SUMMARY
Current models imply that the FERM domain protein Merlin, encoded by the tumor suppressor NF2, inhibits mitogenic signaling at or near the plasma membrane. Here, we show that the closed, growth inhibitory form of Merlin accumulates in the nucleus, binds to the E3 ubiquitin ligase CRL4DCAF1, and suppresses its activity. Depletion of DCAF1 blocks the promitogenic effect of inactivation of Merlin. Conversely, enforced expression of a Merlin-insensitive mutant of DCAF1 counteracts the antimitogenic effect of Merlin. Re-expression of Merlin and silencing of DCAF1 induce a similar, tumor-suppressive program of gene expression. Tumor-derived mutations invariably disrupt Merlin’s ability to interact with or inhibit CRL4DCAF1. Finally, depletion of DCAF1 inhibits the hyperproliferation of Schwannoma cells from NF2 patients and suppresses the oncogenic potential of Merlin-deficient tumor cell lines. We propose that Merlin suppresses tumorigenesis by translocating to the nucleus to inhibit CRL4DCAF1
INTRODUCTION
The tumor suppressor NF2, which is inactivated in the familial cancer syndrome Neurofibromatosis type 2, encodes for Merlin, a member of the Ezrin/Radixin/Moesin (ERM) family of proteins. The prevalence of NF2 mutations in sporadic tumors, especially Schwannomas, meningiomas, and malignant mesotheliomas, and the predisposition of heterozygous Nf2 mutant mice to develop multiple malignant tumor types suggest that Merlin has a relatively broad tumor suppressor function (McClatchey and Giovannini, 2005; Okada et al., 2007). Despite extensive research on Merlin’s role as a tumor suppressor, its biological function remains poorly understood.
Classical ERM proteins, which consist of an N-terminal FERM domain, a coiled-coil domain, and a C-terminal segment containing an actin-binding motif, switch between a closed conformation, presumed dormant, and an open conformation, which mediates linkage of certain cell adhesion receptors to the actin cytoskeleton (Bretscher et al., 2002). The sequence homology of Merlin to ERM proteins and its apparently prevalent localization to the cortical cytoskeleton have led to the hypothesis that Merlin functions at or near the plasma membrane to inhibit the transmission of promitogenic signals (McClatchey and Fehon, 2009). However, Merlin does not contain a canonical actin-binding motif. In addition, it is the closed form of Merlin that suppresses tumorigenesis and is thereby considered “active”. In fact, many tumor-derived missense mutations are predicted to disrupt the closed conformation (Okada et al., 2007).
The interconversion between the closed and the open form of Merlin is promoted by the p21-activated kinase PAK, which phosphorylates Merlin at S518, disrupting the intramolecular C-terminal tail-FERM interaction, which maintains the protein in the closed conformation (Kissil et al., 2002; Xiao et al., 2002). Cadherin-mediated adhesion inhibits PAK, causing an accumulation of the de-phosphorylated, growth inhibitory form of Merlin. Conversely, integrin-dependent adhesion to the matrix activates PAK, causing inactivation of Merlin and, thereby, presumably removing a block to cell cycle progression (Okada et al., 2005). Accordingly, inactivation of Merlin induces exit from contact inhibition (Lallemand et al., 2003; Lallemand et al., 2009; Okada et al., 2005) and accelerates progression through the G1 phase of the cell cycle (Lopez-Lago et al., 2009). These results suggest that Merlin integrates antithetic signals from cadherins and integrins to regulate cell cycle progression.
Merlin exerts inhibitory effects on multiple mitogenic signaling pathways. The closed form of Merlin suppresses the recruitment of Rac to the plasma membrane and the activation of PAK (Kaempchen et al., 2003; Kissil et al., 2003; Okada et al., 2005) and inhibits the activation of mTORC1 independently of Akt (James et al., 2009; Lopez-Lago et al., 2009). Furthermore, Merlin inhibits the Ras-ERK and PI-3K-Akt pathways and Focal Adhesion Kinase (FAK)-Src signaling (Ammoun et al., 2008; Jin et al., 2006a; Poulikakos et al., 2006; Rong et al., 2004). Finally, Merlin cooperates with the related ERM protein Expanded to activate the Hippo tumor suppressor pathway in drosophila (Cho et al., 2006; Hamaratoglu et al., 2006). However, the contribution of each of the identified pathways to NF2-dependent tumorigenesis has not been defined.
Although yeast two hybrid screens and other approaches have led to the identification of several Merlin-binding proteins, the biochemical function of Merlin and, hence, the mechanism through which it suppresses tumorigenesis have remained elusive. This study shows that the closed form of Merlin enters the nucleus, binds to the E3 ubiquitin ligase CRL4DCAF1, and inhibits its activity. Genetic evidence indicates that this inhibitory interaction is essential for tumor suppression.
RESULTS
The Closed Form of Merlin Binds to DCAF1, the Substrate Receptor of the E3 Ubiquitin Ligase CRL4DCAF1
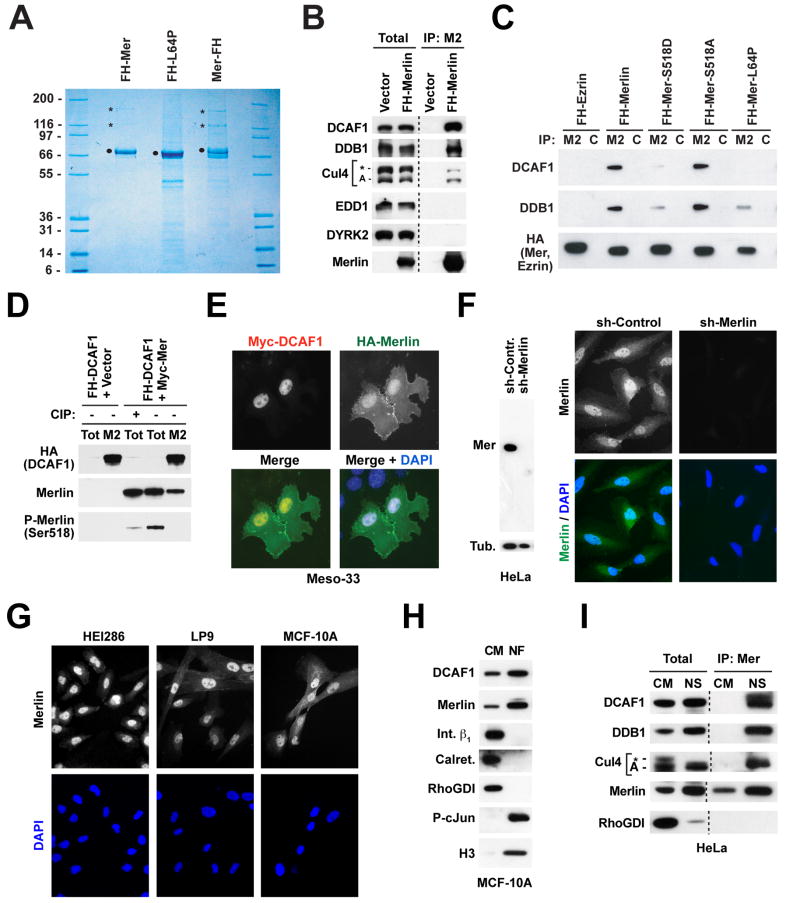
Tandem Affinity Purification (TAP) revealed that Merlin, tagged at either its N- or its C-terminus, associates with proteins displaying an apparent molecular weight of 169 kD and 127 kD (Figure 1A). The two proteins were recovered in similar amounts, each at an apparent ratio of ~ 1:5 to Merlin. Importantly, the tumor-derived mutant L64P, which lacks tumor suppressor activity (Gutmann et al., 2001), did not associate with these proteins (Figure 1A). MALDI-reTOF-MS followed by peptide mass fingerprinting and mass spectrometric sequencing unequivocally identified the 127kD protein as DDB1 (DNA Damage Binding protein 1) and the 169 kD protein as DCAF1 (DDB1-and Cul4-Associated Factor 1) (Figure 1A). NanoLC-MS/MS indicated that wild type Merlin, but not Merlin-L64P, interacts also with Cul4B, albeit at a lower apparent stoichiometry (Table S1).
Figure 1. Merlin Associates with CRL4DCAF1 in the Nucleus.
(A) Cos7 cells were transfected with the indicated FH-tagged (FH; FLAG-HA) forms of Merlin, lysed in RIPA, and subjected to TAP. Proteins were separated by SDS-PAGE and stained with Colloidal Blue. Dots point to the FH-tagged baits and asterisks to the 169 and 127 kD bands identified by mass spectrometry as DCAF1 and DDB1, respectively.
(B) 293T cells were transfected with FH-DCAF1 or empty vector. Total lysates and Flag-immunoprecipitates (M2) were subjected to immunoblotting with antibodies to the indicated proteins.
(C) As in (B), with transfection of the indicated constructs and immunoprecipitation with anti-FLAG (M2) or control (C) antibodies.
(D) As in (B), with cotransfection of Myc-Merlin or empty vector and immunoprecipitation with anti-FLAG (M2) or control (C). To control for the specificity of the anti-P-Merlin antibodies, the indicated sample was treated with calf intestinal alkaline phosphatase (CIP) prior to SDS-PAGE. Endogenous Merlin was detected upon longer exposure.
(E) Meso-33 cells were transiently transfected with the indicated constructs and subjected to double immunofluorescent staining with anti-HA and anti-Myc followed by DAPI.
(F) HeLa cells were infected with lentiviral vectors encoding a sh-RNA targeting Merlin or a control sh-RNA and subjected to immunoblotting (left) or staining with affinity-purified antibodies to the N-terminal segment of Merlin followed by DAPI (right).
(G) The indicated cells were stained as in panel F.
(H) MCF-10A cells were subjected to subcellular fractionation. Equal amounts of proteins from the nuclear (NF) and non-nuclear (CM; cytosol + crude membranes) fraction were subjected to immunoblotting with antibodies to the indicated proteins.
(I) Equal amounts of proteins from the non-nuclear (CM) and nuclear-soluble (NS) fraction of HeLa cells and Merlin-immunoprecipitates were immunoblotted as indicated. Anti-Cul4 recognizes Cul4-A as well as a higher molecular weight band, possibly corresponding to neddylated Cul4-A or Cul4-B (asterisk).
See also Supplemental Figure S1.
Prior studies have established that DDB1 functions as a SKP1-like adaptor to recruit WD40 domain-containing substrate receptors, such as DDB2, CSA (the Cockayne syndrome protein A), and Det1-COP1 to the E3 ligase Cul4-Roc1 (Lee and Zhou, 2007; O’Connell and Harper, 2007). DCAF1, which was originally identified as the cellular receptor for the HIV protein Vpr and designated VprBP, is one of several additional WD40 domain substrate receptors able to combine with DDB1 to mediate assembly of distinct Cul4 (CRL4) ligases (Lee and Zhou, 2007). In agreement with the mass spectrometric analysis, immunoblotting indicated that FH-Merlin combines with DCAF1, DDB1, and Cul4, but not DYRK2 or EDD (Figure 1B), components of the alternative EDD-DYRK2-DDB1-VprBP E3 ligase complex (Maddika and Chen, 2009). These observations indicate that Merlin interacts selectively with the E3 ligase CRL4DCAF1.
Since Merlin formed a stoichiometric complex with DCAF1 but not DDB1 upon cotransfection in Cos7 cells (Figure S1A), we hypothesized that Merlin binds directly to DCAF1 but indirectly to DDB1. To test this hypothesis, we examined the ability of GST-fusion proteins comprising either the FERM or the helical and C-terminal domain of Merlin to bind to in vitro translated DCAF1 or DDB1. As shown in Figure S1B, the FERM domain of Merlin bound directly to DCAF1 but not DDB1, whereas the helical and C-terminal segments did not (Figure S1B). Thus, Merlin binds directly, through its FERM domain, to DCAF1.
To examine if the conformational transition between the open and the closed form regulates Merlin’s binding to CRL4DCAF1, we tested the behavior of two well-characterized phosphorylation mutants. Merlin-S518A, which cannot be phosphorylated at S518 and is stabilized in the closed conformation, associated efficiently with DCAF1 and DDB1 in transfected 293T cells. In contrast, the phosphorylation site mimetic mutant Merlin-S518D, which is thought to maintain an open conformation, displayed dramatically diminished binding to DCAF1 and DDB1 (Figure 1C). As anticipated, Ezrin and Merlin-L64P exhibited essentially no binding in this assay. In agreement with the model that the closed form of Merlin selectively interacts with CRL4DCAF1, immunoblotting with anti-P-Merlin antibodies indicated that the fraction of recombinant Merlin bound to FH-DCAF1 is not detectably phosphorylated at S518 (Figure 1D). Thus, the unphosphorylated, closed form of Merlin interacts specifically with CRL4DCAF1, whereas the open form and a well-characterized tumor-derived mutant do not, raising the possibility that Merlin suppresses cell proliferation and tumorigenesis by binding to CRL4DCAF1.
Merlin Associates with CRL4DCAF1 in the Nucleus
To identify the cellular compartment where Merlin interacts with CRL4DCAF1, we co-transfected Myc-tagged DCAF1 and HA-tagged Merlin into Merlin-deficient Meso-33 mesothelioma cells. These cells undergo growth arrest following re-expression of Merlin and therefore represent an excellent model to study the tumor suppressor function of Merlin (see Figure 5D below). Immunofluorescent staining indicated that Myc-DCAF1 localizes in the nucleus in Meso-33 cells (Figure 1E), suggesting that CRL4DCAF1 functions in this compartment like other CRL4 ligases (Lee and Zhou, 2007; O’Connell and Harper, 2007). Interestingly, in addition to localizing underneath the plasma membrane, HA-Merlin also accumulated in the nucleus (Figure 1E). Subcellular fractionation confirmed these results (Figure S1C).
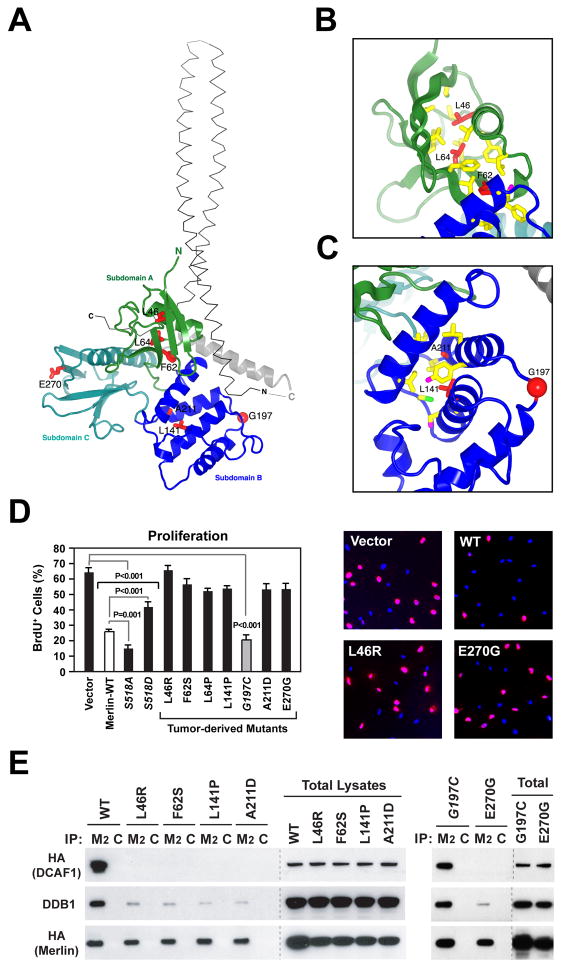
Figure 5. Several Pathogenic Missense Mutations Disrupt Binding of Merlin to CRL4DCAF1 In Vivo.
(A) Tumor derived mutants (red sticks) are shown in the context of the overall structure of the FERM domain of human Merlin (colored ribbons; PDB 1ISN). The α-helical coiled-coil domain from the crystal structure of the closed form of Spodoptera frugiperda Moesin (thin grey αcarbon trace; PDB 2I1K), which is highly homologous to human Merlin, is shown for reference. The coiled-coil domain was positioned by superimposing the FERM domains from the structures of Merlin and Moesin, which have a root mean square deviation for α-carbon atoms of 1.3 Å and share 62% sequence identity.
(B) Close-up view of tumor-derived mutations (red sticks) located in subdomain A (green ribbon) in the crystal structure of human Merlin. Residues in contact with mutated residues are drawn as sticks and colored according to atom type (carbon, yellow; sulfur, green).
(C) Subdomain B represented as in (B), with oxygens as magenta sticks. Red sphere, G197.
(D) Meso-33 cells were transiently transfected with empty vector or vectors encoding wild type Merlin or indicated mutants. Asynchronized cells growing in 10% FCS were subjected to BrdU incorporation assay. The graph indicates the percentage (± SEM) of BrdU-positive cells. Pictures show representative fields of cells transfected with indicated vectors and stained with anti-BrdU (red) and DAPI (blue).
(E) Cos7 cells were transfected with FH-tagged versions of wild type Merlin or indicated mutants. Flag- or control-immunoprecipitates (M2 and C, respectively) and total lysates were immunoblotted as indicated.
See also Supplemental Figure S5.
We reasoned that poor permeabilization of the nuclear membrane or epitope masking may have hampered detection of Merlin in the nucleus in previous immunofluorescence studies (with the exception of Muranen et al., 2005). To study the localization of endogenous Merlin, we designed a fixation and permeabilization protocol able to circumvent these potential problems (see Experimental Procedures). Using this protocol, we found that a monospecific antibody to the N-terminal segment of Merlin generates prominent nuclear staining in multiple cell types, including HeLa squamous carcinoma cells and non-neoplastic HEI286 Schwann cells, LP9 mesothelial cells, and MCF-10A mammary epithelial cells (Figure 1F and 1G). This pattern of staining was specific because it was suppressed by silencing of Merlin (Figure 1F) and could be reproduced by using an antibody to a C-terminal epitope (Figure S1D and E). Finally, biochemical fractionation confirmed that Merlin and DCAF1 accumulate in the nucleus in multiple cell types (Figures 1H and S1F). Thus, Merlin can enter the nucleus and bind to DCAF1.
Many CRL4 ligases associate with DNA or chromatin during the cell cycle or in response to DNA damage (O’Connell and Harper, 2007). However, subnuclear fractionation indicated that both DCAF1 and Merlin accumulate in the soluble nuclear fraction and do not move into the chromatin fraction during cell cycle progression or in response to UV-induced DNA damage (Li and Giancotti, unpublished), consistent with the hypothesis that CRL4DCAF1 performs a function distinct from that of other known CRL4 ligases. Interestingly, coimmunoprecipitation analysis showed that Merlin combines with DCAF1, DDB1, and Cul4 in the soluble nuclear fraction, but not in the non-nuclear fraction despite the presence of abundant amounts of these components (Figure 1I). This suggests that association with a cofactor, dissociation of an inhibitor, or a post-translational modification occurring in the nucleus promote the association of Merlin with CRL4DCAF1. Taken together, these results indicate that the closed, growth inhibitory form of Merlin binds to CRL4DCAF1 in the nucleus.
Merlin is not a Substrate of CRL4DCAF1
Constitutively activated Akt can promote phosphorylation and proteasome-mediated degradation of Merlin (Tang et al., 2007) and overexpression studies have suggested that CRL4DCAF1 mediates this process (Huang and Chen, 2008). However, experiments with Cycloheximide indicated that Merlin exhibits a half-life far exceeding 24 hours in several cell types (Figure S2A). In addition, treatment with the proteasome inhibitor MG132 did not stabilize recombinant Merlin in Cos7 cells, and Merlin-S518D and Merlin-L64P, which do not interact with CRL4DCAF1, did not display increased stability in this assay (Figure S2B). Finally, knock down of DCAF1 did not cause increased accumulation of Merlin in multiple cell types under standard culture conditions as well as in HeLa cells acutely exposed to mitogens (Figure S2C and S2D). In fact, depletion of DCAF1 decreased the levels of Merlin, especially shortly after mitogenic stimulation, suggesting that binding to DCAF1 stabilizes Merlin (Figure S2D). These results argue that Merlin is not targeted for degradation by CRL4DCAF1 and suggest that Merlin functions upstream, not downstream, of CRL4DCAF1.
Merlin Inhibits CRL4DCAF1
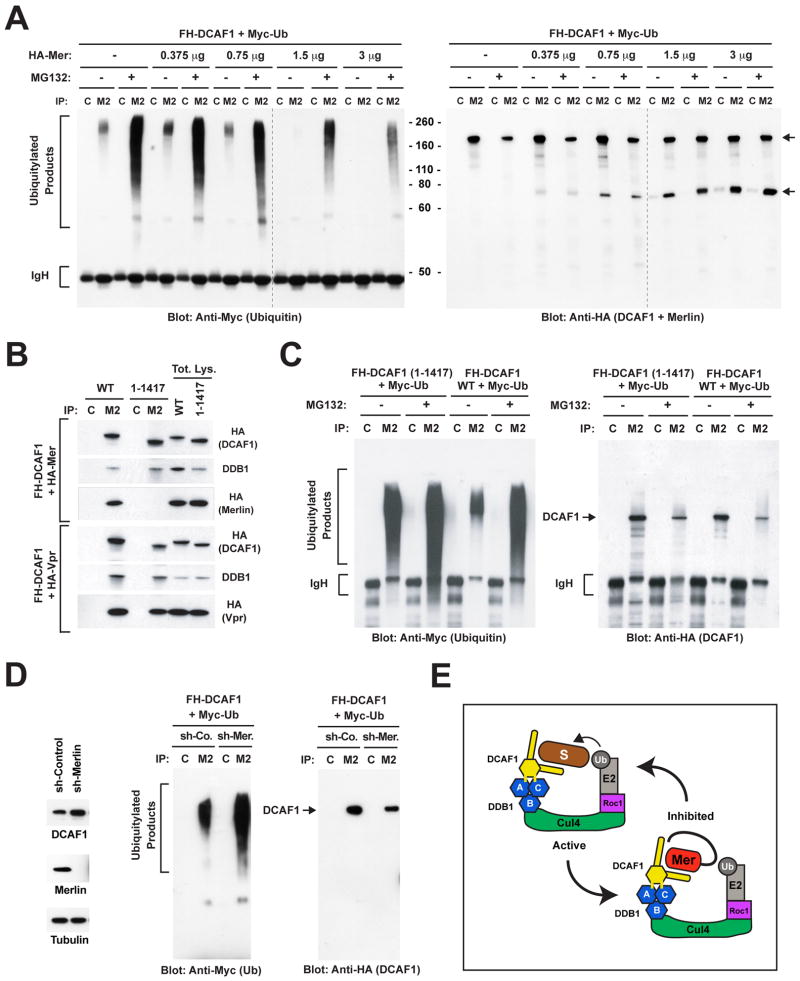
To examine the effect of Merlin on the ubiquitylating activity of CRL4DCAF1, we set up an in vivo ubiquitylation assay. Cos7 cells were transfected with FH-DCAF1 and Myc-ubiquitin, treated with MG132 to stabilize ubiquitylated products, and immunoprecipitated with anti-Flag to isolate CRL4DCAF1 and associated ubiquitylated products. Immunoblotting with anti-Myc antibody revealed that FH-DCAF1 isolated from MG132-treated cells coprecipitates with ubiquitylated species of apparent molecular weight ranging from 55 to 300 kD, suggesting that CRL4DCAF1 ubiquitylates in vivo one or more substrates and that these remain associated with the enzyme throughout immunoprecipitation (Figure 2A, left). In contrast, FH-DCAF1 isolated from untreated cells contained a modest amount of ubiquitylated species of apparent molecular weight ranging from 180 to 250 kD. Co-transfection of increasing concentrations of HA-Merlin caused a dose-dependent suppression of the accumulation of ubiquitylated products, suggesting that Merlin suppresses the ubiquitylating activity of CRL4DCAF1 (Figure 2A left). Immunoblotting with anti-HA antibody indicated that Merlin does not become ubiquitylated upon association with DCAF1, consistent with the notion that Merlin is a regulator and not a substrate of the ligase (Figure 2A right). Immunoblotting of total lysates provided a control for the expression levels of recombinant proteins and indicated that treatment with MG132 partially stabilizes DCAF1 (Figure S2E). These observations suggest that Merlin binds to CRL4DCAF1 and suppresses its ability to mediate ubiquitylation of target proteins.
Figure 2. Merlin Inhibits CRL4DCAF1-dependent Ubiquitylation.
(A) Cos7 cells were transfected with 3 μg of FH-DCAF1 and 2 μg of Myc-Ubiquitin in combination with the indicated amounts of HA-Merlin and treated with MG132 or left untreated. Flag- or control-immunoprecipitates (M2 and C, respectively) were subjected to immunoblotting with anti-Myc to visualize ubiquitylated proteins (left) or anti-HA to visualize DCAF1 and Merlin (right). Upper arrow points to DCAF1 and lower arrow to Merlin.
(B) Cos7 cells were transfected with FH-DCAF1 or FH-DCAF1 (1-1417) in combination with HA-Merlin (top panel) or HA-Vpr (bottom panel). Flag- or control-immunoprecipitates were subjected to immunoblotting. Levels of expression of DCAF1, DDB1 and Merlin or Vpr were verified by immunoblotting aliquots of total lysates.
(C) As in (B), with cotransfection of Myc-Ubiquitin and treatment or not with MG132. Flag- or control-immunoprecipitates were immunoblotted with anti-Myc (left) or anti-HA (right).
(D) Cos7 cells stably infected with lentiviral vectors encoding a control sh-RNA or a sh-RNA targeting Merlin were subjected to immunoblotting (left) or transfected with FH-DCAF1 and Myc-Ubiquitin followed by immunoblotting of Flag- or control-immunoprecipitates with anti-Myc (middle) or anti-HA (right).
(E) Hypothetical model of Merlin-mediated inhibition of CRL4DCAF1. Merlin is proposed to function as a competitive inhibitor of CRL4DCAF1. Blue hexagons, three β propeller folds of DDB1, which contact Cul4 (B) and DCAF1 (A and C). Yellow hexagon, WD40 domain of DCAF1; yellow sticks, double DxR box implicated in binding to DDB1; S, substrate; Ub, ubiquitin; Mer, closed Merlin.
See also Supplemental Figure S2.
To confirm this hypothesis, we examined if a mutant form of DCAF1 unable to bind to Merlin mediates enhanced CRL4DCAF1 ligase activity in cells expressing endogenous Merlin. Deletion mapping indicated that the GST-FERM domain of Merlin binds directly to a 90 amino acid C-terminal segment of DCAF1 in vitro (Figure S2F). Immunoprecipitation experiments confirmed that DCAF1 (1-1417) lacking this C-terminal segment combines with endogenous DDB1 but not with HA-Merlin in Cos7 cells (Figure 2B). Interestingly, this mutant retained the ability to interact with the HIV protein Vpr (Figure 2B), suggesting that it is not grossly misfolded and indicating that Merlin and Vpr bind to distinct sites on DCAF1. In addition, mutation of the double DxR box on the bottom surface of the WD40 propeller fold of DCAF1 (R1247/1283A) disrupted the association of DCAF1 with DDB1, as previously reported (Jin et al., 2006b), but not with Merlin (Figure S2G), indicating that distinct segments of DCAF1 mediate interaction with DDB1 and with Merlin. Notably, in vivo ubiquitylation assays revealed that ligase complexes containing DCAF1 (1-1417) exhibit higher apparent enzymatic activity as compared to those containing wild type DCAF1 in Cos7 cells (Figures 2C). Pretreatment with MG132 caused a large increase in the activity of CRL4 containing wild type DCAF1 but had only a small effect on CRL4 containing DCAF1 (1-1417) (Figures 2C and S2E), suggesting that the latter is a more potent E3 ligase. These results suggest that deletion of the Merlin binding segment of DCAF1 leads to constitutive activation of CRL4DCAF1, consistent with the notion that Merlin inhibits the E3 ligase activity.
Finally, we tested the effect of depletion of Merlin on CRL4DCAF1 activity. Cos7 cells were infected with lentiviruses encoding a control sh-RNA or a sh-RNA targeting Merlin and then subjected to in vivo ubiquitylation assay. The results indicated that depletion of Merlin causes a significant increase in CRL4DCAF1 ligase activity (Figure 2D). The effect of the sh-RNA targeting Merlin was specific, because it was suppressed by overexpression of HA-Merlin but not of HA-Ezrin, which does not associate with FH-DCAF1 (Figure S2H and S2I). Together, the results of positive expression, mutational activation, and depletion provide compelling evidence that Merlin functions as a negative regulator of CRL4DCAF1 (see Figure 2E for a hypothetical model).
CRL4DCAF1 Mediates the Hyperproliferation Caused by Inactivation of Merlin
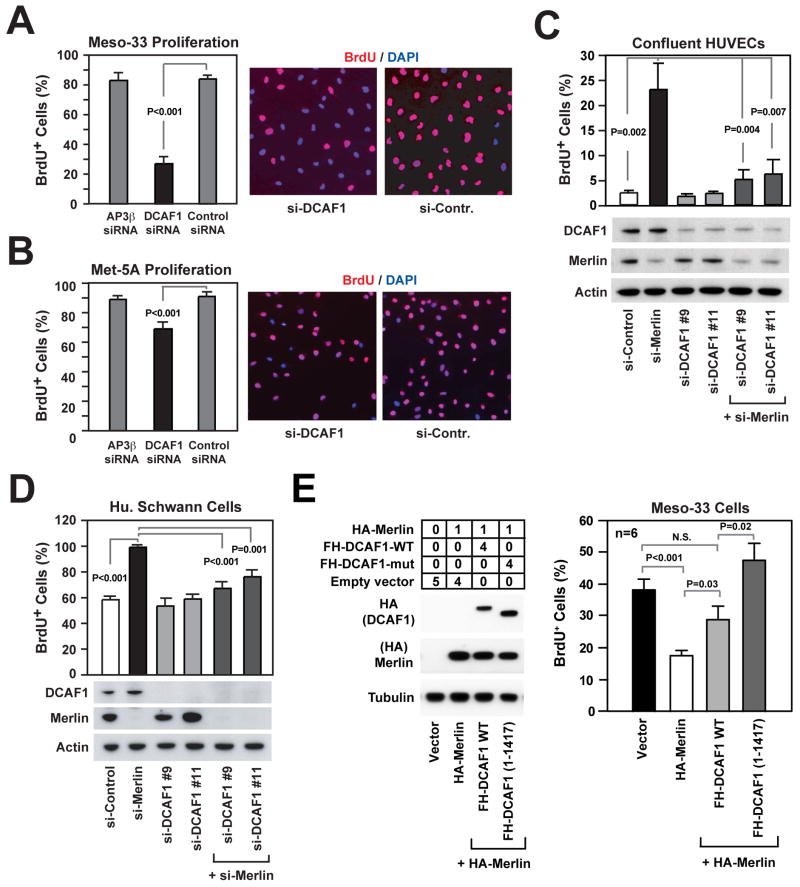
Recent studies have implicated CRL4DCAF1 in DNA replication and progression through the G2 phase of the cell cycle (Hrecka et al., 2007; McCall et al., 2008). To examine if Merlin suppresses cell proliferation by inhibiting CRL4DCAF1, we silenced DCAF1 in NF2 mutant Meso-33 mesothelioma cells using a smart pool of siRNAs (Figure S3A). Since loss of Merlin promotes progression through G1 (Lopez-Lago et al., 2009), we examined if loss of DCAF1 affects this specific phase of the cell cycle. BrdU incorporation experiments revealed that depletion of DCAF1 inhibits the ability of G0 synchronized Meso-33 cells to progress through G1 and enter S phase in response to mitogens (Figure 3A). Deconstruction of the smartpool indicated that each individual siRNA in the pool was able to inhibit expression of DCAF1 and cell proliferation, as expected from specific inhibition (Figure S3B and S3C). In contrast, silencing of DCAF1 exerted a moderate growth inhibitory effect in normal mesothelial Met-5A cells (Figure 3B and S3D), suggesting that Merlin-deficient cells are more sensitive to inactivation of DCAF1 than their normal counterpart.
Figure 3. DCAF1 Mediates Hyperproliferation and Loss of Contact Inhibition in Merlin-deficient Cells.
(A, B) Meso-33 cells (A) and Met-5a cells (B) were transfected with a SMARTpool of siRNAs targeting the adaptor protein AP3β, DCAF1, or a control non-targeting pool, deprived of growth factors for 24 hours, incubated with BrdU in the presence of mitogens for 24 hours, and subjected to anti-BrdU staining followed by counterstaining with DAPI. The graph shows the percentage (± SEM) of BrdU+ cells and the pictures show representative fields.
(C, D) HUVEC cells (C) and HEI 286 human Schwann cells (D) were transfected with a control si-RNA or 2 distinct si-RNAs targeting DCAF1, alone or in combination with a previously validated si-RNA targeting Merlin (Okada et al., 2005). Cells were deprived of growth factors for 24 hours, plated under confluent (C) or sparse (D) conditions on fibronectin, and subjected to BrdU incorporation in the presence of mitogens for 24 hours followed by anti-BrdU staining. The graphs show the percentage (± SEM) of BrdU+ cells. Immunoblotting was used to verify the efficiency of knock down.
(E) Meso-33 cells were transiently transfected with the indicated μg of vectors encoding HA-Merlin, alone or in combination with FH-DCAF1 or FH-DCAF1 (1-1417), and subjected to immunoblotting (left) and BrdU incorporation assay (right). The anti-HA blots including FH-DCAF1 and HA-Merlin were exposed for the same time. The graph shows the percentage (±SEM) of BrdU+ cells.
See also Supplemental Figure S3.
To further explore the role of CRL4DCAF1 in the proliferation of Merlin-deficient tumor cells, we designed a shRNA able to target both human and mouse DCAF1. Silencing of DCAF1 with this shRNA inhibited phosphorylation of Rb and entry into the S phase in Meso-33 cells (Figure S3E). FACS analysis of unsynchronized Meso-33 cells confirmed that depletion of DCAF1 predominantly inhibits progression through G1 (control Meso-33: 23% G1, 68% S, 9% G2/M; DCAF1-depleted Meso-33: 59% G1, 35% S, 6% G2/M). Re-expression of DCAF1 from either a moderate or a strong promoter rescued phosphorylation of Rb and cell cycle progression, confirming the specificity of the shRNA targeting DCAF1 (Figure S3E). We therefore used this shRNA to silence DCAF1 in FC-1801 Schwannoma cells, which were derived from NF2 conditional knock out mice, and their normal counterpart, the FH-912 cells. Notably, inactivation of DCAF1 caused a more profound growth inhibition in Merlin-null FC-1801 cells as compared to Merlin-positive FH-912 cells (Figure S3F), confirming that CRL4DCAF1 is required for the proliferation of Merlin-deficient tumor cells but to a lesser degree for that of normal cells. These observations suggest that CRL4DCAF1 is a key component of the oncogenic signaling pathway activated by loss of Merlin.
To examine if CRL4DCAF1 is required for exit from contact inhibition following inactivation of Merlin, we silenced Merlin in confluent HUVECs. As anticipated, this manipulation induced a significant fraction of confluent HUVECs to enter into S phase (Figure 3C). Notably, depletion of DCAF1 using each of two distinct siRNAs suppressed this unscheduled event (Figure 3C), suggesting that Merlin induces contact inhibition through inhibition of CRL4DCAF1.
To confirm that CRL4DCAF1 functions downstream of Merlin, we conducted an additional genetic epistasis experiment in HEI-286 Schwann cells, which are immortalized but not neoplastic. Silencing of Merlin caused these cells to undergo accelerated progression through G1 and entry into S phase in response to growth factors (Figure 3D), in agreement with the observation that Merlin contributes to restraining G1 progression (Lopez-Lago et al., 2009). Inactivation of DCAF1 reverted the hyperproliferation caused by depletion of Merlin but did not affect the rate of growth of control cells expressing Merlin (Figure 3D), providing additional evidence that CRL4DCAF1 mediates the growth inhibitory effect of Merlin.
If Merlin suppresses cell proliferation by inhibiting CRL4DCAF1, expression of a mutant form of DCAF1 unable to interact with Merlin should counteract the antimitogenic effect of Merlin. To test this prediction, Meso-33 cells were transfected with FH-Merlin, alone or in combination with wild type DCAF1 or DCAF1 (1-1417), and subjected to BrdU incorporation assay. Interestingly, expression of DCAF1 (1-1417) fully rescued Meso-33 cells from undergoing growth arrest upon introduction of Merlin, whereas wild type DCAF1 had only a moderate effect (Figure 3E). We note that, in spite of its higher level of expression, Merlin did not bind stoichiometrically to wild type DCAF1 under these experimental conditions (Figure S3G). Presumably, transient transfection saturates the cellular machinery that modifies Merlin to enable it to bind to CRL4DCAF1 in Meso-33 cells. Taken together, these results provide strong evidence that Merlin suppresses cell proliferation by inhibiting CRL4DCAF1.
Merlin-mediated Inhibition of CRL4DCAF1 Activates a Growth Inhibitory Gene Expression Program
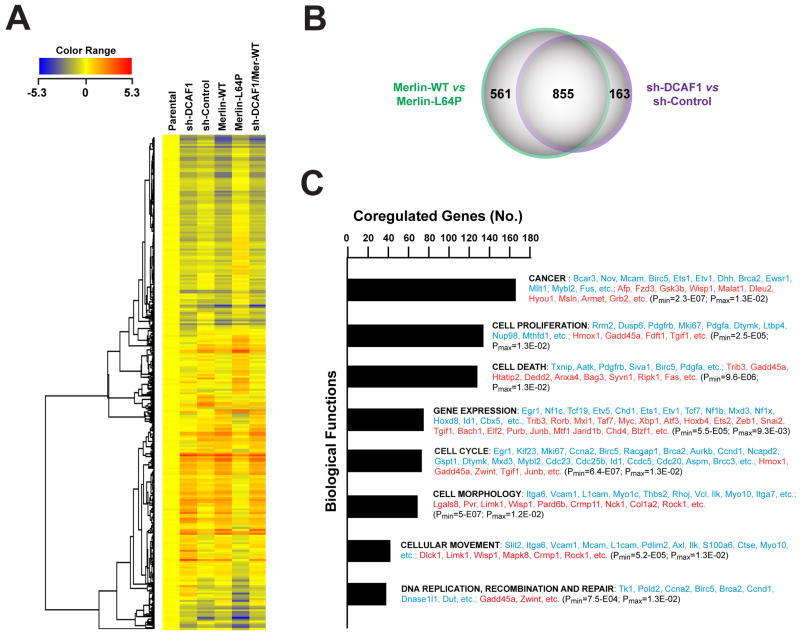
CRL4 ligases can regulate gene expression by promoting ubiquitylation of histones and recruitment of enzymes involved in chromatin remodeling or histone methylation or by inducing proteasome-mediated degradation of transcription factors (O’Connell and Harper, 2007). To examine if Merlin controls gene expression through inhibition of CRL4DCAF1, we compared the gene expression program activated by expression of Merlin or depletion of DCAF1 in Nf2−/− FC-1801 mouse Schwannoma cells. Unsupervised clustering of 1566 probesets differentially expressed upon expression of wild type but not L64P mutant Merlin or upon specific depletion of DCAF1 revealed largely overlapping changes in gene expression (Figure 4A). In fact, expression of Merlin and depletion of DCAF1 induced a concordant upregulation or downregulation of 855 probesets (54%) (Figures 4B and S4A). In spite of this considerable functional overlap, expression of Merlin and knock down of DCAF1 also induced specific effects on gene expression (Figure 4B). These results indicate that Merlin regulates gene expression largely through inhibition of CRL4DCAF1 but also suggest that Merlin has CRL4DCAF1-independent functions.
Figure 4. Expression of Merlin and Depletion of DCAF1 Induce a Largely Overlapping Gene Expression Program.
(A) FC-1801 cells were infected with lentiviral vectors encoding a sh-RNA targeting DCAF1 (sh-DCAF1), a control sh-RNA (sh-Control), wild type Merlin, Merlin-L64P, or sh-DCAF1 in combination with wild type Merlin and subjected to DNA microarray analysis. Unsupervised clustering of 1,566 probesets differentially expressed ≥ 2 fold in at least one of the two major comparisons (sh-DCAF1 versus sh-Control and Merlin-WT versus Merlin-L64P). Values normalized to parental uninfected cells.
(B) Venn Diagram shows the overlap between the list of genes differentially expressed upon expression of Merlin-WT or silencing of DCAF1 (fold change ≥ 2). Coregulated genes (885) are identified as differentially expressed ≥2 fold in one of the two comparisons and displaying a consensual significant change in the other comparison (p ≤ 0.01 by the ANOVA test).
(C). 667 genes coregulated upon expression of Merlin-WT or silencing of DCAF1 (fold change ≥ 2 in both comparisons) was subjected to Ingenuity Pathway analysis. Both down-regulated (353) and upregulated genes (314) are included in the category of Biological Functions. Representative downregulated genes are shown in blue and representative upregulated genes in red in order of decreasing average fold change.
See also Supplemental Figure S4.
Ingenuity pathway analysis revealed that expression of Merlin and inactivation of CRL4DCAF1 cause a concordant downregulation of genes promoting cell cycle progression and upregulation of genes involved in growth arrest and apoptosis (Figure 4C). In addition, these two manipulations downregulated several cell adhesion genes, such as the integrin α subunits Itga6 and Itga7 and the integrin counter-receptor Vcam1, and the integrin-linked kinase Ilk, and upregulated the Rho effectors Rock1 and Limk1. Ingenuity analysis of canonical pathways indicated that expression of Merlin and loss of DCAF1 inhibit integrin and PDGF signaling as well as the signaling pathways jointly regulated by integrins and receptor tyrosine kinases (Figure S4B). Interestingly, these two manipulations also led to downregulation of the integrin signaling component Bcar3, which has been implicated in recruitment of p130CAS to lamellipodia and in activation of Rac (Schrecengost et al., 2007). A survey of the genes coordinately regulated by expression of Merlin or loss of DCAF1 revealed additional features of the gene expression program activated by Merlin through inhibition of CRL4DCAF1. In addition to expanding the category of genes involved in cell adhesion and receptor tyrosine kinase signaling, this analysis revealed that Merlin regulates, through CRL4DCAF1, several genes involved in intracellular trafficking and a fraction of the known Hippo pathway target genes (Figure S4C). These observations suggest that Merlin’s inhibition of CRL4DCAF1 activates a broad tumor suppressive gene expression program.
Intramolecular Association of the FERM Domain with the C-terminal Tail Promotes Nuclear Accumulation of Merlin
In order to obtain insight into the regulation of the association of Merlin with CRL4DCAF1, we studied the mechanism that enables Merlin to accumulate in the nucleus. Preliminary experiments indicated that Merlin’s translocation in the nucleus does not require DCAF1 (Li and Giancotti, unpublished). Since Merlin does not contain a canonical nuclear import sequence, we examined the ability of the FERM domain and of the α-helical and C-terminal domain of the protein to localize to the nucleus. The FERM domain of Merlin (1-341) accumulated in the nucleus of Meso-33 cells to a significantly larger degree as compared to wild type Merlin. In contrast, the α-helical and C-terminal domain of the protein (341–595) failed to localize to the nucleus under the same conditions (Figure S5A and S5B). These results indicate that the FERM domain of Merlin is sufficient for nuclear translocation and suggest that the C-terminal portion of the protein may exert an inhibitory role. Since this portion of Merlin contains a nuclear export signal (Kressel and Schmucker, 2002), it is possible that this signal is masked in the closed conformer.
To examine whether antimitogenic signals, such as cell-to-cell adhesion and growth factor deprivation, regulate the accumulation of Merlin in the nucleus, we monitored the subcellular localization of the unphoshorylated and phosphorylated form of Merlin under various growth conditions. Mobility shift assay and immunoblotting with anti-P-Merlin (S518) indicated that the de-phosphorylated form of Merlin, which was prevalent in confluent cells especially if they were not exposed to mitogens, accumulated predominantly in the nucleus, whereas Merlin phosphorylated at S518 remained largely confined to the non-nuclear fraction under all conditions (Figure S5C and S5D). To confirm that the closed, growth inhibitory form of Merlin accumulates preferentially in the nucleus, we examined the subcellular distribution of Merlin-S518A, which cannot be phosphorylated by PAK, and Merlin-S518D, which mimics the phosphorylated form of the protein. Notably, Merlin-S518A accumulated in the nucleus to a significantly higher extent as compared to wild type Merlin (Figure S5E). In contrast, Merlin-S518D displayed defective nuclear accumulation, suggesting that PAK-mediated phosphorylation inhibits nuclear localization of Merlin. In agreement with this hypothesis, a dominant negative form of PAK enhanced nuclear accumulation of Merlin (Li and Giancotti, unpublished). Thus, intramolecular association of the FERM domain with the C-terminal tail promotes nuclear accumulation of Merlin, whereas phosphorylation of S518 inhibits this process.
Tumor-derived Mutations Prevent Merlin’s Ability to Interact with or to Inhibit CRL4DCAF1
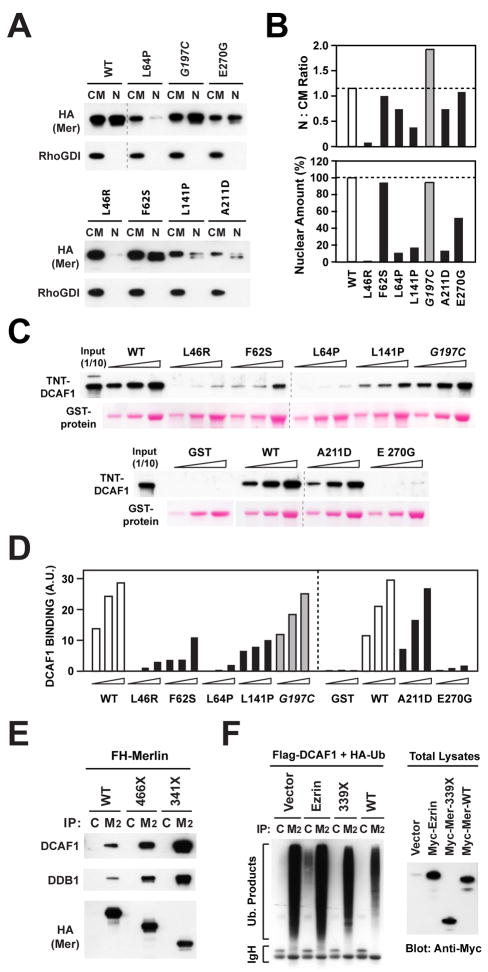
Most NF2 missense mutations map to the FERM domain (Ahronowitz et al., 2007). To further study the relevance of Merlin’s interaction with CRL4DCAF1 for tumor suppression, we examined several of these mutations. L46R, F62S, L64P, L141P, A211D, and E270G have been found in two or more unrelated NF2 patients or are known to track with disease in individual families and are therefore bona fide pathogenic mutations. In contrast, G197C, which was identified in a single patient affected by mild bilateral Schwannoma, may represent a polymorphism or a passenger mutation (Ahronowitz et al., 2007). Structural considerations suggest that L46R, F62S, L64P, L141P, and A211D disrupt the hydrophobic core of subdomain A or B, whereas E270G simply removes a surface charge from subdomain C (Figure 5A–C). In contrast, G197C can be accommodated by a slight adjustment of the surface loop in which it resides, without overt effects on the overall structure of subdomain B, and does not introduce or delete a charge (Figure 5C).
To evaluate their growth suppressive function, Merlin and mutants thereof were introduced into Meso-33 cells. As anticipated, wild type Merlin and Merlin-S518A suppressed progression through G1 and entry in S-phase in these cells, whereas Merlin-S518D inhibited proliferation to a much smaller degree (Figure 5D). Notably, all the bona fide pathogenic mutants were found to be devoid of growth inhibitory activity in Meso-33 cells. In contrast, Merlin G197C suppressed proliferation as effectively as wild type Merlin, suggesting that this is not a pathogenic mutation (Figure 5D). To examine the ability of tumor-derived mutants of Merlin to interact with CRL4DCAF1 in cells, Cos7 cells expressing FH-Merlin and mutants thereof were subjected to coimmunoprecipitation. Remarkably, all the bona fide tumor-derived mutant forms of Merlin did not interact with CRL4DCAF1 in vivo, whereas the control mutant G197C associated with the ligase as efficiently as wild type Merlin under the same conditions (Figure 5E, see Fig. 1C for L64P), providing evidence that Merlin needs to combine with CRL4DCAF1 to suppress tumorigenesis.
To obtain insight into the mechanism through which each of these pathogenic missense mutations impairs association of Merlin with CRL4DCAF1 in vivo, we first assessed their effect on nuclear accumulation of the protein. Biochemical fractionation indicated that L46R, L64P, L141P, and A211D accumulate in the nucleus to a much smaller degree than wild type Merlin or G197C (Figure 6A and 6B). Defective nuclear translocation and decreased level of expression, presumably arising from deficient protein stability, contributed to varying degrees to inhibit accumulation of each of these mutants into the nucleus. At the two opposite ends of the spectrum, L46R and L141 did not accumulate in the nucleus in spite of near wild type levels of expression, suggesting that these two mutations affect predominantly nuclear translocation, whereas A211D nuclear levels were low largely as a consequence of reduced expression (Figure 6A and B). In contrast, F62S and E270 translocated into the nucleus as efficiently as wild type Merlin, although the amounts of the latter in the nucleus appeared to be reduced. These results suggest that defective stability and nuclear translocation contribute to impair the interaction of certain tumor-derived mutant forms of Merlin with CRL4DCAF1.
Figure 6. Pathogenic Mutations Disrupt the Ability of Merlin to Accumulate in the Nucleus, to Bind to DCAF1, or to Inhibit CRL4DCAF1.
(A) Meso-33 cells were transiently transfected with FH-tagged versions of wild type Merlin or the indicated mutants and subjected to subcellular fractionation. Equal amounts of proteins from the non-nuclear (CM) and nuclear (NF) fractions were subjected to immunoblotting.
(B) The amount of Merlin or indicated mutants present in nuclear and non-nuclear fractions was estimated by densitometry of the blot in panel A. (Top) ratio of Merlin or indicated mutants present in nuclear vs. non-nuclear fractions; (Bottom) amount of each Merlin mutant present in the nucleus as a percentage over the wild type value.
(C) In vitro translated and TNT-labeled DCAF1 was incubated with 0.5, 1, and 2 μg of GST fusion proteins comprising the FERM domain of wild type Merlin or indicated mutants. GST-bound DCAF1 was detected by blotting with streptavidin.
(D) Densitometric analysis of (C). The graph shows the binding values in arbitrary units.
(E) Cos7 cells were transfected with the indicated FH-tagged forms of Merlin and subjected to immunoprecipitation with control (C) or anti-Flag Mab (M2) followed by immunoblotting.
(F) Cos7 cells were transfected with FH-DCAF1 and HA-Ubiquitin in combination with the indicated Myc-tagged constructs or empty vector and treated with MG132. Flag- or control-immunoprecipitates were immunoblotted with anti-HA (left). Total lysates were immunoblotted with anti-Myc (right).
To examine the ability of tumor-derived missense mutants to interact with CRL4DCAF1 in vitro, we engineered each mutation into a fusion protein comprising the isolated FERM domain of Merlin, which we had shown binds efficiently to in vitro translated DCAF1. As shown in Figure 6C and 6D, GST-FERM-A211D bound to DCAF1 in vitro almost as efficiently as wild type GST-FERM or GST-FERM-G197C, suggesting that the A211D mutant does not interact in vivo with CRL4DCAF1 because it does not accumulate in the nucleus to sufficient levels (Figure 6A and B). All the other mutant FERM domains displayed decreased ability to bind to recombinant DCAF1: L46R, L64P, and E270G did not bind at all under our experimental conditions, whereas F62S and L141P displayed defective binding (Figure 6C and D). Since the F62S and E270G were able to accumulate in the nucleus to normal or near normal levels (Figure 6A and B), we infer that they do not combine in vivo with CRL4DCAF1 because they are unable to interact directly with DCAF1 with sufficient affinity. Notably, the E270G mutation simply removes a charge from a surface exposed loop on subdomain C (Figure 5A), implicating this loop in interaction with DCAF1 and confirming the high specificity of the interaction of Merlin with DCAF1. In contrast, both defective nuclear accumulation and deficient binding to DCAF1 contribute to the inability of L46R, L64P, and L141P to interact with CRL4DCAF1 in vivo. Together, these experiments indicate that the six tumor-derived missense mutants that we have tested do not combine with CRL4DCAF1 in vivo, owing to defective accumulation in the nucleus, deficient binding to DCAF1, or a combination of the two mechanisms.
While mutations that introduce stop codons within the FERM domain are likely to give rise to misfolded and unstable proteins products, certain nonsense mutations delete only the C-terminal domain – a structural perturbation that may affect the ability of Merlin to maintain a closed conformation. To examine if tumor-derived nonsense mutations affect the ability of Merlin to interact with or to suppress CRL4DCAF1, we examined two prevalent truncation mutants: Merlin 466X, which lacks the C-terminal portion of the α helical domain as well as the C-terminal domain, and Merlin 341X, which retains the FERM domain but lacks both the α helical and the C-terminal domain (Ahronowitz et al., 2007). Coimmunoprecipitation indicated that these two Merlin mutants retain the ability to interact with CRL4DCAF1 in vivo (Figure 6E). In fact, progressive deletion from the C-terminus resulted in increased, as opposed to decreased, binding of Merlin to the ligase. Since the isolated FERM domain of Merlin accumulates in the nucleus to a larger extent than the wild type protein, whereas the C-terminal portion of Merlin including the α helical and C-terminal domain remains in the cytoplasm (Figure S5B), it is possible that increased nuclear accumulation facilitates the interaction of tumor-derived truncations mutants with CRL4DCAF1. Interestingly, in spite of its increased ability to bind to CRL4DCAF1 in vivo, the FERM domain of Merlin did not suppress CRL4DCAF1 ligase activity in vivo as efficiently as wild type Merlin (Figure 6F), indicating that the α helical and C-terminal domain contribute to Merlin’s inhibitory activity. These results suggest that the tumor-derived truncation mutants of Merlin that retain the FERM domain are able to interact with CRL4DCAF1 but fail to suppress its activity. The observation that several distinct tumor-derived mutations impair Merlin’s ability to translocate into the nucleus, to bind to DCAF1, or to inhibit CRL4DCAF1 activity provides strong genetic evidence that Merlin suppresses tumorigenesis by inhibiting CRL4DCAF1.
Inactivation of DCAF1 Suppresses the Tumorigenic Potential of Mesothelioma and Schwannoma Cells
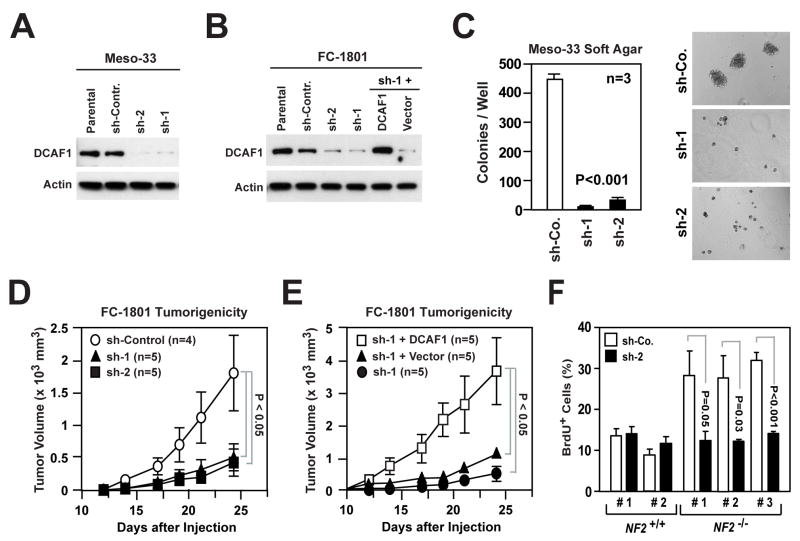
To test if CRL4DCAF1 is required for NF2-dependent tumorigenesis, we silenced DCAF1 in Meso-33 and FC-1801 cells (Figure 7A and 7B). Knock down of DCAF1 inhibited the ability of both mesothelioma and Schwannoma cells to grow in soft agar (Figures 7C and S6A). In contrast, depletion of DCAF1 did not affect the ability of Ras-V12 or v-Src transformed cells to express elevated levels of Cyclin D1 and p-Rb and to proliferate under adherent conditions or in soft agar (Figure S6B and S6C), suggesting that inactivation of CRL4DCAF1 specifically inhibits the malignant properties of Merlin-deficient tumor cells.
Figure 7. Silencing of DCAF1 Suppresses the Tumorigenic Potential of Merlin-deficient Cells.
(A) Meso-33 cells were infected with lentiviral vectors encoding two distinct sh-RNAs targeting DCAF1 (sh-1 and sh-2) or a control sh-RNA (sh-Contr.) and subjected to immunoblotting.
(B) FC-1801 cells treated as in (A). Cells infected with sh-1 were also infected with a retroviral vector encoding a sh-RNA-resistant version of DCAF1 or with empty vector. Immunoblotting was as indicated.
(C) Meso-33 cells infected with the indicated lentiviral vectors were subjected to soft agar assay. The graph illustrates the total number (± SEM) of colonies present in each well after 2 weeks of culture. Representative images shown.
(D, E) 2 × 106 FC-1801 cells infected with the indicated lentiviral vectors were injected subcutaneously in nude mice. (D) shows the increase in volume (± SEM) over time of tumors generated by cells infected with lentiviruses encoding a control sh-RNA (sh-Control) or two distinct sh-RNAs targeting DCAF1 (sh-1 and sh-2). (E) shows the increase in volume (± SEM) over time of tumors generated by cells infected with sh-1 and either a retroviral vector encoding a sh-RNA-resistant version of DCAF1 or empty vector. Data were obtained from the same experiment. Note the different scale of the Y axes of the two graphs.
(F) Primary Schwann cells from 2 normal individuals (NF2+/+) and Schwannoma cells from 2 NF2 patients (NF2−/−) were infected with lentiviruses encoding a sh-RNA targeting DCAF1 (sh-2) or a control sh-RNA (sh-Co.), deprived of growth factors, restimulated with mitogens in the presence of BrdU, and subjected to anti-BrdU staining. The graph shows the percentage (± SEM) of BrdU+ cells.
See also Supplemental Figure S6.
Meso-33 cells as well as other Merlin-deficient mesothelioma cell lines did not give rise to tumors upon subcutaneous or intrapleural injection in mice. In contrast, the FC-1801 cells, which express robust levels of DCAF1, gave rise to sizeable tumors in less than 4 weeks upon subcutaneous injection in nude mice (Figure 7D). Silencing of DCAF1 with two distinct shRNAs inhibited tumor growth in vivo by approximately 75% at day 24. This effect was highly specific, as moderate overexpression of an shRNA-resistant form of DCAF1 rescued the ability of FC-1801 cells expressing one of the two shRNAs targeting DCAF1 to give rise to subcutaneous tumors (Figure 7E). In fact, the tumors generated by these cells grew at a much faster pace as compared to those generated by parental FC-1801 cells (by approximately 2 fold, compare Figure 7D and 7E, data are from the same experiment), suggesting that elevated levels of DCAF1 can promote tumorigenesis.
To explore the relevance of our findings to the pathogenesis of NF2, we examined if DCAF1 affects the ability of primary human Schwannoma cells derived from NF2 patients to proliferate in vitro. Silencing of DCAF1 suppressed the ability of these cells to progress through G1 and enter into S phase in response to mitogens. In contrast, knock down of DCAF1 did not significantly affect the proliferation of control human Schwann cells (Figure 7F). Together, these results indicate that CRL4DCAF1 is necessary for NF2-dependent hyperproliferation, anchorage-independent growth, and tumorigenesis.
DISCUSSION
The mechanism by which Merlin suppresses tumorigenesis has remained elusive since the discovery of the NF2 gene. This study reveals that the closed, growth inhibitory form of Merlin translocates to the nucleus, binds to the E3 ubiquitin ligase CRL4DCAF1, and suppresses its ability to ubiquitylate target proteins. Multiple converging lines of evidence indicate that Merlin’s inhibition of CRL4DCAF1 activity is required to induce growth arrest and suppress tumorigenesis. Notably, analysis of several tumor-derived mutants indicates that some display impaired nuclear translocation, others fail to interact with DCAF1, and still others bind to DCAF1 but do not suppress CRL4DCAF1 E3 ligase activity. Furthermore, silencing of DCAF1 reverses the consequences of Merlin inactivation in normal cells and suppresses the ability of Merlin-deficient tumor cells to grow in soft agar and in nude mice. These findings indicate that entry into the nucleus and inhibition of CRL4DCAF1 are integral components of the mechanism by which Merlin suppresses tumorigenesis.
The mechanisms through which anti-mitogenic signals induce Merlin to inhibit CRL4DCAF1 are likely to be complex. Our results suggest that these signals promote, largely through inhibition of PAK, accumulation of the closed form of Merlin, which translocates into the nucleus. Merlin does not contain a classical importin-α nuclear localization sequence but has a C-terminal CRM1-dependent nuclear export signal (Kressel and Schmucker, 2002). Mutational analysis suggests that the FERM domain is necessary and sufficient for nuclear entry, whereas the C-terminal coiled-coil domain inhibits this process unless it is folded back and interacts with the FERM domain. In addition or instead, unfolding of the C-terminal helical domain may expose the nuclear export signal and thereby facilitate Merlin’s exit from the nucleus. Interestingly, other FERM domain proteins, such as the isoforms H and R of band 4.1 and FAK, appear to shuttle in and out of the nucleus through similar conformational transitions (Gascard et al., 1999; Lim et al., 2008; Luque et al., 1998).
Upon entering the nucleus, the closed form of Merlin binds to the C-terminal segment of DCAF1. Our results indicate that Merlin is not a substrate of CRL4DCAF1 but exerts its antimitogenic function by blocking DCAF1-dependent recruitment or ubiquitylation of CRL4DCAF1 substrates. Since subdomain A of the FERM domain of Merlin consists of an ubiquitin-like fold (Shimizu et al., 2002), Merlin may function as an inhibitory pseudosubstrate for CRL4DCAF1. Reconstitution of CRL4DCAF1 ligase activity in vitro will be necessary to directly test this model. Irrespective of the specific mechanism by which Merlin inhibits CRL4DCAF1, several lines of evidence support the conclusion that this inhibitory interaction restrains cell proliferation. First, inactivation of Merlin enables normal cells to exit from contact inhibition and to undergo cell cycle progression, while depletion of DCAF1 suppresses both effects. Second, enforced expression of a Merlin-insensitive mutant of DCAF1 counteracts the antimitogenic effect of Merlin. Third, depletion of DCAF1 suppresses hyperproliferation in Merlin-deficient Schwannoma and mesothelioma cells but inhibits their normal counterpart to a lesser extent, consistent with the hypothesis that loss of Merlin activates oncogenic signaling through CRL4DCAF1. Finally, silencing of DCAF1 suppresses the ability of primary human Schwannoma cells from NF2 patients to proliferate in vitro, suggesting that deregulated CRL4DCAF1 activity contributes to the pathogenesis of NF2.
CRL4 ligases have been implicated in chromatin remodeling, DNA replication, and the response to DNA damage (Lee and Zhou, 2007; O’Connell and Harper, 2007). For example, CRL4DDB2 promotes the ubiquitylation of several histones and the DNA repair components DDB2 and XPC, restricting the threshold of DNA damage response and inhibiting UV-induced skin carcinogenesis, CRL4CSA mediates ubiquitylation of CSB, a SWI2/SNF2 ATP-dependent chromatin remodeling enzyme, and CRL4DET1-Cop1 promotes ubiquitylation and proteasome–mediated degradation of the transcription factor c-Jun (reviewed in Lee and Zhou, 2007 and in O’Connell and Harper, 2007; Liu et al., 2009). Although the physiological substrates of CRL4DCAF1 have not been identified, our results suggest that CRL4DCAF1 broadly regulates gene expression. In fact, expression of Merlin and depletion of DCAF1 cause similar changes in the expression of several hundred genes, suggesting that Merlin controls a wide gene expression program through inhibition of CRL4DCAF1. Notably, this program includes the inhibition of genes involved in promoting progression through each phase of the cell cycle as well as the upregulation of genes involved in growth arrest and apoptosis. Parenthetically, we speculate that knock down of DCAF1 may cause an accumulation of Merlin-deficient cells in G1 because loss of Merlin specifically deregulates this phase of the cell cycle. In addition, we have observed that Merlin controls, through CRL4DCAF1, the expression of genes involved in receptor tyrosine kinase and adhesion signaling, intracellular trafficking, and a subset of Hippo pathway target genes. The ability of CRL4DCAF1 to regulate these genes may explain some of the effects that Merlin has been reported to exert on receptor tyrosine kinase and adhesion signaling (Ammoun et al., 2008; Curto et al., 2007; Kissil, 2003; Lallemand et al., 2009; Okada et al., 2005; Poulikakos et al., 2006; Shaw et al., 2001). The observation that re-expression of Merlin and depletion of DCAF1 induce a gene expression program of similar breadth, specificity, and scope reinforces the model that Merlin suppresses tumorigenesis through inhibition of CRL4DCAF1.
The analysis of tumor-derived mutations, in particular missense mutations, has provided considerable insight into the function of important tumor suppressors, such as P53, RB, PTEN, and APC. We have studied 6 pathogenic missense mutations mapping to the FERM domain of Merlin and found that each of them impairs the ability of Merlin to accumulate in the nucleus and/or bind to DCAF1. Both types of defects disrupt the association of Merlin with CRL4DCAF1. In contrast, severe truncation mutants, such as 341X, display increased accumulation in the nucleus and interaction with CRL4DCAF1 in vivo but fail to suppress ligase activity. Amongst the missense mutations, L64R, F62S, and L64P, located in subdomain A, disrupt binding to DCAF1 in vitro, whereas L141P and A211D, mapping to subdomain B, do not. Furthermore, E270G, which removes a single charge from the surface of subdomain C, drastically reduces binding to DCAF1 in vitro. These results implicate subdomain A and C in interaction with DCAF1 and suggest that mutations in other portions of the protein, such as subdomain B and the helical segment, may affect nuclear localization determinants or disrupt the closed conformation that is required for nuclear accumulation. The observation that many pathogenic NF2 mutations disrupt, and several others are predicted to disrupt, the ability of Merlin to interact with or to suppress CRL4DCAF1 provides strong genetic evidence that Merlin suppresses tumorigenesis by inhibiting this E3 ligase.
In conclusion, our results indicate that NF2-dependent tumorigenesis arises from the inability of mutant Merlin to enter into the nucleus and to suppress CRL4DCAF1-dependent gene expression. This newly delineated tumor suppressor pathway bears antithetic resemblance to the Wnt/β-catenin pathway, where an intercellular signal hijacks the cytoskeletal component β-catenin to promote oncogenic gene expression in the nucleus (Clevers, 2006). Identification of the substrate(s) of CRL4DCAF1 and elucidation of the mechanism through which this ligase exerts its pro-proliferative and anti-apoptotic role should help to generate targeted therapies for NF2 mutant tumors.
EXPERIMENTAL PROCEDURES
Materials and Standard Protocols
Cell lines, antibodies, vectors, and other materials are described in Supplemental Data. The protocols used for immunoprecipitation, GST pull down, subcellular fractionation, and immunoblotting are outlined in Supplemental Data.
TAP and Mass Spectrometry
Cos7 cells transiently expressing FH-tagged Merlin or Merlin-L64P were lysed in RIPA buffer and subjected to TAP using the FLAG HA Tandem Affinity Purification Kit (Sigma TP0010). Proteins were separated by SDS-PAGE, visualized with Gelcode (Pierce), and subjected to MALDI-reTOF-MS/MS. Alternatively, gel stacks were subjected to nanoLC-MS/MS. See details in Supplemental Data.
Immunofluorescent Staining
Cells were fixed with 4% paraformaldehyde in PBS for 20 min and incubated in permeabilization buffer (PDT: 0.3% sodium deoxycholate, 0.3% Triton X-100 in PBS) for 30 min on ice. Fixation and permeabilization with organic solvents, such as methanol or acetone, or fixation with paraformaldehyde followed by permeabilization with 0.5% Triton X-100 alone for 20 minutes did not allow efficient detection of Merlin in the nucleus. After fixation and permeabilization, cells were incubated overnight at 4°C with primary antibodies diluted in PDT. After washing with PDT, cells were incubated with secondary antibodies diluted in PDT for 2 hours at 4°C. Cells were washed with PDT, rinsed with PBS, and mounted in Gelvatol. When indicated, nuclei were stained with DAPI.
Ubiquitylation Assay
Cos7 cells were transfected with indicated plasmids and, when indicated, treated with 25 μM MG132 for 6 hours. Lysates were incubated with either anti-Flag beads or control mouse IgG beads. Eluted products were subjected to immunoblotting. Detailed procedures are described in Supplemental Data.
BrdU Incorporation Assay
Cells were synchronized in G0 by growth factor deprivation or growth to high density and then stimulated with mitogens for 24 hours in the presence of BrdU. Anti-BrdU staining was used to measure progression through G1 and entry in S phase. See details in Supplemental Data.
Gene Expression and Silencing
Retroviral and lentiviral vectors encoding recombinant proteins or shRNAs were generated as previously described (Pylayeva et al., 2007). Sequences for shRNAs and detailed procedures are described in Supplemental Data.
DNA Microarray Analysis
FC-1801 cells were left untreated or transduced with lentiviruses encoding sh-control, sh-DCAF1, Merlin-WT, Merlin-L64P, or a combination of viruses encoding sh-DCAF1 and Merlin-WT and were plated in triplicate at subconfluent density in complete medium. cDNAs were hybridized to GeneChip Mouse Genome 430a 2.0 Array chips (Affymetrix). Raw data are available in Gene Expression Omnibus (accession no. GSE19305). Data analysis is described in Supplemental Data.
Soft Agar and Tumorigenicity Assay
For soft agar assay, cells were trypsinized, resuspended in complete medium, and plated in 0.34% low melting temperature agarose (FMC Bioproducts) in complete medium. Meso-33 cells were seeded at 2 × 104 and FC-1801 at 6 × 104 per well in 6-well Ultra Low Cluster Plates (Costar). For tumorigenicity assay, 2 × 106 FC-1801 cells were suspended in calcium/magnesium-free PBS and injected subcutaneously into the right flank of nude mice. Tumor volumes were determined by caliper measurement.
Supplementary Material
Acknowledgments
We thank J. Brugge, S. Jhanwar and D. Lim for cell lines, M. Pagano for constructs, L. Fabrizio and M. Yaneva for help with mass spectrometry, and members of the Giancotti laboratory for discussions. This work was supported by the Baker Street Foundation and NIH Cancer Center Support Grant P30 CA08748. W.L. is recipient of a Young Investigator Award from the Children’s Tumor Foundation. P.Z. was supported by NIH grant R01 CA098210.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahronowitz I, Xin W, Kiely R, Sims K, MacCollin M, Nunes FP. Mutational spectrum of the NF2 gene: a meta-analysis of 12 years of research and diagnostic laboratory findings. Hum Mutat. 2007;28:1–12. doi: 10.1002/humu.20393. [DOI] [PubMed] [Google Scholar]
- Ammoun S, Flaiz C, Ristic N, Schuldt J, Hanemann CO. Dissecting and targeting the growth factor-dependent and growth factor-independent extracellular signal-regulated kinase pathway in human schwannoma. Cancer Res. 2008;68:5236–5245. doi: 10.1158/0008-5472.CAN-07-5849. [DOI] [PubMed] [Google Scholar]
- Bretscher A, Edwards K, Fehon RG. ERM proteins and merlin: integrators at the cell cortex. Nat Rev Mol Cell Biol. 2002;3:586–599. doi: 10.1038/nrm882. [DOI] [PubMed] [Google Scholar]
- Cho E, Feng Y, Rauskolb C, Maitra S, Fehon R, Irvine KD. Delineation of a Fat tumor suppressor pathway. Nat Genet. 2006;38:1142–1150. doi: 10.1038/ng1887. [DOI] [PubMed] [Google Scholar]
- Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- Curto M, Cole BK, Lallemand D, Liu CH, McClatchey AI. Contact-dependent inhibition of EGFR signaling by Nf2/Merlin. J Cell Biol. 2007;177:893–903. doi: 10.1083/jcb.200703010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascard P, Nunomura W, Lee G, Walensky LD, Krauss SW, Takakuwa Y, Chasis JA, Mohandas N, Conboy JG. Deciphering the nuclear import pathway for the cytoskeletal red cell protein 4.1R. Mol Biol Cell. 1999;10:1783–1798. doi: 10.1091/mbc.10.6.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutmann DH, Hirbe AC, Haipek CA. Functional analysis of neurofibromatosis 2 (NF2) missense mutations. Hum Mol Genet. 2001;10:1519–1529. doi: 10.1093/hmg/10.14.1519. [DOI] [PubMed] [Google Scholar]
- Hamaratoglu F, Willecke M, Kango-Singh M, Nolo R, Hyun E, Tao C, Jafar-Nejad H, Halder G. The tumour-suppressor genes NF2/Merlin and Expanded act through Hippo signalling to regulate cell proliferation and apoptosis. Nat Cell Biol. 2006;8:27–36. doi: 10.1038/ncb1339. [DOI] [PubMed] [Google Scholar]
- Hrecka K, Gierszewska M, Srivastava S, Kozaczkiewicz L, Swanson SK, Florens L, Washburn MP, Skowronski J. Lentiviral Vpr usurps Cul4-DDB1[VprBP] E3 ubiquitin ligase to modulate cell cycle. Proc Natl Acad Sci USA. 2007;104:11778–11783. doi: 10.1073/pnas.0702102104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Chen J. VprBP targets Merlin to the Roc1-Cul4A-DDB1 E3 ligase complex for degradation. Oncogene. 2008;27:4056–4064. doi: 10.1038/onc.2008.44. [DOI] [PubMed] [Google Scholar]
- James MF, Han S, Polizzano C, Plotkin SR, Manning BD, Stemmer-Rachamimov AO, Gusella JF, Ramesh V. NF2/merlin is a novel negative regulator of mTOR complex 1, and activation of mTORC1 is associated with meningioma and schwannoma growth. Mol Cell Biol. 2009;29:4250–4261. doi: 10.1128/MCB.01581-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Sperka T, Herrlich P, Morrison H. Tumorigenic transformation by CPI-17 through inhibition of a merlin phosphatase. Nature. 2006a;442:576–579. doi: 10.1038/nature04856. [DOI] [PubMed] [Google Scholar]
- Jin J, Arias EE, Chen J, Harper JW, Walter JC. A family of diverse Cul4-Ddb1-interacting proteins includes Cdt2, which is required for S phase destruction of the replication factor Cdt1. Mol Cell. 2006b;23:709–721. doi: 10.1016/j.molcel.2006.08.010. [DOI] [PubMed] [Google Scholar]
- Kaempchen K, Mielke K, Utermark T, Langmesser S, Hanemann CO. Upregulation of the Rac1/JNK signaling pathway in primary human schwannoma cells. Hum Mol Genet. 2003;12:1211–1221. doi: 10.1093/hmg/ddg146. [DOI] [PubMed] [Google Scholar]
- Kissil JL, Johnson KC, Eckman MS, Jacks T. Merlin phosphorylation by p21-activated kinase 2 and effects of phosphorylation on merlin localization. J Biol Chem. 2002;277:10394–10399. doi: 10.1074/jbc.M200083200. [DOI] [PubMed] [Google Scholar]
- Kissil JL, Wilker EW, Johnson KC, Eckman MS, Yaffe MB, Jacks T. Merlin, the product of the Nf2 tumor suppressor gene, is an inhibitor of the p21-activated kinase, Pak1. Mol Cell. 2003;12:841–849. doi: 10.1016/s1097-2765(03)00382-4. [DOI] [PubMed] [Google Scholar]
- Kressel M, Schmucker B. Nucleocytoplasmic transfer of the NF2 tumor suppressor protein merlin is regulated by exon 2 and a CRM1-dependent nuclear export signal in exon 15. Hum Mol Genet. 2002;11:2269–2278. doi: 10.1093/hmg/11.19.2269. [DOI] [PubMed] [Google Scholar]
- Lallemand D, Curto M, Saotome I, Giovannini M, McClatchey AI. NF2 deficiency promotes tumorigenesis and metastasis by destabilizing adherens junctions. Genes Dev. 2003;17:1090–1100. doi: 10.1101/gad.1054603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lallemand D, Manent J, Couvelard A, Watilliaux A, Siena M, Chareyre F, Lampin A, Niwa-Kawakita M, Kalamarides M, Giovannini M. Merlin regulates transmembrane receptor accumulation and signaling at the plasma membrane in primary mouse Schwann cells and in human schwannomas. Oncogene. 2009;28:854–865. doi: 10.1038/onc.2008.427. [DOI] [PubMed] [Google Scholar]
- Lee J, Zhou P. DCAFs, the missing link of the CUL4-DDB1 ubiquitin ligase. Mol Cell. 2007;26:775–780. doi: 10.1016/j.molcel.2007.06.001. [DOI] [PubMed] [Google Scholar]
- Lim ST, Chen XL, Lim Y, Hanson DA, Vo TT, Howerton K, Larocque N, Fisher SJ, Schlaepfer DD, Ilic D. Nuclear FAK promotes cell proliferation and survival through FERM-enhanced p53 degradation. Mol Cell. 2008;29:9–22. doi: 10.1016/j.molcel.2007.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Lee S, Zhang J, Peters SB, Hannah J, Zhang Y, Yin Y, Koff A, Ma L, Zhou P. CUL4A abrogation augments DNA damage response and protection against skin carcinogenesis. Mol Cell. 2009;34:451–460. doi: 10.1016/j.molcel.2009.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Lago MA, Okada T, Murillo MM, Socci N, Giancotti FG. Loss of the tumor suppressor gene NF2, encoding merlin, constitutively activates integrin-dependent mTORC1 signaling. Mol Cell Biol. 2009;29:4235–4249. doi: 10.1128/MCB.01578-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luque CM, Lallena MJ, Alonso MA, Correas I. An alternative domain determines nuclear localization in multifunctional protein 4.1. J Biol Chem. 1998;273:11643–11649. doi: 10.1074/jbc.273.19.11643. [DOI] [PubMed] [Google Scholar]
- Maddika S, Chen J. Protein kinase DYRK2 is a scaffold that facilitates assembly of an E3 ligase. Nat Cell Biol. 2009;11:409–419. doi: 10.1038/ncb1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCall CM, Miliani de Marval PL, Chastain PD, 2nd, Jackson SC, He YJ, Kotake Y, Cook JG, Xiong Y. Human immunodeficiency virus type 1 Vpr-binding protein VprBP, a WD40 protein associated with the DDB1-CUL4 E3 ubiquitin ligase, is essential for DNA replication and embryonic development. Mol Cell Biol. 2008;28:5621–5633. doi: 10.1128/MCB.00232-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClatchey AI, Fehon RG. Merlin and the ERM proteins--regulators of receptor distribution and signaling at the cell cortex. Trends Cell Biol. 2009;19:198–206. doi: 10.1016/j.tcb.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClatchey AI, Giovannini M. Membrane organization and tumorigenesis--the NF2 tumor suppressor, Merlin. Genes Dev. 2005;19:2265–2277. doi: 10.1101/gad.1335605. [DOI] [PubMed] [Google Scholar]
- Muranen T, Gronholm M, Renkema GH, Carpen O. Cell cycle-dependent nucleocytoplasmic shuttling of the neurofibromatosis 2 tumour suppressor merlin. Oncogene. 2005;24:1150–1158. doi: 10.1038/sj.onc.1208283. [DOI] [PubMed] [Google Scholar]
- O’Connell BC, Harper JW. Ubiquitin proteasome system (UPS): what can chromatin do for you? Curr Opin Cell Biol. 2007;19:206–214. doi: 10.1016/j.ceb.2007.02.014. [DOI] [PubMed] [Google Scholar]
- Okada T, Lopez-Lago M, Giancotti FG. Merlin/NF-2 mediates contact inhibition of growth by suppressing recruitment of Rac to the plasma membrane. J Cell Biol. 2005;171:361–371. doi: 10.1083/jcb.200503165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada T, You L, Giancotti FG. Shedding light on Merlin’s wizardry. Trends Cell Biol. 2007;17:222–229. doi: 10.1016/j.tcb.2007.03.006. [DOI] [PubMed] [Google Scholar]
- Pan D. Hippo signaling in organ size control. Genes Dev. 2007;21:886–897. doi: 10.1101/gad.1536007. [DOI] [PubMed] [Google Scholar]
- Poulikakos PI, Xiao GH, Gallagher R, Jablonski S, Jhanwar SC, Testa JR. Re-expression of the tumor suppressor NF2/merlin inhibits invasiveness in mesothelioma cells and negatively regulates FAK. Oncogene. 2006;25:5960–5968. doi: 10.1038/sj.onc.1209587. [DOI] [PubMed] [Google Scholar]
- Pylayeva Y, Guo W, Giancotti FG. Analysis of integrin signaling in genetically engineered mouse models of mammary tumor progression. Methods Enzymol. 2007;426:439–461. doi: 10.1016/S0076-6879(07)26019-7. [DOI] [PubMed] [Google Scholar]
- Rong R, Tang X, Gutmann DH, Ye K. Neurofibromatosis 2 (NF2) tumor suppressor merlin inhibits phosphatidylinositol 3-kinase through binding to PIKE-L. Proc Natl Acad Sci USA. 2004;101:18200–18205. doi: 10.1073/pnas.0405971102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrecengost RS, Riggins RB, Thomas KS, Guerrero MS, Bouton AH. Breast cancer antiestrogen resistance-3 expression regulates breast cancer cell migration through promotion of p130Cas membrane localization and membrane ruffling. Cancer Res. 2007;67:6174–6182. doi: 10.1158/0008-5472.CAN-06-3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw RJ, Paez JG, Curto M, Yaktine A, Pruitt WM, Saotome I, O’Bryan JP, Gupta V, Ratner N, Der CJ, et al. The Nf2 tumor suppressor, merlin, functions in Rac-dependent signaling. Dev Cell. 2001;1:63–72. doi: 10.1016/s1534-5807(01)00009-0. [DOI] [PubMed] [Google Scholar]
- Shimizu T, Seto A, Maita N, Hamada K, Tsukita S, Hakoshima T. Structural basis for neurofibromatosis type 2. Crystal structure of the merlin FERM domain. J Biol Chem. 2002;277:10332–10336. doi: 10.1074/jbc.M109979200. [DOI] [PubMed] [Google Scholar]
- Tang X, Jang SW, Wang X, Liu Z, Bahr SM, Sun SY, Brat D, Gutmann DH, Ye K. Akt phosphorylation regulates the tumour-suppressor merlin through ubiquitination and degradation. Nat Cell Biol. 2007;9:1199–1207. doi: 10.1038/ncb1641. [DOI] [PubMed] [Google Scholar]
- Xiao GH, Beeser A, Chernoff J, Testa JR. p21-activated kinase links Rac/Cdc42 signaling to merlin. J Biol Chem. 2002;277:883–886. doi: 10.1074/jbc.C100553200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.