few years have established microscopy as a high-content screening method for a broad spectrum of biological questions. Both microscope hardware and analysis software are now available to perform imaging-based screening in standard cell biology laboratories. Yet, advanced imaging modalities like time-lapse imaging require vast resources for screening on a genome-wide scale. The increasing number of large-scale systems biology data resources provides a basis to define meaningful candidate gene sets for hypothesis-based screening, for example, by focusing on genes of a certain functional class or intracellular compartment.
The design of new assays and workflows is still tedious, often limited by rigid software user interfaces and proprietary data formats. Improved modular software and databases will be needed to transform automated microscopy into a tool for hypothesis-driven research projects with daily changing assay needs. The integration of machine learning methods into microscope-controlling software will be an important next step for increasing assay content in screening. Based on classification of an image object or phenotypic event, the microscope could be automatically reconfigured, for example, to alter spatial or temporal resolution. This will also enable the implementation of new imaging modalities into screening applications like fluorescence correlation spectroscopy or photobleaching/photoactivation assays or provide event-driven feedback on robotic drug addition. Once the hurdle of hardware and software integration is overcome, imaging-based assays will open new options for systems biology. In the long run, we may envision completely autonomous “robot-scientists” (King et al., 2009) executing cycles of experimentation and hypotheses generation on automated microscopes.
Acknowledgments
We thank Dimitry Kotlyar, Gabor Csucs, Wolfram H. Gerlich, Thomas Walter, Beate Neumann, and Jean-Karim Heriche for critical comments on the manuscript.
This work was supported by Swiss National Science Foundation research grant 3100A0-114120 and a European Young Investigator award of the European Science Foundation to D.W. Gerlich.
Footnotes
Abbreviations used in this paper:
- dsRNA
- double-stranded RNA
- esiRNA
- endo-RNase–prepared siRNA
References
- Aye-Han N.N., Ni Q., Zhang J. 2009. Fluorescent biosensors for real-time tracking of post-translational modification dynamics. Curr. Opin. Chem. Biol. 13:392–397 10.1016/j.cbpa.2009.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakal C., Aach J., Church G., Perrimon N. 2007. Quantitative morphological signatures define local signaling networks regulating cell morphology. Science. 316:1753–1756 10.1126/science.1140324 [DOI] [PubMed] [Google Scholar]
- Bartz F., Kern L., Erz D., Zhu M., Gilbert D., Meinhof T., Wirkner U., Erfle H., Muckenthaler M., Pepperkok R., Runz H. 2009. Identification of cholesterol-regulating genes by targeted RNAi screening. Cell Metab. 10:63–75 10.1016/j.cmet.2009.05.009 [DOI] [PubMed] [Google Scholar]
- Birmingham A., Anderson E.M., Reynolds A., Ilsley-Tyree D., Leake D., Fedorov Y., Baskerville S., Maksimova E., Robinson K., Karpilow J., et al. 2006. 3′ UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat. Methods. 3:199–204 10.1038/nmeth854 [DOI] [PubMed] [Google Scholar]
- Birmingham A., Selfors L.M., Forster T., Wrobel D., Kennedy C.J., Shanks E., Santoyo-Lopez J., Dunican D.J., Long A., Kelleher D., et al. 2009. Statistical methods for analysis of high-throughput RNA interference screens. Nat. Methods. 6:569–575 10.1038/nmeth.1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boland M.V., Murphy R.F. 2001. A neural network classifier capable of recognizing the patterns of all major subcellular structures in fluorescence microscope images of HeLa cells. Bioinformatics. 17:1213–1223 10.1093/bioinformatics/17.12.1213 [DOI] [PubMed] [Google Scholar]
- Boutros M., Kiger A.A., Armknecht S., Kerr K., Hild M., Koch B., Haas S.A., Paro R., Perrimon N. 2004. Genome-wide RNAi analysis of growth and viability in Drosophila cells. Science. 303:832–835 10.1126/science.1091266 [DOI] [PubMed] [Google Scholar]
- Boutros M., Brás L.P., Huber W. 2006. Analysis of cell-based RNAi screens. Genome Biol. 7:R66 10.1186/gb-2006-7-7-r66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brass A.L., Dykxhoorn D.M., Benita Y., Yan N., Engelman A., Xavier R.J., Lieberman J., Elledge S.J. 2008. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 319:921–926 10.1126/science.1152725 [DOI] [PubMed] [Google Scholar]
- Brazma A., Hingamp P., Quackenbush J., Sherlock G., Spellman P., Stoeckert C., Aach J., Ansorge W., Ball C.A., Causton H.C., et al. 2001. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat. Genet. 29:365–371 10.1038/ng1201-365 [DOI] [PubMed] [Google Scholar]
- Buchholz F., Kittler R., Slabicki M., Theis M. 2006. Enzymatically prepared RNAi libraries. Nat. Methods. 3:696–700 10.1038/nmeth912 [DOI] [PubMed] [Google Scholar]
- Ciruela F. 2008. Fluorescence-based methods in the study of protein-protein interactions in living cells. Curr. Opin. Biotechnol. 19:338–343 10.1016/j.copbio.2008.06.003 [DOI] [PubMed] [Google Scholar]
- Conrad C., Erfle H., Warnat P., Daigle N., Lörch T., Ellenberg J., Pepperkok R., Eils R. 2004. Automatic identification of subcellular phenotypes on human cell arrays. Genome Res. 14:1130–1136 10.1101/gr.2383804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DasGupta R., Nybakken K., Booker M., Mathey-Prevot B., Gonsalves F., Changkakoty B., Perrimon N. 2007. A case study of the reproducibility of transcriptional reporter cell-based RNAi screens in Drosophila. Genome Biol. 8:R203 10.1186/gb-2007-8-9-r203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doil C., Mailand N., Bekker-Jensen S., Menard P., Larsen D.H., Pepperkok R., Ellenberg J., Panier S., Durocher D., Bartek J., et al. 2009. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell. 136:435–446 10.1016/j.cell.2008.12.041 [DOI] [PubMed] [Google Scholar]
- Draviam V.M., Stegmeier F., Nalepa G., Sowa M.E., Chen J., Liang A., Hannon G.J., Sorger P.K., Harper J.W., Elledge S.J. 2007. A functional genomic screen identifies a role for TAO1 kinase in spindle-checkpoint signalling. Nat. Cell Biol. 9:556–564 10.1038/ncb1569 [DOI] [PubMed] [Google Scholar]
- Echard A., Hickson G.R., Foley E., O’Farrell P.H. 2004. Terminal cytokinesis events uncovered after an RNAi screen. Curr. Biol. 14:1685–1693 10.1016/j.cub.2004.08.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echeverri C.J., Beachy P.A., Baum B., Boutros M., Buchholz F., Chanda S.K., Downward J., Ellenberg J., Fraser A.G., Hacohen N., et al. 2006. Minimizing the risk of reporting false positives in large-scale RNAi screens. Nat. Methods. 3:777–779 10.1038/nmeth1006-777 [DOI] [PubMed] [Google Scholar]
- Elbashir S.M., Harborth J., Lendeckel W., Yalcin A., Weber K., Tuschl T. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 411:494–498 10.1038/35078107 [DOI] [PubMed] [Google Scholar]
- Erfle H., Pepperkok R. 2007. Production of siRNA- and cDNA-transfected cell arrays on noncoated chambered coverglass for high-content screening microscopy in living cells. Methods Mol. Biol. 360:155–161 [DOI] [PubMed] [Google Scholar]
- Erfle H., Simpson J.C., Bastiaens P.I., Pepperkok R. 2004. siRNA cell arrays for high-content screening microscopy. Biotechniques. 37:454–458: 460: 462 [DOI] [PubMed] [Google Scholar]
- Erfle H., Neumann B., Liebel U., Rogers P., Held M., Walter T., Ellenberg J., Pepperkok R. 2007. Reverse transfection on cell arrays for high content screening microscopy. Nat. Protoc. 2:392–399 10.1038/nprot.2006.483 [DOI] [PubMed] [Google Scholar]
- Erfle H., Neumann B., Rogers P., Bulkescher J., Ellenberg J., Pepperkok R. 2008. Work flow for multiplexing siRNA assays by solid-phase reverse transfection in multiwell plates. J. Biomol. Screen. 13:575–580 10.1177/1087057108320133 [DOI] [PubMed] [Google Scholar]
- Fire A., Xu S., Montgomery M.K., Kostas S.A., Driver S.E., Mello C.C. 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 391:806–811 10.1038/35888 [DOI] [PubMed] [Google Scholar]
- Flockhart I., Booker M., Kiger A., Boutros M., Armknecht S., Ramadan N., Richardson K., Xu A., Perrimon N., Mathey-Prevot B. 2006. FlyRNAi: the Drosophila RNAi screening center database. Nucleic Acids Res. 34:D489–D494 10.1093/nar/gkj114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlich D., Ellenberg J. 2003. 4D imaging to assay complex dynamics in live specimens. Nat. Cell Biol. Suppl:S14–S19 [PubMed] [Google Scholar]
- Gerlich D., Beaudouin J., Gebhard M., Ellenberg J., Eils R. 2001. Four-dimensional imaging and quantitative reconstruction to analyse complex spatiotemporal processes in live cells. Nat. Cell Biol. 3:852–855 10.1038/ncb0901-852 [DOI] [PubMed] [Google Scholar]
- Giepmans B.N., Adams S.R., Ellisman M.H., Tsien R.Y. 2006. The fluorescent toolbox for assessing protein location and function. Science. 312:217–224 10.1126/science.1124618 [DOI] [PubMed] [Google Scholar]
- Glory E., Murphy R.F. 2007. Automated subcellular location determination and high-throughput microscopy. Dev. Cell. 12:7–16 10.1016/j.devcel.2006.12.007 [DOI] [PubMed] [Google Scholar]
- Goldberg I.G., Allan C., Burel J.M., Creager D., Falconi A., Hochheiser H., Johnston J., Mellen J., Sorger P.K., Swedlow J.R. 2005. The Open Microscopy Environment (OME) Data Model and XML file: open tools for informatics and quantitative analysis in biological imaging. Genome Biol. 6:R47 10.1186/gb-2005-6-5-r47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gönczy P., Echeverri C., Oegema K., Coulson A., Jones S.J., Copley R.R., Duperon J., Oegema J., Brehm M., Cassin E., et al. 2000. Functional genomic analysis of cell division in C. elegans using RNAi of genes on chromosome III. Nature. 408:331–336 10.1038/35042526 [DOI] [PubMed] [Google Scholar]
- Gordon A., Colman-Lerner A., Chin T.E., Benjamin K.R., Yu R.C., Brent R. 2007. Single-cell quantification of molecules and rates using open-source microscope-based cytometry. Nat. Methods. 4:175–181 10.1038/nmeth1008 [DOI] [PubMed] [Google Scholar]
- Goshima G., Wollman R., Goodwin S.S., Zhang N., Scholey J.M., Vale R.D., Stuurman N. 2007. Genes required for mitotic spindle assembly in Drosophila S2 cells. Science. 316:417–421 10.1126/science.1141314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunsalus K.C., Yueh W.C., MacMenamin P., Piano F. 2004. RNAiDB and PhenoBlast: web tools for genome-wide phenotypic mapping projects. Nucleic Acids Res. 32:D406–D410 10.1093/nar/gkh110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haralick R.M., Shanmugam K., Dinstein I. 1973. Textural features for image classification. IEEE Trans. Syst. Man Cybern. 3:610–621 10.1109/TSMC.1973.4309314 [DOI] [Google Scholar]
- Harder N., Mora-Bermúdez F., Godinez W.J., Wünsche A., Eils R., Ellenberg J., Rohr K. 2009. Automatic analysis of dividing cells in live cell movies to detect mitotic delays and correlate phenotypes in time. Genome Res. 19:2113–2124 10.1101/gr.092494.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L.H., Hopfield J.J., Leibler S., Murray A.W. 1999. From molecular to modular cell biology. Nature. 402:C47–C52 10.1038/35011540 [DOI] [PubMed] [Google Scholar]
- Haustein E., Schwille P. 2007. Fluorescence correlation spectroscopy: novel variations of an established technique. Annu. Rev. Biophys. Biomol. Struct. 36:151–169 10.1146/annurev.biophys.36.040306.132612 [DOI] [PubMed] [Google Scholar]
- Jackson A.L., Burchard J., Schelter J., Chau B.N., Cleary M., Lim L., Linsley P.S. 2006. Widespread siRNA “off-target” transcript silencing mediated by seed region sequence complementarity. RNA. 12:1179–1187 10.1261/rna.25706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen L.J., Kuhn M., Stark M., Chaffron S., Creevey C., Muller J., Doerks T., Julien P., Roth A., Simonovic M., et al. 2009. STRING 8—a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 37:D412–D416 10.1093/nar/gkn760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang D., Zhao L., Clapham D.E. 2009. Genome-wide RNAi screen identifies Letm1 as a mitochondrial Ca2+/H+ antiporter. Science. 326:144–147 10.1126/science.1175145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsson K. 2009. Visualizing biochemical activities in living cells. Nat. Chem. Biol. 5:63–65 10.1038/nchembio0209-63 [DOI] [PubMed] [Google Scholar]
- Jones T.R., Kang I.H., Wheeler D.B., Lindquist R.A., Papallo A., Sabatini D.M., Golland P., Carpenter A.E. 2008. CellProfiler Analyst: data exploration and analysis software for complex image-based screens. BMC Bioinformatics. 9:482 10.1186/1471-2105-9-482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones T.R., Carpenter A.E., Lamprecht M.R., Moffat J., Silver S.J., Grenier J.K., Castoreno A.B., Eggert U.S., Root D.E., Golland P., Sabatini D.M. 2009. Scoring diverse cellular morphologies in image-based screens with iterative feedback and machine learning. Proc. Natl. Acad. Sci. USA. 106:1826–1831 10.1073/pnas.0808843106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanda T., Sullivan K.F., Wahl G.M. 1998. Histone-GFP fusion protein enables sensitive analysis of chromosome dynamics in living mammalian cells. Curr. Biol. 8:377–385 10.1016/S0960-9822(98)70156-3 [DOI] [PubMed] [Google Scholar]
- King R.D., Rowland J., Oliver S.G., Young M., Aubrey W., Byrne E., Liakata M., Markham M., Pir P., Soldatova L.N., et al. 2009. The automation of science. Science. 324:85–89 10.1126/science.1165620 [DOI] [PubMed] [Google Scholar]
- Kittler R., Pelletier L., Heninger A.K., Slabicki M., Theis M., Miroslaw L., Poser I., Lawo S., Grabner H., Kozak K., et al. 2007. Genome-scale RNAi profiling of cell division in human tissue culture cells. Nat. Cell Biol. 9:1401–1412 10.1038/ncb1659 [DOI] [PubMed] [Google Scholar]
- König R., Chiang C.Y., Tu B.P., Yan S.F., DeJesus P.D., Romero A., Bergauer T., Orth A., Krueger U., Zhou Y., Chanda S.K. 2007. A probability-based approach for the analysis of large-scale RNAi screens. Nat. Methods. 4:847–849 10.1038/nmeth1089 [DOI] [PubMed] [Google Scholar]
- Krishnan M.N., Ng A., Sukumaran B., Gilfoy F.D., Uchil P.D., Sultana H., Brass A.L., Adametz R., Tsui M., Qian F., et al. 2008. RNA interference screen for human genes associated with West Nile virus infection. Nature. 455:242–245 10.1038/nature07207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott-Schwartz J., Altan-Bonnet N., Patterson G.H. 2003. Photobleaching and photoactivation: following protein dynamics in living cells. Nat. Cell Biol. Suppl:S7–S14 [PubMed] [Google Scholar]
- Liu T., Sims D., Baum B. 2009. Parallel RNAi screens across different cell lines identify generic and cell type-specific regulators of actin organization and cell morphology. Genome Biol. 10:R26 10.1186/gb-2009-10-3-r26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo L.H., Wu L.F., Altschuler S.J. 2007. Image-based multivariate profiling of drug responses from single cells. Nat. Methods. 4:445–453 [DOI] [PubMed] [Google Scholar]
- Megason S.G., Fraser S.E. 2007. Imaging in systems biology. Cell. 130:784–795 10.1016/j.cell.2007.08.031 [DOI] [PubMed] [Google Scholar]
- Meister G., Tuschl T. 2004. Mechanisms of gene silencing by double-stranded RNA. Nature. 431:343–349 10.1038/nature02873 [DOI] [PubMed] [Google Scholar]
- Moore J., Allan C., Burel J.M., Loranger B., MacDonald D., Monk J., Swedlow J.R. 2008. Open tools for storage and management of quantitative image data. Methods Cell Biol. 85:555–570 10.1016/S0091-679X(08)85024-8 [DOI] [PubMed] [Google Scholar]
- Mukherji M., Bell R., Supekova L., Wang Y., Orth A.P., Batalov S., Miraglia L., Huesken D., Lange J., Martin C., et al. 2006. Genome-wide functional analysis of human cell-cycle regulators. Proc. Natl. Acad. Sci. USA. 103:14819–14824 10.1073/pnas.0604320103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller P., Kuttenkeuler D., Gesellchen V., Zeidler M.P., Boutros M. 2005. Identification of JAK/STAT signalling components by genome-wide RNA interference. Nature. 436:871–875 10.1038/nature03869 [DOI] [PubMed] [Google Scholar]
- Neumann B., Held M., Liebel U., Erfle H., Rogers P., Pepperkok R., Ellenberg J. 2006. High-throughput RNAi screening by time-lapse imaging of live human cells. Nat. Methods. 3:385–390 10.1038/nmeth876 [DOI] [PubMed] [Google Scholar]
- Neumann B., Walter T., Heriché J.-K., Bulkescher J., Erfle H., Conrad C., Rogers P., Poser I., Held M., Liebe U., et al. 2010. Phenotypic profiling of the human genome by time-lapse microscopy reveals cell division genes. Nature. In Press [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelkmans L., Fava E., Grabner H., Hannus M., Habermann B., Krausz E., Zerial M. 2005. Genome-wide analysis of human kinases in clathrin- and caveolae/raft-mediated endocytosis. Nature. 436:78–86 10.1038/nature03571 [DOI] [PubMed] [Google Scholar]
- Pepperkok R., Ellenberg J. 2006. High-throughput fluorescence microscopy for systems biology. Nat. Rev. Mol. Cell Biol. 7:690–696 10.1038/nrm1979 [DOI] [PubMed] [Google Scholar]
- Perlman Z.E., Slack M.D., Feng Y., Mitchison T.J., Wu L.F., Altschuler S.J. 2004. Multidimensional drug profiling by automated microscopy. Science. 306:1194–1198 10.1126/science.1100709 [DOI] [PubMed] [Google Scholar]
- Plouffe D., Brinker A., McNamara C., Henson K., Kato N., Kuhen K., Nagle A., Adrián F., Matzen J.T., Anderson P., et al. 2008. In silico activity profiling reveals the mechanism of action of antimalarials discovered in a high-throughput screen. Proc. Natl. Acad. Sci. USA. 105:9059–9064 10.1073/pnas.0802982105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poser I., Sarov M., Hutchins J.R., Hériché J.K., Toyoda Y., Pozniakovsky A., Weigl D., Nitzsche A., Hegemann B., Bird A.W., et al. 2008. BAC TransgeneOmics: a high-throughput method for exploration of protein function in mammals. Nat. Methods. 5:409–415 10.1038/nmeth.1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokop R.J., Reeves A.P. 1992. A survey of moment-based techniques for unoccluded object representation and recognition. CVGIP: Graphical Models and Image Processing. 54:438–460 10.1016/1049-9652(92)90027-U [DOI] [Google Scholar]
- Rämö P., Sacher R., Snijder B., Begemann B., Pelkmans L. 2009. CellClassifier: supervised learning of cellular phenotypes. Bioinformatics. 25:3028–3030 10.1093/bioinformatics/btp524 [DOI] [PubMed] [Google Scholar]
- Root D.E., Hacohen N., Hahn W.C., Lander E.S., Sabatini D.M. 2006. Genome-scale loss-of-function screening with a lentiviral RNAi library. Nat. Methods. 3:715–719 10.1038/nmeth924 [DOI] [PubMed] [Google Scholar]
- Schmitz M.H., Gerlich D.W. 2009. Automated live microscopy to study mitotic gene function in fluorescent reporter cell lines. Methods Mol. Biol. 545:113–134 10.1007/978-1-60327-993-2_7 [DOI] [PubMed] [Google Scholar]
- Shen F., Hodgson L., Hahn K. 2006. Digital autofocus methods for automated microscopy. Methods Enzymol. 414:620–632 10.1016/S0076-6879(06)14032-X [DOI] [PubMed] [Google Scholar]
- Sigal A., Milo R., Cohen A., Geva-Zatorsky N., Klein Y., Alaluf I., Swerdlin N., Perzov N., Danon T., Liron Y., et al. 2006. Dynamic proteomics in individual human cells uncovers widespread cell-cycle dependence of nuclear proteins. Nat. Methods. 3:525–531 10.1038/nmeth892 [DOI] [PubMed] [Google Scholar]
- Skop A.R., Liu H., Yates J., III, Meyer B.J., Heald R. 2004. Dissection of the mammalian midbody proteome reveals conserved cytokinesis mechanisms. Science. 305:61–66 10.1126/science.1097931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder B., Sacher R., Rämö P., Damm E.M., Liberali P., Pelkmans L. 2009. Population context determines cell-to-cell variability in endocytosis and virus infection. Nature. 461:520–523 10.1038/nature08282 [DOI] [PubMed] [Google Scholar]
- Snøve O., Jr., Rossi J.J. 2006. Expressing short hairpin RNAs in vivo. Nat. Methods. 3:689–695 10.1038/nmeth927 [DOI] [PubMed] [Google Scholar]
- Sönnichsen B., Koski L.B., Walsh A., Marschall P., Neumann B., Brehm M., Alleaume A.M., Artelt J., Bettencourt P., Cassin E., et al. 2005. Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature. 434:462–469 10.1038/nature03353 [DOI] [PubMed] [Google Scholar]
- Steigemann P., Wurzenberger C., Schmitz M.H., Held M., Guizetti J., Maar S., Gerlich D.W. 2009. Aurora B-mediated abscission checkpoint protects against tetraploidization. Cell. 136:473–484 10.1016/j.cell.2008.12.020 [DOI] [PubMed] [Google Scholar]
- Swedlow J.R., Goldberg I., Brauner E., Sorger P.K. 2003. Informatics and quantitative analysis in biological imaging. Science. 300:100–102 10.1126/science.1082602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanEngelenburg S.B., Palmer A.E. 2008. Fluorescent biosensors of protein function. Curr. Opin. Chem. Biol. 12:60–65 10.1016/j.cbpa.2008.01.020 [DOI] [PubMed] [Google Scholar]
- Walker R.F., Jackway P.T. 1996. Statistical geometric features-extensions for cytological texture analysis. Proceedings of the 13th International Conference on Pattern Recognition. 2:790–794 [Google Scholar]
- Walter T., Held M., Neumann B., Hériché J.K., Conrad C., Pepperkok R., Ellenberg J. 2009. Automatic identification and clustering of chromosome phenotypes in a genome wide RNAi screen by time-lapse imaging. J. Struct. Biol. In press 10.1016/j.jsb.2009.10.004 [DOI] [PubMed] [Google Scholar]
- Wiznerowicz M., Szulc J., Trono D. 2006. Tuning silence: conditional systems for RNA interference. Nat. Methods. 3:682–688 10.1038/nmeth914 [DOI] [PubMed] [Google Scholar]
- Yang D., Buchholz F., Huang Z., Goga A., Chen C.Y., Brodsky F.M., Bishop J.M. 2002. Short RNA duplexes produced by hydrolysis with Escherichia coli RNase III mediate effective RNA interference in mammalian cells. Proc. Natl. Acad. Sci. USA. 99:9942–9947 10.1073/pnas.152327299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Z., Zhou X., Bakal C., Li F., Sun Y., Perrimon N., Wong S.T. 2008. Using iterative cluster merging with improved gap statistics to perform online phenotype discovery in the context of high-throughput RNAi screens. BMC Bioinformatics. 9:264 10.1186/1471-2105-9-264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.H., Chung T.D., Oldenburg K.R. 1999. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 4:67–73 10.1177/108705719900400206 [DOI] [PubMed] [Google Scholar]
- Zhang X.D., Kuan P.F., Ferrer M., Shu X., Liu Y.C., Gates A.T., Kunapuli P., Stec E.M., Xu M., Marine S.D., et al. 2008. Hit selection with false discovery rate control in genome-scale RNAi screens. Nucleic Acids Res. 36:4667–4679 10.1093/nar/gkn435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Li F., Yan J., Wong S.T. 2009. A novel cell segmentation method and cell phase identification using Markov model. IEEE Trans. Inf. Technol. Biomed. 13:152–157 10.1109/TITB.2008.2007098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziauddin J., Sabatini D.M. 2001. Microarrays of cells expressing defined cDNAs. Nature. 411:107–110 10.1038/35075114 [DOI] [PubMed] [Google Scholar]