Abstract
The tumor oncoproteins HRAS, KRAS, and NRAS are the founding members of a larger family of at least 35 related human proteins. Using a somewhat broader definition of sequence similarity reveals a more extended superfamily of more than 170 RAS-related proteins. The RAS superfamily of GTP (guanosine triphosphate) hydrolysis–coupled signal transduction relay proteins can be subclassified into RAS, RHO, RAB, and ARF families, as well as the closely related Gα family. The members of each family can, in turn, be arranged into evolutionarily conserved branches. These groupings reflect structural, biochemical, and functional conservation. Recent findings have provided insights into the signaling characteristics of representative members of most RAS superfamily branches. The analysis presented here may serve as a guide for predicting the function of numerous uncharacterized superfamily members. Also described are guanosine triphosphatases (GTPases) distinct from members of the RAS superfamily. These related proteins employ GTP binding and GTPase domains in diverse structural contexts, expanding the scope of their function in humans.
Introduction
GTPases, together with their associated regulators and effectors, participate as central control elements in signal transduction pathways that touch on virtually every aspect of cell biology. Most of these proteins fall within a superfamily named for the RAS oncoprotein. Research into the biochemistry and function of RAS-related GTPases has focused on a relatively small subset of proteins. Genome analysis and gene expression results from multiple sources were used to create an extensive accounting of the genes and proteins that constitute the human RAS superfamily and some more distantly related GTPases (1). Sequence comparison analysis (2) revealed insights into the relationship among members of this signal transduction superfamily.
RAS Biochemistry and Function
RAS superfamily proteins share a basic biochemical activity: GTP (guanosine triphosphate) binding and hydrolysis (Fig. 1). This commonality is directly reflected in the presence in each protein of several characteristic “G box” sequences (3, 4). The G1 box [aaaaGxxxxGK(S or T), where a = L or I or V or M, and x = any amino acid], also known as a P-loop or Walker A motif (5), is a purine nucleotide binding signature. The G3 box (blbbDxxGl, where l = hydrophilic and b = hydrophobic), which overlaps with the Walker B motif at the invariant aspartic acid residue, is involved in binding a nucleotide-associated Mg2+ ion and is also well conserved among superfamily members. Residues of the G4 box [bbbb(N or T)(K or Q)xD] make hydrogen bond contact with the guanine ring (conferring specificity to GTP over ATP) and provide stabilizing interactions with G1 box residues. Amino acids in the G5 box [bbE(A or C or S or T)SA(K or L)] primarily make indirect associations with the guanine nucleotide and are less well conserved among supergroup members. The G2 box (YDPTIEDSY for HRAS and several other RAS subfamily members) is located in one of two segments that reorient as a function of GDP or GTP binding and provide major components of the effector binding surface. Of all RAS superfamily G2 box sequences, only the threonine residue is highly conserved, but several other residues recur within subfamilies. Mutations in this domain can block association of HRAS with one or more of its downstream effectors (6–8).
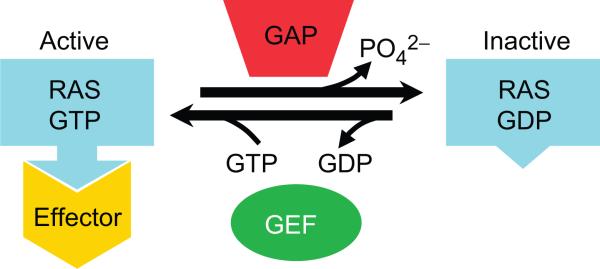
Fig. 1.
RAS proteins exist in equilibrium between GTP- and GDP-bound forms. GEFs and GAPs regulate the relative amounts of each form. The GTP-bound conformation of RAS shows high-affinity interactions with effector proteins that propagate downstream signaling.
RAS proteins share a common mechanism of operation that is tied to nucleotide-regulated conformational shifts [reviewed in (9)]. In the GTP-bound state, they display a binding surface with high affinity for downstream effector proteins [for example, HRAS-GTP has a Kd (dissociation constant) of 18 nM for the protein kinase RAF1 (10)]. The structural changes are confined primarily to two loop regions called switch 1 and switch 2 (11). However, the high-affinity effector-binding conformation of RAS proteins is transient; GTP hydrolysis and release of the γ-phosphate leads to reorientation of effector binding residues, the release of effector proteins (due to reduced affinity), and attenuation of downstream signaling.
The rate-limiting step in RAS protein activation is the exchange of bound GDP for GTP. In most cases this is a slow step (3.4 × 10−4 s−1 for HRAS) (12), favoring an inactive steady-state conformation of RAS even in the presence of a high cellular GTP/GDP ratio [~10-fold (13), although this may not be uniform throughout the cell]. This kinetic limitation is the basis for stimulus-induced mechanisms of RAS protein regulation. Guanine nucleotide exchange factors (GEFs, also called guanine nucleotide–dissociation stimulators or GDSs) catalyze the release of GDP (Fig. 1), thus promoting GTP loading and activation of RAS (~sixfold stimulation for HRAS by the exchange factor SOS1) (14). Several GEFs may act on a particular RAS protein [reviewed in (15)], with each GEF responding to distinct upstream stimuli (for example, growth factor receptor phosphorylation or diacylglycerol production), providing multiple avenues for signal regulation. GEF-mediated regulation is also a point of vulnerability for RAS function: RAS mutants that bind GEFs unproductively (e.g., HRASS17N) can dominantly block the activation of endogenous RAS (16, 17).
Guanine nucleotide dissociation inhibitors (GDIs) act in opposition to exchange factors. GDIs bind specifically to GDP-bound GTPases and inhibit the release of GDP (18), thus prolonging the inactive state. GDI binding also serves to emulsify some lipid-modified GTPases, allowing them to dissociate from membrane surfaces. Multiple GDIs have been identified for RHO and RAB proteins (19, 20) but, to date, not for other subfamilies. GDI-bound, cytoplasmic RHO and RAB proteins are effectively sequestered from membrane-associated effectors as well as regulators.
The intrinsic GTPase activity of RAS-related proteins is typically low (4.2 × 10−4 s−1 for HRAS) (12), which would tend to prolong signal transduction. GTP hydrolysis is greatly enhanced, however, by the intervention of GTPase activating proteins (GAPs) (Fig. 1) (21). As with GEFs, there are often multiple GAPs that function on a given RAS protein (22), allowing for a variety of input sources at this stage of regulation.
Many RAS family proteins are subject to multiple lipid modifications (23), which promote association with cellular membranes. Covalent posttranslational modif ication of C-terminal cysteine residues by isoprenylation (attachment of a farnesyl or geranylgeranyl group) is observed for most RAS, RHO, and RAB family members. This modification has also been implicated in determining subcellular membrane localization, which in turn can influence effector binding or activation and regulatory protein interactions (24–29). Cysteine palmitoylation (covalent attachment of a palmitate fatty acid) also occurs near the C terminus of some RAS and RHO proteins.
At the N terminus of many ARF (ADP ribosylation factor) and Gα subfamily proteins, cotranslational modification of glycine by myristoylation occurs. For some Gα proteins, myristoylation is combined with palmitoylation of a neighboring cysteine (30, 31). In other cases, palmitoylation of cysteines near the N terminus appears to be independent of other modifications (32). As with the modifications at the C terminus, the N-terminal lipid additions likely play a role in membrane localization but may well contribute in other ways to RAS protein structure or function.
RAS Proteins and Cancer
RAS (rat sarcoma) genes were first identified and characterized as transduced oncogenes in the Harvey and Kirsten strains of acutely transforming retroviruses (33, 34) (note: early publications use the name p21src for these genes). Mutationally activated forms of HRAS (also called H-Ras), KRAS (also called K-Ras), and NRAS (also called N-Ras) were subsequently isolated from human tumor cells using transfection-based assays (35–37). Tumor-derived RAS mutations, such as HRASG12V, disable GTPase function and GAP responsiveness (38). Mutations that enhance guanine nucleotide exchange (e.g., HRASN116H) also enhance the basal activation state of RAS proteins (39, 40).
High rates of KRAS-activating missense mutations have been detected in non–small cell lung cancer (15 to 20% of tumors) (41), colon adenomas (40%) (42), and pancreatic adenocarcinomas (95%) (43), making it the single most common mutationally activated human oncoprotein. In some tumors, HRAS- or NRAS-activating mutations are also seen. More than half of the most malignant thyroid tumors, characterized as poorly differentiated or undifferentiated, harbor a mutation in KRAS, HRAS, or NRAS (44). In addition to mutational activation, RAS genes are amplified or overexpressed in some tumors (45). In the case of breast cancer, the incidence of RAS-activating mutations is low, but RAS activity is elevated due in part to increased upstream signaling from the receptor tyrosine kinase ERBB2 (also called Her2) (46). Other mechanisms leading to RAS overactivation in tumor cells include the deletion of genes encoding negative regulators (for example NF1, a GAP for RAS, in neurological tumors) (47–50) and overexpression of positive regulators (such as SOS1, a GEF for RAS, in renal cancer cells) (51). Taken together, these data illustrate the critical and pervasive role played by RAS in cell transformation.
Activating mutations in other members of the RAS superfamily are much less common in human tumors, and the known examples are generally restricted to neoplasias of relatively low frequency. In vitro systems, however, provide compelling evidence that several members of the RAS superfamily, aside from KRAS, HRAS, and NRAS, can enhance or facilitate cell transformation.
RAS Protein Subfamily (35 members)
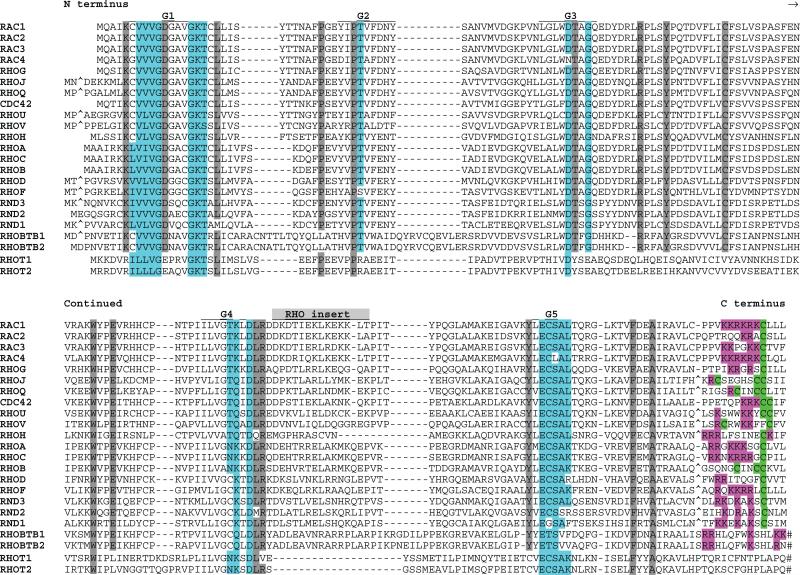
RAS subfamily members show high conservation within the G1, G3, G4, and G5 boxes (Fig. 2). Most proteins in this group are relatively small (183 to 340 amino acids in length) and show no prominent functional motifs outside of those defining their RAS relatedness.
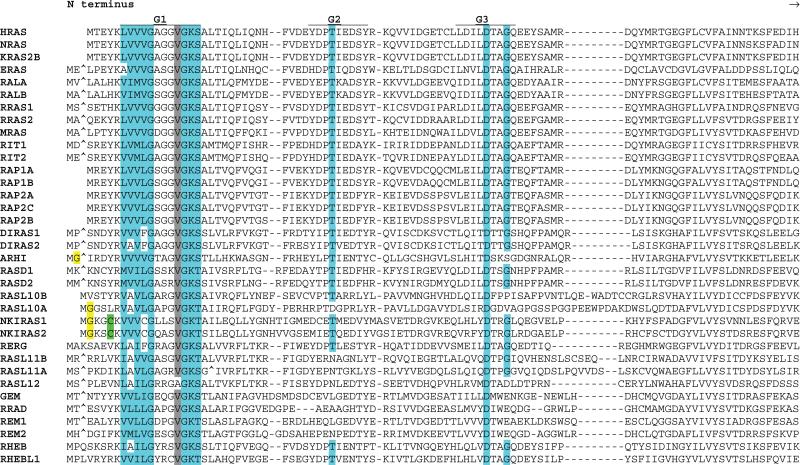
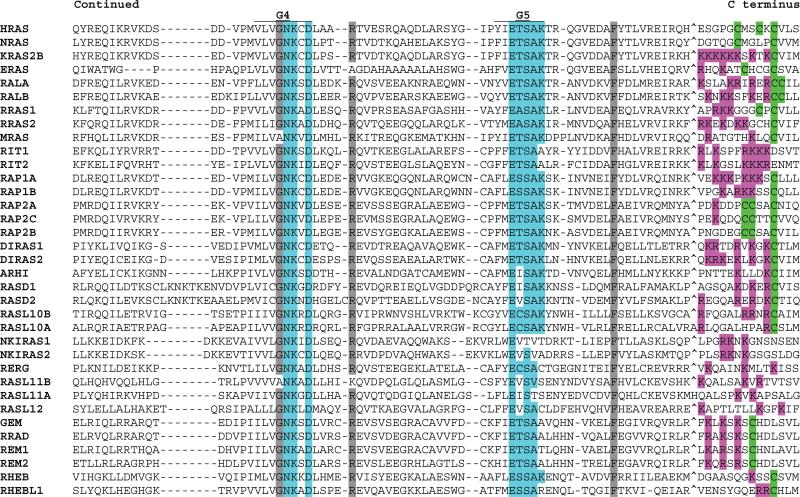
Fig 2.
Alignment of human RAS subfamily members. G box consensus residues are highlighted in blue. N- and C-terminal region cysteines, some of which are substrates for prenylation or fatty acid modification, are highlighted in green. N-terminal glycines in positions favoring myristoylation are highlighted in yellow. C-terminal basic residues are highlighted in pink. Gray highlighting indicates residues that are highly conserved in 90% of members. Amino acids omitted for optimum alignment are indicated with the “^” symbol. For KRAS, the KRAS2B isoform sequence is presented. See Table 1 for alternate gene symbols.
Most of the RAS subfamily proteins localize predominantly to the plasma membrane. Membrane localization results in part from C-terminal prenylation. Prenylation signals mostly conform to the Caax (a = aliphatic, x = terminal amino acid) motif that directs cysteine farnesylation (except when x = L or F, which instructs geranylgeranylation as occurs on RRAS proteins and some RAP proteins). The prenylation reaction is followed by proteolysis of the three C-terminal residues (aax) and methylation of the lipid-modified cysteine [reviewed in (23, 52)]. The later two posttranslational processing steps take place in the endoplasmic reticulum (ER) before transport to the plasma membrane (53). RAL proteins contain the geranylgeranylation signal CCaa. Some RAS subfamily members lack either type of isoprenylation motif and are not subject to any known lipid modification.
Some RAS subfamily proteins contain fatty acid acylation signals. Notably, HRAS, NRAS, ERAS, RRAS1 and RAP2A, RAP2B, and RAP2C have palmitoylated cysteine residues proximal to their C-terminal prenylated cysteines. This modification requires transit through the Golgi compartment (54). Endomembrane localization of RAS proteins may be more than just a posttranslational modification detour, however. Several lines of evidence suggest that RAS proteins are functional signal transducers in the ER-Golgi complex (55–57).
N-terminal lipidations may contribute to the localization of other RAS subfamily members such as ARHI (Ras homolog member I), which has a potential myristoylation site (MetGly) at its N terminus, and NKIRAS1 and NKIRAS2 (NF-κB inhibitor–interacting Ras-like 1 and 2) proteins, which have putative myristoylation or palmitoylation modification signals (MGxxCxxxxC) .
An additional factor in RAS protein trafficking and localization is the presence of a C-terminal polybasic region, as seen on the predominant KRAS splice variant KRAS2B. This protein lacks a palmitoylation site but has a strong polybasic region immediately upstream of the C-terminal farnesylation site. In contrast, HRAS and NRAS have palmitoylation sites but no polybasic regions. These differences are believed to underlie the distinct membrane localization characteristics (58) and signaling properties (59) of these otherwise close paralogs. In the 2A isoform of KRAS (N terminus = KTPGCVKIKKCIIM), the polybasic sequence is replaced with a palmitoylation site. As a result, the KRAS2A isoform may be more similar to HRAS and NRAS in its subcellular localization. Interestingly, RAP1 (prenylation + polybasic) and RAP2 (prenylation + fatty acylation) proteins appear to have a relationship similar to that of KRAS2B and HRAS. The RIT1 and RIT2 proteins encode C-terminal polybasic sequences (6 of 10 residues are R or K) but lack both prenylation and fatty acylation signals. RERG (Ras-related and estrogen-regulated growth inhibitor) appears to be devoid of all standard lipid membrane localization signals and displays cytosolic localization, suggesting that it functions outside the context of cellular membranes (60).
Other posttranslational modifications have been described for RAS subfamily proteins. These include serine phosphorylation (61) and nitrosylation (62–64) of HRAS and tyrosine phosphorylation of RRAS1 (65, 66); the functions of these modifications are still under investigation.
Variation at the level of alternative splicing has been described for some RAS genes. For KRAS (67, 68) and HRAS (69), the primary function of alternate splicing may be to generate isoforms with distinct subcellular localizations.
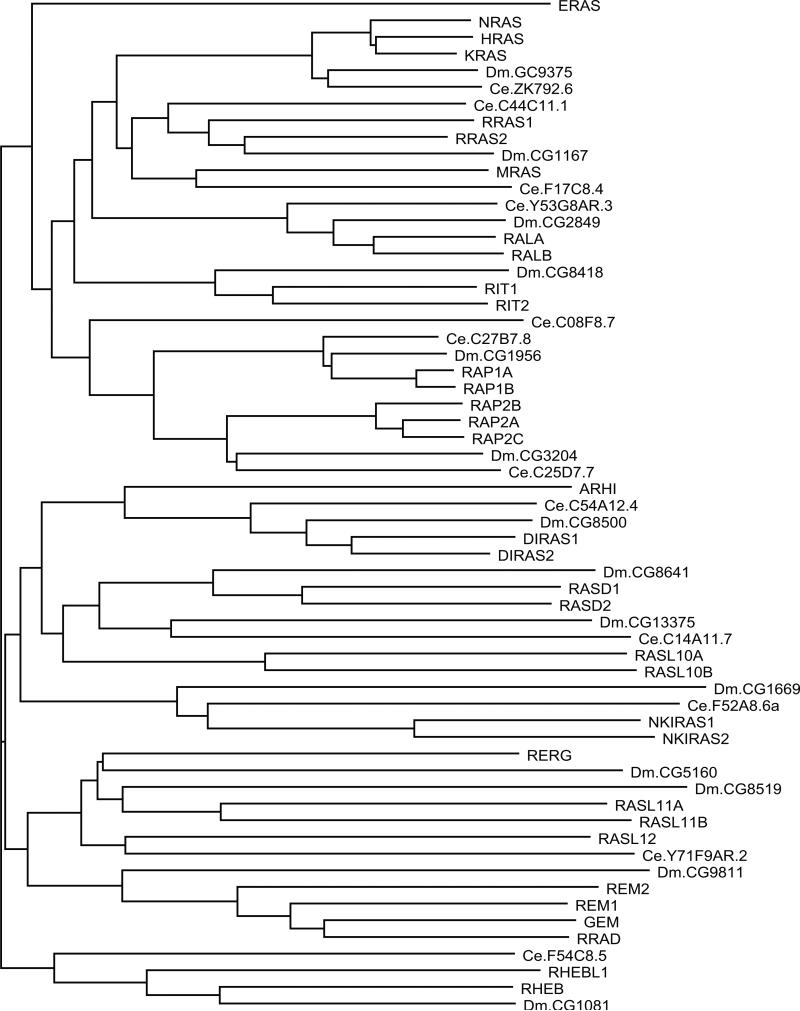
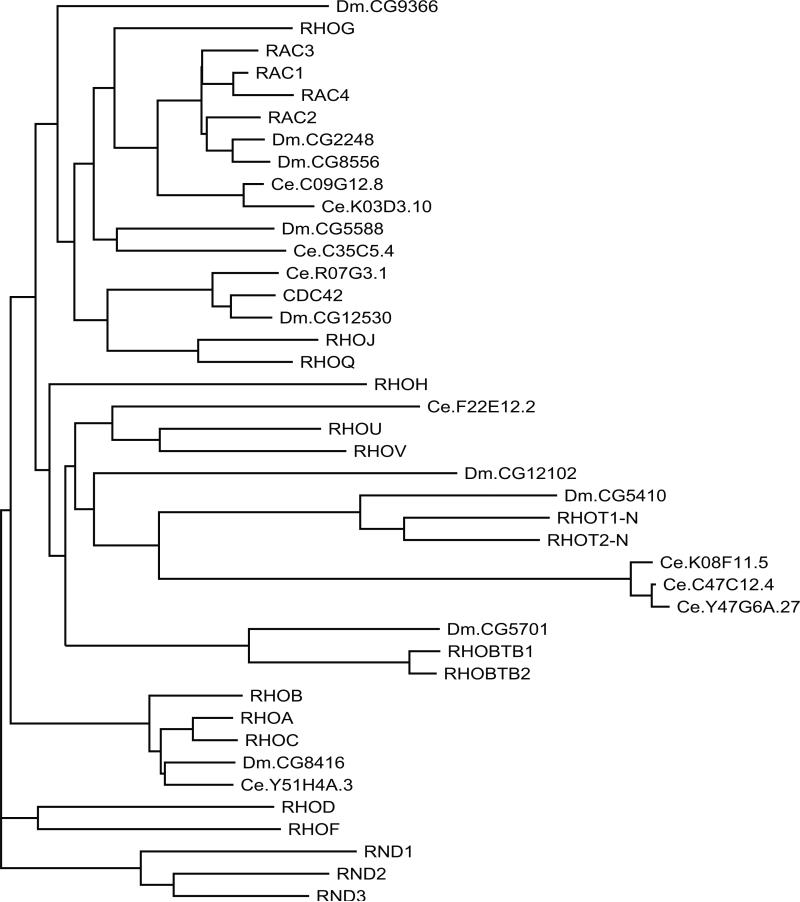
A comparison of RAS subfamily sequences from Homo sapiens, Drosophila Melanogaster, and Caenorhabditis elegans (Fig. 3) shows strong conservation through evolution, with most branches of the dendrogram containing representatives from each species. This analysis also illustrates a notable expansion of RAS subfamily proteins (human = 35, fly = 14, worm = 12) and suggests 12 structural or functional branches.
Fig. 3.
Dendrogram of RAS subfamily members from H. sapiens, D. melanogaster (Dm), and C. elegans (Ce). Human protein names are in uppercase letters. Branch lengths are directly proportional to the number of differences between sequences compared. See Table 1 for alternate names for human protein.
RAS oncoprotein branch (HRAS, KRAS, and NRAS)
HRAS, KRAS, and NRAS (H, K, NRAS) proteins are perhaps best known for their mitogenic properties. As discussed above, mutationally activated forms of these proteins can efficiently transform cells in vitro and in vivo, and such mutations are common in a broad spectrum of human tumors. There is also strong evidence from cell culture experiments (70) and model organisms (71, 72) that H, K, NRAS proteins contribute to cell differentiation and organ development. These same proteins have more recently been implicated in neuronal plasticity in the central nervous system (73–78).
The protein kinase RAF1 (also called c-Raf) was the first identified RAS effector (79–83) and, together with the closely related ARAF (also called A-Raf) and BRAF (also called BRaf), has been the most intensively studied [reviewed in (84)]. Activated RAS binds with high affinity to the “Raf-like Ras–binding domain” (Interpro IPR003116), as well as an adjacent cysteine-rich domain, and leads to activation of the kinase activity of RAF and initiation of the MEK-ERK mitogen-activated protein kinase cascade, which affects transcription and other cellular functions. The precise mechanism of RAS-mediated activation is complex and not yet fully elucidated, but seems to involve enhanced membrane association, as well as allosteric derepression (deletion of the RAS binding domain results in constitutive kinase activity) (85) and promotion of RAF phosphorylation by serine-threonine and tyrosine kinases. Hyperactivation of the RAF effector pathway alone can transform immortalized rodent fibroblast cells, but appears to be insufficient for transformation of some other cell types (86, 87). The frequent occurrence of dominant BRAF mutations in some human cancers (88) further suggests that this effector pathway has a major role in tumorigenesis. Other human proteins with “Raf-like Ras–binding domains” include TIAM1, which functions as a RAS-controlled GEF-type activator of RAC (a member of the Rho subfamily) (89). Mice deficient in TIAM1 function develop normally but are impaired in carcinogen-induced, RAS-mediated, tumorigenesis (90), consistent with a role for this effector in RAS-mediated growth regulation.
The catalytic subunits of phosphotidylinositol 4,5 bisphosphate 3-kinase (PI3K) constitute another well-established class of RAS effectors (91). RAS binds to a consensus “phosphoinositide 3-kinase Ras binding domain” (Interpro IPR000341) found in seven distinct human proteins (PIK3CG, PIK3C2A, PIK3C2G, PIK3CB, PIK3CA, PIK3C2B, and PIK3CD). This interaction promotes PI3K catalytic activity (92), resulting in increased production of membrane-associated PIP3 (phosphatidylinositol 3,4,5-trisphosphate) and the subsequent plasma membrane recruitment of PIP3-binding PH domain proteins such as the protein kinases AKT1 and PDPK1 (3-phosphoinositide–dependent protein kinase 1, also called PDK1). RAS-mediated activation of PI3K is also an important component of cell transformation (8).
Several GEFs for RAL proteins are RAS effectors (93–97). RALGDS, RGL1 (Ral GDP dissociation stimulator–like; also called ARHGAP9), RGL2 (also called Rab2L), and RGL3 each encode a Ras association (RA) domain (Interpro IPR000159), a third type of RAS-effector interaction motif. RAS proteins stimulate the nucleotide-exchange activity of RALGDS (98), and this appears to have a critical role in human cell transformation (99, 100).
RIN1 is another RA domain–containing RAS effector protein (101, 102). The RIN1 protein functions as a RAS-responsive GEF for RAB5 (103) and also stimulates the catalytic activity of the ABL tyrosine kinase (104, 105). RIN1 has a restricted expression pattern (78) and, because of its high-affinity binding to RAS proteins (101), may function in part as a physiological competitor of other effectors. The related proteins RIN2 and RIN3 have discernable RA domains but have not been functionally connected to any RAS protein. Another RAS effector, NORE1 (novel Ras effector 1; also called RASSF5 and RapL), is a positive regulator of cell death through association with the proapoptotic kinase STK4 (106). NORE1 is itself part of a family of related proteins (RASSF1 through RASSF6) that all contain RA domains but have not all been functionally connected to RAS. The RA domain–containing enzyme phospholipase C epsilon (PLCE1; also called PLCε) has also been described as a RAS effector (107). However, RA domains show affinity for RAP as well as RAS proteins. In the case of the RA protein MLLT4 (also called AF6), Rap1 proteins may be the preferred physiological binding partners (108). Finally, another RA domain–containing protein, RASIP1 (Ras-interacting protein 1, also called RAIN), is an effector of RAS and RAP (109). Systematic analysis of RAS family GTPases and multiple effectors has demonstrated binding specificity that often correlates with biochemical and biological activation (110).
BRAP (also called IMP, impedes mitogenic signal propagation) is another protein that binds specifically to activated RAS (111), although BRAP has no RA or other recognizable RAS-interaction domain. BRAP appears to function as a dedicated inhibitor of signaling between RAF and MEK.
RRAS (Related to RAS) branch
RRAS1, RRAS2 (also called TC21), and MRAS (also called RRas3) appear to be involved in control of mitogenesis and the cytoskeleton. RRAS1 localizes to focal adhesions where it promotes cell adhesion and activates integrins (112, 113). Activating (GTPase-defective) mutants of all the RRAS proteins can transform cultured fibroblast cells, with RRAS2 being the most potently transforming (114–117). Activating mutations and overexpression of RRAS2 are found in some human tumors (118–120). Effectors implicated in the function of RRAS family members include PI3K (121, 122), RALGDS and related proteins (97, 122, 123), and RAF kinases (124, 125), but RRAS1 appears to work primarily through PI3K (121). This overlap with effectors of the H, K, NRAS family likely reflects the complete conservation of G2 box (switch 1) sequences among members of both branches. The differences between the physiological consequences of RRAS activation versus that of H, K, NRAS activation may reflect quantitative differences in effector engagement, as well as the contribution of some unique effectors for each protein.
RAP (Ras-Proximal) branch
RAP proteins are activated by mitogenic stimuli and function as regulators of integrin-mediated cell adhesion and cell spreading (126, 127). In cultured cells, RAP proteins do not show transforming activity. Rather, overexpression of RAP1A inhibits RAS-mediated transformation (128). However, RAP1A has been reported to bind and activate BRAF (129), suggesting that it has the capacity to promote mitogenesis and perhaps transformation in some contexts but not others (130). Two observations suggest contributions of RAP proteins in tumorigenesis, but with possible tissue-type specificity. Activation of a RAP-directed GEF (131) or inactivation of a RAP-directed GAP (132) promotes hematopoietic tumor formation. Conversely, the loss of an activator of RAP1 proteins has been found in a mouse osteosarcoma and in several nonhematopoietic human cancer cell lines (133).
RAP proteins may function through activation of RALGDS and related proteins, but not in the same way that RAS does (134), and through associations with PLCE1 (135). In lymphoid cells, RAP1 proteins promote integrin activation through NORE1 (136).
RAL (RAS-Like) branch
RALA and RALB have been implicated in a broad spectrum of functions including mitogenic responses, differentiation, protein trafficking, and cytoskeleton dynamics [reviewed in (137)]. As discussed above, H, K, NRAS, RRAS2, MRAS, and RAP proteins all appear to work in part through RALGDS-type effectors that are expected to stimulate RAL functions. Although mutationally activated RAL proteins are not themselves oncogenic, they can enhance transformation of cultured cells by RAS and EGFR (epidermal growth factor receptor) (98, 138). The two RAL proteins appear to have distinct and complementary roles in cell transformation; RALB is required for tumor cell survival, whereas RALA promotes anchorage-independent cell proliferation (139). Each RAL also has a distinct role in epithelial cell polarization (140).
Several RAL effectors have been identified but, to date, these do not include members of the RAF-PI3K-RALGDS triumvirate. This may seem surprising because RAL proteins show high overall relatedness to H, K, N-RAS proteins. However, the two subfamilies diverge appreciably in their Switch 1 regions (Fig. 2). The sequence YDPTIED is completely conserved in H, K, N-RAS proteins as well as in RRAS1 RRAS2, MRAS, and all RAP proteins, all of which share many effectors. In RALA and RALB the equivalent sequence is YEPTKAD.
The RAL effector RALBP1 (also called RLIP), which has a RAC- and CDC42-directed GAP domain (141–143), regulates endocytosis (144–146). RAL is also a component of the exocyst complex. RAL directly binds to both SEC5L1 (also called Sec5) and EXOC8 (also called EXO84), promoting exocyst complex assembly and membrane trafficking (147–149).
RIT (RAS-like Protein in All Tissues) branch
RIT1 and RIT2 (also called Rin) are positive factors for neuronal cell survival as well as for the initiation, elongation, and branching of neu-rites in culture (150–152). The enhanced expression of RIT1 and RIT2 in developing and mature neurons (153) supports the biological relevance of these properties. RIT2 includes a Ca2+-calmodulin binding site (153), which appears to be required for its neurite outgrowth function (150). Although an activated (GTPase-deficient) mutant of RIT1 can transform a fibroblast cell line (154, 155), there is no evidence that either RIT gene functions in tumorigenesis.
On the basis of protein interaction experiments, RALGDS (and related proteins) and AF6 are potential effectors of RIT1 and RIT2 (156), but no RIT-specific effectors have been characterized. Several lines of evidence indicate that RAF and PI3K are not direct effectors of RIT proteins (150, 154, 156).
ERAS (Embyonic Stem Cell–Expressed Ras) branch
ERAS is an unusual subfamily member in several respects. As indicated in Fig. 3, ERAS occupies a branch with no human paralogs and no fly or worm orthologs. ERAS expression is restricted to undifferentiated embryonic stem (ES) cells (157).
Ectopic expression of wild-type ERAS transforms cultured fibroblast cells (157). This unusual property likely reflects the effect of sequence differences at residues that regulate the GTP/GDP binding equilibrium in other RAS proteins (that is, the amino acid corresponding to Gly12 in H, K, NRAS). ERAS may be an important factor in the propensity of ES cells to form teratomas. A strong candidate effector of ERAS is PI3K (157).
DIRAS (Distinct Subgroup of RAS) and ARHI branches
The DIRAS1 (also called Rig) and DIRAS2 proteins, like RHEBs, show reduced GTPase activity compared to that of most RAS superfamily GTPases, and DIRAS proteins remain predominantly in the GTP-bound state (158). DIRAS and ARHI proteins may have tumor suppressor functions. Overexpression of DIRAS1 antagonizes Ras-mediated signaling and transformation, and DIRAS1 is silenced or down-regulated in many neural tumors and tumor-derived cell lines (159). The ARHI (also called Noey2) protein has been implicated as a tumor suppressor in breast and ovarian cancer (160, 161).
Ectopic expression of DIRAS1 or DIRAS2 can induce the formation of large vacuolar structures (158), but downstream effectors have not been identified for these proteins.
RASD (Ras Induced by Dexamethasone) branch
RASD1. (also called dexRas) was identified as a transcript that shows strong, rapid, and transient induction after treatment of cells with dexamethasone (162), and RASD2 (also called RHES, for Ras homolog enriched in striatum) was identified as a protein expressed in pancreatic beta cells in response to efaroxan, an imidazoline that functions as an α2-adrenergic receptor antagonist and insulin secretagogue (163). There is no evidence to support involvement of RASD1 or RASD2 in transformation or tumorigenesis. RASD1 appears to function as a negative regulator of peptide hormone secretion (164) and as a cell growth suppressor (165). RASD2 has the capacity to activate PI3K and may interfere with G protein–coupled receptor signaling (166).
The uncharacterized gene products RASL10A and RASL10B are the closest related proteins to RASD1 and RASD2.
NKIRAS (NFKB Inhibitor–interacting RAS-like, also called kB-Ras) branch
NKIRAS1 and NKIRAS2 were discovered as proteins that interact with NFKBI (usually called IκB), an inhibitor of the transcription factor NFKB (usually called NF-κB) (167). Binding of NKIRAS to NFKBI-NFKB complexes prevents nuclear translocation of the complex in resting cells, suggesting that NKIRAS proteins participate in the negative regulation of NFKB (168).
REM (Rad and Gem–related) branch
REM1, REM2, RRAD (also called Rad), and GEM (also called Kir) were identified primarily on the basis of their restrictive and regulated expression patterns (169–172). They share a conserved C-terminal cysteine (position –7), but this is not within a context recognized for lipid modification. REM subfamily proteins show no transforming or tumorigenic properties. REM1, RRAD, and GEM function in part as negative regulators of calcium currents through a direct interaction with the β subunit of a voltage-gated Ca2+ channel (173, 174). Overexpression of GEM produces cytoskeletal changes marked by cellular processes. These changes may result from a direct interaction of GEM with the kinesin-like protein KIF9 (175) and RHOA inactivation [reviewed in (176)], perhaps through GMIP (Gem interacting protein), a RHOGAP (177).
RERG (RAS-related and Estrogen-Regulated Growth inhibitor) branch
RERG was identified during a search for genes whose expression in breast tumors correlates with prolonged survival (60). As its name implies, transcription of the RERG gene is increased in response to estrogen, perhaps through direct estrogen receptor binding to the RERG gene promoter. RERG shows no binding to H, K, NRAS effectors tested (RAF, RALGDS, PI3K, and RIN1), and RERG neither transformed cultured fibroblasts nor enhanced HRAS-mediated transformation (60). Ectopic expression of RERG actually blocked transformation and tumorigenesis in a breast tumor cell line (60).
Three gene products
RASL11A, RASL11B, and RASL12—show relatedness to REM. Abundance of RASL11A transcripts is decreased in some prostate tumors (178), but its function is uncharacterized. The RASL11A, RASL11B, and RASL12 gene products have no lipid modification signals, suggesting functions that are not restricted to membrane surfaces. Further analysis of these proteins will determine if they are best considered as a separate branch of RAS proteins.
RHEB (Ras Homolog Enriched in Brain) branch
RHEB proteins are involved in the control of cell cycle and cell growth (179). Although early studies found that RHEB proteins block MAPK (mitogen-activated protein kinase) signaling and inhibit RAS-mediated transformation of cultured fibroblasts (180, 181), it is not yet clear whether these observations represent physiological activities.
RHEB proteins have low intrinsic GTPase activity and exist predominantly in the GTP-bound form. RHEBs are subject to negative regulation, however, by the GAP activity of a TSC1-TSC2 complex. The best-characterized downstream effector of RHEB is the Ser-Thr kinase FRAP1 (also called mTOR, target of rapamycin) (179, 182–186), which in turn regulates translation through its substrates RPS6K (ribosomal protein S6 kinase 1) and EIF4EBP1 (eukaryotic initiation factor 4E–binding protein). Loss-of-function mutations in TSC genes are associated with tuberous sclerosis complex, a benign tumor syndrome, suggesting that RHEB may have tumor promoter functions in vivo.
RHO Protein Subfamily (23 members)
The RHO (Ras homolog) subfamily of proteins is closely related to the RAS subgroup [reviewed in (187–188)], and members of this family show strong conservation among their G1 to G5 boxes (Fig. 4). However, most members of this subfamily have an insert sequence that is not found in other RAS superfamily GTPases. Mounting evidence supports the involvement of RHO proteins in cancer (189). They appear to be primarily collaborators in, rather than initiators of, cell transformation. The unconventional RHO protein RHOBTB2 [named after the BTB (Broad-complex Tramtrack, and Bric a brac) domain; and also called DBC2] is reported as a potential tumor suppressor (87), and overexpression of a RAC1 splice varient, RAC1B, has been reported in colorectal tumors (190).
Fig 4.
Alignment of human RHO subfamily members. Highlighting and symbols are as in Fig. 2. Large C-terminal sequence extensions for RHOBTB and RHOT proteins have been removed (indicated by the “#” symbol). See Table 1 for alternate gene symbols.
RHO subfamily proteins segregate into six branches based on sequence similarity (Fig. 5). Effectors for RHOA, B, and C branch proteins include ROCK1 (Rho-associated protein kinase 1) (and probably ROCK2) (191) and the formin protein DIAPH (also called mDia, mammalian homolog of Diaphinous) (192). RHO proteins bind to and activate PKN proteins, serinethreonine kinases that have been implicated in cell stress responses (193, 194). PLCE is another demonstrated effector of RHO proteins (195). The RHO-binding proteins Rhophilin (194), Rhophilin2 (196), and Rhotekin (197) may also serve as effectors. RAC [Ras-related C3 botulinum toxin substrate (198)] branch proteins appear to function primarily through direct activation of PAK (p21-associated protein kinase) family kinases (199). Other RAC effectors include phospholipase C–β (PLCB) (200). Some effects of CDC42 branch proteins are mediated by WAS (Wiscott-Aldrich syndrome protein; also called WASP) (201–203) together with TOCA1 (transducer of Cdc42-dependent actin assembly), another CDC42 effector (204). CDC42 proteins also participate in the function of a multiprotein complex that includes PAR6, PAR3, and atypical protein kinase C isoforms (205). RND proteins are RHO family members that are constitutively active due to extremely low GTPase activity. They have been reported to antagonize RHOA function, in part through the activation of RHOGAPs (206) or through blocking ROCK activity (207). No effectors have been identified for members of the more distal branches of the RHO family, RHOT1 and RHOT2 (also called Miro for mitochondrial Rho), and RHOBTB1 and RHOBTB2.
Fig. 5.
Dendrogram of RHO subfamily members from H. sapiens, D. melanogaster, and C. elegans. RHOT1-N and RHOT2-N represent the N-terminal GTPase domains of RHOT1 and RHOT2. Human protein names are in uppercase letters.
RHO proteins have been directly implicated in multiple aspects of cytoskeletal remodeling and cell polarity (208, 209), and activated forms of representative RHO proteins demonstrate that certain aspects of remodeling segregate with particular branches of this subfamily (208, 210). The cytoskeletal changes reported include formation of lamellipodia (RAC1 to RAC3 and RHOG), filopodia (CDC42, RHOJ, RHOQ), or stress fibers (RHOA, RHOB, and RHOC) and the disruption of stress fibers (RND1 to RND3).
The majority of RHO subfamily proteins are subject to the same Caax-signaled prenylation and posttranslational modifications as those seen on RAS subfamily members. Some RHO proteins (most notably RAC1, RAC4, RHOA, and RHOC) also include C-terminal polybasic sequences, whereas others are modified by palmitoylation.
RHOT1 and RHOT2 are distinct in several ways from members of the RHO family and might best be considered as a unique subfamily. First, the RHOT proteins show notable sequence divergence from most RHO family members. This includes a lack of C-terminal cysteines, implying that RHOT proteins do not undergo lipid modification, and an absence of the “RHO insert” sequence following the G4 box. Second, RHOT1 and RHOT2 are more than twice as large as other RHO family members. Third, RHOT proteins contain two EF hand (EFh) motifs that may confer calcium binding, a function not associated with other family members. Each RHOT protein also includes a putative GTP-binding motif at the C terminus, but no functional significance has been assigned to these sequences. RHOT proteins localize to mitochondria, and expression of mutationally activated RHOT1 or RHOT2 leads to disruption of the mitochondrial network and to increased rates of apoptosis (209, 210).
An interspecies comparison of RHO subfamily proteins (Fig. 5) shows that they have been highly conserved through evolution and that, as with the RAS subfamily, there has been a notable expansion of the RHO protein subfamily (human = 23, fly = 8, worm = 10).
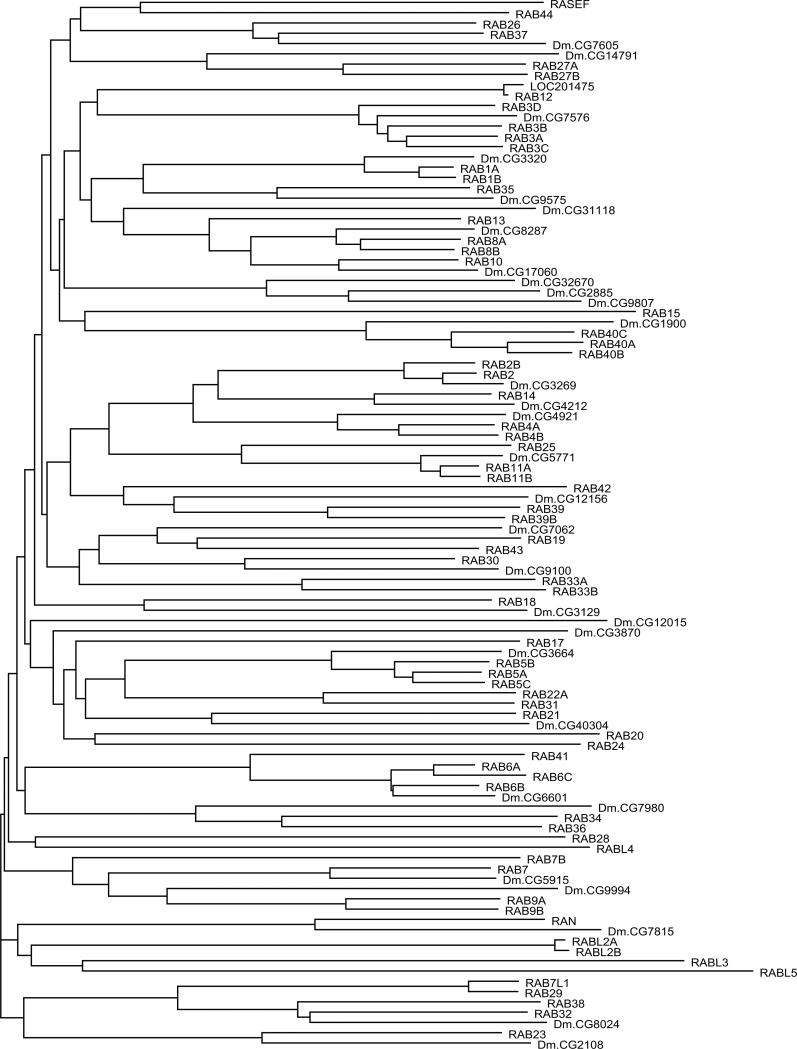
RAB and RAN Protein Subfamily (71 members)
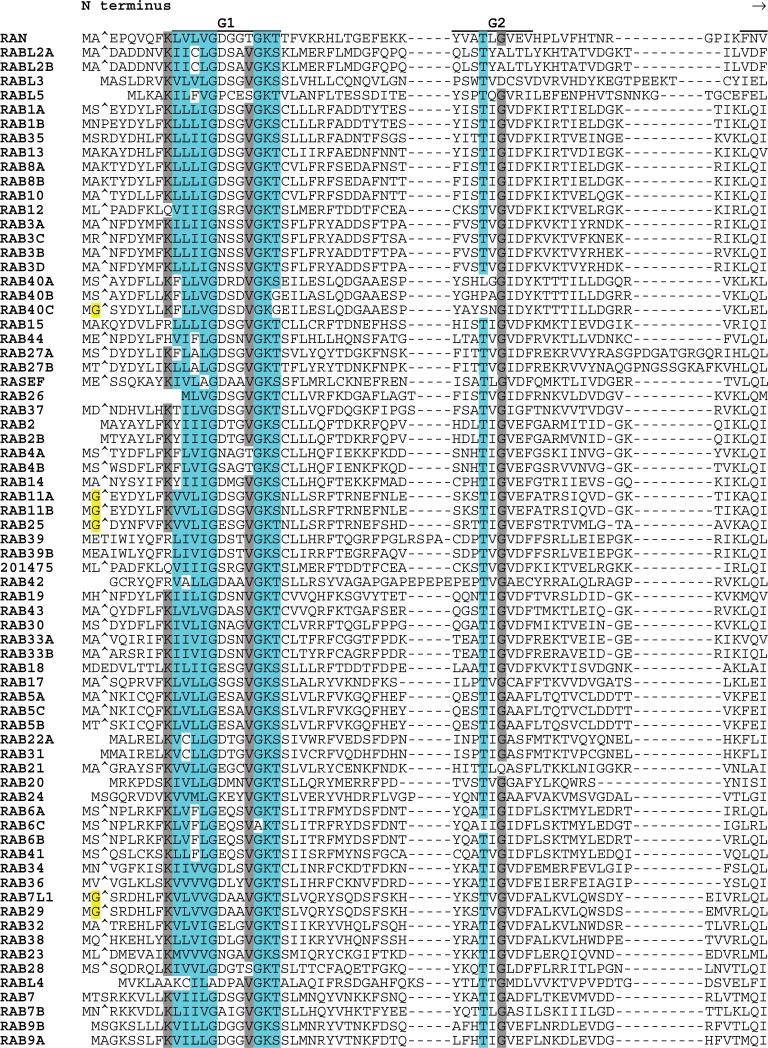
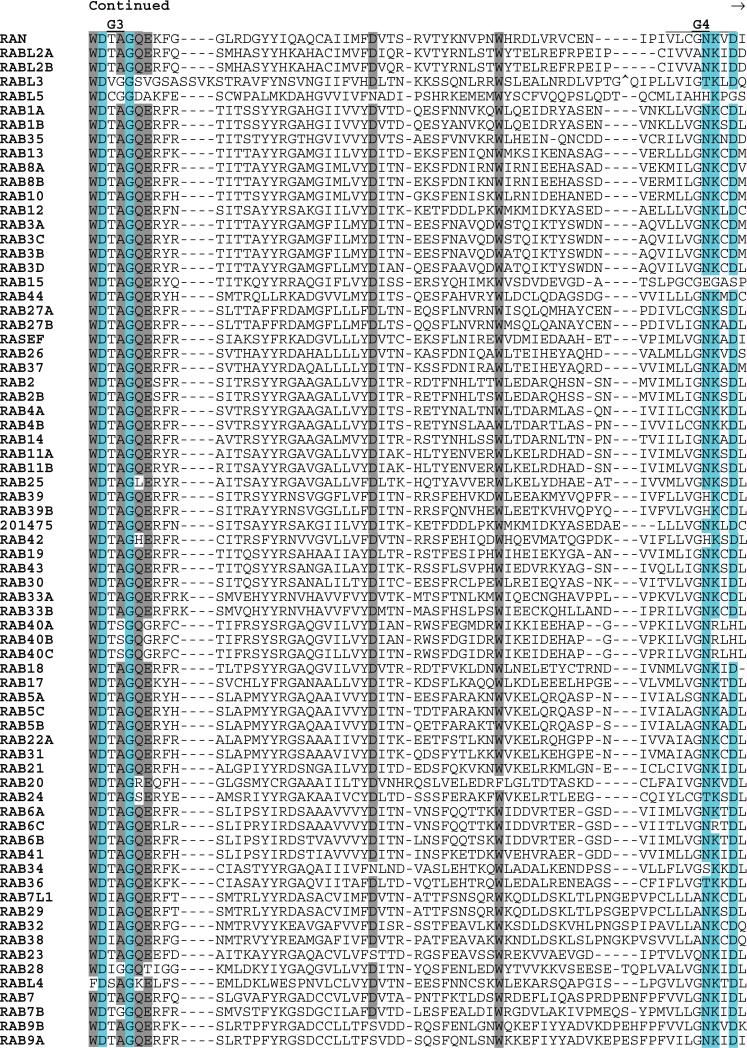
The RAB proteins were first identified as Ras-related genes expressed in rat brain (211). RABs represent the largest subfamily of the RAS superfamily, and the close relatedness of some members (RABL2A to RABL2B; RAB1A to RAB1B; and RAB11A to RAB11B) suggests that recent duplications may have occurred (Fig. 6). Although most RAB proteins include C-terminal prenylation signals, these are distinct from those found in RAS and RHO GTPases. One subfamily member (RAB35) has an adjacent polybasic motif. Several RABs have potential N-terminal myristoylation sites. Sequences of two RABs (RAB6C and RABL4) diverge from the G1 box consensus (otherwise universal in the RAS superfamily) and may not be functional GTPases. RAB44 and RASEF are the largest of the RAB subfamily proteins (predicted molecular masses of 108 and 83 kD, respectively). Each encodes a calcium binding EF hand motif (Interpro IPR002048). In addition, RAB44 encodes a spectrin repeat motif (Interpro IPR002017) and a proline-rich motif (Interpro IPR00064). The RAS domains of RAB44 and RASEF are located at their C termini.
Fig. 6.
Alignment of human RAB subfamily members. Highlighting and symbols are as in Fig. 2. See Table 1 for alternate gene symbols. 201475 (LOC201475, gi41150884) represents an unannotated gene and matching cDNA. The RAB42 sequence is derived from an N-terminal truncated message.
Only one member of the RAB family has been reported to exhibit transforming potential; the RAB8A (also called Mel) gene was first isolated in an NIH3T3 cell transformation assay (212).
RAB proteins function in protein trafficking pathways, regulating vesicle formation, movement, and fusion [reviewed in (213–215)]. Several RAB effectors have been identified. These include rabphilin (also called RPH3A), an effector for the exocytosis function of RAB3 proteins (216). RAB5 proteins regulate endosomal vesicle transport through EEA1 (early endosome antigen 1) (217), early endosome fusion through RBEP1 (rabaptin) (218), and affect nuclear functions through APPL1 (adaptor protein containing PH domain, PTB domain, and leucine zipper motif; also called DIP13α) and APPL2 (also called DIP13β) proteins (219). The participation of RAB7 in lysosomal transport has been attributed in part to the effector protein RILP (Rab7-interacting lysosomal protein) (220, 221). RAB9 proteins interact with the effector M6PRBP1 (also called TIP47) to mediate receptor recognition and cargo selection (222). Rabphilin-11 serves as an effector for RAB11 proteins in their vesicle recycling function (223). RAB27A works through MLPH (melanophilin) to regulate the movement of secretory vesicles along actin filaments (224).
Although the RAN protein has been considered to define a separate family, comparative sequence analysis suggests that RAN resides on a branch of the RAB subfamily. RAN is a regulator of nuclear import and export [reviewed in (225)]. This function is tied directly to the guanine nucleotide status of RAN, with both RAN-GTP and RAN-GDP showing specific interactions with nuclear transport factors. Nuclear RAN is maintained in the GTP-bound state through sequestration of GAP and GEF regulators. RAN-GTP binds importins and exportins but with opposite consequences (promoting import into the nucleus with the former or export from the nucleus with the latter), leading to directed protein transport. RAN binding to importins has also been implicated as a requisite step in mitotic spindle assembly (226, 227). The RAN branch of the RAB subfamily includes four less well-characterized proteins: RABL2A, RABBL2B, RABL3, and RABL5. It remains to be determined if these proteins are functionally similar to RAN.
Most branches of the RAB subfamily appear to be well conserved through evolution (Fig. 7) and show notable expansion (human = 71, fly = 32).
Fig. 7.
Dendrogram of RAB subfamily members from H. sapiens and D. melanogaster. Human protein names are in uppercase letters.
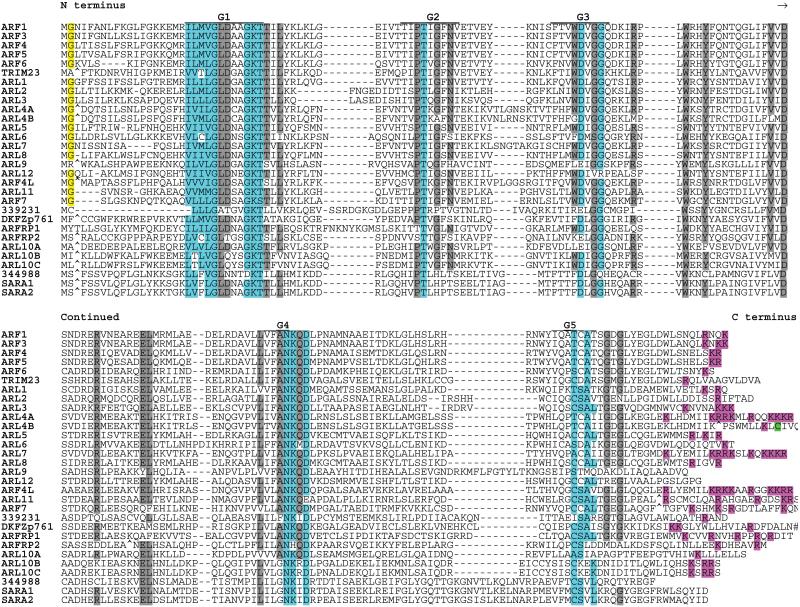
ARF (and SARA) Protein Subfamily (30 members)
The first identified proteins in this subfamily were named for their role as ADP ribosylation factors (228–230). ARFs and related proteins are regulators of trafficking of intracellular proteins and membranes and of cytoskeletal remodeling [reviewed in (231)]. ARF proteins lack C-terminal lipid modification signals (Fig. 8) but in most cases are subject to N-terminal myristoylation.
Fig. 8.
Alignment of human ARF subfamily members. Highlighting and symbols are as in Fig. 2. See Table 1 for alternate gene symbols. The following are unannotated genes with matching cDNAs: 339231 (LOC339231, gi 42661282), 344988 (LOC344988, gi 37539816), and DKFZp761 (DKFZp761H079, gi 33598955).
Effectors mediating ARF functions include the Arfaptins (ARFIPs) (232) and Arfophilin (RAB11FIPs) (233). ARF1 interacts directly with the vesicle coat protein COPI (234) and regulates disassembly (235). At the plasma membrane, ARF6 can regulate endocytic recycling through direct interaction with SEC10L1 (also called Sec10), a subunit of the exocyst complex (236).
ARF proteins also function in part through their ability to regulate phospholipid metabolism by directly activating phosphatidylinositol 4-phosphate 5-kinase (237, 238). ARF1 and ARF3 also bind to GGA (Golgi-localized, gamma-adaptinear–containing, Arf-binding) proteins (239, 240) and function in trans-Golgi network membrane trafficking (241).
The SARA proteins (note that the HGNC name for these proteins overlaps with the common acronym for Smad anchor for receptor activation, a distinct signaling protein) are an off-shoot of the main ARF subfamily. SARA1 (also called Sar1) functions as a component of the COPII complex that mediates export from the ER [reviewed in (242)]. Eight additional ARF-related proteins are located on the same branch as SARA1 and SARA2. There are currently no data, however, on the function of ARL9, ARL10A, ARL10B, ARL10C, ARFRP2, LOC339231, LOC344988, and DKFZp761H0.
Two additional proteins (ENSG00000127917, GI:37538730; ENSG00000185829, GI:7706177) show similarity with ARFs but have major G box motif disruptions that would likely compromise G protein function.
Analysis of ARF and related proteins from human, fly, and worm (Fig. 9) indicates that this is a well-conserved family with a degree of expansion similar to that of other RAS subfamilies (human 30, fly 12, worm 13).
Fig. 9.
Dendrogram of ARF subfamily members from H. sapiens, D. melanogaster, and C. elegans. Human protein names are in uppercase letters.
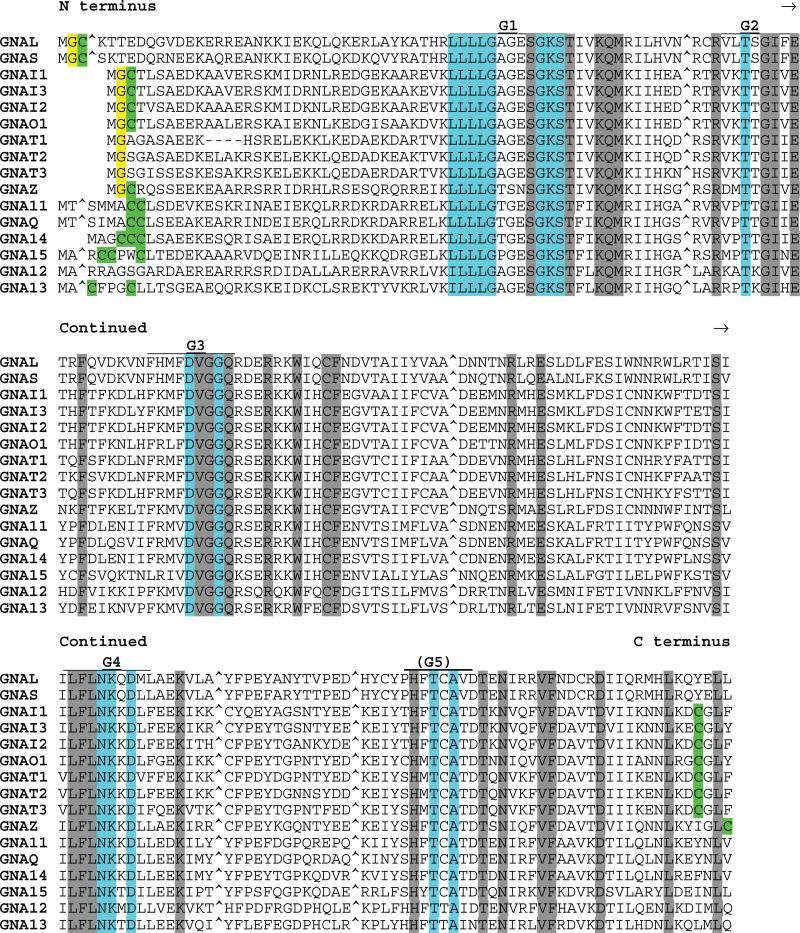
Gα Protein Subfamily (16 members)
The α subunits of G proteins were among the first well-characterized mammalian GTPases. When aligned with other RAS superfamily proteins, the 16 Gα proteins show multiple sequence insertions (Fig. 10) [reviewed in (243)]. The largest of these additional sequence elements, located between the G1 and G2 boxes, is believed to account for the high intrinsic GTPase activity and low intrinsic nucleotide exchange rate of Gα subunits relative to those of RAS [(244), and reviewed in (243)]. Most Gα proteins undergo N-terminal lipid modification by myristate and/or palmitate fatty acids.
Fig. 10.
Alignment of human Gα subfamily members. Highlighting and symbols are as in Fig. 2. Insert sequences (relative to RAS subfamily proteins) have been removed and are indicated with “^”. See Table 1 for alternate gene symbols.
Mutant Gα proteins are associated with several diseases including cancers. Activating mutations in Gαs are found in some pituitary tumors, and Gαi mutations have been reported in tumors of the adrenal cortex (245).
The functions of Gα-type G proteins are inextricably linked to their association with βγ heterodimer subunits and with proteins of the large family of G protein coupled receptors (GPCRs). The inactive (GDP-bound) G protein heterotrimers are typically “parked” on the C-terminal domains of GPCRs. Receptor activation leads to a conformational change that facilitates both GTP loading and reduced affinity for βγ dimers [in some cases, however, the heterotrimer remains intact (246)]. Downstream effectors of Gα include multiple adenylyl cyclase isoforms, several ion channels and transporters, and various other cell regulatory components [reviewed in (247)].
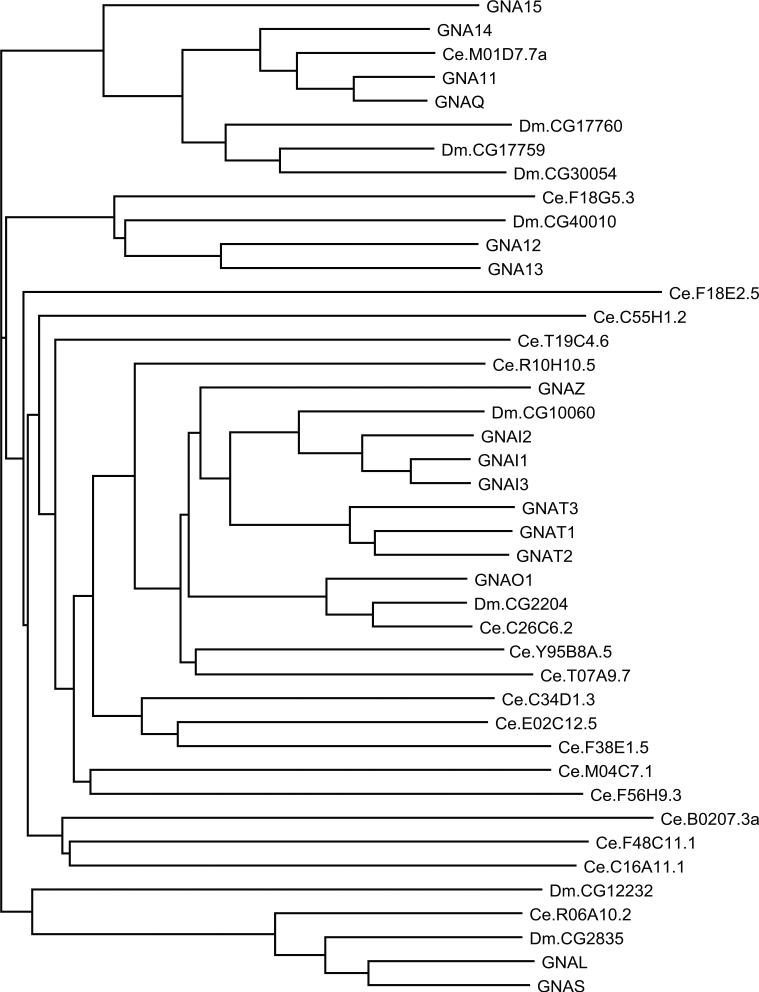
The Gα subfamily of proteins is well conserved in evolution (Fig. 11). The main branches of the Gα tree (the branches containing Gs and Gl, Gi and Gt, G12 and G13, and Gq and G11) are represented in mammals, flies, and worms, but there is an expansion of C. elegans genes in the branch containing Gi, Gt, and Go.
Fig. 11.
Dendrogram of Gα subfamily members from H. sapiens, D. melanogaster, and C. elegans. Human protein names are in uppercase letters.
The RAS Superfamily of Proteins
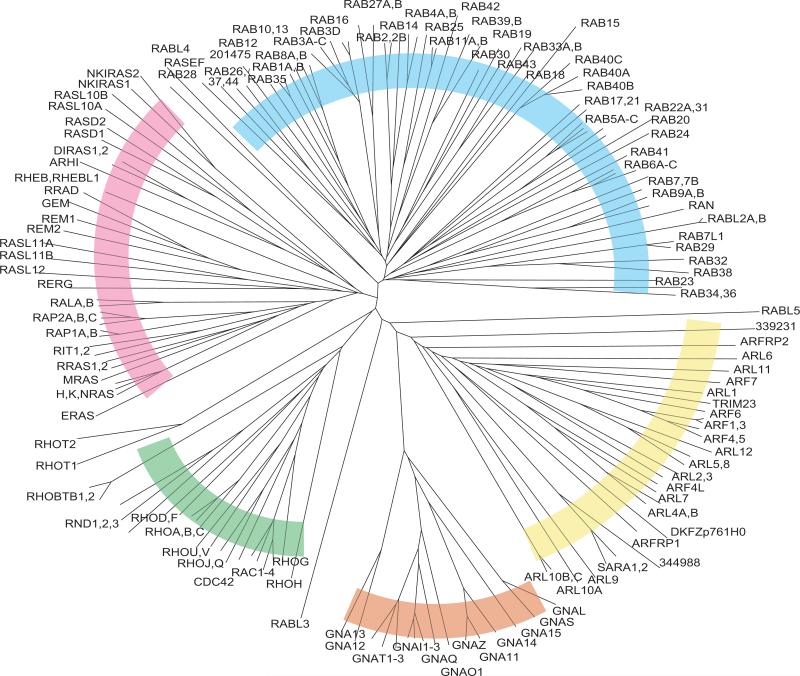
Sequence comparison analysis of all RAS superfamily members (GTPase domains only) highlights the relationship among the subfamilies (Fig. 12). The RAS, RHO, RAB, ARF, and Gα groupings are apparent, and the proximity of the RAS subfamily to the RHO and RAB subfamilies is noteworthy. The close relationship of the ARF and Gα subfamilies is also revealed. The RABL3 and RABL5 proteins segregate outside of the RAB subfamily and are not clearly positioned within any of the other subfamilies. This may reflect an unusual evolutionary origin for these sequences, which also do not have any apparent orthologs in Drosophila (Fig. 7). RABL4 and RAB28 fall slightly outside the main RAB cluster. In addition, the RHOT proteins appear to be only marginally within the RHO family.
Fig. 12.
Unrooted tree of human RAS superfamily members. As with dendrograms in previous figures, branch lengths are directly proportional to the number of differences between sequences compared. Subfamilies of proteins are indicated by colored arcs: RAS (red), RHO (green), Gα (orange), ARF (yellow), and RAB (blue).
Other Human GTPases
GTPases outside the RAS superfamily
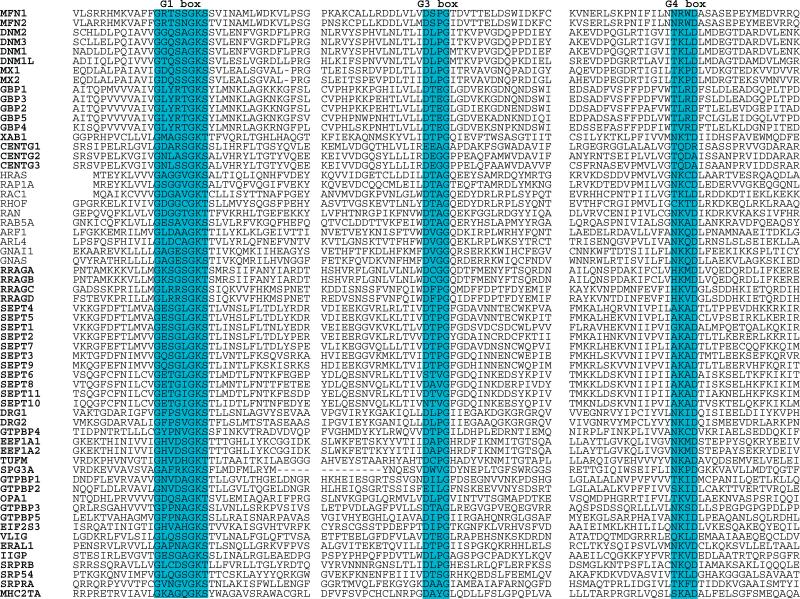
There are at least 50 additional proteins that have demonstrated or predicted GTPase function, but that fall outside the RAS superfamily. These proteins were, in many cases, identified on the basis of genetic or biochemical analyses of function and only subsequently revealed to be GTPases. Each protein includes recognizable G1, G3, and G4 boxes (Fig. 13). They are generally larger than the RAS superfamily proteins, due to the presence of additional functional domains. These “distant” GTPases also do not include the lipid modification signals seen in most RAS subfamily proteins, and they function in multiple subcellular regions.
Fig. 13.
Alignment of human G proteins with representative members of the RAS superfamily. The G1, G3, and G4 box motifs and surrounding sequences are presented. See Table 1 for alternate gene symbols.
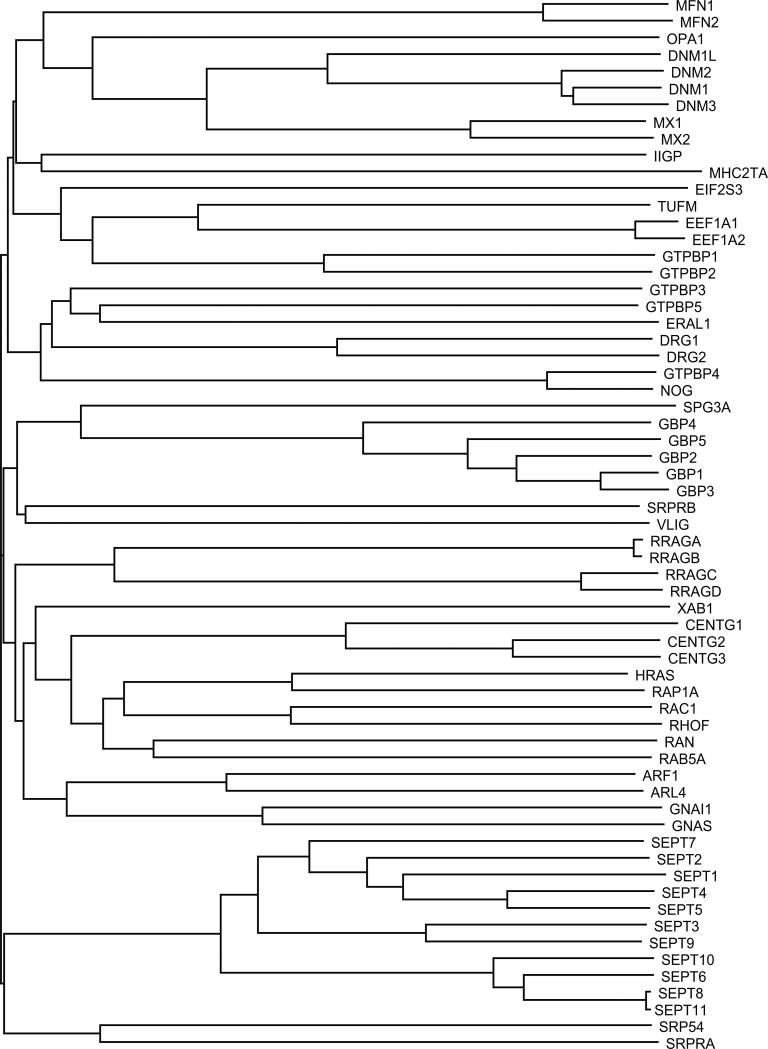
GTPases outside the RAS superfamily are diverse in structure and function. Sequence alignment of their GTPase domains, however, reveals the existence of subfamilies that may also reflect functional characteristics (Fig. 14). One of the largest groups is composed of the septins (SEPT1 to SEPT11), which play a critical role in the constricting ring structure required for cytokinesis. Septins have also been implicated in the formation of focal adhesion complexes and in cell polarity [reviewed in (248)]. Association of septin with the plasma membrane is guanine nucleotide regulated. Specifically, GDP enhances binding to the membrane lipid PIP2 (249).
Fig. 14.
Dendrogram of distant G proteins (uppercase letters) with representative members of the RAS superfamily.
The dynamin family members DNM1 to DNM3 and DNM1L regulate vesicle and organelle dynamics through their participation in a constricting ring structure that requires GTP [reviewed in (250)]. OPA1 (optic atropy 1), MX1 (myxovirus resistance 1), MX2, and the closely related mitofusin proteins (MFN1 and MFN2) are also members of the dynamin family, but their biological functions are not well understood.
Initiation and elongation factors (EIF2S3, EEF1A1, EEF1A2, and TUFM) are perhaps the first class of protein for which GTP binding and hydrolysis were studied in structural and biochemical terms [reviewed in (251, 252)]. During initiation, GTP hydrolysis is coupled to the stepwise assembly of the mRNA-ribosome-tRNA Meti complex. For elongation, conformational changes associated with GTP binding or hydrolysis are used to drive cycles of recruitment and release of aminoacylated elongator tRNAs.
Signal recognition peptide receptor complex components SRPRA (also called SRPR, SRPRα), SRPRB (also called SRPRβ), and SRP54 also utilize GTP hydrolysis to regulate the formation and function of a protein translocation complex (253). However, although the GTPase domains of SRPRΑ and SRP54 are closely related, the sequence of SRPRB is sufficiently divergent to be placed in a distinct branch.
The centaurin gamma proteins CENTG1 (also called GGAP2), CENTG2 (also called GGAP1), and CENTG3 (also called MRIP1) have GTPase domains that are closely related to those of RAS, RHO, and RAB proteins, but do not fit well into any of these RAS subfamilies. CENTGs are unusual because they also include a GAP domain that appears to function intramolecularly to promote GTP hydrolysis (254). Each CENTG protein also encodes a PH (pleckstrin homology) domain (Interpro IPR001849) and an ANK (ankyrin) domain (Interpro IPR002110). The XAB1 GTPase, implicated in the nuclear translocation of a DNA repair factor (255), is structurally similar to members of this branch, raising questions about possible functional similarities.
The RRAG proteins (RRAGA, RRAGB, RRAGC, and RRAGD) are sometimes described as members of the RAS superfamily because they show some similarity with the ARF and Gα proteins. RRAGs have been implicated as factors in the nuclear import and export functions of RAN (256).
Three groups of GTPases were identified because their transcription is induced after treatment of cells with IFNG (also called interferon gamma or IFN-γ) (257). The proteins share an IIGTP (interferon-inducible GTPase) domain (Interpro IPR007743). The first group is represented by IIGP5 in humans but appears to have undergone notable expansion in mice, where they were first discovered and characterized (mouse orthologs are Irg47, Tgtp, Iigp, Lrg47, Igtp, and Gtpi). Some evidence suggests a critical role for IIGTP proteins in normal immune responses, perhaps through participation in vacuolar trafficking (258). Members of the second IFN-inducible group (the guanylate-binding proteins GBP1 through GBP5) are relatively large G proteins. They show unusual GTP binding characteristics (259) that may explain their capacity to generate both GDP and GMP. A closely related gene product is SPG3A (atlastin), mutations in which are associated with hereditary spastic paraplegia (260). The third group of IFN-inducible G proteins are represented by VLIG (very large inducible GTPase 1) and, in mice, by several VLIG paralogs (261).
GTPBP proteins (GTPBP1 to GTPBP5) show some relatedness to the initation/elongation factor branch proteins (262, 263). These genes also show IFN-inducible expression. Although their physiological function remains unclear, GTPBP4 may be identical to NGB (also called Nog1), a nucleolar protein involved in ribosome biogenesis (264). ERAL1 (Era-like 1, also called H-Era, for Escherichia coli Ras-like protein) is named for its structural relationship with Era, an essential bacterial GTPase (265). The ERAL1 protein also includes a KH domain (Interpro IPR004087) involved in RNA binding. ERAL1 is a potential regulator of apoptosis (266).
DRG genes (DRG1 and DRG2) were identified as Developmentally Regulated G proteins (267, 268). Xenopus laevis orthologs of DRG have RNA binding activity (269), but little else is known about their function.
The major histocompatibility complex class II transactivator, MHC2TA (also called CIITA) functions as a master coactivator of MHC class II gene expression. MHC2TA has only weak intrinsic GTPase activity, but GTP binding regulates its nuclear localization (270, 271). The nucleotide-binding domain of MHC2TA has been grouped in the NACHT family (named for founding members NAIP, CIIA, HETE, and TP1) (272). Other mammalian proteins in this large family—which includes BIRC1 (baculovial IAP repeat-containing 1); CIAS1 (cold autoinflamitory syndrome 1); CARD (caspase recruitment domain family) 4, 6, 12, and 15; and NALP (NACHT, leucine rich repeat, and PYD containing) 1, 2, and 4 to 14—that have been tested show greater binding affinity for ATP than for GTP.
TUBB, the β subunit of tubulin, is an established GTP-binding and GTP-hydrolyzing protein, but its structure diverges to such a large extent from those of the other GTPases (273), that it has not been included in sequence comparisons. The identification of other human proteins with GTPase function but relatively low G box sequence conservation will require a deeper understanding of the structure and function relationships for these versatile enzymes. A database of human GTPases can be found at http://www.doe-mbi.ucla.edu/~sievers/gproteins.
GTP-binding proteins
The human genome encodes many proteins with demonstrated or predicted GTP-binding properties, but no apparent GTPase enzymatic function. These include GNL1 (guanine nucleotide binding protein–like 1, also called HSR1), which encodes a likely GTP binding domain that, together with a mouse ortholog (Mmr1) and several bacterial proteins, appears to form a structural subclass of proteins (PFAM domain PF01926) (274). The brain-enriched RING finger protein ZNF179 (also called BFP, brain finger protein) shows close relatedness to the GBPs, but no guanine nucleotide biochemistry has yet been described for BFP.
The dozen or more human RHOGAP protein family members include several with putative GTP-binding domains. RHOGAP5 (also called p190-B or ARHGAP5) and GRLF1 (also called p190A) have the most conserved such domains (275), but even in these cases they have not been demonstrated to have GTPase activity or to play any role in RHO regulatory function. Two other RHOGAP proteins, ARHGAP21 and CHN1 (also called RHOGAP2), have putative GTP-binding domains that are more divergent.
A GTP-binding domain also appears within DAPK1 (death-associated protein kinase), a cell death–associated protein kinase with ankyrin repeats and a death domain (276, 277). Two other DAPK proteins do not include recognizable GTPase domains. Other enzymes with reported GTP-binding domains include transglutaminase, phosphoenolpyruvate carboxy kinase, and glutamate dehydrogenase.
AGTPBP1 (also called NNA1) has an unusual binding site that accommodates either GTP or ATP (278). Other reported GTP-binding proteins of interest include TSN (also called translin or TB-RBP, testis brain RNA-binding protein) (279), ANXA6 (human annexin A6) (280), and MEN1 (multiple endocrine neoplasia 1) (281).
Each of the RHOT proteins has a potential GTP binding site downstream of their RHO-type GTPase domains. The uncharacterized gene product MGC10731 (ENSG00000132881; GI:34147392) also includes a putative GTP binding site.
Conclusions
A survey of all human RAS superfamily GTPases, and analysis of structural and evolutionary relatedness among family members, represents only the end of the beginning of our efforts to understand these pervasive signal transduction regulators. It is likely, for instance, that we have only scratched the surface of variation resulting from alternate splicing and posttranslational modifications of GTPases. Much attention has also turned to determining the specificity of GTPase regulators such as the large family of GAPs and GEFs, as well as continuing efforts to identify downstream effectors. Continued research in these areas should provide a more accurate picture of human GTPase functions in normal and pathological conditions.
Footnotes
Citation: J. Colicelli, Human RAS superfamily proteins and related GTPases. Sci. STKE 2004, re13 (2004).
References and Notes
- 1.Human Genome Nomenclature Committee All prediced proteins discussed in this review are supported by both genomic and cDNA sequences. (HGNC, http://www.gene.ucl.ac.uk/nomenclature) symbols are used throughout (Table 1 provides a list of some common gene names corresponding to HGNC symbols)
- 2.ClustalW was used for all sequence alignments and for the generation of dendrograms. Only GTPase domains were used in the analysis (i.e., extraneous sequences were removed).
- 3.Dever TE, Glynias MJ, Merrick WC. GTP-binding domain: Three consensus sequence elements with distinct spacing. Proc. Natl. Acad. Sci. U.S.A. 1987;84:1814–1818. doi: 10.1073/pnas.84.7.1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bourne HR, Sanders DA, McCormick F. The GTPase superfamily: Conserved structure and molecular mechanism. Nature. 1991;349:117–127. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- 5.Walker JE, Saraste M, Runswick MJ, Gay NJ. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.White MA, Nicolette C, Minden A, Polverino A, Van Aelst L, Karin M, Wigler MH. Multiple Ras functions can contribute to mammalian cell transformation. Cell. 1995;80:533–541. doi: 10.1016/0092-8674(95)90507-3. [DOI] [PubMed] [Google Scholar]
- 7.Joneson T, White MA, Wigler MH, Bar-Sagi D. Stimulation of membrane ruffling and MAP kinase activation by distinct effectors of RAS. Science. 1996;271:810–812. doi: 10.1126/science.271.5250.810. [DOI] [PubMed] [Google Scholar]
- 8.Rodriguez-Viciana P, Warne PH, Khwaja A, Marte BM, Pappin D, Das P, Waterfield MD, Ridley A, Downward J. Role of phosphoinositide 3-OH kinase in cell transformation and control of the actin cytoskeleton by Ras. Cell. 1997;89:457–467. doi: 10.1016/s0092-8674(00)80226-3. [DOI] [PubMed] [Google Scholar]
- 9.Vetter IR, Wittinghofer A. The guanine nucleotide-binding switch in three dimensions. Science. 2001;294:1299–1304. doi: 10.1126/science.1062023. [DOI] [PubMed] [Google Scholar]
- 10.Herrmann C, Horn G, Spaargaren M, Wittinghofer A. Differential interaction of the ras family GTP-binding proteins H-Ras, Rap1A, and R-Ras with the putative effector molecules Raf kinase and Ral-guanine nucleotide exchange factor. J. Biol. Chem. 1996;271:6794–6800. doi: 10.1074/jbc.271.12.6794. [DOI] [PubMed] [Google Scholar]
- 11.Milburn MV, Tong L, deVos AM, Brunger A, Yamaizumi Z, Nishimura S, Kim SH. Molecular switch for signal transduction: Structural differences between active and inactive forms of protooncogenic ras proteins. Science. 1990;247:939–945. doi: 10.1126/science.2406906. [DOI] [PubMed] [Google Scholar]
- 12.Neal SE, Eccleston JF, Hall A, Webb MR. Kinetic analysis of the hydrolysis of GTP by p21N-ras. The basal GTPase mechanism. J. Biol. Chem. 1988;263:19718–19722. [PubMed] [Google Scholar]
- 13.Van Dyke K, Robinson R, Urquilla P, Smith D, Taylor M, Trush M, Wilson M. An analysis of nucleotides and catecholamines in bovine medullary granules by anion exchange high pressure liquid chromatography and fluorescence. Evidence that most of the catecholamines in chromaffin granules are stored without associated ATP. Pharmacology. 1977;15:377–391. doi: 10.1159/000136714. [DOI] [PubMed] [Google Scholar]
- 14.Chardin P, Camonis JH, Gale NW, van Aelst L, Schlessinger J, Wigler MH, Bar-Sagi D. Human Sos1: A guanine nucleotide exchange factor for Ras that binds to GRB2. Science. 1993;260:1338–1343. doi: 10.1126/science.8493579. [DOI] [PubMed] [Google Scholar]
- 15.Quilliam LA, Rebhun JF, Castro AF. A growing family of guanine nucleotide exchange factors is responsible for activation of Ras-family GTPases. Prog. Nucleic Acid Res. Mol. Biol. 2002;71:391–444. doi: 10.1016/s0079-6603(02)71047-7. [DOI] [PubMed] [Google Scholar]
- 16.Feig LA, Cooper GM. Inhibition of NIH 3T3 cell proliferation by a mutant ras protein with preferential affinity for GDP. Mol. Cell. Biol. 1988;8:3235–3243. doi: 10.1128/mcb.8.8.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen SY, Huff SY, Lai CC, Der CJ, Powers S. Ras-15A protein shares highly similar dominant-negative biological properties with Ras-17N and forms a stable, guanine-nucleotide resistant complex with CDC25 exchange factor. Oncogene. 1994;9:2691–2698. [PubMed] [Google Scholar]
- 18.Rak A, Pylypenko O, Durek T, Watzke A, Kushnir S, Brunsveld L, Waldmann H, Goody RS, Alexandrov K. Structure of Rab GDP-dissociation inhibitor in complex with prenylated YPT1 GTPase. Science. 2003;302:646–650. doi: 10.1126/science.1087761. [DOI] [PubMed] [Google Scholar]
- 19.Olofsson B. Rho guanine dissociation inhibitors: Pivotal molecules in cellular signalling. Cell. Signal. 1999;11:545–554. doi: 10.1016/s0898-6568(98)00063-1. [DOI] [PubMed] [Google Scholar]
- 20.Wu SK, Zeng K, Wilson IA, Balch WE. Structural insights into the function of the Rab GDI superfamily. Trends Biochem. Sci. 1996;21:472–476. doi: 10.1016/s0968-0004(96)10062-1. [DOI] [PubMed] [Google Scholar]
- 21.McCormick F. Going for the GAP. Curr. Biol. 1998;8:R673–R674. doi: 10.1016/s0960-9822(98)70431-2. [DOI] [PubMed] [Google Scholar]
- 22.Boguski MS, McCormick F. Proteins regulating Ras and its relatives. Nature. 1993;366:643–654. doi: 10.1038/366643a0. [DOI] [PubMed] [Google Scholar]
- 23.Tamanoi F, Sigman DS, editors. Protein Lipidation. Vol. 21. Academic Press; San Diego, CA: 2001. [Google Scholar]
- 24.Okada T, Masuda T, Shinkai M, Kariya K, Kataoka T. Post-translational modification of H-Ras is required for activation of, but not for association with, B-Raf. J. Biol. Chem. 1996;271:4671–4678. doi: 10.1074/jbc.271.9.4671. [DOI] [PubMed] [Google Scholar]
- 25.Porfiri E, Evans T, Chardin P, Hancock JF. Prenylation of Ras proteins is required for efficient hSOS1-promoted guanine nucleotide exchange. J. Biol. Chem. 1994;269:22672–22677. [PubMed] [Google Scholar]
- 26.McGeady P, Kuroda S, Shimizu K, Takai Y, Gelb MH. The farnesyl group of H-Ras facilitates the activation of a soluble upstream activator of mitogen-activated protein kinase. J. Biol. Chem. 1995;270:26347–26351. doi: 10.1074/jbc.270.44.26347. [DOI] [PubMed] [Google Scholar]
- 27.Rubio I, Wittig U, Meyer C, Heinze R, Kadereit D, Waldmann H, Downward J, Wetzker R. Farnesylation of Ras is important for the interaction with phosphoinositide 3-kinase gamma. Eur. J. Biochem. 1999;266:70–82. doi: 10.1046/j.1432-1327.1999.00815.x. [DOI] [PubMed] [Google Scholar]
- 28.Williams JG, Drugan JK, Yi GS, Clark GJ, Der CJ, Campbell SL. Elucidation of binding determinants and functional consequences of Ras/Raf-cysteine-rich domain interactions. J. Biol. Chem. 2000;275:22172–22179. doi: 10.1074/jbc.M000397200. [DOI] [PubMed] [Google Scholar]
- 29.Gotoh T, Tian X, Feig LA. Prenylation of target GTPases contributes to signaling specificity of Ras-guanine nucleotide exchange factors. J. Biol. Chem. 2001;276:38029–38035. doi: 10.1074/jbc.M104658200. [DOI] [PubMed] [Google Scholar]
- 30.Degtyarev MY, Spiegel AM, Jones TL. The G protein alpha s subunit incorporates [3H]palmitic acid and mutation of cysteine-3 prevents this modification. Biochemistry. 1993;32:8057–8061. doi: 10.1021/bi00083a001. [DOI] [PubMed] [Google Scholar]
- 31.Linder ME, Middleton P, Hepler JR, Taussig R, Gilman AG, Mumby SM. Lipid modifications of G proteins: Alpha subunits are palmitoylated. Proc. Natl. Acad. Sci. U.S.A. 1993;90:3675–3679. doi: 10.1073/pnas.90.8.3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Veit M, Nurnberg B, Spicher K, Harteneck C, Ponimaskin E, Schultz G, Schmidt MF. The alpha-subunits of G-proteins G12 and G13 are palmitoylated, but not amidically myristoylated. FEBS Lett. 1994;339:160–164. doi: 10.1016/0014-5793(94)80406-0. [DOI] [PubMed] [Google Scholar]
- 33.Kirsten WH, Mayer LA. Malignant lymphomas of extrathymic origin induced in rats by murine erythroblastosis virus. J. Natl. Cancer Inst. 1969;43:735–746. [PubMed] [Google Scholar]
- 34.Harvey JJ. An unidentified virus which causes the rapid production of tumours in mice. Nature. 1964;204:1104–1105. doi: 10.1038/2041104b0. [DOI] [PubMed] [Google Scholar]
- 35.Perucho M, Goldfarb M, Shimizu K, Lama C, Fogh J, Wigler M. Human-tumor-derived cell lines contain common and different transforming genes. Cell. 1981;27:467–476. doi: 10.1016/0092-8674(81)90388-3. [DOI] [PubMed] [Google Scholar]
- 36.Krontiris TG, Cooper GM. Transforming activity of human tumor DNAs. Proc. Natl. Acad. Sci. U.S.A. 1981;78:1181–1184. doi: 10.1073/pnas.78.2.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shih C, Padhy LC, Murray M, Weinberg RA. Transforming genes of carcinomas and neuroblastomas introduced into mouse fibroblasts. Nature. 1981;290:261–264. doi: 10.1038/290261a0. [DOI] [PubMed] [Google Scholar]
- 38.Zhang K, DeClue JE, Vass WC, Papageorge AG, McCormick F, Lowy DR. Suppression of c-ras transformation by GTPase-activating protein. Nature. 1990;346:754–756. doi: 10.1038/346754a0. [DOI] [PubMed] [Google Scholar]
- 39.Feig LA, Cooper GM. Relationship among guanine nucleotide exchange, GTP hydrolysis, and transforming potential of mutated ras proteins. Mol. Cell. Biol. 1988;8:2472–2478. doi: 10.1128/mcb.8.6.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Patel G, MacDonald MJ, Khosravi-Far R, Hisaka MM, Der CJ. Alternate mechanisms of ras activation are complementary and favor and formation of ras-GTP. Oncogene. 1992;7:283–288. [PubMed] [Google Scholar]
- 41.Mitsuuchi Y, Testa JR. Cytogenetics and molecular genetics of lung cancer. Am. J. Med. Genet. 2002;115:183–188. doi: 10.1002/ajmg.10692. [DOI] [PubMed] [Google Scholar]
- 42.Grady WM, Markowitz SD. Genetic and epigenetic alterations in colon cancer. Annu. Rev. Genomics Hum. Genet. 2002;3:101–128. doi: 10.1146/annurev.genom.3.022502.103043. [DOI] [PubMed] [Google Scholar]
- 43.Jaffee EM, Hruban RH, Canto M, Kern SE. Focus on pancreas cancer. Cancer Cell. 2002;2:25–28. doi: 10.1016/s1535-6108(02)00093-4. [DOI] [PubMed] [Google Scholar]
- 44.Garcia-Rostan G, Zhao H, Camp RL, Pollan M, Herrero A, Pardo J, Wu R, Carcangiu ML, Costa J, Tallini G. ras mutations are associated with aggressive tumor phenotypes and poor prognosis in thyroid cancer. J. Clin. Oncol. 2003;21:3226–3235. doi: 10.1200/JCO.2003.10.130. [DOI] [PubMed] [Google Scholar]
- 45.Vageli D, Kiaris H, Delakas D, Anezinis P, Cranidis A, Spandidos DA. Transcriptional activation of H-ras, K-ras and N-ras proto-oncogenes in human bladder tumors. Cancer Lett. 1996;107:241–247. doi: 10.1016/0304-3835(96)04372-8. [DOI] [PubMed] [Google Scholar]
- 46.von Lintig FC, Dreilinger AD, Varki NM, Wallace AM, Casteel DE, Boss GR. Ras activation in human breast cancer. Breast Cancer Res. Treat. 2000;62:51–62. doi: 10.1023/a:1006491619920. [DOI] [PubMed] [Google Scholar]
- 47.Xu GF, O'Connell P, Viskochil D, Cawthon R, Robertson M, Culver M, Dunn D, Stevens J, Gesteland R, White R, et al. The neurofibromatosis type 1 gene encodes a protein related to GAP. Cell. 1990;62:599–608. doi: 10.1016/0092-8674(90)90024-9. [DOI] [PubMed] [Google Scholar]
- 48.Xu GF, Lin B, Tanaka K, Dunn D, Wood D, Gesteland R, White R, Weiss R, Tamanoi F. The catalytic domain of the neurofibromatosis type 1 gene product stimulates ras GTPase and complements ira mutants of S. cerevisiae. Cell. 1990;63:835–841. doi: 10.1016/0092-8674(90)90149-9. [DOI] [PubMed] [Google Scholar]
- 49.Martin GA, Viskochil D, Bollag G, McCabe PC, Crosier WJ, Haubruck H, Conroy L, Clark R, O'Connell P, Cawthon RM, et al. The GAP-related domain of the neurofibromatosis type 1 gene product interacts with ras p21. Cell. 1990;63:843–849. doi: 10.1016/0092-8674(90)90150-d. [DOI] [PubMed] [Google Scholar]
- 50.Ballester R, Marchuk D, Boguski M, Saulino A, Letcher R, Wigler M, Collins F. The NF1 locus encodes a protein functionally related to mammalian GAP and yeast IRA proteins. Cell. 1990;63:851–859. doi: 10.1016/0092-8674(90)90151-4. [DOI] [PubMed] [Google Scholar]
- 51.Shinohara N, Ogiso Y, Tanaka M, Sazawa A, Harabayashi T, Koyanagi T. The significance of Ras guanine nucleotide exchange factor, son of sevenless protein, in renal cell carcinoma cell lines. J. Urol. 1997;158:908–911. doi: 10.1097/00005392-199709000-00070. [DOI] [PubMed] [Google Scholar]
- 52.Silvius JR. Mechanisms of Ras protein targeting in mammalian cells. J. Membr. Biol. 2002;190:83–92. doi: 10.1007/s00232-002-1026-4. [DOI] [PubMed] [Google Scholar]
- 53.Choy E, Chiu VK, Silletti J, Feoktistov M, Morimoto T, Michaelson D, Ivanov IE, Philips MR. Endomembrane trafficking of ras: The CAAX motif targets proteins to the ER and Golgi. Cell. 1999;98:69–80. doi: 10.1016/S0092-8674(00)80607-8. [DOI] [PubMed] [Google Scholar]
- 54.Apolloni A, Prior IA, Lindsay M, Parton RG, Hancock JF. H-ras but not K-ras traffics to the plasma membrane through the exocytic pathway. Mol. Cell. Biol. 2000;20:2475–2487. doi: 10.1128/mcb.20.7.2475-2487.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roy S, Wyse B, Hancock JF. H-Ras signaling and K-Ras signaling are differentially dependent on endocytosis. Mol. Cell. Biol. 2002;22:5128–5140. doi: 10.1128/MCB.22.14.5128-5140.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jiang X, Sorkin A. Coordinated traffic of Grb2 and Ras during epidermal growth factor receptor endocytosis visualized in living cells. Mol. Biol. Cell. 2002;13:1522–1535. doi: 10.1091/mbc.01-11-0552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chiu VK, Bivona T, Hach A, Sajous JB, Silletti J, Wiener H, Johnson II RL, Cox AD, Philips MR. Ras signalling on the endoplasmic reticulum and the Golgi. Nat. Cell Biol. 2002;4:343–350. doi: 10.1038/ncb783. [DOI] [PubMed] [Google Scholar]
- 58.Prior IA, Harding A, Yan J, Sluimer J, Parton RG, Hancock JF. GTP-dependent segregation of H-ras from lipid rafts is required for biological activity. Nat. Cell Biol. 2001;3:368–375. doi: 10.1038/35070050. [DOI] [PubMed] [Google Scholar]
- 59.Hancock JF. Ras proteins: Different signals from different locations. Nat. Rev. Mol. Cell Biol. 2003;4:373–384. doi: 10.1038/nrm1105. [DOI] [PubMed] [Google Scholar]
- 60.Finlin BS, Gau CL, Murphy GA, Shao H, Kimel T, Seitz RS, Chiu YF, Botstein D, Brown PO, Der CJ, Tamanoi F, Andres DA, Perou CM. RERG is a novel ras-related, estrogen-regulated and growth-inhibitory gene in breast cancer. J. Biol. Chem. 2001;276:42259–42267. doi: 10.1074/jbc.M105888200. [DOI] [PubMed] [Google Scholar]
- 61.Ballester R, Furth ME, Rosen OM. Phorbol ester- and protein kinase C-mediated phosphorylation of the cellular Kirsten ras gene product. J. Biol. Chem. 1987;262:2688–2695. [PubMed] [Google Scholar]
- 62.Mallis RJ, Buss JE, Thomas JA. Oxidative modification of H-ras: S-thiolation and S-nitrosylation of reactive cysteines. Biochem. J. 2001;355:145–153. doi: 10.1042/0264-6021:3550145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Baker TL, Booden MA, Buss JE. S-Nitrosocysteine increases palmitate turnover on Ha-Ras in NIH 3T3 cells. J. Biol. Chem. 2000;275:22037–22047. doi: 10.1074/jbc.M001813200. [DOI] [PubMed] [Google Scholar]
- 64.Williams JG, Pappu K, Campbell SL. Structural and biochemical studies of p21Ras S-nitrosylation and nitric oxide-mediated guanine nucleotide exchange. Proc. Natl. Acad. Sci. U.S.A. 2003;100:6376–6381. doi: 10.1073/pnas.1037299100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zou JX, Wang B, Kalo MS, Zisch AH, Pasquale EB, Ruoslahti E. An Eph receptor regulates integrin activity through R-Ras. Proc. Natl. Acad. Sci. U.S.A. 1999;96:13813–13818. doi: 10.1073/pnas.96.24.13813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zou JX, Liu Y, Pasquale EB, Ruoslahti E. Activated SRC oncogene phosphorylates R-ras and suppresses integrin activity. J. Biol. Chem. 2002;277:1824–1827. doi: 10.1074/jbc.M103133200. [DOI] [PubMed] [Google Scholar]
- 67.Shimizu K, Birnbaum D, Ruley MA, Fasano O, Suard Y, Edlund L, Taparowsky E, Goldfarb M, Wigler M. Structure of the Ki-ras gene of the human lung carcinoma cell line Calu-1. Nature. 1983;304:497–500. doi: 10.1038/304497a0. [DOI] [PubMed] [Google Scholar]
- 68.McGrath JP, Capon DJ, Smith DH, Chen EY, Seeburg PH, Goeddel DV, Levinson AD. Structure and organization of the human Kiras proto-oncogene and a related processed pseudogene. Nature. 1983;304:501–506. doi: 10.1038/304501a0. [DOI] [PubMed] [Google Scholar]
- 69.Guil S, de La Iglesia N, Fernandez-Larrea J, Cifuentes D, Ferrer JC, Guinovart JJ, Bach-Elias M. Alternative splicing of the human protooncogene c-H-ras renders a new Ras family protein that trafficks to cytoplasm and nucleus. Cancer Res. 2003;63:5178–5187. [PubMed] [Google Scholar]
- 70.Bar-Sagi D, Feramisco JR. Microinjection of the ras oncogene protein into PC12 cells induces morphological differentiation. Cell. 1985;42:841–848. doi: 10.1016/0092-8674(85)90280-6. [DOI] [PubMed] [Google Scholar]
- 71.Simon MA, Bowtell DD, Dodson GS, Laverty TR, Rubin GM. Ras1 and a putative guanine nucleotide exchange factor perform crucial steps in signaling by the sevenless protein tyrosine kinase. Cell. 1991;67:701–716. doi: 10.1016/0092-8674(91)90065-7. [DOI] [PubMed] [Google Scholar]
- 72.Han M, Sternberg PW. let-60, a gene that specifies cell fates during C. elegans vulval induction, encodes a ras protein. Cell. 1990;63:921–931. doi: 10.1016/0092-8674(90)90495-z. [DOI] [PubMed] [Google Scholar]
- 73.Brambilla R, Gnesutta N, Minichiello L, White G, Roylance AJ, Herron CE, Ramsey M, Wolfer DP, Cestari V, Rossi-Arnaud C, Grant SG, Chapman PF, Lipp HP, Sturani E, Klein R. A role for the Ras signalling pathway in synaptic transmission and long-term memory. Nature. 1997;390:281–286. doi: 10.1038/36849. [DOI] [PubMed] [Google Scholar]
- 74.Chen HJ, Rojas-Soto M, Oguni A, Kennedy MB. A synaptic Ras-GTPase activating protein (p135 SynGAP) inhibited by CaM kinase II. Neuron. 1998;20:895–904. doi: 10.1016/s0896-6273(00)80471-7. [DOI] [PubMed] [Google Scholar]
- 75.Kim JH, Liao D, Lau LF, Huganir RL. SynGAP: A synaptic RasGAP that associates with the PSD-95/SAP90 protein family. Neuron. 1998;20:683–691. doi: 10.1016/s0896-6273(00)81008-9. [DOI] [PubMed] [Google Scholar]
- 76.Komiyama NH, Watabe AM, Carlisle HJ, Porter K, Charlesworth P, Monti J, Strathdee DJ, O'Carroll CM, Martin SJ, Morris RG, O'Dell TJ, Grant SG. SynGAP regulates ERK/MAPK signaling, synaptic plasticity, and learning in the complex with postsynaptic density 95 and NMDA receptor. J. Neurosci. 2002;22:9721–9732. doi: 10.1523/JNEUROSCI.22-22-09721.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kim JH, Lee HK, Takamiya K, Huganir RL. The role of synaptic GTPase-activating protein in neuronal development and synaptic plasticity. J. Neurosci. 2003;23:1119–1124. doi: 10.1523/JNEUROSCI.23-04-01119.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dhaka A, Costa RM, Hu H, Irvin DK, Patel A, Kornblum HI, Silva AJ, O'Dell TJ, Colicelli J. The RAS effector RIN1 modulates the formation of aversive memories. J. Neurosci. 2003;23:748–757. doi: 10.1523/JNEUROSCI.23-03-00748.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Van Aelst L, Barr M, Marcus S, Polverino A, Wigler M. Complex formation between RAS and RAF and other protein kinases. Proc. Natl. Acad. Sci. U.S.A. 1993;90:6213–6217. doi: 10.1073/pnas.90.13.6213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vojtek AB, Hollenberg SM, Cooper JA. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell. 1993;74:205–214. doi: 10.1016/0092-8674(93)90307-c. [DOI] [PubMed] [Google Scholar]
- 81.Warne PH, Viciana PR, Downward J. Direct interaction of Ras and the amino-terminal region of Raf-1 in vitro. Nature. 1993;364:352–355. doi: 10.1038/364352a0. [DOI] [PubMed] [Google Scholar]
- 82.Zhang XF, Settleman J, Kyriakis JM, Takeuchi-Suzuki E, Elledge SJ, Marshall MS, Bruder JT, Rapp UR, Avruch J. Normal and oncogenic p21ras proteins bind to the amino-terminal regulatory domain of c-Raf-1. Nature. 1993;364:308–313. doi: 10.1038/364308a0. [DOI] [PubMed] [Google Scholar]
- 83.Koide H, Satoh T, Nakafuku M, Kaziro Y. GTP-dependent association of Raf-1 with Ha-Ras: Identification of Raf as a target downstream of Ras in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 1993;90:8683–8686. doi: 10.1073/pnas.90.18.8683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kolch W. Meaningful relationships: The regulation of the Ras/Raf/MEK/ERK pathway by protein interactions. Biochem. J. 2000;351:289–305. [PMC free article] [PubMed] [Google Scholar]
- 85.Morrison DK, Cutler RE. The complexity of Raf-1 regulation. Curr. Opin. Cell Biol. 1997;9:174–179. doi: 10.1016/s0955-0674(97)80060-9. [DOI] [PubMed] [Google Scholar]
- 86.Oldham SM, Clark GJ, Gangarosa LM, Coffey RJ, Jr., Der CJ. Activation of the Raf-1/MAP kinase cascade is not sufficient for Ras transformation of RIE-1 epithelial cells. Proc. Natl. Acad. Sci. U.S.A. 1996;93:6924–6928. doi: 10.1073/pnas.93.14.6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hamaguchi M, Meth JL, von Klitzing C, Wei W, Esposito D, Rodgers L, Walsh T, Welcsh P, King MC, Wigler MH. DBC2, a candidate for a tumor suppressor gene involved in breast cancer. Proc. Natl. Acad. Sci. U.S.A. 2002;99:13647–13652. doi: 10.1073/pnas.212516099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, Davis N, Dicks E, Ewing R, Floyd Y, Gray K, Hall S, Hawes R, Hughes J, Kosmidou V, Menzies A, Mould C, Parker A, Stevens C, Watt S, Hooper S, Wilson R, Jayatilake H, Gusterson BA, Cooper C, Shipley J, Hargrave D, Pritchard-Jones K, Maitland N, Chenevix-Trench G, Riggins GJ, Bigner DD, Palmieri G, Cossu A, Flanagan A, Nicholson A, Ho JW, Leung SY, Yuen ST, Weber BL, Seigler HF, Darrow TL, Paterson H, Marais R, Marshall CJ, Wooster R, Stratton MR, Futreal PA. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 89.Lambert JM, Lambert QT, Reuther GW, Malliri A, Siderovski DP, Sondek J, Collard JG, Der CJ. Tiam1 mediates Ras activation of Rac by a PI(3)K-independent mechanism. Nat. Cell Biol. 2002;4:621–625. doi: 10.1038/ncb833. [DOI] [PubMed] [Google Scholar]
- 90.Malliri A, van der Kammen RA, Clark K, van der Valk M, Michiels F, Collard JG. Mice deficient in the Rac activator Tiam1 are resistant to Ras-induced skin tumours. Nature. 2002;417:867–871. doi: 10.1038/nature00848. [DOI] [PubMed] [Google Scholar]
- 91.Rodriguez-Viciana P, Warne PH, Dhand R, Vanhaesebroeck B, Gout I, Fry MJ, Waterfield MD, Downward J. Phosphatidylinositol-3-OH kinase as a direct target of Ras. Nature. 1994;370:527–532. doi: 10.1038/370527a0. [DOI] [PubMed] [Google Scholar]
- 92.Rodriguez-Viciana P, Warne PH, Vanhaesebroeck B, Waterfield MD, Downward J. Activation of phosphoinositide 3-kinase by interaction with Ras and by point mutation. EMBO J. 1996;15:2442–2451. [PMC free article] [PubMed] [Google Scholar]
- 93.Spaargaren M, Bischoff JR. Identification of the guanine nucleotide dissociation stimulator for Ral as a putative effector molecule of R-ras, H-ras, K-ras, and Rap. Proc. Natl. Acad. Sci. U.S.A. 1994;91:12609–12613. doi: 10.1073/pnas.91.26.12609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kikuchi A, Demo SD, Ye ZH, Chen YW, Williams LT. ralGDS family members interact with the effector loop of ras p21. Mol. Cell. Biol. 1994;14:7483–7491. doi: 10.1128/mcb.14.11.7483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hofer F, Fields S, Schneider C, Martin GS. Activated Ras interacts with the Ral guanine nucleotide dissociation stimulator. Proc. Natl. Acad. Sci. U.S.A. 1994;91:11089–11093. doi: 10.1073/pnas.91.23.11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Peterson SN, Trabalzini L, Brtva TR, Fischer T, Altschuler DL, Martelli P, Lapetina EG, Der CJ, White GC. Identification of a novel RalGDS-related protein as a candidate effector for Ras and Rap1. J. Biol. Chem. 1996;271:29903–29908. doi: 10.1074/jbc.271.47.29903. [DOI] [PubMed] [Google Scholar]
- 97.Ehrhardt GR, Korherr C, Wieler JS, Knaus M, Schrader JW. A novel potential effector of M-Ras and p21 Ras negatively regulates p21 Ras-mediated gene induction and cell growth. Oncogene. 2001;20:188–197. doi: 10.1038/sj.onc.1204053. [DOI] [PubMed] [Google Scholar]
- 98.Urano T, Emkey R, Feig LA. Ral-GTPases mediate a distinct downstream signaling pathway from Ras that facilitates cellular transformation. EMBO J. 1996;15:810–816. [PMC free article] [PubMed] [Google Scholar]
- 99.Ward Y, Wang W, Woodhouse E, Linnoila I, Liotta L, Kelly K. Signal pathways which promote invasion and metastasis: Critical and distinct contributions of extracellular signal-regulated kinase and Ral-specific guanine exchange factor pathways. Mol. Cell. Biol. 2001;21:5958–5969. doi: 10.1128/MCB.21.17.5958-5969.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hamad NM, Elconin JH, Karnoub AE, Bai W, Rich JN, Abraham RT, Der CJ, Counter CM. Distinct requirements for Ras oncogenesis in human versus mouse cells. Genes Dev. 2002;16:2045–2057. doi: 10.1101/gad.993902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wang Y, Waldron RT, Dhaka A, Patel A, Riley MM, Rozengurt E, Colicelli J. The RAS effector RIN1 directly competes with RAF and is regulated by 14-3-3 proteins. Mol. Cell. Biol. 2002;22:916–926. doi: 10.1128/MCB.22.3.916-926.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Han L, Wong D, Dhaka A, Afar D, White M, Xie W, Herschman H, Witte O, Colicelli J. Protein binding and signaling properties of RIN1 suggest a unique effector function. Proc. Natl. Acad. Sci. U.S.A. 1997;94:4954–4959. doi: 10.1073/pnas.94.10.4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tall GG, Barbieri MA, Stahl PD, Horazdovsky BF. Ras-activated endocytosis is mediated by the Rab5 guanine nucleotide exchange activity of RIN1. Dev. Cell. 2001;1:73–82. doi: 10.1016/s1534-5807(01)00008-9. [DOI] [PubMed] [Google Scholar]
- 104.Afar DE, Han L, McLaughlin J, Wong S, Dhaka A, Parmar K, Rosenberg N, Witte ON, Colicelli J. Regulation of the oncogenic activity of BCR-ABL by a tightly bound substrate protein RIN1. Immunity. 1997;6:773–782. doi: 10.1016/s1074-7613(00)80452-5. [DOI] [PubMed] [Google Scholar]
- 105.Hu H, Colicelli J. unpublished.
- 106.Khokhlatchev A, Rabizadeh S, Xavier R, Nedwidek M, Chen T, Zhang XF, Seed B, Avruch J. Identification of a novel Ras-regulated proapoptotic pathway. Curr. Biol. 2002;12:253–265. doi: 10.1016/s0960-9822(02)00683-8. [DOI] [PubMed] [Google Scholar]
- 107.Kelley GG, Reks SE, Ondrako JM, Smrcka AV, Phospholipase C. (epsilon): A novel Ras effector. EMBO J. 2001;20:743–754. doi: 10.1093/emboj/20.4.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Boettner B, Govek EE, Cross J, Van Aelst L. The junctional multidomain protein AF-6 is a binding partner of the Rap1A GTPase and associates with the actin cytoskeletal regulator profilin. Proc. Natl. Acad. Sci. U.S.A. 2000;97:9064–9069. doi: 10.1073/pnas.97.16.9064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mitin NY, Ramocki MB, Zullo AJ, Der CJ, Konieczny SF, Taparowsky EJ. Identification and characterization of rain, a novel Ras-interacting protein with a unique subcellular localization. J. Biol. Chem. 2004;279:22353–22361. doi: 10.1074/jbc.M312867200. [DOI] [PubMed] [Google Scholar]
- 110.Rodriguez-Viciana P, Sabatier C, McCormick F. Signaling specificity by Ras family GTPases is determined by the full spectrum of effectors they regulate. Mol. Cell. Biol. 2004;24:4943–4954. doi: 10.1128/MCB.24.11.4943-4954.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Matheny SA, Chen C, Kortum RL, Razidlo GL, Lewis RE, White MA. Ras regulates assembly of mitogenic signalling complexes through the effector protein IMP. Nature. 2004;427:256–260. doi: 10.1038/nature02237. [DOI] [PubMed] [Google Scholar]
- 112.Kwong L, Wozniak MA, Collins AS, Wilson SD, Keely PJ. R-Ras promotes focal adhesion formation through focal adhesion kinase and p130(Cas) by a novel mechanism that differs from integrins. Mol. Cell. Biol. 2003;23:933–949. doi: 10.1128/MCB.23.3.933-949.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Furuhjelm J, Peranen J. The C-terminal end of R-Ras contains a focal adhesion targeting signal. J. Cell Sci. 2003;116:3729–3738. doi: 10.1242/jcs.00689. [DOI] [PubMed] [Google Scholar]
- 114.Chan AM, Miki T, Meyers KA, Aaronson SA. A human oncogene of the RAS superfamily unmasked by expression cDNA cloning. Proc. Natl. Acad. Sci. U.S.A. 1994;91:7558–7562. doi: 10.1073/pnas.91.16.7558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Graham SM, Cox AD, Drivas G, Rush MG, D'Eustachio P, Der CJ. Aberrant function of the Ras-related protein TC21/R-Ras2 triggers malignant transformation. Mol. Cell. Biol. 1994;14:4108–4115. doi: 10.1128/mcb.14.6.4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ehrhardt GR, Leslie KB, Lee F, Wieler JS, Schrader JW. M-Ras, a widely expressed 29-kD homologue of p21 Ras: Expression of a constitutively active mutant results in factor-independent growth of an interleukin-3-dependent cell line. Blood. 1999;94:2433–2444. [PubMed] [Google Scholar]
- 117.Cox AD, Brtva TR, Lowe DG, Der CJ. R-Ras induces malignant, but not morphologic, transformation of NIH3T3 cells. Oncogene. 1994;9:3281–3288. [PubMed] [Google Scholar]
- 118.Huang Y, Saez R, Chao L, Santos E, Aaronson SA, Chan AM. A novel insertional mutation in the TC21 gene activates its transforming activity in a human leiomyosarcoma cell line. Oncogene. 1995;11:1255–1260. [PubMed] [Google Scholar]
- 119.Clark GJ, Kinch MS, Gilmer TM, Burridge K, Der CJ. Overexpression of the Ras-related TC21/R-Ras2 protein may contribute to the development of human breast cancers. Oncogene. 1996;12:169–176. [PubMed] [Google Scholar]
- 120.Barker KT, Crompton MR. Ras-related TC21 is activated by mutation in a breast cancer cell line, but infrequently in breast carcinomas in vivo. Br. J. Cancer. 1998;78:296–300. doi: 10.1038/bjc.1998.490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Marte BM, Rodriguez-Viciana P, Wennstrom S, Warne PH, Downward J. R-Ras can activate the phosphoinositide 3-kinase but not the MAP kinase arm of the Ras effector pathways. Curr. Biol. 1997;7:63–70. doi: 10.1016/s0960-9822(06)00028-5. [DOI] [PubMed] [Google Scholar]
- 122.Rosario M, Paterson HF, Marshall CJ. Activation of the Ral and phosphatidylinositol 3′ kinase signaling pathways by the ras-related protein TC21. Mol. Cell. Biol. 2001;21:3750–3762. doi: 10.1128/MCB.21.11.3750-3762.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Gao X, Satoh T, Liao Y, Song C, Hu CD, Kariya Ki K, Kataoka T. Identification and characterization of RA-GEF-2, a Rap guanine nucleotide exchange factor that serves as a downstream target of M-Ras. J. Biol. Chem. 2001;276:42219–42225. doi: 10.1074/jbc.M105760200. [DOI] [PubMed] [Google Scholar]
- 124.Spaargaren M, Martin GA, McCormick F, Fernandez-Sarabia MJ, Bischoff JR. The Ras-related protein R-ras interacts directly with Raf-1 in a GTP-dependent manner. Biochem. J. 1994;300:303–307. doi: 10.1042/bj3000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Rosario M, Paterson HF, Marshall CJ. Activation of the Raf/MAP kinase cascade by the Ras-related protein TC21 is required for the TC21-mediated transformation of NIH 3T3 cells. EMBO J. 1999;18:1270–1279. doi: 10.1093/emboj/18.5.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Reedquist KA, Ross E, Koop EA, Wolthuis RM, Zwartkruis FJ, van Kooyk Y, Salmon M, Buckley CD, Bos JL. The small GTPase, Rap1, mediates CD31-induced integrin adhesion. J. Cell Biol. 2000;148:1151–1158. doi: 10.1083/jcb.148.6.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.McLeod SJ, Shum AJ, Lee RL, Takei F, Gold MR. The Rap GTPases regulate integrin-mediated adhesion, cell spreading, actin polymerization, and Pyk2 tyrosine phosphorylation in B lymphocytes. J. Biol. Chem. 2003 doi: 10.1074/jbc.M313098200. [DOI] [PubMed] [Google Scholar]
- 128.Kitayama H, Sugimoto Y, Matsuzaki T, Ikawa Y, Noda M. A ras-related gene with transformation suppressor activity. Cell. 1989;56:77–84. doi: 10.1016/0092-8674(89)90985-9. [DOI] [PubMed] [Google Scholar]
- 129.Vossler MR, Yao H, York RD, Pan MG, Rim CS, Stork PJ. cAMP activates MAP kinase and Elk-1 through a B-Raf- and Rap1-dependent pathway. Cell. 1997;89:73–82. doi: 10.1016/s0092-8674(00)80184-1. [DOI] [PubMed] [Google Scholar]
- 130.Enserink JM, Christensen AE, de Rooij J, van Triest M, Schwede F, Genieser HG, Doskeland SO, Blank JL, Bos JL. A novel Epac-specific cAMP analogue demonstrates independent regulation of Rap1 and ERK. Nat. Cell Biol. 2002;4:901–906. doi: 10.1038/ncb874. [DOI] [PubMed] [Google Scholar]
- 131.Dupuy AJ, Morgan K, von Lintig FC, Shen H, Acar H, Hasz DE, Jenkins NA, Copeland NG, Boss GR, Largaespada DA. Activation of the Rap1 guanine nucleotide exchange gene, CalDAG-GEF I, in BXH-2 murine myeloid leukemia. J. Biol. Chem. 2001;276:11804–11811. doi: 10.1074/jbc.M008970200. [DOI] [PubMed] [Google Scholar]
- 132.Ishida D, Kometani K, Yang H, Kakugawa K, Masuda K, Iwai K, Suzuki M, Itohara S, Nakahata T, Hiai H, Kawamoto H, Hattori M, Minato N. Myeloproliferative stem cell disorders by deregulated Rap1 activation in SPA-1-deficient mice. Cancer Cell. 2003;4:55–65. doi: 10.1016/s1535-6108(03)00163-6. [DOI] [PubMed] [Google Scholar]
- 133.Yajnik V, Paulding C, Sordella R, McClatchey AI, Saito M, Wahrer DC, Reynolds P, Bell DW, Lake R, van den Heuvel S, Settleman J, Haber DA. DOCK4, a GTPase activator, is disrupted during tumorigenesis. Cell. 2003;112:673–684. doi: 10.1016/s0092-8674(03)00155-7. [DOI] [PubMed] [Google Scholar]
- 134.Mirey G, Balakireva M, L'Hoste S, Rosse C, Voegeling S, Camonis J. A Ral guanine exchange factor-Ral pathway is conserved in Drosophila melanogaster and sheds new light on the connectivity of the Ral, Ras, and Rap pathways. Mol. Cell. Biol. 2003;23:1112–1124. doi: 10.1128/MCB.23.3.1112-1124.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Song C, Satoh T, Edamatsu H, Wu D, Tadano M, Gao X, Kataoka T. Differential roles of Ras and Rap1 in growth factor-dependent activation of phospholipase C epsilon. Oncogene. 2002;21:8105–8113. doi: 10.1038/sj.onc.1206003. [DOI] [PubMed] [Google Scholar]
- 136.Katagiri K, Maeda A, Shimonaka M, Kinashi T. RAPL, a Rap1-binding molecule that mediates Rap1-induced adhesion through spatial regulation of LFA-1. Nat. Immunol. 2003;4:741–748. doi: 10.1038/ni950. [DOI] [PubMed] [Google Scholar]
- 137.Feig LA. Ral-GTPases: Approaching their 15 minutes of fame. Trends Cell Biol. 2003;13:419–425. doi: 10.1016/s0962-8924(03)00152-1. [DOI] [PubMed] [Google Scholar]
- 138.Lu Z, Hornia A, Joseph T, Sukezane T, Frankel P, Zhong M, Bychenok S, Xu L, Feig LA, Foster DA. Phospholipase D and RalA cooperate with the epidermal growth factor receptor to transform 3Y1 rat fibroblasts. Mol. Cell. Biol. 2000;20:462–467. doi: 10.1128/mcb.20.2.462-467.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Chien Y, White MA. RAL GTPases are linchpin modulators of human tumour-cell proliferation and survival. EMBO Rep. 2003;4:800–806. doi: 10.1038/sj.embor.embor899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Shipitsin M, Feig LA. RalA but not RalB enhances polarized delivery of membrane proteins to the basolateral surface of epithelial cells. Mol. Cell. Biol. 2004;24:5746–5756. doi: 10.1128/MCB.24.13.5746-5756.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Park SH, Weinberg RA. A putative effector of Ral has homology to Rho/Rac GTPase activating proteins. Oncogene. 1995;11:2349–2355. [PubMed] [Google Scholar]
- 142.Jullien-Flores V, Dorseuil O, Romero F, Letourneur F, Saragosti S, Berger R, Tavitian A, Gacon G, Camonis JH. Bridging Ral GTPase to Rho pathways. RLIP76, a Ral effector with CDC42/Rac GTPase-activating protein activity. J. Biol. Chem. 1995;270:22473–22477. doi: 10.1074/jbc.270.38.22473. [DOI] [PubMed] [Google Scholar]
- 143.Cantor SB, Urano T, Feig LA. Identification and characterization of Ral-binding protein 1, a potential downstream target of Ral GTPases. Mol. Cell. Biol. 1995;15:4578–4584. doi: 10.1128/mcb.15.8.4578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Jullien-Flores V, Mahe Y, Mirey G, Leprince C, Meunier-Bisceuil B, Sorkin A, Camonis JH. RLIP76, an effector of the GTPase Ral, interacts with the AP2 complex: Involvement of the Ral pathway in receptor endocytosis. J. Cell Sci. 2000;113:2837–2844. doi: 10.1242/jcs.113.16.2837. [DOI] [PubMed] [Google Scholar]
- 145.Nakashima S, Morinaka K, Koyama S, Ikeda M, Kishida M, Okawa K, Iwamatsu A, Kishida S, Kikuchi A. Small G protein Ral and its downstream molecules regulate endocytosis of EGF and insulin receptors. EMBO J. 1999;18:3629–3642. doi: 10.1093/emboj/18.13.3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Rosse C, L'Hoste S, Offner N, Picard A, Camonis J. RLIP, an effector of the Ral GTPases, is a platform for Cdk1 to phosphorylate epsin during the switch off of endocytosis in mitosis. J. Biol. Chem. 2003;278:30597–30604. doi: 10.1074/jbc.M302191200. [DOI] [PubMed] [Google Scholar]
- 147.Moskalenko S, Henry DO, Rosse C, Mirey G, Camonis JH, White MA. The exocyst is a Ral effector complex. Nat. Cell Biol. 2002;4:66–72. doi: 10.1038/ncb728. [DOI] [PubMed] [Google Scholar]
- 148.Moskalenko S, Tong C, Rosse C, Mirey G, Formstecher E, Daviet L, Camonis J, White MA. Ral GTPases regulate exocyst assembly through dual subunit interactions. J. Biol. Chem. 2003;278:51743–51748. doi: 10.1074/jbc.M308702200. [DOI] [PubMed] [Google Scholar]
- 149.Sugihara K, Asano S, Tanaka K, Iwamatsu A, Okawa K, Ohta Y. The exocyst complex binds the small GTPase RalA to mediate filopodia formation. Nat. Cell Biol. 2002;4:73–78. doi: 10.1038/ncb720. [DOI] [PubMed] [Google Scholar]
- 150.Hoshino M, Nakamura S. Small GTPase Rin induces neurite outgrowth through Rac/Cdc42 and calmodulin in PC12 cells. J. Cell Biol. 2003;163:1067–1076. doi: 10.1083/jcb.200308070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Hynds DL, Spencer ML, Andres DA, Snow DM. Rit promotes MEK-independent neurite branching in human neuroblastoma cells. J. Cell Sci. 2003;116:1925–1935. doi: 10.1242/jcs.00401. [DOI] [PubMed] [Google Scholar]
- 152.Spencer ML, Shao H, Andres DA. Induction of neurite extension and survival in pheochromocytoma cells by the Rit GTPase. J. Biol. Chem. 2002;277:20160–20168. doi: 10.1074/jbc.M201092200. [DOI] [PubMed] [Google Scholar]
- 153.Lee CH, Della NG, Chew CE, Zack DJ. Rin, a neuron-specific and calmodulin-binding small G-protein, and Rit define a novel subfamily of ras proteins. J. Neurosci. 1996;16:6784–6794. doi: 10.1523/JNEUROSCI.16-21-06784.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Rusyn EV, Reynolds ER, Shao H, Grana TM, Chan TO, Andres DA, Cox AD. Rit, a non-lipid-modified Ras-related protein, transforms NIH3T3 cells without activating the ERK, JNK, p38 MAPK or PI3K/Akt pathways. Oncogene. 2000;19:4685–4694. doi: 10.1038/sj.onc.1203836. [DOI] [PubMed] [Google Scholar]
- 155.Sakabe K, Teramoto H, Zohar M, Behbahani B, Miyazaki H, Chikumi H, Gutkind JS. Potent transforming activity of the small GTP-binding protein Rit in NIH 3T3 cells: Evidence for a role of a p38gamma-dependent signaling pathway. FEBS Lett. 2002;511:15–20. doi: 10.1016/s0014-5793(01)03264-1. [DOI] [PubMed] [Google Scholar]
- 156.Shao H, Kadono-Okuda K, Finlin BS, Andres DA. Biochemical characterization of the Ras-related GTPases Rit and Rin. Arch. Biochem. Biophys. 1999;371:207–219. doi: 10.1006/abbi.1999.1448. [DOI] [PubMed] [Google Scholar]
- 157.Takahashi K, Mitsui K, Yamanaka S. Role of ERas in promoting tumour-like properties in mouse embryonic stem cells. Nature. 2003;423:541–545. doi: 10.1038/nature01646. [DOI] [PubMed] [Google Scholar]
- 158.Kontani K, Tada M, Ogawa T, Okai T, Saito K, Araki Y, Katada T. Di-Ras, a distinct subgroup of ras family GTPases with unique biochemical properties. J. Biol. Chem. 2002;277:41070–41078. doi: 10.1074/jbc.M202150200. [DOI] [PubMed] [Google Scholar]
- 159.Ellis CA, Vos MD, Howell H, Vallecorsa T, Fults DW, Clark GJ. Rig is a novel Ras-related protein and potential neural tumor suppressor. Proc. Natl. Acad. Sci. U.S.A. 2002;99:9876–9881. doi: 10.1073/pnas.142193799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Yu Y, Xu F, Peng H, Fang X, Zhao S, Li Y, Cuevas B, Kuo WL, Gray JW, Siciliano M, Mills GB, Bast RC., Jr. NOEY2 (ARHI), an imprinted putative tumor suppressor gene in ovarian and breast carcinomas. Proc. Natl. Acad. Sci. U.S.A. 1999;96:214–219. doi: 10.1073/pnas.96.1.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Wang L, Hoque A, Luo RZ, Yuan J, Lu Z, Nishimoto A, Liu J, Sahin AA, Lippman SM, Bast RC, Jr., Yu Y. Loss of the expression of the tumor suppressor gene ARHI is associated with progression of breast cancer. Clin. Cancer Res. 2003;9:3660–3666. [PubMed] [Google Scholar]
- 162.Kemppainen RJ, Behrend EN. Dexamethasone rapidly induces a novel ras superfamily member-related gene in AtT-20 cells. J. Biol. Chem. 1998;273:3129–3131. doi: 10.1074/jbc.273.6.3129. [DOI] [PubMed] [Google Scholar]
- 163.Chan SL, Monks LK, Gao H, Deaville P, Morgan NG. Identification of the monomeric G-protein, Rhes, as an efaroxan-regulated protein in the pancreatic beta-cell. Br. J. Pharmacol. 2002;136:31–36. doi: 10.1038/sj.bjp.0704680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Graham TE, Key TA, Kilpatrick K, Dorin RI. Dexras1/AGS-1, a steroid hormone-induced guanosine triphosphate-binding protein, inhibits 3′,5′-cyclic adenosine monophosphate-stimulated secretion in AtT-20 corticotroph cells. Endocrinology. 2001;142:2631–2640. doi: 10.1210/endo.142.6.8209. [DOI] [PubMed] [Google Scholar]
- 165.Vaidyanathan G, Cismowski MJ, Wang G, Vincent TS, Brown KD, Lanier SM. The Ras-related protein AGS1/RASD1 suppresses cell growth. Oncogene. 2004;23:5858–5863. doi: 10.1038/sj.onc.1207774. [DOI] [PubMed] [Google Scholar]
- 166.Vargiu P, De Abajo R, Garcia-Ranea JA, Valencia A, Santisteban P, Crespo P, Bernal J. The small GTP-binding protein, Rhes, regulates signal transduction from G protein-coupled receptors. Oncogene. 2004;23:559–568. doi: 10.1038/sj.onc.1207161. [DOI] [PubMed] [Google Scholar]
- 167.Fenwick C, Na SY, Voll RE, Zhong H, Im SY, Lee JW, Ghosh S. A subclass of Ras proteins that regulate the degradation of IkappaB. Science. 2000;287:869–873. doi: 10.1126/science.287.5454.869. [DOI] [PubMed] [Google Scholar]
- 168.Chen Y, Wu J, Ghosh G. KappaB-Ras binds to the unique insert within the ankyrin repeat domain of IkappaBbeta and regulates cytoplasmic retention of IkappaBbeta × NF-kappaB complexes. J. Biol. Chem. 2003;278:23101–23106. doi: 10.1074/jbc.M301021200. [DOI] [PubMed] [Google Scholar]
- 169.Finlin BS, Andres DA. Rem is a new member of the Rad- and Gem/Kir Ras-related GTP-binding protein family repressed by lipopolysaccharide stimulation. J. Biol. Chem. 1997;272:21982–21988. doi: 10.1074/jbc.272.35.21982. [DOI] [PubMed] [Google Scholar]
- 170.Finlin BS, Shao H, Kadono-Okuda K, Guo N, Andres DA. Rem2, a new member of the Rem/Rad/Gem/Kir family of Ras-related GTPases. Biochem. J. 2000;347:223–231. [PMC free article] [PubMed] [Google Scholar]
- 171.Reynet C, Kahn CR. Rad: A member of the Ras family overexpressed in muscle of type II diabetic humans. Science. 1993;262:1441–1444. doi: 10.1126/science.8248782. [DOI] [PubMed] [Google Scholar]
- 172.Maguire J, Santoro T, Jensen P, Siebenlist U, Yewdell J, Kelly K. Gem: An induced, immediate early protein belonging to the Ras family. Science. 1994;265:241–244. doi: 10.1126/science.7912851. [DOI] [PubMed] [Google Scholar]
- 173.Finlin BS, Crump SM, Satin J, Andres DA. Regulation of voltage-gated calcium channel activity by the Rem and Rad GTPases. Proc. Natl. Acad. Sci. U.S.A. 2003;100:14469–14474. doi: 10.1073/pnas.2437756100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Beguin P, Nagashima K, Gonoi T, Shibasaki T, Takahashi K, Kashima Y, Ozaki N, Geering K, Iwanaga T, Seino S. Regulation of Ca2+ channel expression at the cell surface by the small G-protein kir/Gem. Nature. 2001;411:701–706. doi: 10.1038/35079621. [DOI] [PubMed] [Google Scholar]
- 175.Piddini E, Schmid JA, de Martin R, Dotti CG. The Ras-like GTPase Gem is involved in cell shape remodelling and interacts with the novel kinesin-like protein KIF9. EMBO J. 2001;20:4076–4087. doi: 10.1093/emboj/20.15.4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Chardin P. GTPase regulation: Getting aRnd Rock and Rho inhibition. Curr. Biol. 2003;13:R702–R704. doi: 10.1016/j.cub.2003.08.042. [DOI] [PubMed] [Google Scholar]
- 177.Aresta S, de Tand-Heim MF, Beranger F, de Gunzburg J. A novel Rho GTPase-activating-protein interacts with Gem, a member of the Ras superfamily of GTPases. Biochem. J. 2002;367:57–65. doi: 10.1042/BJ20020829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Louro R, Nakaya HI, Paquola AC, Martins EA, da Silva AM, Verjovski-Almeida S, Reis EM. RASL11A, member of a novel small monomeric GTPase gene family, is down-regulated in prostate tumors. Biochem. Biophys. Res. Commun. 2004;316:618–627. doi: 10.1016/j.bbrc.2004.02.091. [DOI] [PubMed] [Google Scholar]
- 179.Patel PH, Thapar N, Guo L, Martinez M, Maris J, Gau CL, Lengyel JA, Tamanoi F. Drosophila Rheb GTPase is required for cell cycle progression and cell growth. J. Cell Sci. 2003;116:3601–3610. doi: 10.1242/jcs.00661. [DOI] [PubMed] [Google Scholar]
- 180.Clark GJ, Kinch MS, Rogers-Graham K, Sebti SM, Hamilton AD, Der CJ. The Ras-related protein Rheb is farnesylated and antagonizes Ras signaling and transformation. J. Biol. Chem. 1997;272:10608–10615. doi: 10.1074/jbc.272.16.10608. [DOI] [PubMed] [Google Scholar]
- 181.Im E, von Lintig FC, Chen J, Zhuang S, Qui W, Chowdhury S, Worley PF, Boss GR, Pilz RB. Rheb is in a high activation state and inhibits B-Raf kinase in mammalian cells. Oncogene. 2002;21:6356–6365. doi: 10.1038/sj.onc.1205792. [DOI] [PubMed] [Google Scholar]
- 182.Castro AF, Rebhun JF, Clark GJ, Quilliam LA. Rheb binds tuberous sclerosis complex 2 (TSC2) and promotes S6 kinase activation in a rapamycin- and farnesylation-dependent manner. J. Biol. Chem. 2003;278:32493–32496. doi: 10.1074/jbc.C300226200. [DOI] [PubMed] [Google Scholar]
- 183.Inoki K, Li Y, Xu T, Guan KL. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 2003;17:1829–1834. doi: 10.1101/gad.1110003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Garami A, Zwartkruis FJ, Nobukuni T, Joaquin M, Roccio M, Stocker H, Kozma SC, Hafen E, Bos JL, Thomas G. Insulin activation of Rheb, a mediator of mTOR/S6K/4E-BP signaling, is inhibited by TSC1 and 2. Mol. Cell. 2003;11:1457–1466. doi: 10.1016/s1097-2765(03)00220-x. [DOI] [PubMed] [Google Scholar]
- 185.Tee AR, Manning BD, Roux PP, Cantley LC, Blenis J. Tuberous sclerosis complex gene products, Tuberin and Hamartin, control mTOR signaling by acting as a GTPase-activating protein complex toward Rheb. Curr. Biol. 2003;13:1259–1268. doi: 10.1016/s0960-9822(03)00506-2. [DOI] [PubMed] [Google Scholar]
- 186.Zhang Y, Gao X, Saucedo LJ, Ru B, Edgar BA, Pan D. Rheb is a direct target of the tuberous sclerosis tumour suppressor proteins. Nat. Cell Biol. 2003;5:578–581. doi: 10.1038/ncb999. [DOI] [PubMed] [Google Scholar]
- 187.Burridge K, Wennerberg K. Rho and rac take center stage. Cell. 2004;116:167–179. doi: 10.1016/s0092-8674(04)00003-0. [DOI] [PubMed] [Google Scholar]
- 188.Wennerberg K, Der CJ. Rho-family GTPases: It's not only Rac and Rho (and I like it). J. Cell Sci. 2004;117:1301–1312. doi: 10.1242/jcs.01118. [DOI] [PubMed] [Google Scholar]
- 189.Sahai E, Marshall CJ. RHO-GTPases and cancer. Nat. Rev. Cancer. 2002;2:133–142. doi: 10.1038/nrc725. [DOI] [PubMed] [Google Scholar]
- 190.Jordan P, Brazao R, Boavida MG, Gespach C, Chastre E. Cloning of a novel human Rac1b splice variant with increased expression in colorectal tumors. Oncogene. 1999;18:6835–6839. doi: 10.1038/sj.onc.1203233. [DOI] [PubMed] [Google Scholar]
- 191.Kimura K, Ito M, Amano M, Chihara K, Fukata Y, Nakafuku M, Yamamori B, Feng J, Nakano T, Okawa K, Iwamatsu A, Kaibuchi K. Regulation of myosin phosphatase by Rho and Rho-associated kinase (Rho-kinase). Science. 1996;273:245–248. doi: 10.1126/science.273.5272.245. [DOI] [PubMed] [Google Scholar]
- 192.Watanabe N, Kato T, Fujita A, Ishizaki T, Narumiya S. Cooperation between mDia1 and ROCK in Rho-induced actin reorganization. Nat. Cell Biol. 1999;1:136–143. doi: 10.1038/11056. [DOI] [PubMed] [Google Scholar]
- 193.Amano M, Mukai H, Ono Y, Chihara K, Matsui T, Hamajima Y, Okawa K, Iwamatsu A, Kaibuchi K. Identification of a putative target for Rho as the serine-threonine kinase protein kinase N. Science. 1996;271:648–650. doi: 10.1126/science.271.5249.648. [DOI] [PubMed] [Google Scholar]
- 194.Watanabe G, Saito Y, Madaule P, Ishizaki T, Fujisawa K, Morii N, Mukai H, Ono Y, Kakizuka A, Narumiya S. Protein kinase N (PKN) and PKN-related protein rhophilin as targets of small GTPase Rho. Science. 1996;271:645–648. doi: 10.1126/science.271.5249.645. [DOI] [PubMed] [Google Scholar]
- 195.Wing MR, Snyder JT, Sondek J, Harden TK. Direct activation of phospholipase C-epsilon by Rho. J. Biol. Chem. 2003;278:41253–41258. doi: 10.1074/jbc.M306904200. [DOI] [PubMed] [Google Scholar]
- 196.Peck JW, Oberst M, Bouker KB, Bowden E, Burbelo PD. The RhoA-binding protein, rhophilin-2, regulates actin cytoskeleton organization. J. Biol. Chem. 2002;277:43924–43932. doi: 10.1074/jbc.M203569200. [DOI] [PubMed] [Google Scholar]
- 197.Reid T, Furuyashiki T, Ishizaki T, Watanabe G, Watanabe N, Fujisawa K, Morii N, Madaule P, Narumiya S. Rhotekin, a new putative target for Rho bearing homology to a serine/threonine kinase, PKN, and rhophilin in the rho-binding domain. J. Biol. Chem. 1996;271:13556–13560. doi: 10.1074/jbc.271.23.13556. [DOI] [PubMed] [Google Scholar]
- 198.Didsbury J, Weber RF, Bokoch GM, Evans T, Snyderman R. rac, a novel ras-related family of proteins that are botulinum toxin substrates. J. Biol. Chem. 1989;264:16378–16382. [PubMed] [Google Scholar]
- 199.Bagrodia S, Taylor SJ, Creasy CL, Chernoff J, Cerione RA. Identification of a mouse p21Cdc42/Rac activated kinase. J. Biol. Chem. 1995;270:22731–22737. doi: 10.1074/jbc.270.39.22731. [DOI] [PubMed] [Google Scholar]
- 200.Snyder JT, Singer AU, Wing MR, Harden TK, Sondek J. The pleckstrin homology domain of phospholipase C-beta2 as an effector site for Rac. J. Biol. Chem. 2003;278:21099–21104. doi: 10.1074/jbc.M301418200. [DOI] [PubMed] [Google Scholar]
- 201.Aspenstrom P, Lindberg U, Hall A. Two GTPases, Cdc42 and Rac, bind directly to a protein implicated in the immunodeficiency disorder Wiskott-Aldrich syndrome. Curr. Biol. 1996;6:70–75. doi: 10.1016/s0960-9822(02)00423-2. [DOI] [PubMed] [Google Scholar]
- 202.Symons M, Derry JM, Karlak B, Jiang S, Lemahieu V, McCormick F, Francke U, Abo A. Wiskott-Aldrich syndrome protein, a novel effector for the GTPase CDC42Hs, is implicated in actin polymerization. Cell. 1996;84:723–734. doi: 10.1016/s0092-8674(00)81050-8. [DOI] [PubMed] [Google Scholar]
- 203.Kolluri R, Tolias KF, Carpenter CL, Rosen FS, Kirchhausen T. Direct interaction of the Wiskott-Aldrich syndrome protein with the GTPase Cdc42. Proc. Natl. Acad. Sci. U.S.A. 1996;93:5615–5618. doi: 10.1073/pnas.93.11.5615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Ho HY, Rohatgi R, Lebensohn AM, Le M, Li J, Gygi SP, Kirschner MW. Toca-1 mediates Cdc42-dependent actin nucleation by activating the N-WASP-WIP complex. Cell. 2004;118:203–216. doi: 10.1016/j.cell.2004.06.027. [DOI] [PubMed] [Google Scholar]
- 205.Garrard SM, Capaldo CT, Gao L, Rosen MK, Macara IG, Tomchick DR. Structure of Cdc42 in a complex with the GTPase-binding domain of the cell polarity protein, Par6. EMBO J. 2003;22:1125–1133. doi: 10.1093/emboj/cdg110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Wennerberg K, Forget MA, Ellerbroek SM, Arthur WT, Burridge K, Settleman J, Der CJ, Hansen SH. Rnd proteins function as RhoA antagonists by activating p190 RhoGAP. Curr. Biol. 2003;13:1106–1115. doi: 10.1016/s0960-9822(03)00418-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Riento K, Guasch RM, Garg R, Jin B, Ridley AJ. RhoE binds to ROCK I and inhibits downstream signaling. Mol. Cell. Biol. 2003;23:4219–4229. doi: 10.1128/MCB.23.12.4219-4229.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Heo WD, Meyer T. Switch-of-function mutants based on morphology classification of Ras superfamily small GTPases. Cell. 2003;113:315–328. doi: 10.1016/s0092-8674(03)00315-5. [DOI] [PubMed] [Google Scholar]
- 209.Fransson A, Ruusala A, Aspenstrom P. Atypical Rho GTPases have roles in mitochondrial homeostasis and apoptosis. J. Biol. Chem. 2003;278:6495–6502. doi: 10.1074/jbc.M208609200. [DOI] [PubMed] [Google Scholar]
- 210.Aspenstrom P, Fransson A, Saras J. Rho GTPases have diverse effects on the organization of the actin filament system. Biochem. J. 2004;377:327–337. doi: 10.1042/BJ20031041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Touchot N, Chardin P, Tavitian A. Four additional members of the ras gene superfamily isolated by an oligonucleotide strategy: Molecular cloning of YPT-related cDNAs from a rat brain library. Proc. Natl. Acad. Sci. U.S.A. 1987;84:8210–8214. doi: 10.1073/pnas.84.23.8210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Nimmo ER, Sanders PG, Padua RA, Hughes D, Williamson R, Johnson KJ. The MEL gene: A new member of the RAB/YPT class of RAS-related genes. Oncogene. 1991;6:1347–1351. [PubMed] [Google Scholar]
- 213.Segev N. Ypt/rab gtpases: Regulators of protein trafficking. Sci. STKE. 2001;2001:re11. doi: 10.1126/stke.2001.100.re11. [DOI] [PubMed] [Google Scholar]
- 214.Stenmark H, Olkkonen VM. The Rab GTPase family. Genome Biol. 2001;2:REVIEWS3007. doi: 10.1186/gb-2001-2-5-reviews3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Pereira-Leal JB, Seabra MC. Evolution of the Rab family of small GTP-binding proteins. J. Mol. Biol. 2001;313:889–901. doi: 10.1006/jmbi.2001.5072. [DOI] [PubMed] [Google Scholar]
- 216.Stahl B, Chou JH, Li C, Sudhof TC, Jahn R. Rab3 reversibly recruits rabphilin to synaptic vesicles by a mechanism analogous to raf recruitment by ras. EMBO J. 1996;15:1799–1809. [PMC free article] [PubMed] [Google Scholar]
- 217.Christoforidis S, McBride HM, Burgoyne RD, Zerial M. The Rab5 effector EEA1 is a core component of endosome docking. Nature. 1999;397:621–625. doi: 10.1038/17618. [DOI] [PubMed] [Google Scholar]
- 218.Stenmark H, Vitale G, Ullrich O, Zerial M. Rabaptin-5 is a direct effector of the small GTPase Rab5 in endocytic membrane fusion. Cell. 1995;83:423–432. doi: 10.1016/0092-8674(95)90120-5. [DOI] [PubMed] [Google Scholar]
- 219.Miaczynska M, Christoforidis S, Giner A, Shevchenko A, Uttenweiler-Joseph S, Habermann B, Wilm M, Parton RG, Zerial M. APPL proteins link Rab5 to nuclear signal transduction via an endosomal compartment. Cell. 2004;116:445–456. doi: 10.1016/s0092-8674(04)00117-5. [DOI] [PubMed] [Google Scholar]
- 220.Jordens I, Fernandez-Borja M, Marsman M, Dusseljee S, Janssen L, Calafat J, Janssen H, Wubbolts R, Neefjes J. The Rab7 effector protein RILP controls lysosomal transport by inducing the recruitment of dynein-dynactin motors. Curr. Biol. 2001;11:1680–1685. doi: 10.1016/s0960-9822(01)00531-0. [DOI] [PubMed] [Google Scholar]
- 221.Cantalupo G, Alifano P, Roberti V, Bruni CB, Bucci C. Rab-interacting lysosomal protein (RILP): The Rab7 effector required for transport to lysosomes. EMBO J. 2001;20:683–693. doi: 10.1093/emboj/20.4.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Carroll KS, Hanna J, Simon I, Krise J, Barbero P, Pfeffer SR. Role of Rab9 GTPase in facilitating receptor recruitment by TIP47. Science. 2001;292:1373–1376. doi: 10.1126/science.1056791. [DOI] [PubMed] [Google Scholar]
- 223.Mammoto A, Ohtsuka T, Hotta I, Sasaki T, Takai Y. Rab11BP/Rabphilin-11, a downstream target of rab11 small G protein implicated in vesicle recycling. J. Biol. Chem. 1999;274:25517–25524. doi: 10.1074/jbc.274.36.25517. [DOI] [PubMed] [Google Scholar]
- 224.Desnos C, Schonn JS, Huet S, Tran VS, El-Amraoui A, Raposo G, Fanget I, Chapuis C, Menasche G, de Saint Basile G, Petit C, Cribier S, Henry JP, Darchen F. Rab27A and its effector MyRIP link secretory granules to F-actin and control their motion towards release sites. J. Cell Biol. 2003;163:559–570. doi: 10.1083/jcb.200302157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Weis K. Regulating access to the genome: Nucleocytoplasmic transport throughout the cell cycle. Cell. 2003;112:441–451. doi: 10.1016/s0092-8674(03)00082-5. [DOI] [PubMed] [Google Scholar]
- 226.Carazo-Salas RE, Gruss OJ, Mattaj IW, Karsenti E. Ran-GTP co-ordinates regulation of microtubule nucleation and dynamics during mitotic-spindle assembly. Nat. Cell Biol. 2001;3:228–234. doi: 10.1038/35060009. [DOI] [PubMed] [Google Scholar]
- 227.Wilde A, Lizarraga SB, Zhang L, Wiese C, Gliksman NR, Walczak CE, Zheng Y. Ran stimulates spindle assembly by altering microtubule dynamics and the balance of motor activities. Nat. Cell Biol. 2001;3:221–227. doi: 10.1038/35060000. [DOI] [PubMed] [Google Scholar]
- 228.Kahn RA, Gilman AG. Purification of a protein cofactor required for ADP-ribosylation of the stimulatory regulatory component of adenylate cyclase by cholera toxin. J. Biol. Chem. 1984;259:6228–6234. [PubMed] [Google Scholar]
- 229.Sewell JL, Kahn RA. Sequences of the bovine and yeast ADP-ribosylation factor and comparison to other GTP-binding proteins. Proc. Natl. Acad. Sci. U.S.A. 1988;85:4620–4624. doi: 10.1073/pnas.85.13.4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Price SR, Nightingale M, Tsai SC, Williamson KC, Adamik R, Chen HC, Moss J, Vaughan M. Guanine nucleotide-binding proteins that enhance choleragen ADP-ribosyltransferase activity: Nucleotide and deduced amino acid sequence of an ADP-ribosylation factor cDNA. Proc. Natl. Acad. Sci. U.S.A. 1988;85:5488–5491. doi: 10.1073/pnas.85.15.5488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Randazzo PA, Nie Z, Miura K, Hsu VW. Molecular aspects of the cellular activities of ADP-ribosylation factors. Sci. STKE. 2000;2000:re1. doi: 10.1126/stke.2000.59.re1. [DOI] [PubMed] [Google Scholar]
- 232.Kanoh H, Williger BT, Exton JH. Arfaptin 1, a putative cytosolic target protein of ADP-ribosylation factor, is recruited to Golgi membranes. J. Biol. Chem. 1997;272:5421–5429. doi: 10.1074/jbc.272.9.5421. [DOI] [PubMed] [Google Scholar]
- 233.Shin OH, Ross AH, Mihai I, Exton JH. Identification of arfophilin, a target protein for GTP-bound class II ADP-ribosylation factors. J. Biol. Chem. 1999;274:36609–36615. doi: 10.1074/jbc.274.51.36609. [DOI] [PubMed] [Google Scholar]
- 234.Zhao L, Helms JB, Brunner J, Wieland FT. GTP-dependent binding of ADP-ribosylation factor to coatomer in close proximity to the binding site for dilysine retrieval motifs and p23. J. Biol. Chem. 1999;274:14198–14203. doi: 10.1074/jbc.274.20.14198. [DOI] [PubMed] [Google Scholar]
- 235.Bigay J, Gounon P, Robineau S, Antonny B. Lipid packing sensed by ArfGAP1 couples COPI coat disassembly to membrane bilayer curvature. Nature. 2003;426:563–566. doi: 10.1038/nature02108. [DOI] [PubMed] [Google Scholar]
- 236.Prigent M, Dubois T, Raposo G, Derrien V, Tenza D, Rosse C, Camonis J, Chavrier P. ARF6 controls post-endocytic recycling through its downstream exocyst complex effector. J. Cell Biol. 2003;163:1111–1121. doi: 10.1083/jcb.200305029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Honda A, Nogami M, Yokozeki T, Yamazaki M, Nakamura H, Watanabe H, Kawamoto K, Nakayama K, Morris AJ, Frohman MA, Kanaho Y. Phosphatidylinositol 4-phosphate 5-kinase alpha is a downstream effector of the small G protein ARF6 in membrane ruffle formation. Cell. 1999;99:521–532. doi: 10.1016/s0092-8674(00)81540-8. [DOI] [PubMed] [Google Scholar]
- 238.Jones DH, Morris JB, Morgan CP, Kondo H, Irvine RF, Cockcroft S. Type I phosphatidylinositol 4-phosphate 5-kinase directly interacts with ADP-ribosylation factor 1 and is responsible for phosphatidylinositol 4,5-bisphosphate synthesis in the golgi compartment. J. Biol. Chem. 2000;275:13962–13966. doi: 10.1074/jbc.c901019199. [DOI] [PubMed] [Google Scholar]
- 239.Dell'Angelica EC, Puertollano R, Mullins C, Aguilar RC, Vargas JD, Hartnell LM, Bonifacino JS. GGAs: A family of ADP ribosylation factor-binding proteins related to adaptors and associated with the Golgi complex. J. Cell Biol. 2000;149:81–94. doi: 10.1083/jcb.149.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Boman AL, Zhang C, Zhu X, Kahn RA. A family of ADP-ribosylation factor effectors that can alter membrane transport through the trans-Golgi. Mol. Biol. Cell. 2000;11:1241–1255. doi: 10.1091/mbc.11.4.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Puertollano R, Randazzo PA, Presley JF, Hartnell LM, Bonifacino JS. The GGAs promote ARF-dependent recruitment of clathrin to the TGN. Cell. 2001;105:93–102. doi: 10.1016/s0092-8674(01)00299-9. [DOI] [PubMed] [Google Scholar]
- 242.Haucke V. Vesicle budding: A coat for the COPs. Trends Cell Biol. 2003;13:59–60. doi: 10.1016/s0962-8924(02)00011-9. [DOI] [PubMed] [Google Scholar]
- 243.Conklin BR, Bourne HR. Structural elements of G alpha subunits that interact with G beta gamma, receptors, and effectors. Cell. 1993;73:631–641. doi: 10.1016/0092-8674(93)90245-l. [DOI] [PubMed] [Google Scholar]
- 244.Markby DW, Onrust R, Bourne HR. Separate GTP binding and GTPase activating domains of a G alpha subunit. Science. 1993;262:1895–1901. doi: 10.1126/science.8266082. [DOI] [PubMed] [Google Scholar]
- 245.Lyons J, Landis CA, Harsh G, Vallar L, Grunewald K, Feichtinger H, Duh QY, Clark OH, Kawasaki E, Bourne HR, et al. Two G protein oncogenes in human endocrine tumors. Science. 1990;249:655–659. doi: 10.1126/science.2116665. [DOI] [PubMed] [Google Scholar]
- 246.Bunemann M, Frank M, Lohse MJ. From The Cover: Gi protein activation in intact cells involves subunit rearrangement rather than dissociation. Proc. Natl. Acad. Sci. U.S.A. 2003;100:16077–16082. doi: 10.1073/pnas.2536719100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Neves SR, Ram PT, Iyengar R. G protein pathways. Science. 2002;296:1636–1639. doi: 10.1126/science.1071550. [DOI] [PubMed] [Google Scholar]
- 248.Faty M, Fink M, Barral Y. Septins: A ring to part mother and daughter. Curr. Genet. 2002;41:123–131. doi: 10.1007/s00294-002-0304-0. [DOI] [PubMed] [Google Scholar]
- 249.Zhang J, Kong C, Xie H, McPherson PS, Grinstein S, Trimble WS. Phosphatidylinositol polyphosphate binding to the mammalian septin H5 is modulated by GTP. Curr. Biol. 1999;9:1458–1467. doi: 10.1016/s0960-9822(00)80115-3. [DOI] [PubMed] [Google Scholar]
- 250.Danino D, Hinshaw JE. Dynamin family of mechanoenzymes. Curr. Opin. Cell Biol. 2001;13:454–460. doi: 10.1016/s0955-0674(00)00236-2. [DOI] [PubMed] [Google Scholar]
- 251.Andersen GR, Nissen P, Nyborg J. Elongation factors in protein biosynthesis. Trends Biochem. Sci. 2003;28:434–441. doi: 10.1016/S0968-0004(03)00162-2. [DOI] [PubMed] [Google Scholar]
- 252.Pestova TV, Hellen CU. Functions of eukaryotic factors in initiation of translation. Cold Spring Harb. Symp. Quant. Biol. 2001;66:389–396. doi: 10.1101/sqb.2001.66.389. [DOI] [PubMed] [Google Scholar]
- 253.Nagai K, Oubridge C, Kuglstatter A, Menichelli E, Isel C, Jovine L. Structure, function and evolution of the signal recognition particle. EMBO J. 2003;22:3479–3485. doi: 10.1093/emboj/cdg337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Xia C, Ma W, Stafford LJ, Liu C, Gong L, Martin JF, Liu M. GGAPs, a new family of bifunctional GTP-binding and GTPase-activating proteins. Mol. Cell. Biol. 2003;23:2476–2488. doi: 10.1128/MCB.23.7.2476-2488.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Nitta M, Saijo M, Kodo N, Matsuda T, Nakatsu Y, Tamai H, Tanaka K. A novel cytoplasmic GTPase XAB1 interacts with DNA repair protein XPA. Nucleic Acids Res. 2000;28:4212–4218. doi: 10.1093/nar/28.21.4212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Sekiguchi T, Hirose E, Nakashima N, Ii M, Nishimoto T. Novel G proteins, Rag C and Rag D, interact with GTP-binding proteins, Rag A and Rag B. J. Biol. Chem. 2001;276:7246–7257. doi: 10.1074/jbc.M004389200. [DOI] [PubMed] [Google Scholar]
- 257.Boehm U, Guethlein L, Klamp T, Ozbek K, Schaub A, Futterer A, Pfeffer K, Howard JC. Two families of GTPases dominate the complex cellular response to IFN-gamma. J. Immunol. 1998;161:6715–6723. [PubMed] [Google Scholar]
- 258.MacMicking JD, Taylor GA, McKinney JD. Immune control of tuberculosis by IFN-gamma-inducible LRG-47. Science. 2003;302:654–659. doi: 10.1126/science.1088063. [DOI] [PubMed] [Google Scholar]
- 259.Prakash B, Renault L, Praefcke GJ, Herrmann C, Wittinghofer A. Triphosphate structure of guanylate-binding protein 1 and implications for nucleotide binding and GTPase mechanism. EMBO J. 2000;19:4555–4564. doi: 10.1093/emboj/19.17.4555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Zhao X, Alvarado D, Rainier S, Lemons R, Hedera P, Weber CH, Tukel T, Apak M, Heiman-Patterson T, Ming L, Bui M, Fink JK. Mutations in a newly identified GTPase gene cause autosomal dominant hereditary spastic paraplegia. Nat. Genet. 2001;29:326–331. doi: 10.1038/ng758. [DOI] [PubMed] [Google Scholar]
- 261.Klamp T, Boehm U, Schenk D, Pfeffer K, Howard JC. A giant GTPase, very large inducible GTPase-1, is inducible by IFNs. J. Immunol. 2003;171:1255–1265. doi: 10.4049/jimmunol.171.3.1255. [DOI] [PubMed] [Google Scholar]
- 262.Kudo H, Senju S, Mitsuya H, Nishimura Y. Mouse and human GTPBP2, newly identified members of the GP-1 family of GTPase. Biochem. Biophys. Res. Commun. 2000;272:456–465. doi: 10.1006/bbrc.2000.2763. [DOI] [PubMed] [Google Scholar]
- 263.Senju S, Nishimura Y. Identification of human and mouse GP-1, a putative member of a novel G-protein family. Biochem. Biophys. Res. Commun. 1997;231:360–364. doi: 10.1006/bbrc.1997.6103. [DOI] [PubMed] [Google Scholar]
- 264.Jensen BC, Wang Q, Kifer CT, Parsons M. The NOG1 GTP-binding protein is required for biogenesis of the 60 S ribosomal subunit. J. Biol. Chem. 2003;278:32204–32211. doi: 10.1074/jbc.M304198200. [DOI] [PubMed] [Google Scholar]
- 265.Britton RA, Chen SM, Wallis D, Koeuth T, Powell BS, Shaffer LG, Largaespada D, Jenkins NA, Copeland NG, Court DL, Lupski JR. Isolation and preliminary characterization of the human and mouse homologues of the bacterial cell cycle gene era. Genomics. 2000;67:78–82. doi: 10.1006/geno.2000.6243. [DOI] [PubMed] [Google Scholar]
- 266.Akiyama T, Gohda J, Shibata S, Nomura Y, Azuma S, Ohmori Y, Sugano S, Arai H, Yamamoto T, Inoue J. Mammalian homologue of E. coli Ras-like GTPase (ERA) is a possible apoptosis regulator with RNA binding activity. Genes Cells. 2001;6:987–1001. doi: 10.1046/j.1365-2443.2001.00480.x. [DOI] [PubMed] [Google Scholar]
- 267.Li B, Trueb B. DRG represents a family of two closely related GTP-binding proteins. Biochim. Biophys. Acta. 2000;1491:196–204. doi: 10.1016/s0167-4781(00)00025-7. [DOI] [PubMed] [Google Scholar]
- 268.Kumar S, Tomooka Y, Noda M. Identification of a set of genes with developmentally down-regulated expression in the mouse brain. Biochem. Biophys. Res. Commun. 1992;185:1155–1161. doi: 10.1016/0006-291x(92)91747-e. [DOI] [PubMed] [Google Scholar]
- 269.Ishikawa K, Azuma S, Ikawa S, Morishita Y, Gohda J, Akiyama T, Semba K, Inoue J. Cloning and characterization of Xenopus laevis drg2, a member of the developmentally regulated GTP-binding protein subfamily. Gene. 2003;322:105–112. doi: 10.1016/j.gene.2003.08.016. [DOI] [PubMed] [Google Scholar]
- 270.Harton JA, Cressman DE, Chin KC, Der CJ, Ting JP. GTP binding by class II transactivator: Role in nuclear import. Science. 1999;285:1402–1405. doi: 10.1126/science.285.5432.1402. [DOI] [PubMed] [Google Scholar]
- 271.Raval A, Weissman JD, Howcroft TK, Singer DS. The GTP-binding domain of class II transactivator regulates its nuclear export. J. Immunol. 2003;170:922–930. doi: 10.4049/jimmunol.170.2.922. [DOI] [PubMed] [Google Scholar]
- 272.Koonin EV, Aravind L. The NACHT family - a new group of predicted NTPases implicated in apoptosis and MHC transcription activation. Trends Biochem. Sci. 2000;25:223–224. doi: 10.1016/s0968-0004(00)01577-2. [DOI] [PubMed] [Google Scholar]
- 273.Nogales E, Downing KH, Amos LA, Lowe J. Tubulin and FtsZ form a distinct family of GTPases. Nat. Struct. Biol. 1998;5:451–458. doi: 10.1038/nsb0698-451. [DOI] [PubMed] [Google Scholar]
- 274.Vernet C, Ribouchon MT, Chimini G, Pontarotti P. Structure and evolution of a member of a new subfamily of GTP-binding proteins mapping to the human MHC class I region. Mamm. Genome. 1994;5:100–105. doi: 10.1007/BF00292335. [DOI] [PubMed] [Google Scholar]
- 275.Burbelo PD, Miyamoto S, Utani A, Brill S, Yamada KM, Hall A, Yamada Y. p190-B, a new member of the Rho GAP family, and Rho are induced to cluster after integrin cross-linking. J. Biol. Chem. 1995;270:30919–30926. doi: 10.1074/jbc.270.52.30919. [DOI] [PubMed] [Google Scholar]
- 276.Deiss LP, Feinstein E, Berissi H, Cohen O, Kimchi A. Identification of a novel serine/threonine kinase and a novel 15-kD protein as potential mediators of the gamma interferon-induced cell death. Genes Dev. 1995;9:15–30. doi: 10.1101/gad.9.1.15. [DOI] [PubMed] [Google Scholar]
- 277.Inbal B, Bialik S, Sabanay I, Shani G, Kimchi A. DAP kinase and DRP-1 mediate membrane blebbing and the formation of autophagic vesicles during programmed cell death. J. Cell Biol. 2002;157:455–468. doi: 10.1083/jcb.200109094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Harris A, Morgan JI, Pecot M, Soumare A, Osborne A, Soares HD. Regenerating motor neurons express Nna1, a novel ATP/GTP-binding protein related to zinc carboxypeptidases. Mol. Cell. Neurosci. 2000;16:578–596. doi: 10.1006/mcne.2000.0900. [DOI] [PubMed] [Google Scholar]
- 279.Chennathukuzhi VM, Kurihara Y, Bray JD, Yang J, Hecht NB. Altering the GTP binding site of the DNA/RNA-binding protein, Translin/TB-RBP, decreases RNA binding and may create a dominant negative phenotype. Nucleic Acids Res. 2001;29:4433–4440. doi: 10.1093/nar/29.21.4433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Bandorowicz-Pikula J, Kirilenko A, van Deursen R, Golczak M, Kuhnel M, Lancelin JM, Pikula S, Buchet R. A putative consensus sequence for the nucleotide-binding site of annexin A6. Biochemistry. 2003;42:9137–9146. doi: 10.1021/bi034359m. [DOI] [PubMed] [Google Scholar]
- 281.Yaguchi H, Ohkura N, Tsukada T, Yamaguchi K. Menin, the multiple endocrine neoplasia type 1 gene product, exhibits GTP-hydrolyzing activity in the presence of the tumor metastasis suppressor nm23. J. Biol. Chem. 2002;277:38197–38204. doi: 10.1074/jbc.M204132200. [DOI] [PubMed] [Google Scholar]
- 282.I thank R. Lovering and other members of the HUGO Nomenclature Committee (H. Wain, E. Bruford, M. Lush, V. Khodiyar, C. Talbot, M. Wright, and S. Povey) for extensive assistance in determining gene symbols; A. Bernards for help in identifying some GTPase sequences; and C. Der, F. Taminoi, A. van der Bliek, and J. Lengyel for insightful comments and criticisms.
Table 1.
Protein nomenclature. Human Genome Nomenclature Committee (HGNC) symbols for genes discussed in this review are shown (left) with common names and aliases (right).
| HGNC name Common names and aliases | |
| AGTPBP1 | ATP/GTP-binding protein 1, Nna1 |
| AKT1 | Akt, PKB |
| ANXA6 | annexin 6 |
| APPL1 | DIP13α, adaptor protein with PH, PTB and Leucine zipper domains |
| APPL2 | DIP13β |
| ARAF | A-Raf |
| ARFIP | arfaptin |
| ARHGAP21 | ArhGAP10 |
| ARHI | Noey2, Arhi |
| BIRC1 | baculovirus IAP repeat containing 1, NAIP |
| BRAF | B-Raf |
| BRAP | BRap2, RNF52, IMP |
| CARD | caspase recruitment domain family protein |
| CDC42 | cell division cycle 42-like, Cdc42 |
| CENTG1 | GGAP2 |
| CENTG2 | GGAP1 |
| CENTG3 | MRIP1 |
| CHN1 | RhoGAP2 |
| CIAS1 | cold autoinflamitory syndrome 1, NALP3 |
| DAPK1 | death-associated protein kinase, DAPK |
| DIAPH | diaphanous, mDia |
| DIRAS1 | Di-Ras1, Rig |
| DIRAS2 | Di-Ras2 |
| DNM1-3 | dynamin 1-3, Dnm1-3 |
| DNM1L | dynamin 1-like, Dnm1L |
| DRG1 and 2 | developmentally regulated GTP-binding protein |
| EEA1 | early endosome antigen |
| EEF1A1 | eukaryotic translation elongation factor 1 α1, eEF1A, eEF1α |
| EEF1A2 | eukaryotic translation elongation factor 1 α2 |
| EGFR | epidermal growth factor receptor, EGF-R |
| EIF2S3 | eukaryotic translation initiation factor 2 subunit 3, Eif2 γ |
| EIF4EBP1 | eukaryotic translation inititation factor 4E binding protein 1 |
| ERAL1 | Era G protein-like 1, Hera-B |
| ERAS | ERas, Eras |
| ERBB2 | HER2, neu |
| ERK | mitogen activated protein kinase, MAPK, Erk1, 2 |
| EXO8 | Exo84 |
| FRAP | mTOR, mammalian target of rapamycin |
| GBP1-5 | guanylate-binding protein 1-5 |
| GEM | Kir |
| GGA | Golgi-localized, γ-adaptin-ear-containing, Arf-binding |
| GMIP | GEM interacting protein, Gmip |
| GNA11 | Gα11 |
| GNA12 | Gα12 |
| GNA13 | Gα13 |
| GNA14 | Gα14 |
| GNA15 | Gα15, Gα16 |
| GNAI1 | Gαi-1 |
| GNAI2 | Gαi-2 |
| GNAI3 | Gαi-3 |
| GNAL | Gαl (olfactory) |
| GNAO | Gαo |
| GNAQ | Gαq |
| GNAS | Gαs |
| GNAT1 | transducin 1 |
| GNAT2 | transducin 2 |
| GNAT3 | transducin 3, gustducin |
| GNAZ | Gαz |
| GNL1 | guanine nucleotide binding protein-like 1, HSR1 |
| GRLF1 | p190A RhoGAP |
| GTBP1-5 | GTP binding protein |
| HRAS | H-Ras, c-Ha-ras |
| IFNG | IFN-γ |
| IIGP5 | interferon-inducible GTPase 5 |
| KIF9 | kinesin family member 9 |
| KRAS2B | K-Ras, c-Ki-ras |
| M6PRBP1 | TIP47 |
| MEK | mitogen activated protein kinase kinase, Mek1, 2 |
| MEN1 | multiple endocrine neoplasia 1, menin |
| MFN1 | mitofusin 1, Mfn1 |
| MFN2 | mitofusin 2, Mfn2 |
| MHC2TA | major histocompatibility complex class II transactivator, CIITA |
| MLLT4 | mixed-lineage leukemia translocated to 4, AF6 |
| MLPH | melanophilin |
| MX1 | Myxovirus resistance 1 |
| MX2 | Myxovirus resistance 2 |
| NALP | NACHT, leucine rich repeat and PYD contianing |
| NF1 | neurofibromin 1 |
| NFKB | nuclear factor of κ gene enhancer in B cells, NF-κB |
| NFKBI | nuclear factor of κ gene enhancer in B cells inhibitor, IκB |
| NGB | neuroglobin, Nog1 |
| NKIRAS1 | NF-κB inhibitor interacting Ras 1, κB-ras1 |
| NKIRAS2 | NF-κB inhibitor interacting Ras 2, κB-ras2 |
| NORE1 | Rassf5, RapL |
| NRAS | N-Ras |
| OPA1 | optic atrophy 1, Opa1 |
| PAK1-6 | Pak, p21-accociated kinase |
| PARD3 | par-3, partitioning defective 3 homolog |
| PARD6 | Par-6, partitioning defective 6 homolog |
| PDPK1 | 3-phosphoinositide dependent protein kinase, PDK1 |
| PIK3C2A | phosphoinositide-3-kinase class 2 α |
| PIK3C2B | phosphoinositide-3-kinase class 2 β |
| PIK3C2G | phosphoinositide-3-kinase class 2 γ |
| PIK3CA | phosphoinositide-3-kinase p110α |
| PIK3CB | phosphoinositide-3-kinase p110β |
| PIK3CD | phosphoinositide-3-kinase p110δ |
| PIK3CG | phosphoinositide-3-kinase p110γ |
| PKN1-3 | protein kinase N, PRK, DBK |
| PLCB | PLCβ |
| PLCE1 | PLCε |
| RAB10 | Rab10 |
| RAB11A | Rab11A, Rab11 |
| RAB11B | RAB11B |
| RAB11FIP1 | arfophilin 1, RCP |
| RAB12 | Rab12 |
| RAB13 | Rab13 |
| RAB14 | Rab14 |
| RAB15 | Rab15 |
| RAB17 | Rab17 |
| RAB18 | Rab18 |
| RAB19 | Rab19 |
| RAB1A | Rab1A, Rab1 |
| RAB1B | Rab1B |
| RAB2 | Rab2 |
| RAB20 | Rab20 |
| RAB21 | Rab21 |
| RAB22A | Rab22A, Rab22 |
| RAB23 | Rab23 |
| RAB24 | Rab24 |
| RAB25 | Rab25 |
| RAB26 | Rab26 |
| RAB27A | Rab27A, Rab27, Ram |
| RAB27B | Rab27B |
| RAB28 | Rab28 |
| RAB29 | Rab29 |
| RAB2B | Rab2b |
| RAB30 | Rab30 |
| RAB31 | Rab31, Rab22B |
| RAB32 | Rab32 |
| RAB33A | Rab33A, RabS10 |
| RAB33B | Rab33B |
| RAB34 | Rab34 |
| RAB35 | Rab35 |
| RAB36 | Rab36 |
| RAB37 | Rab37 |
| RAB38 | Rab38 |
| RAB39 | Rab39 |
| RAB39B | Rab39B |
| RAB3A | Rab3A |
| RAB3B | Rab3B |
| RAB3C | Rab3C |
| RAB3D | Rab3D, Rab16 |
| RAB40A | Rab40A, Rar2 |
| RAB40C | Rab40C, RasL8C, RarL |
| RAB40C | Rab40C, RarL |
| RAB41 | Rab41 |
| RAB42 | Rab42 |
| RAB43 | Rab43, Rab41, Rab11B |
| RAB44 | Rab44, RASD3, RASL13 |
| RAB4A | Rab4A, Rab4 |
| RAB4B | Rab4B |
| RAB5A | Rab5A, Rab5 |
| RAB5B | Rab5B |
| RAB5C | Rab5C, RabL |
| RAB6A | Rab6A, Rab6 |
| RAB6B | Rab6B |
| RAB6C | Rab6C |
| RAB7 | Rab7 |
| RAB7B | Rab7B |
| RAB7L1 | Rab7L1, Rab7L |
| RAB8A | Rab8A, Mel |
| RAB8B | Rab8B |
| RAB9A | Rab9A, Rab9 |
| RAB9B | Rab9B, Rab9L |
| RABEP1 | rabaptin-5, neurocrescin |
| RABEP1, 2 | rabaptin 1, 2 |
| RABL2A | RabL2A |
| RABL2B | RabL2B |
| RABL3 | RabL3 |
| RABL4 | RabL4, RayL |
| RABL5 | RabL5 |
| RAC1 | Ras-related C3 botulinum toxin substrate 1, Rac1, TC25 |
| RAC2 | Rac2 |
| RAC3 | Rac3 |
| RAC4 | Rac4 |
| RAF1 | c-Raf, Raf1 |
| RALA | RalA |
| RALB | RalB |
| RALBP1 | RalBP1, RLIP |
| RALGDS | ral guanine nucleotide dissociation stimulator, RalGDS, RalGEF |
| RAN | Ran |
| RAP1A | Rap1-A |
| RAP1B | Rap1-B |
| RAP2A | Rap2-A |
| RAP2B | Rap2-B |
| RAP2C | Rap2-C |
| RASD1 | DexRas |
| RASD2 | Rhes |
| RASEF | RasEF, Rab45 |
| RASIP1 | Ras interacting protein 1, RAIN |
| RASL10A | RasL10A |
| RASL10B | RasL10B |
| RASL11A | RasL11A |
| RASL11B | RasL11B |
| RASL12 | RasL12, RIS |
| RASSF1 | Rassf1 |
| RASSF2 | Rassf2 |
| RASSF3 | Rassf3 |
| RASSF4 | Rassf4 |
| RASSF6 | Rassf6 |
| REM1 | Rem1, rem, ges |
| REM2 | Rem2 |
| RERG | Rerg, rerg |
| RGL1 | ral guanine nucleotide dissociation stimulator-like 1, Rgl |
| RGL2 | ral guanine nucleotide dissociation stimulator-like 1, Rgl2 |
| RGL3 | Rgl3 |
| RHEB | Rheb1 |
| RHEBL1 | Rheb2 |
| RHOA | RhoA |
| RHOB | RhoB |
| RHOBTB1 | RhoBTB1 |
| RHOBTB2 | RhoBTB2, DBC2 |
| RHOC | RhoC |
| RHOD | RhoD |
| RHOF | Rif |
| RHOG | RhoG |
| RHOH | RhoH |
| RHOJ | RhoJ, Tcl |
| RHOQ | RhoQ, Tc10 |
| RHOT1 | Miro1 |
| RHOT2 | Miro2 |
| RHOU | RhoU, wrch-1 |
| RHOV | RhoV, wrch-2, chp |
| RILP | Rab7-interacting lysosomal protein |
| RIN1 | Ras and Rab interacting 1, Rin1 |
| RIN2 | Ras and Rab interacting 2, Rin2 |
| RIN3 | Ras and Rab interacting 3, Rin3 |
| RIT1 | rit |
| RIT2 | rin |
| RND1 | Rho6 |
| RND2 | RhoN, Rho7, ARHE |
| RND3 | RhoE, Rho8, ARHN |
| ROCK1 | Rock1, Rho-associated protein kinase 1 |
| ROCK2 | Rock2, Rho-associated protein kinase 2 |
| RPH11 | rabphilin 11 |
| RPH3A | rabphilin 3A |
| RPS6K | ribosomal protein S6 kinase |
| RRAD | Rad |
| RRAGA | Rag A, Gtr1 |
| RRAGB | Rag B |
| RRAGC | Rag C, GTR2 |
| RRAGD | Rag-D |
| RRAS1 | R-Ras |
| RRAS2 | TC21 |
| RRAS3 | M-Ras |
| SARA1 | Sar1 |
| SARA2 | Sar2 |
| SEC10L1 | Sec10 |
| SEC5L1 | Sec5 |
| SEPT1 | PNUTL3 |
| SEPT10 | Sept10 |
| SEPT11 | Sept11 |
| SEPT2 | NEDD5, DIFF6 |
| SEPT3 | Sept3 |
| SEPT4 | Sept4, PNULT2, CDCREL-2, ARTS |
| SEPT5 | Sept5, PNUTL1, CDCREL-1 |
| SEPT6 | SEP2, Sept6 |
| SEPT7 | Sept7, CDC10 |
| SEPT8 | Sept8 |
| SEPT9 | Sept9, MSF, PNUTL4 |
| SOS1 | son of sevenless, mSOS |
| SPG3A | spastic paraplegia, atlastin |
| SRP54 | Srp54 |
| SRPRA | SRPR, SrpRa, Signal recognition peptide receptor subunit α |
| SRPRB | SrpRb, Signal recognition peptide receptor subunit β |
| STK4 | serine/threonine kianse 4, MST1 |
| TIAM1 | T cell lymphoma invasion and metastsis, Tiam1 |
| TOCA1 | transducer of Cdc42-dependent actin assembly |
| TSC1 | tuberous sclerosis 1, hamartin |
| TSC2 | tuberous sclerosis 2, uberin |
| TSN | translin, TRSLN, TB-RBP |
| TUBB | β-tubulin |
| TUFM | EFTu |
| VLIG1 | very large inducible GTPase 1 |
| WAS | Wiskott-Aldrich syndrome, WASP |
| XAB1 | MBDin |
| ZNF179 | BFP |