Abstract
Elastomers based on poly(dimethylsiloxane) (PDMS) are promising materials for fabrication of a wide range of microanalytical systems due to their mechanical and optical properties and ease of processing. To date, however, quantitative studies that demonstrate reliable and reproducible methods for attachment of binding groups that capture complex receptor proteins of relevance to biomedical applications of PDMS microsystems have not been reported. Herein we describe methods that lead to the reproducible capture of a transmembrane protein, the human epidermal growth factor (EGF) receptor, onto PDMS surfaces presenting covalently immobilized antibodies for EGF receptor, and subsequent isolation of the captured receptor by mechanical transfer of the receptor onto a chemically functionalized surface of a gold film for detection. This result is particularly significant because the physical properties of transmembrane proteins make this class of proteins a difficult one to analyze. We benchmark the performance of antibodies to the human EGF receptor covalently immobilized on PDMS against the performance of the same antibodies physisorbed to conventional surfaces utilized in ELISA assays through the use of EGF receptor that was 32P-radiolabeled in its autophosphorylation domain. These results reveal that two pan-reactive antibodies for the EGF receptor (H11 and 111.6) and one phosphospecific EGF receptor antibody (pY1068) capture the receptor on both PDMS and ELISA plates. When using H11 antibody to capture EGF receptor and subsequent treatment with a stripping buffer (NaOH and sodium dodecylsulfate) to isolate the receptor, the signal-to-background obtained using the PDMS surface was 82:1, exceeding the signal-to-background measured on the ELISA plate (<48:1). We also characterized the isolation of captured EGF receptor by mechanical contact of the PDMS surface with a chemically functionalized gold film. The efficiency of mechanical transfer of the transmembrane protein from the PDMS surface was found to be 75–81%. However, the transfer of non-specifically bound protein was substantially less than 75%, thus leading to the important finding that mechanical transfer of the EGF receptor leads to an approximately four-fold increase in signal-to-background from 20:1 to 88:1. The signal-to-background obtained following mechanical transfer is also better than that obtained using ELISA plates and stripping buffer (<48:1). The EGF receptor is a clinically important protein and the target of numerous anticancer agents and thus these results, when combined, provide guidance for the design of PDMS-based microanalytical systems for the capture and isolation of complex and clinically important transmembrane proteins.
Introduction
Elastomers based on poly(dimethylsiloxane) (PDMS) are emerging as an important class of technological materials for the fabrication of micro-scale systems because they combine properties such as biocompatibility, chemical inertness and optical transparency with ease of processing via replica molding (soft lithography). During the past few years, for example, PDMS has been used in studies of microfluidic devices,1 patterned cell culture systems,2 and DNA3 and protein microanalysis systems.4 In these past reports and many others, a key challenge confronted by the investigators was the control of the interactions of biomolecules with the surfaces of the PDMS. The heterogeneous nature of proteins and their various mechanisms of interaction with surfaces make the engineering of surfaces of PDMS a particular challenge when designing microanalytical systems for use with proteins.5–8 It is this challenge that is addressed herein in the context of the capture and detection of a biomedically important transmembrane protein, the epidermal growth factor (EGF) receptor, on the surface of PDMS.
Past attempts to modify the surface properties of PDMS for use in microsystems can be organized into two categories: (i) physical approaches,7–12 and (ii) covalent approaches.13,14 Physical approaches include the physisorption of serum or extracellular matrix (ECM) proteins or the layer-by-layer deposition of synthetic polyelectrolytes. These approaches have been largely pursued in order to passivate the surface of PDMS or to promote the attachment of mammalian cells to PDMS surfaces (ECM proteins). The second class of approaches used to modify the surface properties of PDMS has involved the chemical activation of the surface of PDMS13 and the use of heterobifunctional cross-linkers to form covalent bonds between biomolecules (e.g., primary amine groups of proteins) and the activated PDMS surface.14 Although this second approach offers the potential advantage of stable and long-lived attachment of specific binding molecules (e.g., antibodies) to the surface of PDMS, there are surprisingly few reports that (i) establish procedures that are validated to lead to reliable and reproducible capture of complex and clinically important proteins such as membrane proteins on PDMS, (ii) quantify the capture of these targeted proteins, and (iii) benchmark the performance of the binding groups on PDMS against that obtained using physisorption of the same binding groups on conventional (polystyrene-based) ELISA plates. In this paper, we report on the covalent attachment of pan-reactive and phosphospecific antibodies for the EGF receptor to the surface of PDMS, and quantify capture of the EGF receptor from solution onto the surface of the PDMS. We also validate a method for isolation of the captured EGF receptor from the surface of the stamp; such an isolation step enables a variety of detection methods (e.g., stains and dyes of proteins) that can be implemented within microsystems. Importantly, as reported below, we have found that the isolation method also leads to a substantial increase in signal-to-background.
The study described in this paper builds from past work by Bernard et al14 at IBM-Zurich who reported on the covalent attachment of anti-mouse IgG to the surfaces of PDMS stamps. These investigators quantified the capture of 125I-labelled mouse IgG onto the surface of the PDMS stamp, and the extent of subsequent transfer of the captured 125I-labelled mouse IgG onto glass by contact of the PDMS with the surface of glass. These studies revealed that >95% of the captured mouse IgG was transferred to the glass. While specific capture of several other antigens to antibodies immobilized on PDMS has been demonstrated qualitatively,15 quantitative data of the type described above for mouse IgG is lacking in the literature dealing with complex proteins such as membrane receptor proteins. In this paper, we provide such data utilizing a biomedically relevant protein associated with the development of human cancers, namely the epidermal growth factor (EGF) receptor. The results reported represent an important step forward in the development of microsystems for analysis of the EGF receptor and other cell-associated molecules of clinical and basic science interest.
EGF is a peptide hormone that can regulate many cellular events, including cell-cycle progression, differentiation and migration, in a variety of tissues. This factor signals through the ubiquitously expressed and widely studied glycoprotein, the EGF receptor.16 The EGF receptor is a 170 kDa transmembrane receptor tyrosine kinase involved in cellular growth and survival that, when over-expressed or mutated, has been linked to the development of many deadly cancers, notably those of the lung, brain, head, neck and pancreas.17 In addition to over-expression, unregulated self-phosphorylation (hyperactivation) as a result of EGF receptor mutation(s) is often associated with the aforementioned malignant conditions. Because of the association of the EGF receptor with cancer development and because of its utility as a prognosticator of tumor severity and treatment response, the development of clinical diagnostics for cancer cases involving EGF receptor mutations or over-expression has become an active area of research. The complexity of the EGF receptor and its signaling network make the development of analytic tools for EGF receptor particularly challenging.
Relative to antibodies, which have been successfully analyzed using surface-based microsystems,8,14,18 the detection of EGF receptor presents a variety of challenges with respect to maintaining phosphorylation status, preventing protease degradation, and preserving structural integrity. In the context of the current study, these issues impact radiolabeling by [γ-32P] MgATP, which is a sensitive means to measure EGF receptor activity and quantity. Thus, in addition to high biological relevance, the EGF receptor represents a challenging model system that captures many of the complexities associated with clinically relevant proteins that are absent when using antibodies as target analytes.
In the study outlined below, we report methods that lead to highly reproducible capture of EGF receptor to antibodies covalently immobilized on the surface of PDMS. The methods are demonstrated to work with three anti-EGF receptor antibodies, including one antibody that is specific for a site of tyrosine phosphorylation (pY1068) that is associated with the activated state of the EGF receptor. We also report on the mechanical transfer and isolation of the EGF receptor from the antibody-functionalized PDMS. An important finding of the current study is that mechanical transfer of the EGF receptor leads to an increase in signal-to-background and thus represents a promising step for implementation within microsystems for analysis of EGF receptor. This finding is discussed in light of measurements of the action of “stripping buffers” that are widely used in conventional solution-phase immunoassays.
Experimental
Materials
All materials were used as received, unless otherwise noted. Fisher’s Finest glass slides were obtained from Fisher Scientific (Pittsburgh, PA). Gold (99.999% purity) was obtained from International Advanced Materials (Spring Valley, NY). Titanium (99.99% purity) was obtained from PureTech (Brewster, NY). Polished silicon wafers were purchased from Silicon Sense (Nashua, NH). 2-aminoethanethiol hydrochloride, N-hydroxysuccinimide (NHS) and PBS, pH 7.6 (for NHS/EDC activation procedure), were obtained from Sigma-Aldrich (Milwaukee, WI). 1-Ethyl-3-(3-dimethylaminopropyl)carbodiimide Hydrochloride (EDC) was obtained from Pierce Biotechnology (Rockford, IL). Anhydrous ethanol containing 5% isopropyl alcohol and 5% methyl alcohol as denaturants was obtained from Sigma-Aldrich and purged with argon gas for 1 hour prior to use. Poly(dimethylsiloxane) (PDMS) elastomeric stamps were prepared using Sylgard 184 silicone elastomer kit obtained from Dow Corning (Midland, MI). IgG-free BSA was purchased from Jackson Immunoresearch (West Grove, PA). Radioactive [γ-32P] ATP was purchased from Amersham/GE Health Care. Monoclonal murine anti-EGF receptor IgG clones H11 and 111.6 were obtained from LabVision (Fremont, CA). Polyclonal rabbit anti-pY1068-EGFR IgG was obtained from BioSource International (Camarillo, CA). Monoclonal rabbit and murine IgG isotype control antibodies were purchased from Caltag/Invitrogen (Carlsbad, CA) and eBioscience (San Diego, CA), respectively. All antibodies were obtained at 1 mg/ml concentration and free of BSA and azide and were used as is or following dilution with phosphate buffered saline (PBS, 138 mM NaCl, 8 mM Na2HPO4, 2.8 mM KCl, 1.5 mM KH2PO4, Sigma-Aldrich, Milwaukee, WI), pH 7.4.
Purification of the Epidermal Growth Factor Receptor
The human EGF receptor was purified by immunoaffinity chromatography as previously described.19 Briefly, A431 squamous carcinoma cells were harvested by lifting in PBS/EDTA/EGTA (10 mM Na2HPO4, 140 mM NaCl, 2.5 mM EDTA, 2.5 mM EGTA), pH 7.2 and homogenized by sonication in the presence of phosphatase and protease inhibitors. The homogenate was then incubated overnight with sepharose beads functionalized with anti-EGF receptor monoclonal antibody 528. Bound EGF receptor was washed and eluted by competitive interaction with recombinant human EGF. The eluted fraction was subsequently concentrated and quantified. We note that the EGF receptor is stable in aqueous solution in the absence of surfactants and other amphiphillic stabilizers.20
Radiolabeling of Epidermal Growth Factor Receptor
EGF receptor was labeled via a modification of autophosphorylation assays previously described.19 Microcon YM50 spin columns (Millipore, Billerica, MA) were passivated with 0.5 mg/ml BSA in 20 mM HEPES (Sigma-Aldrich, St. Louis, MO, Catalog # H-0891), pH 7.4 for 2 hours. Purified EGF receptor was incubated on ice for 15 min in the presence of 40–80 µCi [γ-32P] ATP and a reaction buffer composed of 20 mM HEPES, pH 7.4, 2 mM MnCl2, 5 mM MgCl2, 100 µM Leupeptin, 0.5% Aprotinin, 50 µM Na3VO4, and 50 µM unlabeled ATP (Buffer A). The receptor was diluted, added to the passivated YM50 ultrafiltration chamber, and centrifuged at 4 °C and 14,000 × g in order to remove non-bound [γ-32P] ATP. The filter was washed twice with Buffer A without ATP (Buffer B) and the concentrated 32P-radiolabeled EGF receptor was recovered and subsequently brought up to a final concentration of 5.2 nM (unless otherwise indicated) in Buffer B supplemented with 10 mM EDTA and 20 mM MnCl2 (Buffer C). Receptor labeling and the absence of free [γ-32P] ATP were confirmed by autoradiography.
Preparation and Incubation of Polystyrene ELISA Plates
Antibody diluted to 50 µg/ml in coating buffer, pH 9.6 (70 ml 100 mM Na2CO3, 170 ml 100 mM NaHCO3) was incubated in Costar 96-well half-area polystyrene ELISA plates (Corning Inc., Corning, NY) overnight at 4 °C in a humidified chamber. Antibody was then removed and wells were washed four times with PBS, pH 7.4 containing 0.05% Tween-20 with 5 mM ATP (Buffer D). Plates were then blocked for 2 hr at room temperature with 1% IgG-free BSA in Buffer D. The wells were washed four times with Buffer D, and EGF receptor was incubated for 2 hours at room temperature. Each well was washed four times with Buffer D and two times with water. The receptor was then removed from the wells using 100 µl of pre-warmed stripping buffer (50 °C, 0.1 N NaOH/0.1% SDS) and deposited into liquid scintillation cocktail (LSC, ScintiSafe from Fisher Scientific) for one minute of scintillation counting.
Cleaning of Glass Substrates
Glass microscope slides were cleaned sequentially in piranha solution (70% H2SO4, 30% H2O2) and alkaline solution (70% KOH, 30% H2O2) for 1 h at ~80 °C. Warning: Piranha solution should be handled with extreme caution; in some circumstances, most probably when it has been mixed with significant quantities of an oxidizable organic material, it has detonated unexpectedly. The slides were then rinsed thoroughly with deionized water (18.2 MΩ*cm), ethanol, and methanol and dried under a stream of nitrogen gas. The cleaned slides were stored in an oven at 110 °C.
Preparation of Gold Substrates
Glass slides were placed within the chamber of an electron beam evaporator. All metal films were deposited at chamber pressures < 2 × 10−6 torr at deposition rates of 0.2 Å/s. A thin film of titanium (thickness of 60Å) was deposited onto the glass substrate to serve as an adhesion layer. Next, semitransparent films of gold (140Å) were deposited onto the titanium-coated substrate. All gold substrates were used within 1 day of removal from the evaporator chamber.
Formation of Self-Assembled Monolayers (SAMs)
Gold films were functionalized with 2-aminoethanethiol by immersion of the glass slides bearing the gold films in a 2 mM ethanolic solution of 2-aminoethanethiol for 18 h. After incubation, the slides were rinsed sequentially with 70% EtOH in water, water, and 70% EtOH in water. The slides were then dried under a stream of nitrogen gas and immersed in 0.01 M HCl for 30 s followed by drying with a stream of nitrogen gas. The slides were used immediately for affinity contact printing.
Fabrication of PDMS Affinity Capture Surfaces
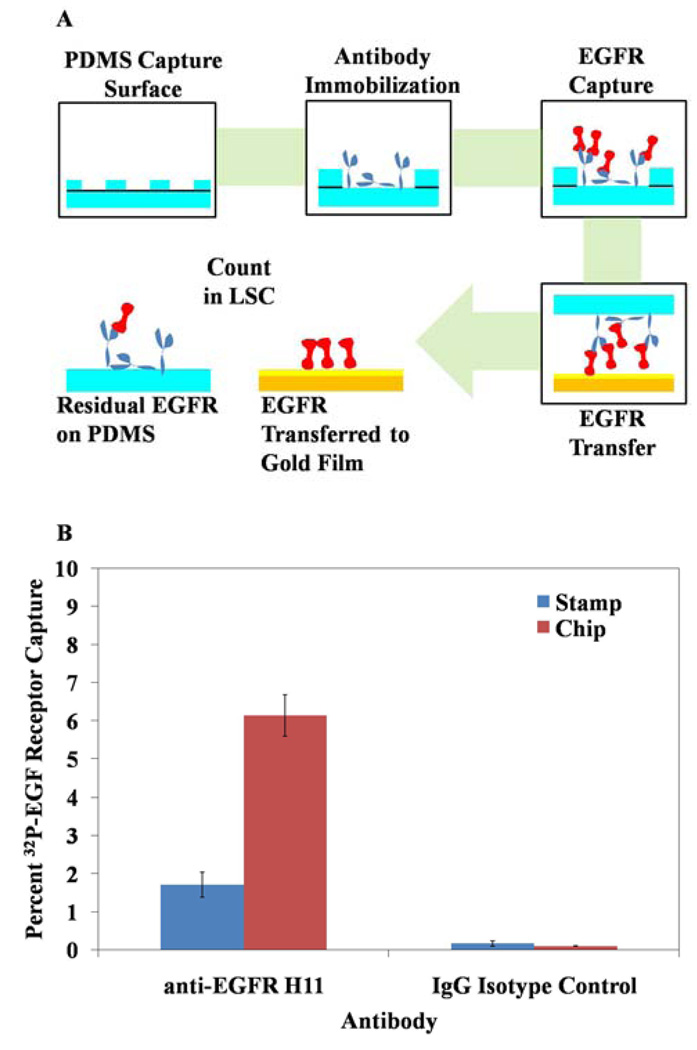
PDMS surfaces for affinity contact printing were formed by casting PDMS against the surface of silicon wafers. Alternatively, hemispherical PDMS wells (4.76 mm diameter, 35.6 mm2 surface area) (Figure 1) were formed by casting against stainless steel ball bearings set into a recessed aluminum surface. After curing the PDMS for at least 24 hr at 65 °C, the stamps were released from the masters and ultrasonicated three times in 2:1 ethanol/water for 10 min. The PDMS was dried under a stream of nitrogen gas and placed in a vacuum chamber for 30 min. The PDMS surface was oxidized using a PlasmaTherm 1441 RIE Instrument (8 sccm O2, 20 s, 100 0W). Care was taken in subsequent steps to avoid organic solvents that may cause swelling of the PDMS.21 Accordingly, the oxidized PDMS was then functionalized with a primary amine by immersion into an aqueous solution of 10% 3-aminopropyltriethoxysilane at 80 °C for 50 min. The surface was rinsed copiously with water, and dried with a stream of nitrogen. Next, the PDMS stamp was reacted with 0.1 M succinic anhydride in N,N-dimethylformamide at room temperature for 10 min to produce a carboxylic acid-terminated surface. Antibodies were covalently attached to the carboxylic acid groups by first incubating the carboxylic acid groups in PBS, pH 7.6 containing 0.05 M NHS and 0.2 M EDC for 10 min. After incubation, the surfaces were rinsed with PBS and dried with a stream of nitrogen gas. The PDMS was then incubated with antibody (1 mg/ml unless otherwise stated) for 40 min to immobilize the antibody via amide linkages formed between primary amines of the antibodies and the NHS-activated PDMS surface. PDMS surfaces were subsequently washed and used to capture EGF receptor, as described for polystyrene ELISA plates. PDMS wells were deposited directly into LSC for scintillation counting (Figure 1) whereas PDMS stamps were blown dry with air prior to immersion into LSC for scintillation counting or for affinity contact printing (Figure 5), as described below.
Fig. 1.
Schematic representation of the procedure used to verify antibody immobilization and EGF receptor capture on both polystyrene-based (PS) ELISA plates and chemically functionalized poly(dimethylsiloxane) (PDMS) wells.
Fig. 5.
Transfer of EGF receptor from PDMS to a chemically functionalized gold film. (A) shows a schematic representation of the procedure. (B) shows the distribution of EGF receptor between the PDMS stamp and capture surfaces after mechanical transfer. n = 12 for all measurements.
Affinity Contact Printing
Flat affinity capture stamps, prepared as described above, were dried under a stream of nitrogen gas and placed into conformal contact with gold films presenting ammoniated 2-aminoethanethiol. The stamps were held in conformal contact by applying minimal pressure (~1 kPa, uniformly distributed) for 7–10 min. The stamps were then peeled away from the gold films and the stamps and gold films were placed subsequently into separate vials of LSC for scintillation counting.
Results and Discussion
Capture of human EGF receptor by using antibody covalently immobilized on PDMS
We first sought to immobilize antibody on PDMS surfaces such that it would retain its capability to specifically capture the EGF receptor. A schematic illustration of the methods used is shown in Figure 1 and described in detail in the Experimental section above. Briefly, PDMS surfaces functionalized with N-hydroxysuccinimide were incubated with antibody (1 mg/ml unless otherwise stated) for 40 min before washing with Buffer D. The surfaces were subsequently blocked for 2 hr at room temperature with 1% BSA dissolved in Buffer D prior to washing and further incubation for two hours with 32P-radiolabeled EGF receptor (5.2 nM unless otherwise stated). Finally, PDMS surfaces were washed with Buffer D and water prior to excision and deposition into liquid scintillation cocktail (LSC) for scintillation counting. Repeated observations from scintillation counting revealed that radiolabeled EGF receptor was captured at high levels when supplemented with both 10 mM EDTA and 20 mM MnCl2. We note that MnCl2 is known to be important for EGF receptor activity.22
Use of murine anti-EGF receptor antibody Clone H11 (1 mg/ml) led to an overall capture of EGF receptor that ranged from 10–30% of the 32P-labeled receptor preparation incubated on the PDMS surface. When using the scintillation counting method described above (excision), the signal-to-background ratios were typically measured to be ~19:1 (a representative example can be seen on the middle bar of Figure 2a). We also investigated several variations on the above-described procedure, such as omission of MnCl2 and EDTA from the receptor preparation, and omission of ATP from the washes performed using Buffer D (data not shown). These variations on the preferred procedure yielded capture of only 5–10% of EGF receptor in the sample and signal-to-background ratios were substantially lower, typically 2:1 to 5:1. While the procedure reported above does not necessarily represent an optimal protocol, it was found to be robust and highly reproducible, with consistent capture of EGFR with signal-to-background of ~19:1.
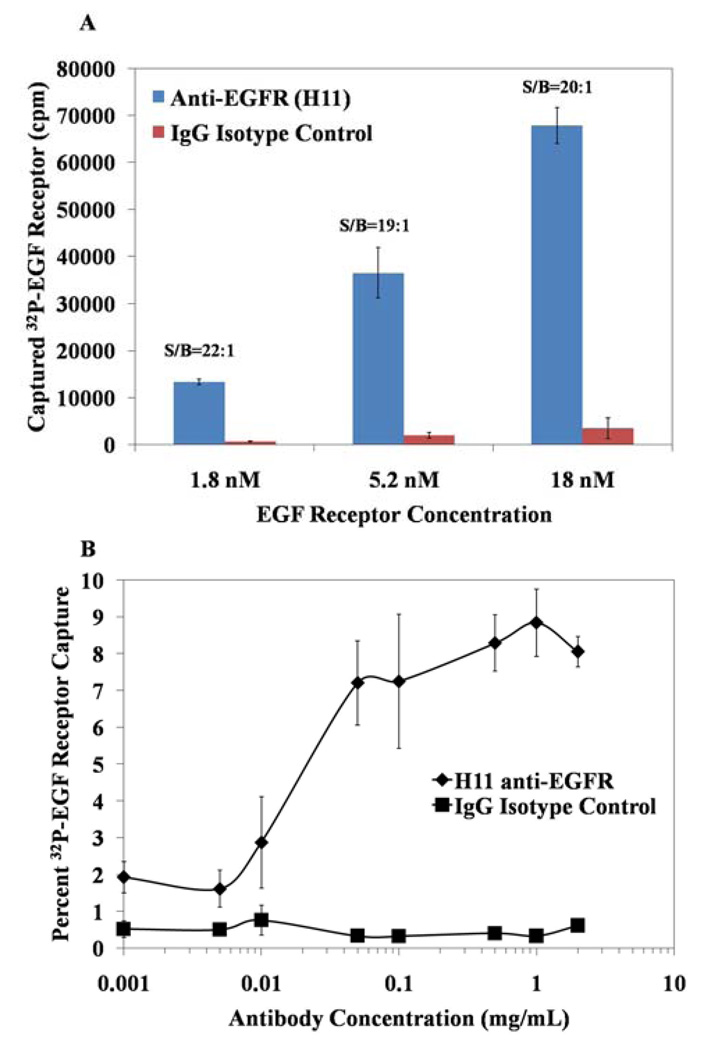
Fig. 2.
The effects of EGF receptor (A) and antibody (B) concentration on capture of EGF receptor using antibody immobilized on PDMS. The concentration of antibody used in (A) was 1 mg/mL and the concentration of EGF receptor used in (B) was 5.2 nM. EGF receptor incubated on the surface in A corresponded to either 260,000 counts (1.8 nM) or 3,000,000 (18nM). n ≥ 3 for all measurements.
The dissociation constant (Kd) of the H11 anti-EGF receptor antibody is approximately 4 nM,23 which is comparable to the EGF receptor concentration of 5.2 nM used in the above experiments. The use of a receptor concentration that is comparable to the antibody Kd has the advantage that it will unmask factors that impact binding of the EGF receptor. However, because antibody affinity and avidity in bulk solution can differ from that measured when the antibody is immobilized on a surface, we also determined the capture efficiency and signal-to-background ratio at three concentrations (1.8 nM, 5.2 nM, 18 nM) of EGF receptor incubated on PDMS surfaces functionalized with 1 mg/ml H11 antibody. As shown in Figure 2a, we measured the scintillation counts-per-minute to increase from 13,000 to 68,000 cpm when the concentration of EGF receptor was increased from 1.8 nM to 18 nM. Interestingly, the signal-to-background ratio was consistently observed to be ~20:1 for all EGF receptor concentrations tested. Thus, while the overall signal is reduced with lower EGF receptor concentration, the EGF receptor was detected with consistent signal-to-background over the log range of concentrations investigated. These data suggest that detection of EGF receptor at concentrations that are relevant to samples prepared from biological specimens is feasible with high selectivity and sensitivity.
As mentioned above, antibodies are known to differ in their structure and function when dissolved in bulk solution as compared to immobilized on a surface.24,25 In particular, because binding affinities of antibodies can vary between solution-phase and solid/liquid interfaces, and because antibody concentrations of 1 mg/ml would be viewed as relatively high for development of microanalytical systems, we evaluated the binding efficacy of a range of antibody concentrations to determine the minimal concentration of antibody required for effective EGF receptor capture (at a constant 5.2 nM concentration of EGF receptor) on PDMS surfaces. The results (Figure 2b) indicate that capture of EGF receptor at concentrations of antibody above 100 µg/ml is approximately constant, but that capture of EGF receptor drops at lower concentrations of antibody. These results lead us to conclude that antibody concentrations of ~100 µg/ml are probably close to optimal for our current procedures. We note that the antibody solutions removed from the PDMS surfaces do not have to be discarded; they can be used for functionalization of additional PDMS surfaces.
Benchmarking the capture of EGF receptor by PDMS-immobilized antibodies against capture on surfaces used in ELISAs
The use of PDMS to immobilize functional antibodies for ligand capture draws its motivation from the well-established technique of Enzyme-Linked Immunosorbent Assays (ELISAs).25 We therefore sought to compare antibody-mediated capture of EGF receptor on PDMS to the gold standard using polystyrene-based (PS) ELISA surfaces. Because ELISAs are known to be effective for a variety of antibody-antigen interactions, we also sought to determine whether the protocol described above for immobilization of H11 antibodies to the PDMS surfaces was one that could be used to successfully immobilize other EGF receptor antibodies to PDMS surfaces in a functional state.
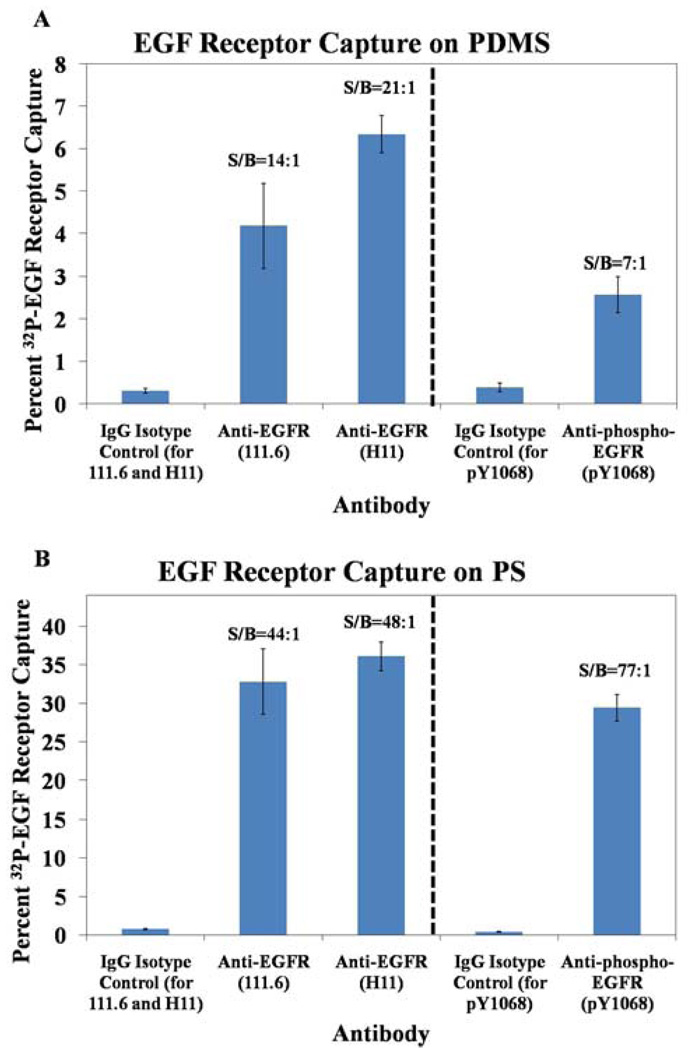
We first compared the capture efficiency and signal-to-background ratios of two pan-reactive, murine anti-EGF receptor antibodies (Clone 111.6 and Clone H11). Because it was not possible to excise the PS wells for direct immersion in the LSC, as described in the experimental section, we extracted the captured EGF receptor from the PS surfaces using the stripping buffer, and placed the extracted EGF receptor into the LSC. In these studies, we observed favorable capture and signal-to-background ratios using both the PDMS (Figure 3a) and the PS (Figure 3b) surfaces, demonstrating that antibody activity is preserved during surface immobilization of both antibodies. We note that the percentage capture of the EGF receptor was consistently higher on PS as compared to PDMS. We speculate that this difference in capture efficiency is due to the covalent attachment of the antibody to the PDMS as opposed to the physisorbed state of antibody on the PS surface. Although the antibodies are likely immobilized on both PS and PDMS in random orientations, the physisorbed state on PS will allow the antibodies to reorganize to accommodate receptor binding. In contrast, multi-point covalent attachment of the antibody to the PDMS surface prevents such reorganization and likely leads to a lower binding capacity at the surface. We also note that the latter form of attachment can cause loss of activity due to denaturation, a result of the strain induced by multiple and permanent attachment sites.25 Finally, the nature of attachment of antibody likely affects the antibody surface packing densities, which is known to affect the activity of the bound antibody.26,27 Inspection of Figure 3 also reveals that a higher signal-to-background ratio was measured in these experiments with the PS capture surface (48:1 and 44:1 for H11 and 111.6, respectively) as compared to PDMS (21:1 and 14:1). We caution, however, that this result is influenced by the different scintillation counting methods used to acquire the data on PS as compared to PDMS. As described above, scintillation counting with the PS surfaces involved extraction of the EGF receptor from the surface into a buffer prior to counting. This extraction step, as reported below, leads to an increase in signal-to-background. We note that we initially adopted the procedure with PDMS in which we immersed the PDMS surfaces into scintillation cocktail to avoid potential changes in signal-to-background associated with the extraction step.
Fig. 3.
Percent capture of EGF receptor on PDMS (A) and PS (B) by pan-reactive anti-EGF receptor antibodies 111.6 and H11, phosphospecific anti-EGF receptor antibody pY1068, and IgG isotype controls. PDMS surface area = 35.6 mm2 and PS surface area = 18.2 mm2. n = 3 for all measurements.
Although quantification of total EGF receptor expression is very useful in biological studies, quantification of the activation of EGF receptor by tyrosine phosphorylation is particularly important for gaining insight into normal physiological processes as well as hyperactivation and over-expression of EGF receptor in the context of cancer. We therefore sought to determine whether we could also detect phosphorylated EGF receptor using phosphospecific receptor antibodies immobilized on PDMS and PS (Figure 3). Using an antibody directed against an activated/phosphorylated form of EGF receptor (pY1068) we detected EGF receptor with favorable signal-to-background ratios. Similar to the pan-reactive antibodies, the PS surface demonstrated both higher levels of EGF receptor capture and signal-to-background, as compared to PDMS when using scintillation counting methods that involved immersion of the PDMS into the scintillation cocktail. We also note that the activity of the phosphospecific antibody immobilized on PS was found to be very similar to the pan-reactive antibodies immobilized on the PS. However, this was not the case for the antibodies immobilized on PDMS; the percentage capture of EGF receptor with the pY1068 antibody was lower than the pan-reactive antibodies. This result suggests that determination of percent EGF receptor phosphorylation using pan-reactive and phosphospecific antibodies immobilized on PDMS will require an analysis that includes consideration of this differential binding capacity. However, these results, taken together, demonstrate a promising array of antibodies that can detect both total and activated amounts of EGF receptor. Such a capability is a necessary foundation for the development of clinically relevant assays for EGF receptor.
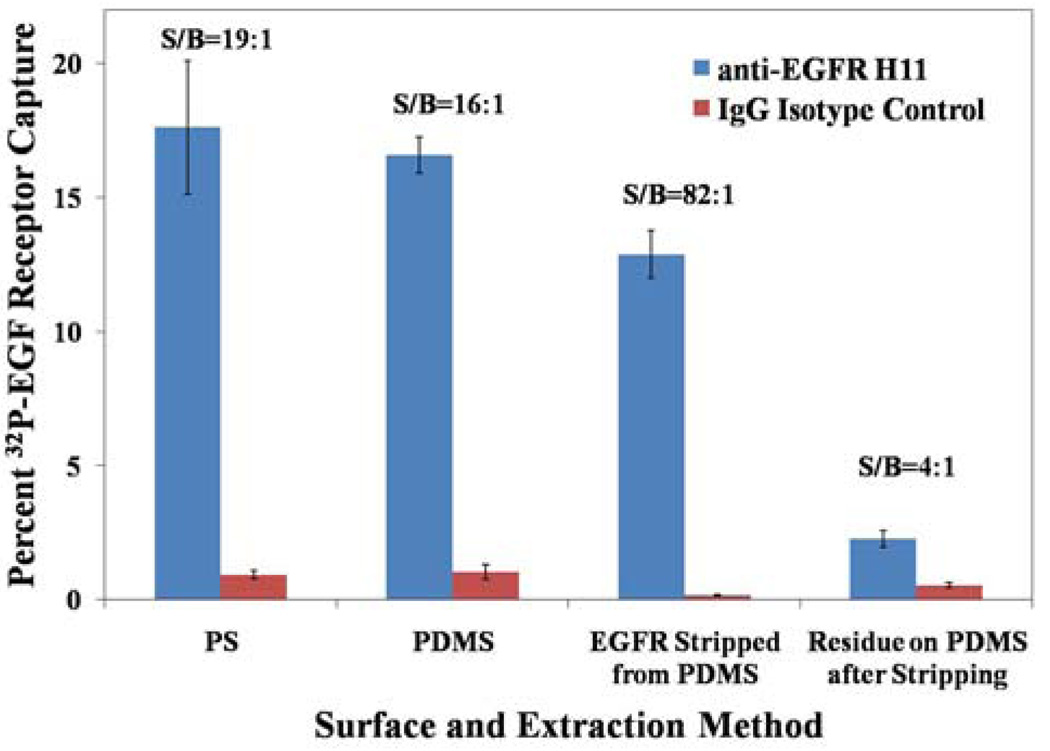
As noted previously, inspection of Figure 3 reveals that the signal-to-background ratio was typically greater when using the H11 anti-EGFR antibody physisorbed on PS as compared to covalently attached to PDMS. Because the method of measurement of EGF receptor differed between the two surfaces (EGF receptor was chemically stripped from the PS surfaces prior to measurement whereas the PDMS with captured EGF receptor was immersed directly into the LSC, see Figure 1), we sought to determine if use of the stripping buffer would lead to higher signal-to-background on PDMS surfaces. To this end, we compared signal-to-background obtained when the captured EGF receptor was quantified by immersion of the PDMS into the scintillation cocktail to the case where the EGF receptor was quantified by stripping the EGF receptor from the surface of the PDMS using the stripping buffer (Figure 4). These experiments revealed that the signal-to-background ratio improved dramatically from ~16:1 to ~82:1 when the stripping buffer was used on PDMS. This signal-to-background substantially exceeded the average signal-to-background ratio obtained in any of our experiments using H11 antibody on PS. We also note that we observed some variability on the PS surfaces, with signal-to-background ranging from 19:1 (Figure 4) to 48:1 (Figure 3). The results obtained with PDMS in Figure 4 indicate that anti-EGF receptor functionalized PDMS is capable of releasing an average of 85% of its counts to the stripping buffer, whereas the IgG isotype control released less than 23% of the counts on its surface. This observation suggests that there is a baseline non-specific adsorption of EGF receptor that is more or less irreversible on the PDMS surface. This result is an important one because it suggests that isolation of the captured EGF receptor surface from the antibody-decorated surface can substantially increase signal-to-background. We note that the data in Figure 3 and Figure 4 reveal some variability in the percentage of EGF receptor captured by antibodies immobilized on PDMS. This variability reflects variation in the concentration of the EGF receptor in solution (due to losses during sample preparation), and the use of concentrations of EGF receptor that are close to the dissociation constant for the antibodies. Below we explore a general and facile separation process that involves the mechanical transfer of the EGF receptor. This procedure is demonstrated to also lead to dramatic improvement of signal-to-background, but without requiring the use of stripping buffer.
Fig. 4.
Comparison of extraction of EGF receptor signal from PDMS capture surfaces by excision and by stripping buffer extraction, compared to PS. PDMS surface area = 35.6 mm2 and PS surface area = 18.2 mm2. n = 6 for all measurements.
Isolation of human EGF receptor by mechanical transfer from PDMS stamps to gold chips
Much of the technological interest in PDMS is based on its elastomeric properties, which enables easy fabrication of microfluidic devices as well as conformal contact with surfaces for mechanical transfer (printing).4,14 In our case, we sought to demonstrate use of PDMS to capture and transfer EGF receptor to a chemically functionalized gold-coated surface for isolation, with the long term goal of developing simple and sensitive microanalytical systems that permit detection of the EGF receptor. To determine the extent of mechanical transfer of EGF receptor during printing, we used the procedure outlined in Figure 5A, in which the PDMS surfaces were contacted with gold-coated glass chips functionalized with 2-aminoethanethiol monolayers. When comparing the anti-EGF receptor antibody (clone H11) to the IgG isotype control (Figure 5B), we consistently found that 75–81% of the EGF receptor captured on the H11-decorated PDMS stamp was transferred from the stamp to the chemically functionalized surface. In contrast, when using IgG control-functionalized stamps, only 37% of counts were transferred. We note that this trend is similar to the results described above that were obtained with stripping buffer (Figure 4). This result suggests that mechanical transfer has likely transferred the maximum amount of EGF receptor that can be released from the PDMS stamp and that a residual amount of EGF receptor that is non-specifically captured on the PDMS is, for practical purposes, irreversibly bound to the PDMS. Importantly, this differential transfer of EGF receptor using H11 versus control IgG functionalized stamps leads to an increase in signal-to-background from ~19:1 (on the PDMS stamp) to 88:1 (on the gold chip, following transfer). We also investigated capture surfaces reported by Bernard et al.,14 by stamping onto non-functionalized, sonicated (5 minutes in 2:1 solution of ethanol:water) glass microscope slides. Using these surfaces, we measured a 47–75% transfer efficiency (compared to a 28–48% transfer efficiency from IgG isotype control). This result indicates that the amine-terminated capture surface used in our experiments leads to higher capture levels of EGF receptor than glass slides. This result also suggests that control of the chemical functionality of the capture surface likely affords an opportunity to improve signal-to-background in microanalytic systems that exploit mechanical transfer of protein analytes.
Conclusion
The main conclusions of the study reported in this paper are threefold. First, we have established protocols that lead to the reproducible capture of the transmembrane protein EGF receptor onto the PDMS surfaces presenting covalently immobilized antibodies for EGF receptor, and subsequent mechanical transfer of the receptor onto a chemically functionalized surface of a gold film. The physical properties of transmembrane proteins make this class of proteins a difficult one to analyze, and thus results reported in this paper provide quantitative data that establish the feasibility of performing assays for transmembrane proteins using PDMS-based microanalytical systems. Second, we have benchmarked the performance of antibodies to EGF receptor covalently immobilized on PDMS against the performance of the same antibodies physisorbed to conventional surfaces used in ELISA assays. These results reveal that two pan-reactive antibodies for EGF receptor (H11 and 111.6) and one phosphospecific antibody (pY1068) directed against activated EGF receptor can capture the receptor on both PDMS and ELISA plates. When using the H11 antibody and identical procedures for extraction of EGF receptor from the PDMS and PS analytic surfaces (stripping buffer), the signal-to-background obtained on the PDMS surface was 82:1, thus exceeding the signal-to-background measured on the PS surface. Third, we characterized the transfer of EGF receptor during mechanical contact of the PDMS surface with a chemically functionalized gold film. The efficiency of mechanical transfer of the transmembrane protein from the PDMS surface was found to be approximately 75–81%. In contrast, the transfer of non-specifically bound protein was substantially less than 81%, thus leading to the conclusion that mechanical transfer of the EGF receptor leads to an increase in signal-to-background from ~19:1 (on PDMS surfaces) to ~88:1 (after printing). This signal-to-background is superior to that obtained using PS-based ELISA plates (<44:1). These results, when combined, provide guidance for the design of microanalytical systems for complex and clinically important proteins such as EGF receptor that employ PDMS.
Acknowledgements
This research was partially supported by the National Institutes of Health (CA108467 and CA122892) and the National Science Foundation (DMR 0520527)
References
- 1.McDonald JC, Whitesides GM. Acc. Chem. Res. 2002;35:491–499. doi: 10.1021/ar010110q. [DOI] [PubMed] [Google Scholar]; Whitesides GM. Nature. 2007;442:368–373. doi: 10.1038/nature05058. [DOI] [PubMed] [Google Scholar]
- 2.Chang JC, Brewer GJ, Wheeler BC. Biomaterials. 2003;24:2863–2870. doi: 10.1016/s0142-9612(03)00116-9. [DOI] [PubMed] [Google Scholar]; Rozkiewicz DI, Kraan Y, Werten MWT, De Wolf FA, Subramaniam V, Ravoo BJ, Reinhoudt DN. Chem. Eur. J. 2006;12:6290–6297. doi: 10.1002/chem.200501554. [DOI] [PubMed] [Google Scholar]; Millet LJ, Stewart ME, Sweedler JV, Nuzzo RG, Gillette MU. Lab Chip. 7:987–994. doi: 10.1039/b705266a. [DOI] [PubMed] [Google Scholar]
- 3.Liu DJ, Perdue RK, Sun L, Crooks RM. Langmuir. 2004;20:5905–5910. doi: 10.1021/la049605p. [DOI] [PubMed] [Google Scholar]; Tan H, Huang S, Yang KL. Langmuir. 2007;23:8607–8613. doi: 10.1021/la701258c. [DOI] [PubMed] [Google Scholar]
- 4.Bernard A, Delamarche E, Schmid H, Michel B, Bosshard HR, Biebuyck H. Langmuir. 1998;14:2225–2229. [Google Scholar]; Tingey ML, Wilyana S, Snodgrass EJ, Abbott NL. Langmuir. 2004;20:6818–6826. doi: 10.1021/la049728+. [DOI] [PubMed] [Google Scholar]; Inerowicz HD, Howell S, Regnier FE, Reifenberger R. Langmuir. 2002;18:5263–5268. [Google Scholar]
- 5.Butler JE, Lu EP, Navarro P, Christiansen B. J. Mol. Recognit. 1997;10:36–51. doi: 10.1002/(SICI)1099-1352(199701/02)10:1<36::AID-JMR353>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 6.Wu DP, Zhao BX, Dai ZP, Qin JH, Lin BC. Lab Chip. 2006;6:942–947. doi: 10.1039/b600765a. [DOI] [PubMed] [Google Scholar]
- 7.Linder V, Verpoorte E, Thormann W, De Rooij NF, Sigrist M. Anal. Chem. 2001;73:4181–4189. doi: 10.1021/ac010421e. [DOI] [PubMed] [Google Scholar]
- 8.Eteshola E, Leckband D. Sens. Actuators B. 2001;72:129–133. [Google Scholar]
- 9.Chen CS, Mrksich M, Huang S, Whitesides GM. Science. 1997;276:1425–1428. doi: 10.1126/science.276.5317.1425. [DOI] [PubMed] [Google Scholar]
- 10.Makamba H, Hsieh Y-Y, Sung W-C, Hui Chen S. Anal. Chem. 2005;77:3971–3978. doi: 10.1021/ac0502706. [DOI] [PubMed] [Google Scholar]
- 11.Huang B, Wu H, Kim S, Kobilka BK, Zare RN. Lab Chip. 2006;6:369–373. doi: 10.1039/b515840k. [DOI] [PubMed] [Google Scholar]
- 12.Cesaro-Tadic S, Dernick G, Juncker D, Buurman G, Kropshofer H, Michel B, Fattinger C, Delamarche E. Lab Chip. 2004;4:563–569. doi: 10.1039/b408964b. [DOI] [PubMed] [Google Scholar]
- 13.Hollahan JR, Carlson GL. J. Appl. Polym. Sci. 1970;14:2499–2508. [Google Scholar]; Chaudhury MK, Whitesides GM. Langmuir. 1991;7:1013–1025. [Google Scholar]; Diaz-Quijada GA, Wayner DDM. Langmuir. 2004;20:9607–9611. doi: 10.1021/la048761t. [DOI] [PubMed] [Google Scholar]; Sui G, Wang J, Lee C-C, Lu W, Lee SP, Leyton JV, Wu AM, Tseng H-R. Anal. Chem. 2006;78:5543–5551. doi: 10.1021/ac060605z. [DOI] [PubMed] [Google Scholar]
- 14.Bernard A, Fitzli D, Sonderegger P, Delamarche E, Michel B, Bosshard HR, Biebuyck H. Nat. Biotech. 2001;19:866–869. doi: 10.1038/nbt0901-866. [DOI] [PubMed] [Google Scholar]
- 15.Lin FYH, Sabri M, Erickson D, Alirezaie J, Li D, Sherman PM. Analyst. 2004;129:823–828. doi: 10.1039/b409222h. [DOI] [PubMed] [Google Scholar]; Jang C-H, Tingey ML, Korpi NL, Luk Y-Y, Bertics PJ, Abbott NL. J. Am. Chem. Soc. 2005;127:8912–8913. doi: 10.1021/ja051079g. [DOI] [PubMed] [Google Scholar]
- 16.Ullrich A, Schlessinger J. Cell. 1990;61:203–212. doi: 10.1016/0092-8674(90)90801-k. [DOI] [PubMed] [Google Scholar]; Irmer D, Funk JO, Blaukat A. Oncogene. 2007;26:5693–5701. doi: 10.1038/sj.onc.1210383. [DOI] [PubMed] [Google Scholar]
- 17.Baselga J. Oncologist. 2002;7:2–8. doi: 10.1634/theoncologist.7-suppl_4-2. [DOI] [PubMed] [Google Scholar]
- 18.Bernard A, Michel B, Delamarche E. Anal. Chem. 2001;73:8–12. doi: 10.1021/ac0008845. [DOI] [PubMed] [Google Scholar]; Kanda V, Kariuki JK, Harrison DJ, McDermott MT. Anal. Chem. 2004;76:7257–7262. doi: 10.1021/ac049318q. [DOI] [PubMed] [Google Scholar]
- 19.Wiepz GJ, Guadaramma AG, Fulgham DL, Bertics PJ. Methods Mol. Biol. 2006;327:25–38. doi: 10.1385/1-59745-012-X:25. [DOI] [PubMed] [Google Scholar]
- 20.Weber W, Bertics PJ, Gill GN. J. Biol. Chem. 1984;259:14631–14636. [PubMed] [Google Scholar]
- 21.Lee JN, Park C, Whitesides GM. Anal. Chem. 2003;75:6544–6554. doi: 10.1021/ac0346712. [DOI] [PubMed] [Google Scholar]
- 22.Koland JG, Cerione RA. Biochim. Biophys. Acta. 1990;1052:489–498. doi: 10.1016/0167-4889(90)90160-f. [DOI] [PubMed] [Google Scholar]; Hubler L, Kher U, Bertics PJ. Biochim. Biophys. Acta. 1992;1133:307–319. doi: 10.1016/0167-4889(92)90052-d. [DOI] [PubMed] [Google Scholar]
- 23.Cochran JR, Kim Y-S, Olsen MJ, Bhandari R, Wittrup KD. J. Immunological Methods. 2004;287:147–158. doi: 10.1016/j.jim.2004.01.024. [DOI] [PubMed] [Google Scholar]
- 24.Xu H, Zhao X, Grant C, Lu JR. Langmuir. 2006;22:6313–6320. doi: 10.1021/la0532454. [DOI] [PubMed] [Google Scholar]
- 25.Peluso P, Wilson DS, Do D, Tran H, Venkatasubbaiah M, Quincy D, Heidecker B, Poindexter K, Tolani N, Phelan M, Witte K, Jung LS, Wagner P, Nock S. Anal. Biochem. 2003;312:113–124. doi: 10.1016/s0003-2697(02)00442-6. [DOI] [PubMed] [Google Scholar]
- 26.Butler JE. Methods. 2000;22:4–23. doi: 10.1006/meth.2000.1031. [DOI] [PubMed] [Google Scholar]
- 27.Xu H, Lu JR, Williams DE. J. Phys. Chem. B. 2006;110:1907–1914. doi: 10.1021/jp0538161. [DOI] [PubMed] [Google Scholar]