We characterized interactions between the human proteins Sec62 and Sec63 as well as the putative interaction of human Sec62 with ribosomes. The data demonstrate evolutionary conservation of Sec62/Sec63 interaction and indicate that in the course of evolution Sec62 of vertebrates has gained the additional function to interact with ribosomes.
Abstract
Because of similarity to their yeast orthologues, the two membrane proteins of the human endoplasmic reticulum (ER) Sec62 and Sec63 are expected to play a role in protein biogenesis in the ER. We characterized interactions between these two proteins as well as the putative interaction of Sec62 with ribosomes. These data provide further evidence for evolutionary conservation of Sec62/Sec63 interaction. In addition, they indicate that in the course of evolution Sec62 of vertebrates has gained an additional function, the ability to interact with the ribosomal tunnel exit and, therefore, to support cotranslational mechanisms such as protein transport into the ER. This view is supported by the observation that Sec62 is associated with ribosomes in human cells. Thus, the human Sec62/Sec63 complex and the human ER membrane protein ERj1 are similar in providing binding sites for BiP in the ER-lumen and binding sites for ribosomes in the cytosol. We propose that these two systems provide similar chaperone functions with respect to different precursor proteins.
INTRODUCTION
In eukaryotic cells, protein secretion begins with the translocation of presecretory proteins across the membrane of the rough endoplasmic reticulum (ER). Translocation is mediated by a protein translocase (also termed translocon), that resides in the ER membrane, and occurs co- or posttranslationally. The posttranslational mechanism is abundant in the yeast Saccharomyces cerevisiae and the human parasite Trypanosoma brucei (Goldshmidt et al., 2008), whereas the cotranslational mechanism dominates in mammalian cells. In uni- as well as multicellular organisms and for both modes of operation, the protein translocase contains the heterotrimeric Sec61 complex as a central pore-forming component. In yeast ER, the cotranslationally operating protein translocase comprises Sec63p (a membrane-integrated heat shock protein [Hsp] 40), Sec71p, Sec72p, and Kar2p (a lumenal Hsp70) as additional components. The posttranslationally operating protein translocase contains these same components, plus Sec62p (Jermy et al., 2006). The Sec61 complex as well as Sec63p and Kar2p are also involved in protein export from the ER (Plemper et al., 1997; Kalies et al., 2005). Two of the subunits of the Sec61 complex as well as Sec62p, Sec63p, and Kar2p are essential proteins in yeast. Insect Sec62 isoform A can complement inactivation of the SEC62 gene in yeast (Noel and Cartwright, 1994). Sec63 is also involved in cotranslational protein transport into the trypanosomal ER and is also essential in trypanosomes (Goldshmidt et al., 2008; Zimmerman and Blatch, 2009). In mammalian cells, the lumenal Hsp70 immunoglobulin heavy-chain binding protein (BiP) and a so far unidentified Hsp40 are also involved in cotranslational protein import into the organelle (Dierks et al., 1996; Hamman et al., 1998; Tyedmers et al., 2003; Alder et al., 2005). Furthermore, in mammalian microsomes orthologues of yeast Sec62p and Sec63p are found in stoichiometric amounts as compared with Sec61α subunits, and in detergent extracts derived from canine pancreatic microsomes they are found in association with each other as well as with the Sec61 complex (Meyer et al., 2000; Tyedmers et al., 2000).
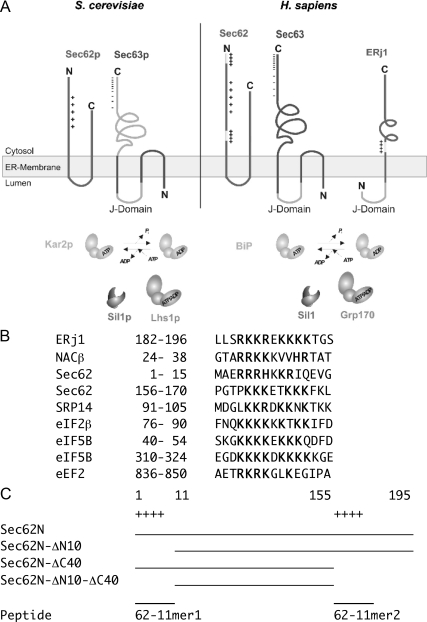
In contrast to the situation in yeast and trypanosomes, the mammalian ER membrane contains the additional Hsp40, ERj1, which is related to Sec63 in providing a lumenal J-domain (Dudek et al., 2002; see Figure 1A). Expression of human ERj1 in yeast can complement inactivation of the SEC63 gene (Kroczynska et al., 2004). The cytosolic domain of ERj1 associates with the ribosomal tunnel exit and (via the lumenal J-domain) recruits BiP to translating ribosomes (Blau et al., 2005; Dudek et al., 2005). In addition, ERj1 may play a dual role in gene expression, i.e., at the level of translation as well as transcription (Dudek et al., 2005). We identified a basic oligopeptide motif in ERj1 that is necessary and sufficient for its association with ribosomes and for its regulatory effect on translation. Similar basic oligopeptide motifs are not only present in the established ribosomal ligands signal recognition particle (SRP) and nascent polypeptide-associated complex (NAC) but also in mammalian Sec62 (see Figure 1B). We proposed that these basic oligopeptide motifs represent a general mechanism for attachment of ribosomal ligands to ribosomes (Dudek et al., 2005) and that mammalian Sec62 also may be able to bind to ribosomes. We note that similar basic oligopeptide motifs are absent from yeast Sec62p (see Figure 1A).
Figure 1.
Structural and functional features of human Sec62 and Sec63. (A) Domain organization of Sec62, Sec63, and ERj1. Sec63 and ERj1 are membrane resident Hsp40s with ER lumenal J-domains and cochaperones for ER-lumenal Hsp70s (BiP and Kar2p). Sil1, Grp170, and Lhs1p act as nucleotide exchange factors for these Hsp70s. We note that loss of Sil1 function is linked to the human neurodegenerative disease Marinesco-Sjögren syndrome and may be nonlethal due to functional redundancy of Sil1 with Grp170 (Zimmermann et al., 2006). (B) Basic oligopeptides that are present in Sec62 and established ribosomal ligands in Homo sapiens. (C) Recombinant derivatives of human Sec62 and synthetic basic oligopeptides that are used throughout this study.
There also is considerable medical interest in human Sec62 and Sec63. Genetic work linked mutations in the SEC63 gene to polycystic liver disease (Davila et al., 2004). According to the two-hit hypothesis for this disease, mutations in the second allele in liver cells of heterozygous carriers of a SEC63 mutation do not result in cell death, but rather lead to cell proliferation and the progressive development of liver cysts. Furthermore, mutations in the SEC63 gene were described for HNPCC-associated small-bowel cancer (Schulmann et al., 2005) and microsatellite-unstable gastric and colorectal cancers (Mori et al., 2002). In addition, overexpression of the SEC62 gene was found to be associated with sporadic colorectal cancer (Eschrich et al., 2005) and prostate cancer (Jung et al., 2006).
Here, we provide direct evidence for conservation of Sec62/Sec63 interaction from yeast to humans and show that in the course of evolution Sec62 of vertebrates has gained a function, i.e., the ability to interact with the ribosomal tunnel exit. Two basic oligopeptide motifs are responsible for this interaction and are absent from yeast Sec62p as well as from Sec62 in invertebrates or plants. Thus, the human Sec62/Sec63 complex and human ERj1 are similar in providing a binding site for ribosomes on the cytosolic face and a binding site for BiP on the lumenal face of the ER membrane, i.e., may both be involved in cotranslational transport of polypeptides into the ER. This is consistent with the observation that Sec62 is protected against externally added antibodies by ribosomes in permeabilized human cells.
MATERIALS AND METHODS
Materials
Aurintricarboxylic acid (ATA) and CHAPS were from Calbiochem (La Jolla, CA). [35S]methionine and enhanced chemiluminescence (ECL) were obtained from GE Healthcare (Waukesha, WI). Sulfo-SMCC was purchased from Pierce (Rockford, IL). Antibodies against calnexin and protein disulfide isomerase (PDI) were from Stressgen (San Diego, CA); peroxidase-coupled anti-penta-histidine antibodies were from Qiagen (Hilden, Germany); peroxidase conjugate of anti-rabbit IgG goat antibodies were from Sigma (St. Louis, MO). Antibodies against yeast Sec62 (CNKKKAINEKAEQN) and human ribosomal proteins L4 (CEKKPTTEEKKPAA) and S3 (CGKPEPPAMPQPV) were raised against the indicated C-terminal oligopeptides.
Peptides were synthesized on a 433A peptide synthesizer (Applied Biosystems, Foster City, CA), cleaved, and deprotected as described previously (Dierks et al., 1996). Oligopeptides 62-11mer 1 and 2 corresponded to amino acid residues 2 through 12 and 157 through 167 of human Sec62, respectively. Peptide iv62-11mer corresponded to amino acid residues 2 through 12 of Sec62 isoform A of Drosophila melanogaster (SEKKRARRRKD).
Ribosomes were purified from canine pancreas by gradient centrifugation and washed with puromycin (0.5 mM) and high salt (500 mM KOAc). The A260/280 of the purified ribosomes was 1.98. Ribosomal subunits were prepared by incubation of washed ribosomes in high salt (1 M KCl) and subsequent sucrose gradient centrifugation according to published procedures (Spedding, 1990). The integrity of 80S ribosomes and ribosomal subunits was confirmed by analysis of molecular mass and hydrodynamic radius by a combination of asymmetric-flow field-flow fractionation (Eclipse 2, Wyatt Technology, Santa Barbara, CA) and light-scattering analysis (miniDAWN Tristar and QELS, Wyatt Technology; Supplemental Figure 1). Pretreatment of ribosomes with RNase A (80–240 μg/ml) was carried out by incubation for 30 min at 30°C.
Recombinant derivatives of human or yeast Sec62 with C-terminal hexa-histidine tag and derivatives of human Sec62 and Sec63, respectively, with N-terminal glutathione-S-transferase (GST) were purified from Escherichia coli. Sec62N corresponded to amino acid residues 1 through 195 of human Sec62, Sec62N-ΔN10 to 11-195, Sec62N-ΔC40 to 1-155 to Sec62N-ΔN10-ΔC40 to 11-155, and Sec62C to 279-399. Sec63C corresponded to amino acid residues 210 through 760 of human Sec63, Sec63C26 corresponded to residues 734-760. Yeast Sec62N corresponded to amino acid residues 1 through 149 and also contained a C-terminal hexa-histidine tag (Willer et al., 2003); “humanized” Sec62N was identical except that the residue 1 of yeast Sec62N was replaced by the 12 aminoterminal amino acid residues from human Sec62. The molecular mass of the purified human Sec62N was determined by a combination of asymmetric-flow field-flow fractionation and light-scattering analysis as 35 kDa (Supplemental Figure 2), which is consistent with a monomer (calculated mass of monomer: 23 kDa).
Pulldown Assay
Purified GST or related hybrid proteins (10 μg) were immobilized on GSH-Sepharose. Then, buffer (20 mM HEPES-KOH, pH 7.5, 150 mM KCl, 2 mM MgCl2, 0.65% CHAPS) or purified hexa-histidine fusion protein (12 μg) in the same buffer was applied to the resin. Where indicated oligopeptides (final concentration: 300 μg/ml) were added simultaneously. After washing, the bound material was eluted and analyzed by SDS-PAGE as followed by either protein staining with Coomassie Brilliant Blue or Western blotting plus immunodetection with anti-penta-histidine antibodies, coupled to peroxidase. The antibodies were visualized by incubation of the blots in ECL and subsequent luminescence imaging (Lumi-Imager F1 with LumiAnalyst software 3.1, Roche Diagnostics, Mannheim, Germany).
Ribosome-Binding Assay
Recombinant proteins (150 pmol) were diluted into 20 mM HEPES-KOH, pH 7.5, 200 mM KCl, 1.5 mM MgCl2, 1 mM EDTA, and 0.65% CHAPS and incubated in the simultaneous absence or presence of ribosomes (50 pmol) for 15 min at 30°C (Dudek et al., 2005). Where indicated oligopeptides (50 nmol) were present. Subsequently the mixture was layered onto a low-salt cushion (0.5 M sucrose in 40 mM HEPES-KOH, pH 7.5, 150 mM KOAc, 5 mM MgOAc, 2 mM DTT) and subjected to centrifugation for 90 min at 356,000 × g and 2°C (Beckman table top ultracentrifuge Optima MAX-E, Beckmann rotor TLA-120.2; Fullerton, CA). The supernatants were subjected to protein precipitation with trichloroacetic acid. The precipitates and the pellets, respectively, were subjected to SDS-PAGE and subsequent protein staining or Western blotting plus immunodetection with peroxidase-coupled anti-penta-histidine antibodies or antibodies against ribosomal proteins plus peroxidase conjugate of anti-rabbit IgG goat antibodies. The antibodies were visualized by incubation of the blots in ECL and subsequent luminescence imaging.
Alternatively, ribosomal complexes were subjected to sucrose gradient centrifugation (linear sucrose gradient between 10 and 60%, wt/vol in low-salt buffer, adjusted to 33 μg/ml BSA) for 60 or 90 min at 280,000 × g and 2°C (Beckman ultracentrifuge Optima L-80, Beckmann rotor SW 55 Ti; Dudek et al., 2002). After fractionation of the gradients, proteins were precipitated and subjected to SDS-PAGE and subsequent protein staining or Western blotting plus immunodetection as described above.
Surface Plasmon Resonance Spectroscopy
Surface plasmon resonance (SPR) spectroscopy was performed in a BIAlite upgrade system (Biacore, Freiburg, Germany). Sensor chip NTA was activated with Ni2+ according to the manufacturer's protocol (Biacore). Hexa-histidine–tagged proteins were immobilized on the chip surface at a flow rate of 10 μl/min in 10 mM HEPES-NaOH, pH 7.4, 150 mM NaCl, 50 μM EDTA, and 0.005% surfactant P20. For interaction analysis, the chip was equilibrated with running buffer (10 mM HEPES-KOH, pH 7.6, 150 mM NaCl, 0.125% digitonin) at a flow rate of 5 μl/min. Subsequently, solutions containing increasing concentrations of analyte were passed over the chip surface. Each analyte application was followed by application of running buffer and eventually with running buffer that was supplemented with high salt. The analysis was carried out by employing BIA evaluation software version 3.1 (Biacore).
In Vitro Translation
Synthesis of firefly luciferase, bovine preprolactin (ppl), or nascent preprolactin (ppl86mer) was carried out in rabbit reticulocyte lysate in the presence of [35S]methionine (translation kit type I or II, Roche Diagnostics; Dudek et al., 2005). The translation reactions contained either buffer (20 mM HEPES-KOH, pH 7.5, 500 mM KCl, 2 mM MgCl2, 2 mM DTT, 0.65% CHAPS), recombinant protein (final concentration: 1 μM) in buffer, DMSO (final concentration: 2%), or oligopeptide (final concentration: 150 μM) in DMSO. After incubation for various times at 30°C, the translation reactions were subjected to SDS-PAGE and phosphorimaging (Molecular Dynamics, Sunnyvale, CA; model SF with Image Quant software).
Chemical Cross-Linking
ppl86-mer was synthesized in reticulocyte lysate in the presence of [35S]methionine for 20 min (Dudek et al., 2005). After adjusting the translation reaction to 500 mM KOAc for 5 min at 0°C, the ribosomes were pelleted by centrifugation through a high-salt cushion. The ribosomes were resuspended in XL-buffer (50 mM HEPES-KOH, pH 7.5, 200 mM KOAc, 2 mM MgOAc, 0.2 M sucrose, 0.65% (wt/vol) CHAPS), divided into various aliquots, and supplemented with buffer or recombinant protein (2 μM). After incubation for 15 min at 30°C, ribosomes were reisolated by centrifugation through a low-salt cushion and incubated with Sulfo-SMCC (0.25 mM) for 20 min at 0°C. All samples were subjected to SDS-PAGE and phosphorimaging.
Yeast Manipulations
Yeast SEC62 ts mutant strain RDM50-94C (leu2-3 leu2-112 his4 ura3-52 sec62-1 MATα) was kindly provided by R. Schekman (Berkeley, CA). The CEN-multicopy vector YEp with URA3 as a selection marker was provided by G. Schlenstedt (Homburg). Cells were transformed with a derivative of YEp that contained the human SEC62 cDNA under control of the GAL1 promoter and were grown on SD medium without uracil at 24 or 37°C in the presence of glucose (2%) or galactose (2%).
Quantitative Fluorescence Microscopy
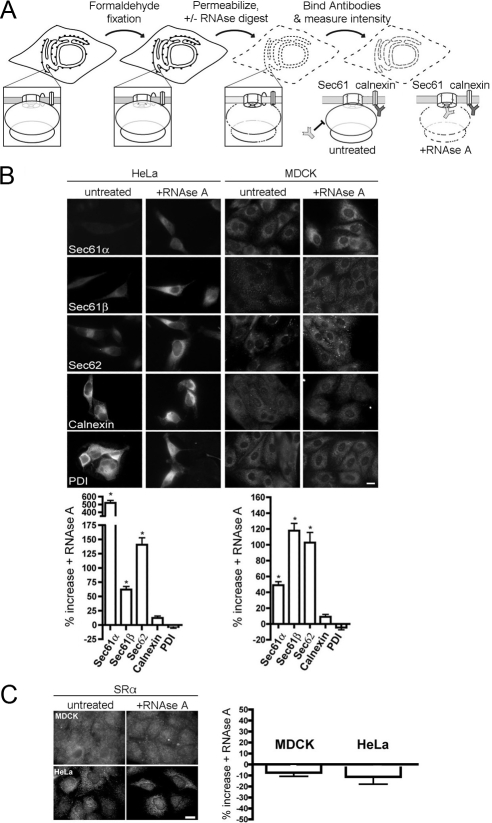
Cells were grown on eight-chambered Lab-Tek glass coverslips (Naperville, IL) in the respective standard media to 40–70% confluence. Cells were fixed with fresh 3.7% formaldehyde in PBS for 15 min at room temperature, permeabilized with PBS with 0.1% Triton X-100, and—where indicated—treated with 50 μg/ml RNase A during the block step in 10% fetal bovine serum/PBS. Subsequently, the cells were labeled with affinity-purified primary antibodies, followed by Alexa 555–conjugated anti-rabbit IgG secondary antibodies. Cells were imaged using fluorescence microscopy with a widefield microscope (Axiovert 200; Carl Zeiss Microimaging, Thornwood, NY; 63× oil 1.4 NA objective, 450–490 excitation/500–550 emission bandpass filter) and a Retiga 2000R camera. Image analysis was performed with ImageJ 1.39i (http://rsb.info.nih.gov/ij/). Figures were prepared using Microsoft Excel 2004 (Redmond, WA) and Adobe Photoshop CS2 and Adobe Illustrator 11.0 (San Jose, CA).
RESULTS
Human Sec62 and Sec63 Interact with Each Other in a Manner Similar to their Yeast Orthologues
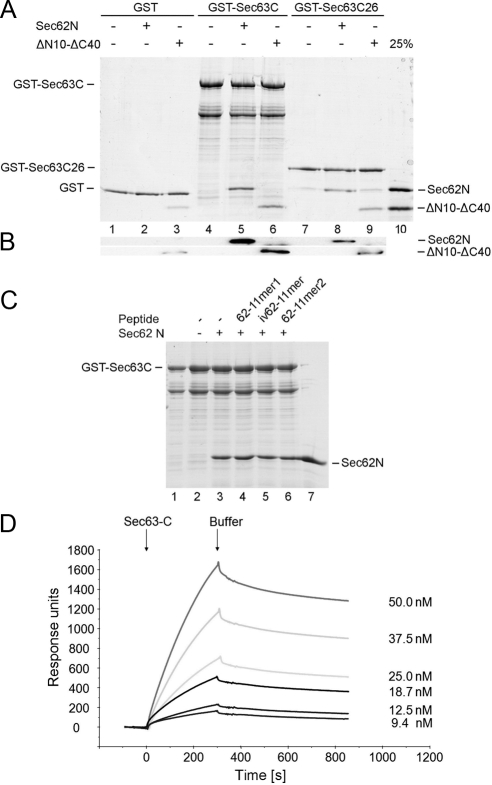
In yeast, the negatively charged carboxyterminus of Sec63p interacts with the overall positively charged aminoterminal (N-terminal) domain of Sec62p (Wittke et al., 2000; Figure 1A). Thus, we asked if this mode of interaction is conserved in the two human proteins. Purified GST hybrid proteins that comprised the complete carboxyterminal (C-terminal) domain of human Sec63 (termed Sec63C) or only its 26 C-terminal amino acid residues (Sec63C26) were immobilized on GSH-Sepharose. GST served as a negative control. Then, buffer or purified hexa-histidine fusion of the N-terminal domain of mammalian Sec62 (Sec62N) were applied to the resin. The bound material was eluted and analyzed by SDS-PAGE, followed by either protein staining (Figure 2A, lanes 1 and 2, 4 and 5, and 7 and 8) or Western blotting plus immunodetection with anti-penta-histidine antibodies (Figure 2B). Although there was no Sec62N detected in the eluate of the immobilized GST (lane 2), there was a significant amount of Sec62N found in the eluate of both GST-Sec63C (lane 5) as well as GST-Sec63C26 (lane 8). Thus, human Sec63 and Sec62 directly interact with each other, and the C-terminal 26 amino acid residues of Sec63C are sufficient for this interaction.
Figure 2.
Human Sec62N interacts with human Sec63C. (A and B) GST or the two GST hybrid proteins, GST-Sec63C or GST-Sec63C26, were immobilized on GSH-Sepharose. Buffer, Sec62N, or Sec62N-ΔN10-ΔC40 (1 μM) was applied to the resin. After washing, the bound material was eluted and analyzed by SDS-PAGE as followed by either protein staining (A) or Western blotting plus immunodetection with anti-penta-histidine antibodies (B). Twenty-five percent of the input of the two Sec62 derivatives were run on the stained gel for comparison (A, lane 10). (C) GST-Sec63C hybrid was immobilized. Buffer, Sec62N (1 μM), or Sec62N in combination with the indicated oligopeptide (225 μM) was applied to the resin. After washing, the bound material was eluted and analyzed by SDS-PAGE followed by protein staining. Aliquots of the input of GST-Sec63 (lane 1) and Sec62 (lane 7) were run on the same gel for comparison. iv62-11mer, oligopeptide from invertebrate Sec62. (D) SPR analysis of the Sec62/Sec63 interaction. Human Sec62N was immobilized on an activated NTA sensor chip in the measuring cell and human Sec62C as a negative control in the reference cell. Increasing concentrations of human Sec63C (as indicated) were passed over the chip and were followed by running buffer. Association and dissociation kinetics were recorded.
We analyzed which part of Sec62N was involved in this interaction: the central and overall positive region or one of the two positively charged oligopeptides at/near the amino- or carboxyterminus (Figure 1, A and B). This was addressed by employing a truncated Sec62N that lacks the two positively charged oligopeptides (termed Sec62N-ΔN10-ΔC40; Figure 1C) or the two positively charged oligopeptides (termed 62-11mer1 and 2; Figure 1C) as potential competitors of Sec62N, respectively. Sec62N-ΔN10-ΔC40 efficiently bound to GST-Sec63C as well as GST-Sec63C26 (Figure 2, A and B, lanes 6 and 9). Furthermore, even at a >100-fold molar excess (that is effective in the competition of ribosome binding; see below) the two basic oligopeptides did not interfere with binding of Sec62N to GST-Sec63C (Figure 2C). Thus the central and overall positive region of Sec62 is involved in interaction with Sec63C. This was confirmed by binding studies that used peptide spot membranes according to Frank (1992) (Supplemental Figures 3 and 4). In this respect, both mammalian proteins behaved like their yeast orthologues.
To further characterize the Sec62/Sec63 interaction, SPR experiments were carried out. Human Sec62N was immobilized in the measuring cell of a NTA sensor chip via its hexa-histidine tag. Human Sec62C served as a negative control and was immobilized in the reference cell. Then increasing concentrations of human Sec63C were passed over the chip and were followed by buffer. Association of the analyte and its dissociation were recorded and analyzed (Figure 2D). We could fit the kinetics with a 1:1 binding model and determined an apparent affinity (Kd) of Sec63C for Sec62N of 4.78 nM. This Kd was consistent with the fact that native Sec62 was coimmunoprecipitated with Sec63 from a microsomal detergent extract (Tyedmers et al., 2000).
Human Sec62 Interacts with 80S Ribosomes
The N-terminal domain of mammalian Sec62 (Sec62N) contains two basic oligopeptide motifs that are reminiscent of similar peptides in established ribosomal tunnel exit ligands (such as NAC and ERj1; Ferbitz et al., 2004; Blau et al., 2005; Figure 1B). Deletion of the basic oligopeptides in the N-terminal domains of mammalian and yeast NACβ (Grallath et al., 2006; Wegrzyn et al., 2006) as well as the cytosolic domain of ERj1 (Dudek et al., 2005) led to loss of ribosome-binding ability. Therefore, Sec62N was analyzed with respect to its ribosome-binding ability by a number of different experimental approaches, and the role of the two basic oligopeptide motifs was characterized. The same experimental strategies were previously used for the characterization of the ribosome interaction of ERj1 (Dudek et al., 2002, 2005).
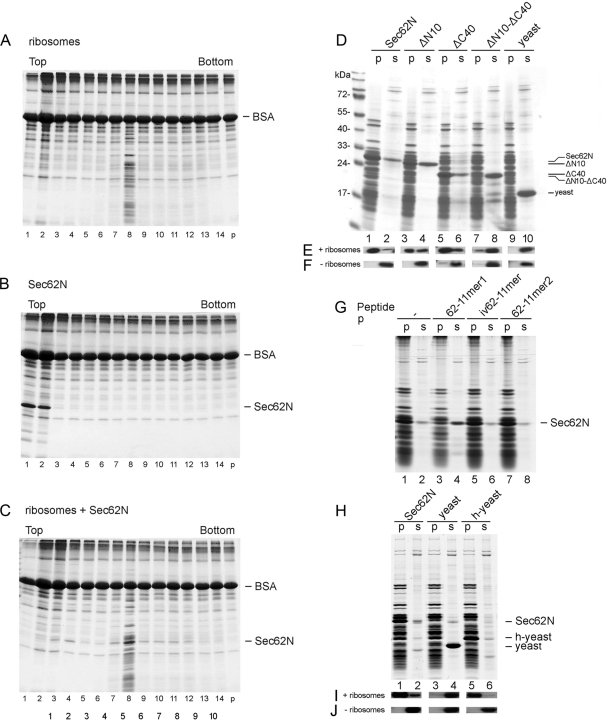
Sec62N was incubated in the presence or absence of nontranslating 80S ribosomes. An aliquot of ribosomes was incubated in the absence of Sec62N and served as reference (Figure 3A). The samples were analyzed by gradient centrifugation and subsequent SDS-PAGE and protein staining. In the absence of ribosomes, Sec62N stayed at the top of the gradient (Figure 3B). After incubation with ribosomes, Sec62N comigrated with ribosomes (Figure 3C). Thus the observed comigration of Sec62N with ribosomes was not due to aggregation, but rather reflected an interaction between the two molecules.
Figure 3.
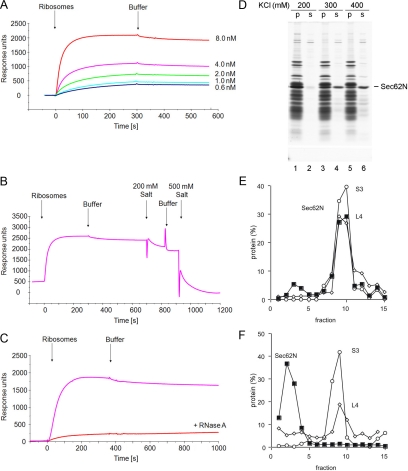
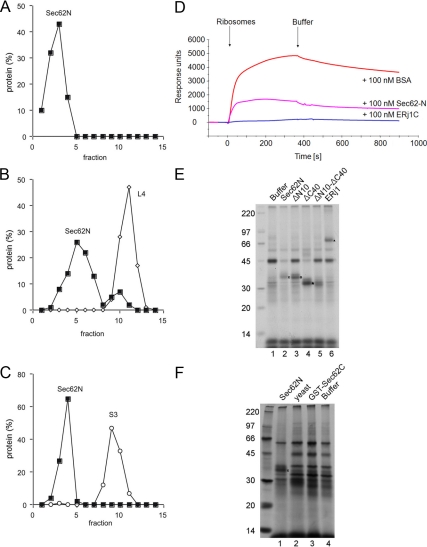
Ribosome binding of human Sec62N and inhibition by basic oligopeptides. (A–C) Sec62N was adjusted to KCl concentration of 100 mM and incubated without (B) or with ribosomes (C). A third mixture contained ribosomes but was free of Sec62N (A). Subsequently the samples were subjected to sucrose gradient centrifugation. After fractionation of the gradients, aliquots of the fractions and the pellets (p) were subjected to SDS-PAGE and protein staining. (D–F) Recombinant proteins were incubated in the presence (D and E) or absence (F) of nontranslating ribosomes. Subsequently, the mixtures were layered onto a sucrose cushion and subjected to ultracentrifugation. The pellets (p) and supernatants (s) were analyzed by SDS-PAGE and subsequent protein staining (D) and Western blotting plus immunodetection with anti-penta-histidine antibodies (E and F). (G) Sec62N (150 pmol) or Sec62N in combination with the indicated oligopeptide (50 nmol) were incubated in the presence of nontranslating ribosomes. Subsequently, the mixtures were layered onto a sucrose cushion and subjected to ultracentrifugation. The pellets and supernatants were analyzed by SDS-PAGE and subsequent protein staining. (H–J) Recombinant proteins were incubated in the presence (H and I) or absence (J) of nontranslating ribosomes. Subsequently, the mixtures were analyzed as in D–F. We note that the recombinant proteins did not pellet in the absence of ribosomes (F and J). yeast, yeast Sec62N; h-yeast, humanized yeast Sec62N.
Next, Sec62N and three truncated derivatives (Figure 1C) were each incubated in the presence or absence of nontranslating ribosomes in order to address the question of which part of Sec62N was involved in this interaction. Yeast Sec62N that lacks similar basic oligopeptide motifs as compared with human Sec62N (Figure 1A) served as negative control. Subsequently, the ribosomes were reisolated by centrifugation and the relative amount of ribosome associated Sec62N was determined by SDS-PAGE and protein staining (Figure 3D) or Western blotting plus immunodetection with anti-penta-histidine antibodies (Figure 3, E and F). All proteins stayed in the supernatant in the absence of ribosomes (Figure 3F). However, Sec62N and constructs that contained at least a single positively charged oligopeptide were pelleted together with ribosomes, i.e., were active in ribosome binding (although to a varying degree), whereas the construct with the double deletion (termed Sec62N-ΔN10-ΔC40) was almost completely inactive in ribosome binding (Figure 3, D and E, lanes 7 and 8). Furthermore, yeast Sec62N that lacks similarly charged oligopeptides was unable to bind ribosomes (Figure 3, D and E, lanes 9 and 10). Although both basic oligopeptides within Sec62N contributed to ribosome binding, the aminoterminal peptide seemed to be more important.
To further substantiate the role of the two highly charged oligopeptides for ribosome binding, synthetic oligopeptides were used as potential competitors of Sec62N in ribosome binding (peptides 62-11mer1 and 2; Figure 1C). The aminoterminal oligopeptide from an invertebrate Sec62 served as negative control (peptide iv62-11mer). Only the aminoterminal peptide from human Sec62 competed with Sec62N for ribosome binding (Figure 3G, lanes 3 and 4).
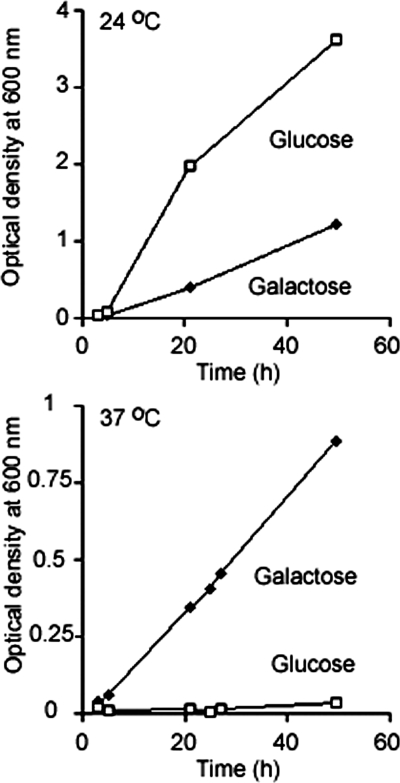
Having observed an interaction of human Sec62N with ribosomes, i.e., a gain of function of the human protein compared with the yeast protein, we asked if yeast Sec62N can be turned into a ribosome-binding protein by the addition of the aminoterminal dodecapeptide from human Sec62 and if human SEC62 is able to rescue the thermosensitive growth phenotype of a translocation-deficient yeast SEC62 mutant. Yeast Sec62N was extended at its aminoterminus by the aminoterminal dodecapeptide from human Sec62 (termed humanized Sec62N) and incubated in the presence or absence of nontranslating ribosomes. Subsequently, the ribosomes were reisolated by centrifugation and the relative amount of ribosome associated humanized Sec62N was determined by SDS-PAGE and protein staining or Western blotting plus immunodetection with anti-penta-histidine antibodies. In contrast to wild-type yeast Sec62N, humanized Sec62N bound to ribosomes (Figure 3, H–J). Thus the aminoterminal peptide that is present in human Sec62 is sufficient for ribosome binding. Because of a protein translocation defect at the nonpermissive temperature, the yeast mutant strain RDM50-94C hardly grows at 37°C (Deshaies and Schekman, 1990). However, when the human SEC62 gene was expressed in this strain after the addition of galactose, the cells grew at 37°C (Figure 4). Thus, the additional positively charged oligopeptides that are present in human Sec62 compared with yeast Sec62p do not interfere with the posttranslational function of yeast Sec62p.
Figure 4.
Human Sec62 complements a corresponding yeast mutant strain. Yeast SEC62 ts mutant strain RDM50-94C was grown at 24 or 37°C in the absence of uracil and in the presence of glucose or galactose as indicated. The cells had been transformed with a multicopy vector that contained URA3 as a selection marker and the human SEC62 cDNA under control of the GAL1 promoter. The optical density (OD) was measured at 600 nm.
To quantitatively characterize the ribosome interaction of mammalian Sec62, SPR experiments were carried out. Human Sec62N was immobilized in the measuring cell of a NTA sensor chip via its hexa-histidine tag. Based on the experiments that are depicted in Figure 3, D and E, yeast Sec62N served as a negative control and was immobilized in the reference cell. Then increasing concentrations of mammalian ribosomes were passed over the chip and were followed by buffer. Association of the analyte to and its dissociation from the ligand were recorded and analyzed (Figure 5A). We determined an apparent affinity (Kd) of ribosomes for Sec62N of 0.13 nM.
Figure 5.
Characterization of the Sec62/ribosome interaction. (A) Human Sec62N was immobilized on an activated NTA sensor chip in the measuring cell and yeast Sec62N as a negative control in the reference cell. Increasing concentrations of nontranslating ribosomes (as indicated) were passed over the chip and were followed by running buffer. Association and dissociation kinetics were recorded. (B) Sensorgram after application of ribosomes, running buffer, and subsequent application of running buffer with 200 and 500 mM KOAc, respectively. (C) Sensorgram after application of ribosomes and the same concentration of ribosomes that had been pretreated with RNase A (240 μg/ml), respectively, for 30 min at 30°C. (D) Sec62N was incubated in the presence of nontranslating ribosomes at the indicated salt concentrations. Subsequently, the mixture was layered onto a sucrose cushion and subjected to ultracentrifugation. The pellets and supernatants were analyzed by SDS-PAGE and subsequent protein staining. (E and F) Sec62N was incubated in the presence of ribosomes that had been pretreated with buffer (E) or RNase A in buffer (80 μg/ml; F) for 30 min at 30°C. Subsequently the samples were subjected to sucrose gradient centrifugation. The fractions were analyzed by SDS-PAGE and subsequent Western blotting plus immunodetection with anti-penta-histidine (■), anti-L4 (◇), and anti-S3 (○) antibodies. The Western blot signals were quantified by luminescence imaging and plotted against the fraction number.
Ribosome Association of Sec62 Is Salt- and RNase-sensitive
The interaction of ERj1 with ribosomes was shown to be salt-sensitive and to involve rRNA (Dudek et al., 2002 and 2005). Here, we asked if this is also true for Sec62N. Thus, ribosome binding assays and SPR experiments were carried out above the standard 100–150 mM salt concentration and by substituting ribosomes by RNase-treated ribosomes, respectively. Human Sec62N was immobilized in the measuring cell of a NTA sensor chip via its hexa-histidine tag, and yeast Sec62N was immobilized in the reference cell. After association of native ribosomes, the dissociation was carried out at 200 and 500 mM KOAc concentration (Figure 5B). According to the sensorgram, the interaction of Sec62N with ribosomes was salt-sensitive. When RNase treated ribosomes were used as analyte the sensorgram suggested that the interaction of Sec62N with ribosomes was RNase-sensitive (Figure 5C). Similar observations were made in the ribosome-binding assays (Figure 5, D and E vs. F). Thus binding of Sec62N to ribosomes most likely involves electrostatic interactions with rRNA. We note that the observed salt sensitivity of ribosome binding of Sec62N may explain why Sec62 was not characterized as ribosome-associated membrane protein after solubilization of microsomes in detergent (salt concentration: 400 mM KCl; Meyer et al., 2000; Tyedmers et al., 2000).
Sec62 Interacts with Ribosomes near or at the Tunnel Exit
Next, we used three independent experimental approaches to assess whether or not the ribosomal exit site of nascent polypeptides is also the binding site for Sec62N, because this site has previously been observed to provide a docking site for several proteins that are involved in protein biogenesis at the ER (SRP, NAC, Sec61 complex, ERj1; Beckmann et al., 2001; Halic et al., 2004; Ferbitz et al., 2004; Blau et al., 2005).
First we assessed if there was a preference of Sec62N for the large ribosomal subunit. Sec62N was incubated in the presence or absence of 60S and 40S ribosomal subunits. The samples were analyzed by gradient centrifugation and subsequent SDS-PAGE as followed by Western blotting plus immunodetection with anti-penta-histidine, anti-L4, or anti-S3 antibodies, respectively. After incubation with 60S subunits, a fraction of the Sec62N comigrated with ribosomes (Figure 6B). In the absence of ribosomes and in the presence of 40S subunits, Sec62N stayed at the top of the gradient (Figure 6, A and C). Thus, Sec62 can only interact with large ribosomal subunits.
Figure 6.
Sec62N binds near the ribosomal tunnel exit. (A–C) Sec62N was incubated in the absence (A) or presence of 60S (B) or 40S (C) ribosomal subunits. Subsequently the samples were subjected to sucrose gradient centrifugation. The fractions were analyzed by SDS-PAGE and subsequent Western blotting plus immunodetection with anti-penta-histidine (■), anti-L4 (◇), and anti-S3 (○) antibodies. The Western blot signals were quantified by luminescence imaging and plotted against the fraction number. (D) Human Sec62N was immobilized on an activated NTA sensor chip in the measuring cell and yeast Sec62N as a negative control in the reference cell. SPR sensorgram after application of ribosomes that had been preincubated with the indicated concentrations of Sec62N, ERj1C, or BSA for 10 min at 30°C. (E and F) ppl86mer was synthesized in reticulocyte lysate. The ribosomes were pelleted by ultracentrifugation and supplemented with buffer or recombinant protein as indicated (ERj1 served as a positive control; Dudek et al., 2005). After incubation and reisolation of ribosomes the cross-linking reagent was added, and the incubation was continued. All samples were subjected to SDS-PAGE. The positions of cross-linking products between ppl86mer and recombinant proteins are indicated. We note that different types of translation kit were used in the two experiments, which may account for the differences in cross-linking efficiencies.
Next, we analyzed the possible overlap in binding sites between Sec62N and ERj1 using SPR experiments (Blau et al., 2005). Sec62N was immobilized in the measuring cell of a NTA sensor chip via its hexa-histidine tag and analyzed with respect to binding of native ribosomes that had been preincubated with ERj1C (Figure 6D). Native ribosomes that had been preincubated with Sec62N served as positive control, and native ribosomes that had been preincubated with BSA as negative control. The results demonstrate that Sec62N and ERj1 compete for an overlapping binding site on ribosomes, most likely at or near the ribosomal tunnel exit.
To directly address this point, Sec62N or its derivatives were allowed to form complexes with ribosomes that contained radiolabeled nascent polypeptide chains of defined length (i.e., peptidyl-tRNAs comprising 86 amino-terminal amino acid residues of preprolactin, ppl86mer). Subsequently, the ribosome/nascent chain/Sec62N complexes were reisolated and subjected to chemical cross-linking. ERj1 served as a positive control (Dudek et al., 2005; Figure 6E, lane 6). In the presence of Sec62N or a construct that contains a single positively charged oligopeptide, cross-linked products of the nascent polypeptide were detected (Figure 6E, lanes 2–4). These were absent when buffer, Sec62N-ΔN10-ΔC40 (Figure 6E, lanes 1 and 5), yeast Sec62N (Figure 6F, lane 2), or human Sec62C were used (Figure 6F, lane 3). Thus, like ERj1, Sec62N binds to the ribosome near the tunnel exit. We note that there is a dominant 45-kDa cross-linking product to be seen in the absence of Sec62N that is hardly seen in the presence of Sec62N. We suggest that this cross-linking product involves a 36-kDa ribosomal protein (possibly L4; Woolhead et al., 2004) and that Sec62N that is bound near the tunnel exit affects the positioning of the nascent polypeptide chain with respect to this ribosomal protein. According to this interpretation, removal of the aminoterminal oligopeptide has a more pronounced effect on binding of Sec62N compared with removal of the carboxyterminal oligopeptide.
Sec62N Does Not Simultaneously Interact with Sec63C and Ribosomes
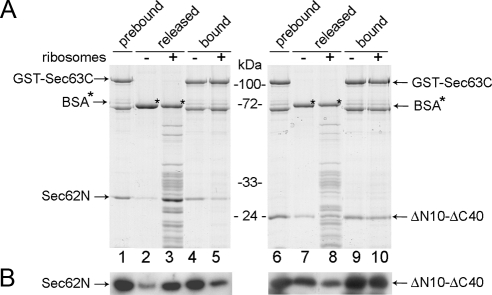
Having seen an interaction of Sec62N with ribosomes (Figure 3) and with Sec63C (Figure 2), we asked if Sec62N can recruit Sec63C to ribosomes. In ribosome binding assays, however, we failed to detect a trimeric complex (data not shown). Therefore, the immobilized complex between GST-Sec63C and Sec62N was used (Figure 7, left panels). In parallel, a complex was formed between GST-Sec63C and Sec62N-ΔN10-ΔC40, i.e., the truncated Sec62N that lacks the two positively charged oligopeptides (Figure 7, right panels). The resins were eluted with buffer or nontranslating ribosomes in the same buffer. Subsequently, the resins were eluted with SDS sample buffer, and all samples were analyzed as described above. On elution with buffer, Sec62N and Sec62N-ΔN10-ΔC40 remained bound to the immobilized Sec63C as expected (Figure 7, lanes 4 and 9). On elution with ribosomes, however, Sec62N eluted together with the ribosomes (lane 3 vs. 5). In contrast, Sec62N-ΔN10-ΔC40 that was unable to bind to ribosomes was not eluted with ribosomes (lane 8 vs. 10). Thus, interaction of Sec62N with Sec63C does not prevent binding of Sec62 to ribosomes, which is consistent with the observed Kd values for the interactions of Sec62N with Sec63C (4.78 nM) and ribosomes (0.13 nM).
Figure 7.
Sec62N but not Sec62N-ΔN10-ΔC40 is displaced from Sec63C by ribosomes. GST-Sec63C was immobilized on GSH-Sepharose. Sec62N (lanes 2–5) or Sec62N-ΔN10-ΔC40 (lanes 7–10) were applied to the resin in the presence of BSA. After washing, the resin was eluted with phosphate buffer (3 mM KCl, 1 mM MgCl2, 155 mM NaCl, 50 mM NaH2PO4, 200 mM Na2HPO4, pH 7.3, 0.65% CHAPS, 10 μg/ml BSA) or nontranslating ribosomes in phosphate buffer. After washing, the bound material was eluted and analyzed by SDS-PAGE followed by either protein staining (A) or Western blotting plus immunodetection with anti-penta-histidine antibodies (B).
Mammalian Sec62 Inhibits Translation at the Level of Initiation
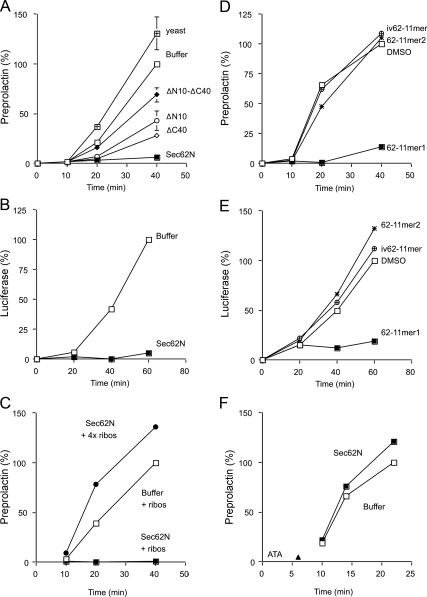
ERj1 was shown to be able to bind to ribosomes as well as to inhibit protein synthesis at the level of initiation (Dudek et al., 2005). Therefore, Sec62N and its derivatives were tested for their ability to inhibit synthesis of firefly luciferase or bovine preprolactin in reticulocyte lysate (Figure 8, A and B). In this assay, Sec62N and constructs that contained at least a single positively charged oligopeptide were active in inhibiting protein synthesis (although to a varying degree), and the construct with the double deletion (termed Sec62N-ΔN10-ΔC40) was less active (Figure 8A). Furthermore, yeast Sec62N that lacks similarly charged oligopeptides was inactive (Figure 8A). We note that a similar inhibitory effect of human Sec62N on the synthesis of luciferase, and preprolactin was observed in the presence of canine pancreatic microsomes (Supplemental Figure 5). A control experiment demonstrated that the translational inhibition activity of Sec62N was specific: the inhibitory effect of Sec62N on translation correlated reciprocally with the ribosome content of the reticulocyte lysate (Figure 8C).
Figure 8.
Inhibition of in vitro protein synthesis by Sec62N and by basic oligopeptides. (A and B) Preprolactin (A) or luciferase (B) were synthesized in reticulocyte lysate in the presence of [35S]methionine. The translation reactions were supplemented with buffer or recombinant protein (1 μM) in the same buffer. After the indicated incubation times, aliquots of the complete translation reactions were subjected to SDS-PAGE and phosphorimaging. The amount of preprolactin or luciferase that was present at the end of the buffer control reaction was set to 100%. Average values and SEMs are presented in A and were based on n = 6 (yeast) and n = 9 (all others). (C) Before translation, the reticulocyte lysate was fractionated into ribosomes and postribosomal supernatant by centrifugation (Dudek et al., 2002). Subsequently, the postribosomal supernatant was combined with the ribosomal pellet that corresponded to the same (□ and ■) or the fourfold (•) volume of reticulocyte lysate. The preprolactin was synthesized in the presence of buffer (□) or recombinant Sec62N (• and ■) and analyzed as described in A. (D and E) Preprolactin (D) or luciferase (E) were synthesized in reticulocyte lysate in the presence of [35S]methionine. The translation reactions were supplemented with DMSO or the indicated oligopeptide (150 μM) in DMSO and analyzed as described in A. (F) Synthesis of preprolactin was initiated for 6 min. Then aurintricarboxylic acid (ATA; 77 μM) was added in order to block further initiation (indicated by arrowhead), and the translation reactions were supplemented with buffer or recombinant protein in buffer and analyzed as described in A.
To further substantiate the role of the two highly charged oligopeptides for translational modulation by Sec62N, the synthetic oligopeptides were used in translation (peptides 62-11mer1 and 2; Figure 1C). Again, the aminoterminal oligopeptide from an invertebrate Sec62 served as negative control (peptide iv62-11mer). Indeed, the aminoterminal peptide from human Sec62N inhibited synthesis of preprolactin as well as luciferase (Figure 8, D and E).
In addition, Sec62N was tested for its ability to inhibit translation after inhibition of initiation (Figure 8F). Sec62N did not affect protein synthesis under these conditions. Thus, Sec62N inhibits synthesis of presecretory as well as nonsecretory proteins at the level of initiation.
Mammalian Sec62 Is Protected from Antibody Access by Ribosomes in Permeabilized Cells
Snapp et al. (2004) established a microscopic method to address the organization of protein translocase in the ER of mammalian cells. The experimental strategy was cell fixation, cell permeabilization, optional ribosome destruction by RNase treatment, and incubation with specific primary and fluorescently labeled secondary antibodies (Figure 9A). In the subsequent fluorescence microscopy and image analysis, the quantitative data from the RNase-treated cells were compared with the minus RNase control. Positive differential effects in fluorescence intensity were taken as an indication of association of the respective protein with ribosomes. Here, two different cell types (Madin-Darby canine kidney [MDCK] and HeLa) were analyzed with respect to Sec62 (Figure 9B). The Sec61α and Sec61β subunits of the translocase served as positive controls, the ER-membrane protein calnexin and the ER-lumenal PDI served as negative controls (Snapp et al., 2004). Although the extents of the differential effects varied between the two cell types, we detected a significant increase in fluorescence intensity after RNase treatment for Sec61α, Sec61β, and Sec62, but not for calnexin and PDI. Similar results were obtained for Cos-7 and HepG2 cells (data not shown). Thus, Sec62 is in the vicinity of ribosomes and, by extrapolation, of Sec61 complexes in the intact ER. This is consistent with our previous findings that Sec61 subunits can be coimmunoprecipitated from microsomal detergent extracts with antibodies against Sec62 and vice versa (Tyedmers et al., 2000).
Figure 9.
Sec62 is associated with ribosomes at the ER of permeabilized mammalian cells. (A) Experimental strategy. (B) HeLa and MDCK cells were fixed, left untreated or treated with RNase, and labeled with the indicated primary antibodies plus Alexa 555–conjugated secondary antibodies as described in Material and Methods. Fluorescence was recorded and quantified as described in Material and Methods. The difference in fluorescence intensity between the RNase-treated and the control sample is given as % change, and the SEM is indicated. Asterisk indicates significance as determined by Student's t test, i.e., p < 0.0001 relative to control (n > 20 cells). (C) HeLa and MDCK cells were fixed, left untreated or treated with RNase A, and labeled with the primary antibodies that were directed against SRa plus Alexa 555–conjugated secondary antibodies. We note that the axis scales are different for B versus C. Scale bar, 20 μm.
To rule out the possibility that the observed epitope protection is due to the ribonucleoprotein particles SRP rather than ribosomes, the α-subunit of the SRP receptor (SRα) was analyzed under identical conditions (Figure 9C). There was no increase in fluorescence intensity after RNase treatment for SRα in either MDCK or HeLa cells. Thus, the observed epitope protection in the case of Sec61α, Sec61β, and Sec62, was indeed due to ribosomes.
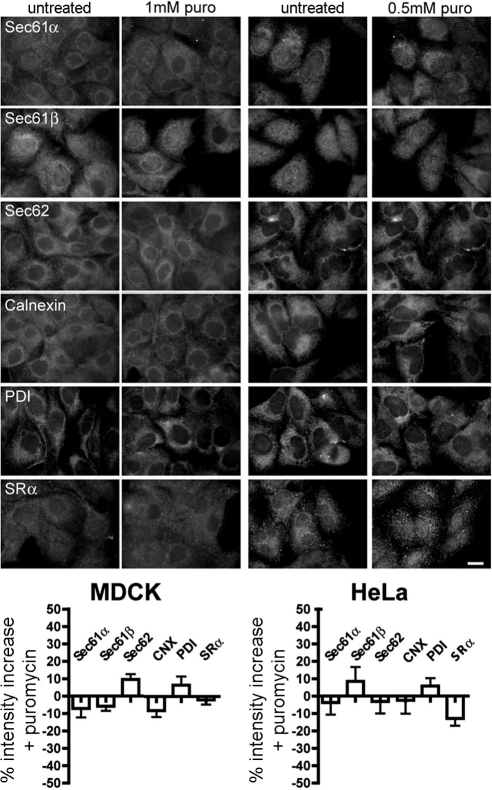
Next we analyzed if the effect of RNase treatment can be mimicked by puromycin treatment of MDCK and HeLa cells, i.e., under conditions where the nascent polypeptides are released from translating ribosomes (Figure 10). There was no intensity increase detected for any of the proteins after addition of puromycin. This supports the notion that many ribosomes or 60S ribosomal subunits do not leave the ER surface after termination of protein synthesis (Potter et al., 2001). Furthermore, these data suggested that the RNase experiments visualized translating as well as nontranslating ribosomes.
Figure 10.
Sec62 is associated with nontranslating ribosomes at the ER of permeabilized mammalian cells. HeLa and MDCK cells were incubated in the absence or presence of the indicated concentrations of puromycin for 1 h at 37°C. Then the cells were fixed and labeled with the indicated primary antibodies plus Alexa 555–conjugated secondary antibodies. We note that 1 mM puromycin is sufficient to inhibit translation within 5 min in MDCK cells and that the axis scales are different compared with Figure 9B. Scale bar, 20 μm.
DISCUSSION
Similarities between Human and Yeast Sec62
Here we demonstrated that the human ER membrane proteins Sec62 and Sec63 interact in the same manner as compared with their yeast orthologues, i.e., involving the conserved oppositely charged regions at the central region of Sec62N and the carboxyterminus of Sec63C (Wittke et al., 2000; Figure 1A). This is consistent with the observations that in T. brucei there is no Sec62 ortholog and that the trypanosomal Sec63 lacks the negatively charged carboxyterminal region (Goldshmidt et al., 2008; Supplemental Figures 6 and 7). Taken together with the fact that the two proteins are associated with each other and with the Sec61 complex in detergent extracts from both yeast (Jermy et al., 2006) and canine pancreatic microsomes (Meyer et al., 2000; Tyedmers et al., 2000), it is very likely that the two mammalian proteins play a similar role in protein biogenesis compared with their yeast orthologues, i.e., are involved in protein transport into the ER. This view is supported by our immunofluorescence analysis (see below).
Differences between Human and Yeast Sec62
Furthermore, we have shown that in the course of evolution Sec62 of vertebrates has gained a function, i.e., the ability to interact with the ribosomal tunnel exit. This is consistent with our observation that human Sec62 is associated with ribosomes at the ER of mammalian cells and with the fact that the majority of transport into the mammalian ER is cotranslational. In the case of Sec62, two basic oligopeptide motifs may be responsible that are absent from yeast Sec62p (Figure 1, A and B) as well as from Sec62 orthologues in invertebrates (such as Caenorhabditis elegans or Drosophila melanogaster) or plants (such as Arabidopsis thaliana or Oryza sativa; Supplemental Figures 6 and 7). Interestingly, this apparent gain of function did not interfere with the original function of the yeast protein because human Sec62 was able to complement the respective yeast ts mutant strain, as had previously been shown for the insect protein (Noel and Cartwright, 1994). Thus Sec62 showed most of the characteristics that were previously observed for ERj1 (Figure 1): 1) high salt– and RNase-sensitive ribosome binding of the cytosolic domain, 2) essential basic oligopeptide(s) in the cytosolic domain, 3) binding near the ribosomal tunnel exit, and 4) modulatory effect on initiation of protein synthesis (Dudek et al., 2002, 2005; Blau et al., 2005). Because Sec63 shares with ERj1 the ability to bind to BiP via its ER-luminal domain (J-domain), the human Sec62/Sec63 complex may have overlapping functions with ERj1. This view is supported by the observations that human ERj1 can complement a yeast Sec63p knockout (Kroczynska et al., 2004) and that loss of Sec63 function in human polycystic liver disease is not lethal (Davila et al., 2004; Zimmermann et al., 2006). We note that in the case of ERj1 the basic oligopeptide motif is also found only in vertebrate orthologues (Supplemental Figure 8).
Possible Function(s) of Human Sec62/Sec63 and ERj1
What are the specific functions of Sec62/Sec63 and ERj1 in the mammalian ER? We are convinced that the properties of the two proteins/complexes suggest a cotranslational function, most likely in protein transport into the ER. Both could serve in recruiting BiP to Sec61 complexes and/or incoming polypeptides (Dierks et al., 1996; Tyedmers et al., 2003; Alder et al., 2005). Furthermore, both proteins/complexes could be involved in the transport of those nascent polypeptides that are synthesized by ribosomes or 60S ribosomal subunits that stay permanently associated with the ER surface (Potter et al., 2001), i.e., do not depend on SRP and its receptor for targeting to the ER (Figure 10). In this case it would make perfect sense to allow initiation of protein synthesis only if BiP is available, i.e., is bound to an ER-lumenal J-domain. Indeed, we observed for ERj1 that binding of BiP to the J-domain still allows binding of the ERj1/BiP complex to the ribosome and that this heterodimeric complex does not inhibit translation (Dudek et al., 2005). We suggest that a similar mechanism may be in operation for the Sec62/Sec63-complex. Here the model is that binding of BiP to the ER-lumenal J-domain of Sec63 induces a Sec63/Sec62-interaction that allows binding of the aminoterminal domain of Sec62 to ribosomes in a way that is compatible with translation.
Why would there be two similar systems? There could be functional specialization: the one system could recruit BiP to ribosomes and the Sec61 complex for sealing of the Sec61 channel (Alder et al., 2005), whereas the other system could recruit BiP as a molecular ratchet for incoming polypeptide chains (Tyedmers et al., 2003). Alternatively, the two systems may have different substrate specificities. A first indication for a potential substrate specificity of ERj1 was reported by S. Y. Blond and coworkers (Kroczynska et al., 2004, 2005). In a two-hybrid screening these authors identified two interaction partners of the SANT-2 domain that is present in the cytosolic domain of human ERj1, the secretory proteins α1-antichymotrypsin (ACT, residues 140-400) and inter-α-trypsin inhibitor heavy chain 4 (ITIH4, residues 588-930). The two proteins have in common that they are secreted in the human liver, are serum protease inhibitors, and are acute-phase proteins. In both cases the signal peptides were not present, indicating that the mature proteins interacted with the SANT-2 domain. This may suggest that ACT and ITIH4 are substrates of ERj1 in the human liver. Therefore, we used the aminoterminal domain of human Sec62 as bait in a two-hybrid screening of a human liver cDNA library (data not shown). We identified four putative interaction partners, two uncharacterized proteins and the precursors of two secretory proteins, haptoglobin (residues 1-296) and complement factor B (residues 1-237). The two secretory proteins have in common that they are secreted in the human liver, are serum proteins with a serine protease domain, and contain complement control protein (CCP)-domains or SUSHI repeats in their aminoterminal regions (two in residues 1-296 of haptoglobin and three in residues 1-237 of complement factor B). In both cases the signal peptides were present, possibly indicating that the signal peptides interacted with Sec62N. A signal peptide receptor function of human Sec62 would not be unexpected because the yeast Sec62p was shown to be part of the signal peptide receptor that is involved in posttranslational transport. Thus, these observations may indicate that haptoglobin and complement factor B may be substrates of Sec62 in the human liver. In future work, this interpretation of the two two-hybrid screenings has to be evaluated after siRNA-mediated silencing of the ERJ1 and the SEC62 gene, respectively.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Dr. R. S. Hegde (National Institute of Child Health and Human Development, Bethesda, MD) for his kind gift of anti-Sec61α antibodies and to Dr. G. Schlenstedt (Homburg) for his advice with respect to the complementation analysis. Furthermore, we acknowledge critical reading of the manuscript by Dr. G. L. Blatch (Rhodes University, Grahamstown, South Africa) and expert technical assistance by Simone Amann and Nicole Heim. E.L.S. is an Ellison Medical Foundation New Scholar in Aging. He is also supported by National Institutes of Health Grant R21 DK074650-01 and a pilot award from the Marion Bessin Liver Center. M.D.deE., C.B., and L.M. were supported by fellowships from the Deutsche Forschungsgemeinschaft (DFG). In addition, this work was supported by grants from Homburger Forschungsförderungsprogramm (HOMFOR) and DFG (SFB 530).
Abbreviations used:
- BiP
immunoglobulin heavy-chain binding protein
- ER
endoplasmic reticulum
- GST
glutathione S-transferase
- Hsp
heat shock protein
- NAC
nascent polypeptide-associated complex
- PDI
protein disulfide isomerase
- SPR
surface plasmon resonance
- SRP
signal recognition particle.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-08-0730) on January 13, 2010.
REFERENCES
- Alder N. N., Shen Y., Brodsky J. L., Hendershot L. M., Johnson A. E. The molecular mechanism underlying BiP-mediated gating of the Sec61 translocon of the endoplasmic reticulum. J. Cell Biol. 2005;168:389–399. doi: 10.1083/jcb.200409174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann R., Spahn C.M.T., Eswar N., Helmers J., Penczek P., Sali A., Frank J., Blobel G. Architecture of the protein-conducting channel associated with the translating 80S ribosome. Cell. 2001;107:361–372. doi: 10.1016/s0092-8674(01)00541-4. [DOI] [PubMed] [Google Scholar]
- Blau M., Mullapudi S., Becker T., Dudek J., Zimmermann R., Penczek P., Beckmann R. ERj1p uses a universal ribosomal adaptor site to coordinate the 80S ribosome at the membrane. Nat. Struct. Mol. Biol. 2005;12:1015–1016. doi: 10.1038/nsmb998. [DOI] [PubMed] [Google Scholar]
- Davila S., et al. Mutations in SEC63 cause autosomal dominant polycystic liver disease. Nat. Genet. 2004;36:575–577. doi: 10.1038/ng1357. [DOI] [PubMed] [Google Scholar]
- Deshaies R. J., Schekman R. Structural and functional dissection of Sec62p, a membrane-bound component of the yeast endoplasmic reticulum protein import machinery. Mol. Cell. Biol. 1990;10:6024–6035. doi: 10.1128/mcb.10.11.6024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dierks T., Volkmer J., Schlenstedt G., Jung C., Sandholzer U., Zachmann K., Schlotterhose P., Neifer K., Schmidt B., Zimmermann R. A microsomal ATP-binding protein involved in efficient protein transport into the mammalian endoplasmic reticulum. EMBO J. 1996;15:6931–6942. [PMC free article] [PubMed] [Google Scholar]
- Dudek J., et al. A novel type of co-chaperone mediates transmembrane recruitment of DnaK-like chaperones to ribosomes. EMBO J. 2002;21:2958–2967. doi: 10.1093/emboj/cdf315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudek J., Greiner M., Müller A., Hendershot L. M., Kopsch K., Nastainczyk W., Zimmermann R. ERj1p has a basic role in protein biogenesis at the endoplasmic reticulum. Nat. Struct. Mol. Biol. 2005;12:1008–1014. doi: 10.1038/nsmb1007. [DOI] [PubMed] [Google Scholar]
- Eschrich S., et al. Molecular staging or survival prediction of colorectal cancer patients. J. Clin. Oncol. 2005;23:3526–3535. doi: 10.1200/JCO.2005.00.695. [DOI] [PubMed] [Google Scholar]
- Ferbitz L., Maier T., Patzelt H., Bukau B., Deuerling E., Ban N. Structure of the trigger factor chaperone complex with the ribosome defines the molecular environment of the emerging nascent protein chain. Nature. 2004;431:590–596. doi: 10.1038/nature02899. [DOI] [PubMed] [Google Scholar]
- Frank R. Spot synthesis: an easy technology for positionally addressable, parallel chemical synthesis on a membrane support. Tetrahedron. 1992;48:9217–9232. [Google Scholar]
- Goldshmidt H., Sheiner L., Bütikofer P., Roditi I., Uliel S., Günzel M., Engstler M., Michaeli S. Role of protein translocation pathways across the ER in Trypanosoma brucei. J. Biol. Chem. 2008;283:32085–32098. doi: 10.1074/jbc.M801499200. [DOI] [PubMed] [Google Scholar]
- Grallath S., Schwarz J. P., Böttcher U.M.K., Bracher A., Hartl F. U., Siegers K. L25 functions as a conserved ribosomal docking site shared by nascent chain-associated complex and signal recognition particle. EMBO Rep. 2006;7:78–84. doi: 10.1038/sj.embor.7400551. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Halic M., Becker T., Pool M. R., Spahn C.M.T., Grassucci R. A., Frank J., Beckmann R. Structure of the signal recognition particle interacting with the elongation-arrested ribosome. Nature. 2004;427:808–814. doi: 10.1038/nature02342. [DOI] [PubMed] [Google Scholar]
- Hamman B. D., Hendershot L. M., Johnson A. E. BiP maintains the permeability barrier of the ER membrane by sealing the lumenal end of the translocon pore before and early in translocation. Cell. 1998;92:747–758. doi: 10.1016/s0092-8674(00)81403-8. [DOI] [PubMed] [Google Scholar]
- Jermy A. J., Willer M., Davis E., Wilkinson B. M., Stirling C. J. The Brl domain of Sec63p is required for assembly of functional ER translocons. J. Biol. Chem. 2006;281:7899–7906. doi: 10.1074/jbc.M511402200. [DOI] [PubMed] [Google Scholar]
- Jung V., Kamradt J., Kindich R., Jung M., Mueller M., Schulz W. A., Engers R., Stoeckle M., Zimmermann R., Wullich B. B. Genomic and expression analysis of the 3q25–q26 amplicon reveals TLOC1/SEC62 as a probable target gene in prostate cancer. Mol. Cancer Res. 2006;4:169–176. doi: 10.1158/1541-7786.MCR-05-0165. [DOI] [PubMed] [Google Scholar]
- Kalies K.-U., Allan S., Sergeyenko T., Kröger H., Römisch K. The protein translocation channel binds proteasomes to the endoplasmic reticulum membrane. EMBO J. 2005;24:2284–2293. doi: 10.1038/sj.emboj.7600731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroczynska B., Evangelista C. M., Samant S. S., Elguindi E. C., Blond S. Y. The SANT2 domain of murine tumor cell DnaJ-like protein 1 human homologue interacts with α1-antichymotrypsin and kinetically interferes with its serpin inhibitory activity. J. Biol. Chem. 2004;279:11432–11443. doi: 10.1074/jbc.M310903200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroczynska B., King-Simmons L., Alloza L., Alava M. A., Elguindi E. C., Blond S. Y. BIP co-chaperone MTJ1/ERDJ1 interacts with inter-α-trypsin inhibitor heavy chain 4. Biochem. Biophys. Res. Comm. 2005;338:1467–1477. doi: 10.1016/j.bbrc.2005.10.101. [DOI] [PubMed] [Google Scholar]
- Meyer H.-A., Grau H., Kraft R., Kostka S., Prehn S., Kalies K.-U., Hartmann E. Mammalian Sec61 is associated with Sec62 and Sec63. J. Biol. Chem. 2000;275:14550–14557. doi: 10.1074/jbc.275.19.14550. [DOI] [PubMed] [Google Scholar]
- Mori Y., et al. Instabilotyping reveals unique mutational spectra in microsatellite-unstable gastric cancers. Cancer Res. 2002;62:3641–3645. [PubMed] [Google Scholar]
- Noel P. J., Cartwright I. L. A Sec62p-related component of the secretory protein translocon from Drosophila displays developmentally complex behavior. EMBO J. 1994;13:5253–5261. doi: 10.1002/j.1460-2075.1994.tb06859.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plemper R. K., Böhmler S., Bordallo J., Sommer T., Wolf D. H. Mutant analysis links the translocon and BiP to retrograde protein transport for ER degradation. Nature. 1997;388:891–895. doi: 10.1038/42276. [DOI] [PubMed] [Google Scholar]
- Potter M. D., Seiser R. M., Nicchitta C. V. Ribosome exchange revisited: a mechanism for translation-coupled ribosome detachment from the ER membrane. Trends Cell Biol. 2001;11:112–115. doi: 10.1016/s0962-8924(00)01905-x. [DOI] [PubMed] [Google Scholar]
- Schulmann K., et al. HNPCC-associated small bowel cancer: clinical and molecular characteristics. Gastroenterology. 2005;128:590–599. doi: 10.1053/j.gastro.2004.12.051. [DOI] [PubMed] [Google Scholar]
- Snapp E. L., Reinhart G. A., Bogert B. A., Lippincott-Schwartz J., Hegde R. S. The organization of engaged and quiescent translocons in the endoplasmic reticulum of mammalian cells. J. Cell Biol. 2004;164:997–1007. doi: 10.1083/jcb.200312079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spedding G. Isolation and analysis of ribosomes from prokaryotes, eukaryotes, and organelles. In: Spedding G., editor. Ribosomes and Protein Synthesis: A Practical Approach. Oxford: IRL Press; 1990. pp. 1–29. [Google Scholar]
- Tyedmers J., Lerner M., Bies C., Dudek J., Skowronek M. H., Haas I. G., Heim N., Nastainczyk W., Volkmer J., Zimmermann R. Homologs of the yeast Sec complex subunits Sec62p and Sec63p are abundant proteins in dog pancreas microsomes. Proc. Natl. Acad. Sci. USA. 2000;97:7214–7219. doi: 10.1073/pnas.97.13.7214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyedmers J., Lerner M., Wiedmann M., Volkmer J., Zimmermann R. Polypeptide-binding proteins mediate completion of cotranslational protein translocation into the mammalian endoplasmic reticulum. EMBO Rep. 2003;4:505–510. doi: 10.1038/sj.embor.embor826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegrzyn R. D., Hofmann D., Merz F., Nikolay R., Rauch T., Graf C., Deuerling E. A conserved motif is prerequisite for the interaction of NAC with ribosomal protein L23 and nascent chains. J. Biol. Chem. 2006;281:2847–2857. doi: 10.1074/jbc.M511420200. [DOI] [PubMed] [Google Scholar]
- Willer M., Jermy A. J., Young B. P., Stirling C. J. Identification of novel protein-protein interactions at the cytosolic surface of the Sec63-complex in the yeast ER membrane. Yeast. 2003;20:133–148. doi: 10.1002/yea.954. [DOI] [PubMed] [Google Scholar]
- Wittke S., Dünnwald M., Johnsson N. Sec62p, a component of the endoplasmic reticulum translocation machinery, contains multiple binding sites for the Sec-complex. Mol. Biol. Cell. 2000;11:3859–3871. doi: 10.1091/mbc.11.11.3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolhead C. A., McCormick P. J., Johnson A. E. Nascent membrane and secretory proteins differ in FRET-detected folding far inside the ribosome and in their exposure to ribosomal proteins. Cell. 2004;116:725–736. doi: 10.1016/s0092-8674(04)00169-2. [DOI] [PubMed] [Google Scholar]
- Zimmermann R., Müller L., Wullich B. Protein transport into the endoplasmic reticulum: mechanisms and pathologies. Trends Mol. Med. 2006;12:567–573. doi: 10.1016/j.molmed.2006.10.004. [DOI] [PubMed] [Google Scholar]
- Zimmermann R., Blatch G. L. A novel twist to protein secretion in eukaryotes. Trends Parasitol. 2009;25:147–150. doi: 10.1016/j.pt.2009.01.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.