The functional significance of nuclear translocation of β-actin remains unclear. Here, we demonstrate that PMA induces β-actin accumulation in the nucleus and binding to various target genes with different functions. We also find that accumulated nuclear β-actin is involved in recruitment of RNA polymerase II and in transcription regulation.
Abstract
Studies have shown that nuclear translocation of actin occurs under certain conditions of cellular stress; however, the functional significance of actin import remains unclear. Here, we demonstrate that during the phorbol 12-myristate 13-acetate (PMA)-induced differentiation of HL-60 cells toward macrophages, β-actin translocates from the cytoplasm to the nucleus and that this process is dramatically inhibited by pretreatment with p38 mitogen-activated protein kinase inhibitors. Using chromatin immunoprecipitation-on-chip assays, the genome-wide maps of β-actin binding to gene promoters in response to PMA treatment is analyzed in HL-60 cells. A gene ontology-based analysis shows that the identified genes belong to a broad spectrum of functional categories such as cell growth and differentiation, signal transduction, response to external stimulus, ion channel activity, and immune response. We also demonstrate a correlation between β-actin occupancy and the recruitment of RNA polymerase II at six selected target genes, and β-actin knockdown decreases the mRNA expression levels of these target genes induced by PMA. We further show that nuclear β-actin is required for PMA-induced transactivation of one target gene, solute carrier family 11 member 1, which is important for macrophage activation. Our data provide novel evidence that nuclear accumulation of β-actin is involved in transcriptional regulation during macrophage-like differentiation of HL-60 cells.
INTRODUCTION
Actin is one of the most abundant proteins in eukaryotic cells. It has been extensively studied as a cytoplasmic cytoskeletal protein that plays important roles in cellular processes such as cell motility, growth, cytokinesis, endocytosis, and intracellular trafficking (Brakebusch and Fassler, 2003; Suetsugu and Takenawa, 2003; Ascough, 2004). For many years, its presence in the nucleus has been questioned. However, in recent years, convincing evidence has clearly demonstrated that actin, actin-related proteins (ARPs), as well as actin-binding proteins, are not only present in the nucleus but also play important roles in diverse nuclear activities such as chromatin remodeling and RNA transcription (Olave et al., 2002; Blessing et al., 2004; Chen and Shen, 2007). The mammalian BAF (BRG1/hBRM-associated factor) complex is one of several SWI/SWF-like chromatin-remodeling complexes that can remodel chromatin in vitro. Purification of the subunits of the BAF complex revealed that the complex consists of actin, ARP BAF53, and nine additional proteins. When the actin subunit is absent from the complex, the activity of BAF is reduced and overall chromatin reorganization is altered (Zhao et al., 1998; Rando et al., 2002). Chromatin-remodeling INO80 complexes lacking actin as well as Arp5 and Arp8 are compromised for ATPase activity and DNA binding, suggesting that actin, Arp5, and Arp8 are necessary for these activities and for ATP-dependent chromatin remodeling (Shen et al., 2003). Involvement of nuclear actin in RNA transcription was reported many years ago (Smith et al., 1979; Egly et al., 1984; Scheer et al., 1984) but was received with skepticism. Recently, Percipalle et al. (2001) demonstrated that actin bound to the heterogeneous nuclear ribonucleoprotein hrp36 is necessary for transcription from Balbiani rings by RNA polymerase (RNAP) II in Chironomus tentants and that actin–hrp65-2 interaction is required for the maintenance of normal transcriptional activity in the cell (Percipalle et al., 2003). Furthermore, several studies have indicated that nuclear actin is associated with transcription by RNA polymerases (Pol) I (Fomproix and Percipalle, 2004; Philimonenko et al., 2004; Sjolinder et al., 2005), II (Hofmann et al., 2004; Kukalev et al., 2005; Sjolinder et al., 2005), and III (Hofmann et al., 2004; Hu et al., 2004; Philimonenko et al., 2004; Kukalev et al., 2005). In addition to chromatin remodeling and transcription, actin has also been implicated in other nuclear processes, including assembly of the nuclear structure (Krauss et al., 2002, 2003), RNA processing (Hofmann et al., 2001; Percipalle et al., 2001), and nuclear export (Hofmann et al., 2001).
Although actin accumulation in the nuclei in response to cellular stress has been reported previously, it is still not clear whether the occurrence of actin in nuclei is universal and what biological significance it has. Sanger et al. (1980a,b) demonstrated that a disappearance of stress fibers from the cytoplasm and a reversible translocation of cytoplasmic actin into the nucleus occurred after treatment of PtK2 and WI-38 cells with 10% dimethyl sulfoxide. Courgeon et al. (1993) showed that heat shock caused actin to accumulate in the nucleus in Drosophila cells. In mast cells, entry of actin into the nucleus was induced by either treatment with latrunculin B, which led to disassembly of F-actin in the cytoplasm, or depletion of ATP (Pendleton et al., 2003). It has been suggested that nuclear actin is in a soluble monomeric form rather than in a filament polymeric form as that in the cytoskeleton (Gonsior et al., 1999; Bettinger et al., 2004). So far, the molecular mechanism by which actin enters into the nucleus in response to cellular stress has not been established, and the potential functions of nuclear accumulation of actin in response to external signals remain unclear. It has been reported that nuclear actin is involved in gene transcription either by direct interaction with the DNA (Ou et al., 2005) or indirectly as a member of a DNA-binding complex such as chromatin remodeling (Zhao et al., 1998). Under stress, β-actin translocates into nuclei to function as a transcriptional modulator, playing an important role in the regulation of gene transcription along with stress-activated transcription factor.
In the present study, we find that when HL-60 cells and human peripheral blood monocytes are differentiated toward macrophages by phorbol 12-myristate 13-acetate (PMA), β-actin translocates from the cytoplasm to the nucleus and accumulates therein. To establish whether the accumulated nuclear β-actin is involved in gene transcription regulation during the differentiation of HL-60 cells toward macrophages, the chromatin immunoprecipitation (ChIP)-on-chip assay was used to identify β-actin target genes. Gene ontology (GO)-based analysis shows a broad spectrum of functional categories of the β-actin–enriched genes. Our data also demonstrate that nuclear translocation of β-actin is involved in regulating gene transcription during macrophage differentiation.
MATERIALS AND METHODS
Plasmid Constructs
pREP4-luc was constructed as described previously (Liu et al., 2001). The promoter regions of the solute carrier family 11 member 1 (SLC11A1) gene and SCG2 gene were amplified using human genomic DNA as a template, prepared from U937 cells, and using the following two pairs of primers: 5′-GAGCTAGCACTCCAGTCTGGGCAACAGAGTAA-3′ and 5′-CAAAGCTTAGTGCCCTGCCTCTTACATCAACA-3′; 5′-GAGCTAGCGTACGAAGCTTCCTTTCGATTGCA-3′ and 5′-CAAAGCTTGGCTCCACAGCATATTCCTCCCGTTCT-3′. The two polymerase chain reaction (PCR) products (one product covering nucleotides −750 to +46 of the human SLC11A1 promoter and showing the highest peak of enrichment by the β-actin antibody, and the other product covering nucleotides −857 to +52 of the human SCG2 promoter) were gel purified and digested with NheI and HindIII and then cloned into pREP4-luc vector. These two constructs were termed pREP4-SLC11A1–Luc and pREP4-SCG2–Luc, respectively.
Preparation of Monocytes and Cell Culture
Human venous blood from healthy medication-free volunteers was collected, and isolation of peripheral blood monocytes was performed as described previously (Xu et al., 2005). Both monocytes and HL-60 cells were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS), 2 mM l-glutamine, and 1% penicillin/streptomycin.
Western Blot Analysis
After the appropriate treatment, total cellular extracts as well as nuclear and cytoplasmic fractions were prepared and Western blot analysis was performed as described previously (Xu et al., 2005). A monoclonal antibody (mAb) against glyceraldehyde-3-phosphate dehydrogenase (GAPDH; Sigma-Aldrich, St. Louis, MO) was used at a 1:10,000 dilution and a mAb against β-actin (clone AC-15; Sigma-Aldrich) was used at a 1:5000 dilution. A polyclonal antibody against histone deacetylase (HDAC)1 (Santa Cruz Biotechnology, Santa Cruz, CA) or green fluorescent protein (GFP; Invitrogen, Carlsbad, CA) was used at a 1:1000 dilution.
Immunoprecipitation Assays
Cells were treated with PMA for 48 h or left untreated; subsequently, the cells were washed once with ice-cold phosphate-buffered saline (PBS) and lysed in EBMK/0.1% NP-40 buffer (25 mM HEPES, pH 7.6, 5 mM MgCl2, 1.5 mM KCl, 75 mM NaCl, 175 mM sucrose, 0.1% NP-40, and protease inhibitors) on ice for 10 min. The nuclear pellet was collected by centrifugation at 500 × g for 4 min and washed three times with EBMK buffer (no NP-40). The nuclei were then lysed in 1 ml of radioimmunoprecipitation assay (RIPA) buffer containing protease inhibitors, passed repeatedly through a 22-gauge needle, and centrifuged at 10,000 × g for 30 min. The supernatants were precleared with protein A/G agarose for 30 min. Immunoprecipitation was performed overnight at 4°C using the antibody against RNAP II (Covance, Berkeley, CA) or GFP. To precipitate the antigen–antibody complex, protein A/G agarose was added and incubated for 1 h at 4°C. After washing with RIPA buffer, the precipitated proteins were eluted by boiling in SDS sample buffer and analyzed by immunoblotting using antibodies to β-actin or RNAP II.
ChIP-on-Chip
HL-60 cells were treated with PMA (10 ng/ml) or left untreated. A complete protocol for chromatin immunoprecipitation and amplification can be found on the website at http://www.vmrf.org/researchcenters/gene-chip/chromatin_immunoprecipitation.pdf. High-density promoter arrays (2006-04-28 HG18_RefSeq_promoter arrays) were created by NimbleGen (Madison, WI) systems and contained 390,000 50–75 mer probes per array that tiled through 2200 base pairs upstream and 500 base pairs downstream of the transcriptional start sites of the selected genes. Promoter arrays were hybridized and data were extracted by NimbleGen system as part of a chromatin immunoprecipitation custom array service.
ChIP-Quantitative (q)PCR
The ChIP assay was performed by using a chromatin immunoprecipitation assay kit (Millipore, Billerica, MA) according to the manufacturer's instructions. HL-60 cells were treated with PMA for 48 h or left untreated and then fixed with 1% formaldehyde for 10 min, washed with ice-cold PBS containing protease inhibitors, and lysed with SDS lysis buffer. The lysate was sonicated to yield DNA fragments between 300 and 1000 base pairs and centrifuged at 13,000 rpm for 10 min. The supernatant was diluted 10-fold with ChIP dilution buffer and precleared with salmon sperm DNA/protein A agarose. Immunoprecipitation was performed overnight at 4°C using either nonspecific mouse immunoglobulin (Ig)G or the antibodies against β-actin (clone AC-15; Sigma-Aldrich) or RNAP II (clone 8WG16; Covance). Immunoprecipitates were washed and eluted, and the cross-links were reversed by adding 20 μl of 5 M NaCl to the eluates and heating for 4 h at 65°C. The precipitated DNA fragments were purified and quantified by Quant-iT dsDNA assay kit (Invitrogen) and were then amplified by real-time qPCR as described in the following Quantitative Real-Time PCR section. Primer sequences used for real-time PCR are listed in Supplemental Table S1.
Small RNA Interference (siRNA) Experiment
HL-60 cells were transiently transfected with either control or β-actin siRNA using the Cell Line Nucleofector kit V (Amaxa Biosystems, Gaithersburg, MD) according to the manufacturer's instructions. In brief, 2 × 106 log-growth cells were suspended in 100 μl of Cell Line Nucleofector solution V and mixed with 2.5 μg of control siRNA or β-actin siRNA. The siRNA duplexes used in the experiment are Silencer β-actin siRNA (Ambion, Austin, TX). Transfection was performed with a Nucleofector II device using the program T-019. Transfected cells were cultured for 24 h at 37°C in 5% CO2 and were then further treated with PMA (10 ng/ml) for 48 h or left untreated, followed by real-time quantitative reverse transcription (RT)-PCR analysis.
Quantitative Real-Time PCR
Total RNA was extracted from cells using TRIzol Reagent (Invitrogen) according to the manufacturer's instructions. Next, 1 μg of total RNA was reverse transcribed with the QuantiTect reverse transcription kit (QIAGEN, Mississauga, ON, Canada). An equal amount of cDNA from each experimental condition or purified DNA fragment from ChIP was amplified by real-time PCR using the Stratagene Mx-4000 and Brilliant SYBR Green QPCR Master Mix. We normalized gene expression to a house-keeping gene (GAPDH), and the relative expression value between the samples was calculated based on the threshold cycle (CT) value using the 2−ΔΔCT method (Thuraisingam et al., 2007).
Luciferase Activity
The HL-60 cells were transfected with either control or β-actin siRNA as described above. Twenty-four hours after transfection, transient transfections of siRNA-transfected or untransfected HL-60 cells with luciferase reporter constructs were performed using HiFect transfection reagent (Amaxa Biosystems) according to manufacturer's instructions. Four hours after transfection, cells were treated with PMA (10 ng/ml) for 48 h or left untreated, and then the cells were harvested. Luciferase reporter assays were performed using the Dual-Luciferase reporter assay system (Promega, Madison, WI), and the luminescence measurements were done with a luminometer (model TD-20/20; Turner Designs, Sunnyvale, CA). Firefly luciferase activity was normalized to Renilla luciferase activity.
Immunofluorescence
HL-60 cells were seeded on four-well culture slides (Becton Dickinson Labware, BD Biosciences, Franklin Lakes, NJ), pretreated with signal inhibitors or dimethyl sulfoxide (DMSO; the vehicle for inhibitors), and then stimulated with PMA. Unstimulated HL-60 cells were collected by centrifugation at 500 × g for 5 min and placed on culture slides. Both treated and untreated cells were fixed for 15 min in PBS containing 3.0% paraformaldehyde and permeabilized for 20 min with 0.18% Triton X-100 in M-buffer (containing 50 mM imidazole, 50 mM KCl, 0.5 mM MgCl2, 1 mM EGTA, 0.1 mM EDTA, 1 mM β-mercaptoethanol, and 4 mM glycerol, pH 6.8). After soaking in blocking buffer (PBS containing 1% goat serum and 1% bovine serum albumin) for 30 min, cells were incubated with a mouse anti-β-actin antibody (clone AC-15; Sigma-Aldrich) at 1:1000 dilution in blocking buffer for 16h at 4°C. After washes with PBS, cells were incubated with an Alexa Fluor 568 goat anti-mouse IgG (1:300; Invitrogen) for 1h at room temperature. After washing in PBS, the cells were mounted in Vectashield with 4,6-diamidino-2-phenylindole (DAPI; Vector Laboratories, Burlingame, CA) and visualized with an AxioVision 3.1 microscope (Carl Zeiss, Jena, Germany) using a 63× oil objective. An AxioCam HR digital camera (Carl Zeiss) was used for photography.
Microinjection
HL-60 cells were plated on one-well Lab-Tek chambers in RPMI 1640 medium with 10% heat-inactivated FBS, 100 U/ml penicillin, and 100 ng/ml streptomycin, and treated with 10 ng/ml PMA. Thirty hours after PMA treatment, the nuclei of PMA-treated HL-60 cells were comicroinjected with tetramethylrhodamine B isothiocyanate (TRITC)-labeled dextran (Sigma-Aldrich) with either anti-β-actin antibody (Sigma-Aldrich) or mouse IgG using a microinjection system (micromanipulator model 5170 and microinjector model 5242; Eppendorf, Hamburg, Germany) linked to a microscope from Olympus (Tokyo, Japan). The pressure injection device was fitted with sterile glass capillaries (Ø 0.5 ± 0.2 μm, Femtotips; Eppendorf). The antibodies were dialyzed in PBS, concentrated, and combined with dextran. The final concentration of β-actin antibody, IgG, or dextran is 5.0 mg/ml. Approximately 40 fl was injected into each nucleus and ∼30 nuclei were microinjected in 30 min for each chamber. Eighteen hours after injection, SLC11A1 mRNA expression was detected by using RNA in situ hybridization.
mRNA In Situ Hybridization
Plasmid PGEM-T-3′ untranslated region (Xu et al., 2005) was linearized with the restriction endonuclease PstI and then used as the template to produce anti-sense biotin-labeled RNA probes by in vitro transcription. mRNA in situ hybridization was performed by using ELF 97 mRNA in situ hybridization kit #2 (Invitrogen). HL-60 cells were fixed with 3.7% formaldehyde and 5% acetic acid in 0.9% NaCl for 15 min and then washed twice in PBS. Samples were rinsed in 100% xylene for 5 min and washed with PBS. Samples were then prehybridized for 1 h and hybridized overnight at 42°C. Prehybridization was done in 25% formamide, 6× SSC, 5× Dehardt's solution, and 500 μg/ml salmon sperm DNA; hybridization was done in the same solution with biotinylated antisense probes. The next day, samples were sequentially washed three times with 2× SSC, three times with 0.1× SSC, and once with wash buffer (1×). Subsequently, the samples were blocked with blocking buffer for 30 min and then incubated with a dilution of 1:50 streptavidin-alkaline phosphatase in the same blocking buffer for 30 min. Samples were then washed and incubated with substrate working solution (1:10 dilution of phosphatase substrate and 1:500 dilution of substrate additives 1 and 2 in the development buffer) for 1 h. After wash, the cells were mounted in mounting medium and viewed with an AxioSkop II microscope (Carl Zeiss).
RESULTS
PMA Treatment, Which Induces Differentiation of HL-60 Cells and Human Monocytes, Stimulates Nuclear Accumulation of β-Actin
When HL-60 cells were induced with PMA to differentiate into macrophage-like cells, we found that actin redistribution occurred. Western blot analysis using whole-cell lysates revealed that total β-actin content increased by 1.48 ± 0.16-fold HL-60 cells treated with PMA for 24 h, and the content of β-actin remained stable after 24 h of treatment. The analysis of subcellular fractions revealed that β-actin was strongly expressed in the cytoplasm and very weakly (scarcely detectable) in the nuclei in untreated HL-60 cells. After treatment of HL-60 cells with PMA, β-actin content was increased by 1.38 ± 0.13-fold in the cytoplasm during the 72 h of differentiation, whereas the content of nuclear actin increased 12.5 ± 2.1-fold after 24 h and increased 32.9 ± 3.8-fold after 48 and 72 h of treatment (Figure 1A). These results demonstrate that the observed nuclear accumulation of β-actin in response to PMA treatment was not only due to an overall increase in β-actin expression but also probably resulted from an elevated import of cytoplasmic β-actin to the nucleus. Western blotting of the same membranes to detect nucleus- and cytoplasm-specific markers (histone deacetylase 1 and GAPDH, respectively) verified that the cytoplasmic protein did not leak into the nuclear fractions during cell treatment or fractionation, and further monitored the equal loading and transfer of the samples. As shown in Figure 1B, similar results were observed when human monocytes were used. PMA-induced nuclear accumulation of β-actin was further confirmed by immunofluorescence analysis using a mouse anti-β-actin antibody. As shown in Figure 1C, in untreated cells, we observed that β-actin was exclusively cytoplasmic, whereas in PMA-treated HL-60 cells, β-actin was intensely stained in the nuclei and the cytoplasmic staining diminished. In control experiments, anti-β-actin antibodies were omitted from the immunostaining, and no fluorescence signals were observed either in the nucleus or cytoplasm (data not shown). To rule out any possibility that some of the cytoplasmic actin might enter into the nucleus during immunostaining, HL-60 cells were transfected with vector pAcGFP1-Actin (Supplemental Materials and Methods) and GFP-β-actin fusion protein expression was then directly monitored by fluorescence microcopy. As shown in Figure 1D, after PMA treatment, the fluorescence of GFP-β-actin fusion protein could also be observed in the nuclei from differentiating HL-60 cells.
Figure 1.
PMA treatment induces β-actin translocation to the nucleus. HL-60 cells (A) and human monocytes (B) were left untreated or treated with PMA (10 ng/ml) for 24, 48, and 72 h. Total cell, cytoplasmic, and nuclear extracts (TE, CE, and NE, respectively) were prepared as described in Materials and Methods. Total and subcellular β-actin expression was analyzed by Western blot analysis. The expression of the cytoplasmic marker GAPDH and nuclear marker HDAC1 were also monitored. (C) HL-60 cells treated with or without PMA (10 ng/ml) for 48 h were then fixed with 3% paraformaldehyde and permeabilized with 0.18% Triton X-100. Subsequently, the cells were labeled with a monoclonal mouse anti-β-actin antibody, followed by Alexa Four 568 goat anti-mouse IgG (red). The nuclei were stained with DAPI (blue). (D) Transfected HL-60 cells expressing GFP-β-actin fusion protein were left untreated or treated with PMA for 48 h, and then the effects of PMA treatment on GFP-β-actin distribution in the transfected cells was observed with an AxioVision 3.1 microscope (Carl Zeiss).
Signaling Events Involved in PMA-induced Nuclear Localization of β-Actin
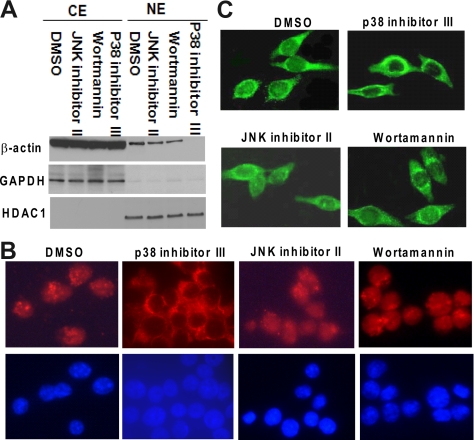
Given the growing evidence showing that actin translocates to the nucleus in response to cellular stress and that nuclear actin is involved in diverse nuclear activities, we sought to identify the signaling pathway(s) influencing the nuclear accumulation of actin. We first tested a series of inhibitors of signaling pathways at two different concentrations to determine their influence on PMA-induced nuclear accumulation of actin. This assessment was carried out by Western blot analysis of actin abundance in the cytoplasmic and nuclear fractions. Our results showed that all mitogen-activated protein kinase kinase (MEK)/extracellular signal-regulated kinase (ERK), p38 mitogen-activated protein kinase (MAPK), and protein kinase C (PKC) inhibitors could block PMA-induced translocation of actin into nuclei, whereas treatment with inhibitors of c-Jun NH2-terminal kinase (JNK) or phosphatidylinositol 3-kinase (PI3-K) had no effect on nuclear translocation (Supplemental Table S2). The PKC and MEK/ERK signal pathway inhibitors could also block PMA-induced morphological changes of HL-60 cells undergoing differentiation such as cellular adherence to plastic and spreading in a manner characteristic for macrophages (data not shown). In contrast, the p38 inhibitor (p38 inhibitor III), JNK inhibitor (JNK inhibitor II), and PI3-K inhibitor (wortmannin) did not prevent the differentiation of HL-60 cells to the macrophage-like phenotype (data not shown), and their effects on PMA-induced differentiation of HL-60 cells were assessed both by nitro blue tetrazolium (NBT) reduction assay (Supplemental Figure S1A) and by monitoring of CD11b expression using flow cytometry analysis (Supplemental Figure S1, B and C). The percentage of NBT-positive cells and CD11b expression were greatly increased after 48 h of PMA treatment, compared with untreated control. Both JNK inhibitor II and wortmannin did not significantly inhibit the HL-60 cell differentiation induced by PMA, as indicated by NBT reduction ability and CD11b expression (both in terms of the percentage of CD11b-positive cells and mean fluorescence intensity). The p38 inhibitor III increased the PMA-induced differentiation of HL-60 cells, but not significantly, as assessed using the same assays. The representative Western blot analysis illustrating the effects of p38 inhibitor III, JNK inhibitor II, and wortmannin on PMA-induced nuclear translocation of actin was shown in Figure 2A. As stated above, the p38 inhibitor III blocked the nuclear accumulation of β-actin, whereas the JNK inhibitor II and wortmannin did not. Indirect immunofluorescence analysis of β-actin distribution in HL-60 cells, illustrated in Figure 2B, confirmed the results of Western blot analysis. Pretreatment with JNK inhibitor II or wortmannin (inhibitor of PI3-K) had no effect on PMA-induced nuclear accumulation of β-actin, whereas β-actin was confined to the cytoplasm after pretreatment with p38 inhibitor III. Similar results were also obtained when using HL-60 cells expressing GFP–β-actin fusion protein (Figure 2C). Our results demonstrate that p38 signal pathway is involved in PMA-induced nuclear translocation of β-actin.
Figure 2.
Effects of JNK, PI3-K, or p38 MAPK inhibitors on PMA-induced translocation of β-actin into nucleus. (A) JNK inhibitor (JNK inhibitor II; 5 μM), PI3-K inhibitor (wortmannin; 1 μM), p38 MAPK inhibitor (p38 inhibitor III; 20 μM), or DMSO (the vehicle for inhibitors) was separately added to HL-60 cells 1 h before treating with PMA (10 ng/ml) for 48 h. (A) β-Actin levels in cytoplasmic fractions and nuclear fractions were monitored using Western blot analysis. The expression of the cytoplasmic marker GAPDH and nuclear marker HDAC1 was also monitored. (B) Effects of signal inhibitors on PMA-induced nuclear translocation of β-actin were detected by indirect immunofluorescence as described in Figure 1C. Red color represents β-actin; blue color depicts nucleus. (C) HL-60 cells expressing GFP-β-actin fusion protein were pretreated with kinase inhibitors or DMSO as described in A for 1 h and then cultured with PMA for 48 h. Fluorescence microscopy was used to observe the effects of kinase inhibitors on PMA-induced GFP-β-actin translocation.
Accumulated Nuclear Actin Is Colocalized with RNA Polymerase II
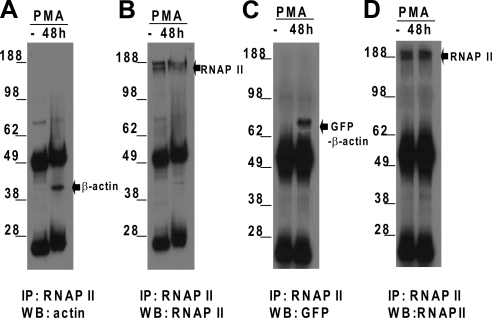
To determine whether the nuclear β-actin accumulation is associated with an interaction with RNAP II, we have performed series of immunoprecipitations and Western blot analyses. Nuclear extracts from HL-60 cells either untreated or treated with PMA for 48 h were immunoprecipitated with anti-RNAP II antibody. The immunocomplexes were detected by immunoblotting with an antibody against β-actin and reprobed with an antibody against RNAP II. As shown in Figure 3A, β-actin was undetectable in RNAP II complexes from untreated HL-60 cells, but it appeared in the complexes from PMA-treated HL-60 cells. In both untreated and PMA-treated HL-60 cells, RNAP II could be detected (Figure 3B). Similar results were obtained when the same experiments were performed using human peripheral blood monocytes (data not shown). To investigate whether exogenous β-actin could incorporate into the RNAP II complex, binding of GFP-tagged β-actin to RNAP II was tested in HL-60 cells transfected with expression vector pAcGFP1-actin. RNAP II complexes were immunoprecipitated from lysates of the transfected HL-60 cells, and the precipitated complexes were then probed with a GFP antibody and reprobed with an antibody against RNAP II. As shown in Figure 3, C and D, GFP-β-actin was associated with RNAP II only when transfected HL-60 cells were treated with PMA, as in wild-type HL-60 cells. Our results demonstrate that PMA treatment promotes interaction between nuclear β-actin and RNAP II.
Figure 3.
Endogenous or exogenous β-actin is associated with RNAP II during the differentiation of HL-60 cells. (A) Nuclear extracts prepared from 5 × 107 HL-60 cells untreated or treated with PMA (10 ng/ml) for 48 h were subjected to immunoprecipitation with an RNAP II antibody. The bound proteins were eluted by boiling in SDS sample buffer and separated on SDS-PAGE gel, and analyzed by Western blotting with a mouse mAb against β-actin. (B) Western blots from A were stripped and reprobed using an anti-RNAP II antibody. (C) Coimmunoprecipitation experiments and Western blot analyses were performed as described in A, except that HL-60 cells transfected with vector pAcGFP1-actin were used. Nuclear extracts were immunoprecipitated with an anti-RNAP II antibody and probed with a GFP antibody. (D) The Western blots from C were stripped and reprobed using the anti-RNAP II antibody. IP, immunoprecipitation; WB, Western blot analysis.
Identification of Genome-wide Binding of β-Actin to the Promoters in HL-60 Cells
To further investigate whether accumulated nuclear β-actin is associated with genome-wide transcriptional regulation, we used an anti-β-actin antibody to perform ChIP-on-chip experiments, searching for genome-wide promoters enriched in β-actin binding. HL-60 cells were untreated or treated with PMA for 48 h, and standard ChIP assays were performed using the β-actin antibody and a nonspecific mouse IgG as a control. The immunoprecipitated DNAs were amplified by two rounds of ligation-mediated (LM)-PCR as described in Materials and Methods. The produced amplicons contained DNA fragments spanning from 200 to 600 base pairs. The amplicons from the β-actin and IgG-immunoprecipitated chromatin samples were labeled with cyanine (Cy)5 dye, and the amplicons from unenriched input chromatin samples were labeled with Cy3 dye. Equal amounts of Cy5-labeled and Cy3-labeled amplicons were cohybridized onto separate HG18_RefSeq_promoter arrays. A representative profile of the β-actin complex along chromosome 2 and a close-up of β-actin ChIP-on-chip hybridization signals around the 5′ end of the SLC11A1 gene are shown in Figure 4, A and B.
Figure 4.
High-resolution ChIP-on-chip analysis of β-actin binding to chromosome 2. Cross-linked and sonicated chromatin was prepared from HL-60 cells with or without PMA treatment for 48 h. ChIP experiments were performed, and then the precipitated DNAs prepared from a β-actin antibody and a nonspecific IgG as well as total input DNA were used to prepare amplicons by LM-PCR. High-density HG18_RefSeq_promoter arrays were hybridized with the amplicons. (A) Representative profile of the β-actin complex along the chromosome 2 was shown. The logarithmic ratio (log2R) means hybridization intensities between β-actin or IgG enriched DNA and input DNA. (B) A close-up view of ChIP-on-chip hybridization signals for β-actin and nonspecific IgG binding around the 5′-end of SLC11A1 gene.
To identify the promoters that were significantly bound by β-actin, we compared the hybridization intensities between the β-actin array and the input array, and we calculated a β-actin/input ratio for each oligonucleotide probe. To eliminate from consideration promoter regions that are enriched nonspecifically during the ChIP procedure, we also calculated an IgG/input ratio for each oligonucleotide probe. We considered a promoter positive for β-actin binding based on data analysis when 1) significant differences were found (p < 0.01 by t test) in hybridization signals between the immunoprecipitation-enriched DNA channel compared with the input control channel; and 2) log2 (β-actin/input ratio) was higher than 1.0, and at the same time, log2 (IgG/input ratio) was <0.5. This was further analyzed using SignalMap1.8 (NimbleGen). In untreated cells, only 25 gene promoters were identified as actin binding targets, whereas at 48 h after PMA treatment, 827 gene promoters were identified (Supplemental Table S3). To gain insights into the functions of the identified target genes, GO enrichment analysis was performed using DAVID (Dennis et al., 2003) and EASE (http://david.abcc.ncifcrf.gov/ease/ease.jsp) This analysis revealed that the 20 GO categories, such as cell growth and/or maintenance, apoptosis, cell surface receptor-linked signal transduction, response to external stimuli, and immune response were significantly enriched after 48 h of PMA treatment (Table 1). It is noteworthy that a given gene product may display one or more functions. Interestingly, we found that some target genes possess the same functions as nuclear actin does and are involved in chromatin remodeling, transcription, RNA splicing, and nucleocytoplasmic trafficking (Supplemental Table S4). We also identified a number of genes that are associated with ion transport, immune response, and apoptosis.
Table 1.
GO categories significantly enriched for β-actin target genes
| GO category | β-Actintarget genes | p valuea(EASE score) |
|---|---|---|
| Apoptosis (GO:0006915) | 17 | 0.028 |
| Cell surface receptor linked | ||
| Signal transduction (GO:0007166) | 60 | 0.0054 |
| Response to external stimulus(GO:0009605) | 64 | 0.013 |
| Chemotaxis (GO:0006935) | 15 | 0.0029 |
| Immune response (GO:0006955) | 36 | 0.0036 |
| Cell cycle (GO:0007049) | 29 | 0.0081 |
| Protein biosynthesis(GO:0006412) | 34 | 0.0041 |
| Protein modification(GO:0006464) | 46 | 0.0086 |
| Catabolism (GO:0009056) | 53 | 0.0018 |
| Biosynthesis (GO:0009058) | 51 | 0.0036 |
| RNA biosynthetic process(GO:0032774) | 28 | 0.0046 |
| RNA processing (GO:0006396) | 23 | 0.0027 |
| DNA repair (GO:0006281) | 11 | 0.0012 |
| DNA replication (GO:0006260) | 14 | 0.0052 |
| Ion channel activity(GO:0005216) | 46 | 0.0025 |
| Ion transport (GO:0006811) | 28 | 0.0093 |
| Cell motility (GO:0048870) | 15 | 0.0053 |
| Cell growth and/or maintenance(GO:0009987) | 87 | 0.0062 |
| Cell differentiation (GO:0043697) | 12 | 0.0039 |
| Cell development (GO:0007275) | 72 | 0.0076 |
a The threshold of EASE score, a modified Fisher exact p value, for gene-enrichment analysis. Usually, the p value is ≤0.05 to be considered strongly enriched in the annotation categories.
The results from these ChIP-on-chip experiments were validated in independent conventional ChIP experiments using PCR primers (Supplemental Table S1) that spanned the regions showing the highest peak of enrichment by the β-actin antibody. Twelve of 48 promoters bound by β-actin in chromosome 2 were randomly selected. As shown in Figure 5A, the results demonstrated that β-actin was not associated with the selected promoters in undifferentiated cells (without PMA treatment); however, after PMA treatment, β-actin was recruited to the promoter regions, which was in agreement with the observations from ChIP-on-chip analysis. The ChIP experiments verified the binding of β-actin to 11 of the selected promoters of β-actin target genes analyzed by ChIP-on-chip (Figure 5B).
Figure 5.
Validation of β-actin target genes identified by ChIP-on-chip. (A) Twelve of 48 targets bound by β-actin on chromosome 2 were randomly selected and were confirmed by conventional ChIP. The immunoprecipitated DNAs prepared as described in Materials and Methods were analyzed by PCR using primers specific to the promoters of the indicated genes. A nonspecific IgG was used as a negative control, and input DNAs were used as positive controls. (B) TreeView depiction of the binding changes of the 12 target genes in HL-60 cells treated or left untreated with PMA analyzed by ChIP-on-chip.
Correlation between β-Actin and RNAP II Occupancy at Target Promoters
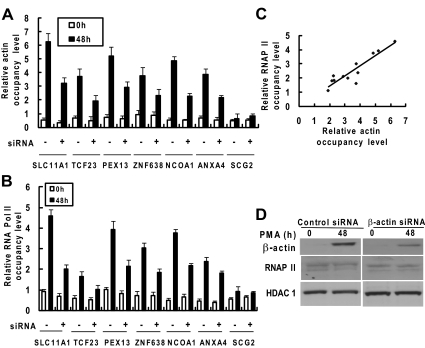
Our results have shown that nuclear β-actin interacts with RNA Pol II in macrophages. To examine potential correlation between β-actin binding and RNAP II recruitment at the promoters of β-actin targeted genes, we performed ChIP-qPCR for β-actin and RNAP II occupancies at six target promoters (SLC11A1, TCF23, PEX13, ZNF638, NCOA1, and ANXA4) and one nontarget promoter (SCG2). As shown in Figure 6, A and B, both the occupancies of β-actin and RNAP II at the selected target promoters increased upon PMA treatment in control siRNA- or β-actin siRNA-treated HL-60 cells. β-Actin knockdown resulted in the decrease of PMA-induced β-actin binding to the promoters accompanied by a diminished recruitment of RNAP II. Both the PMA treatment and β-actin knockdown had no effects on the occupancies of β-actin and RNAP II at the SCG2 promoter. To examine the relationship between β-actin and RNAP II binding at target gene promoters, the results for β-actin occupancies were compared with the results obtained for RNAP II occupancies. Using Spearman rank correlation analysis, we revealed a significant correlation between PMA-induced β-actin occupancy and RNAP II occupancy at the selected promoters (r = 0.83, p = 0.000785 at 48 h; Figure 6C). As shown in Figure 6D, transfection of β-actin siRNA significantly decreased the nuclear translocation of β-actin induced by PMA treatment in HL-60 cells. The β-actin siRNA treatment had no significant effect on the protein levels of RNAP II and nuclear HDAC1. These results demonstrate that nuclear β-actin is involved in recruiting RNAP II to the target genes.
Figure 6.
Effects of β-actin knockdown on β-actin binding and RNAP II recruitment at selected promoters in HL-60 cells as assayed by ChIP-qPCR. HL-60 cells were transiently transfected with either control siRNA (−) or β-actin siRNA (+). Twenty-four hours after transfection, HL-60 cells were left untreated or treated with PMA (10 ng/ml) for 48h. (A) ChIP experiments were performed, and then the precipitated DNAs prepared from a β-actin antibody and a nonspecific mouse IgG as well as total input DNA were quantified and were amplified using real-time PCR with primers that target the selected regions of six target genes. β-Actin occupancy level is represented as the ratio of signal from immunoprecipitation (IP) samples versus that of the input minus background of IgG control. (B) ChIP and real-time PCR experiments were performed and RNAP II occupancy was calculated the same as described in A except for an RNAP II antibody used for IP. (C) Correlation between β-actin and RNAP II occupancy at the six target promoters at 48 h after PMA treatment. The correlation coefficient (r) is 0.83 at 48 h (p = 0.000785). (D) Nuclear extracts from PMA-treated or untreated transfected cells were prepared and analyzed by Western blot analysis using antibodies specific to β-actin and HDAC1.
β-Actin Binding to the Gene Promoters Is Functionally Relevant in Actin-mediated Modulation of Target Gene Expression
To determine the target genes whose proper expression depends, directly or indirectly, on the presence of nuclear β-actin in differentiating HL-60 cells, β-actin was knocked down using a small RNA interference experiment as described in Figure 6A, and cells were treated with PMA for 48 h. We further performed quantitative RT-PCR analysis to measure the time-dependent changes in the expression of the selected six target genes (SLC11A1, TCF23, PEX13, ZNF638, NCOA1, and ANXA4) in HL-60 cells. As shown in Figure 7, mRNA expression levels of SLC11A1, PEX13, NCOA1, ANXA4, and ZNF638 in β-actin siRNA-treated cells decreased significantly compared with those in control siRNA-treated cells after 48 h of PMA treatment. The mRNA levels of these genes correlated with the β-actin occupancies at the promoters (Figures 6A and 7) after PMA treatment. β-Actin knockdown had no effect on the mRNA expression level of one target gene TCF23 and the nontarget gene SCG2. The results demonstrate that the β-actin occupancy level at the promoters is associated with the expression level of β-actin target gene.
Figure 7.
Effects of β-actin knockdown on the expression of six target genes. HL-60 cells were transfected with either control (−) or β-actin (+) siRNA and were then treated with PMA as described in Figure 7. The expression levels of six target genes SLC11A1, TCF23, PEX13, ZNF638, NCOA1, and ANXA4 and one nontarget gene, SCG2, were measured by real-time PCR. Experiments were conducted in quadruplicate and normalized to GAPDH mRNA level (data shown are mean ± SEM, n = 4, unpaired t test, **p < 0.001 compared with mRNA level in β-actin siRNA-treated cells at the corresponding time point of PMA treatment).
Nuclear β-Actin Is Required for Transactivation of SLC11A1 Gene
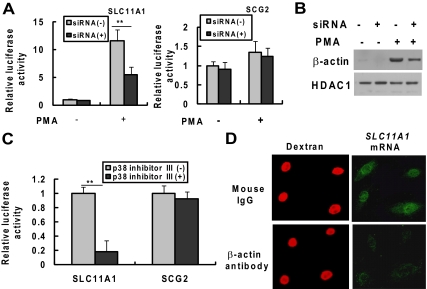
The findings described above about β-actin's binding to the gene promoters and its association with the mRNA expression of the target genes prompted us to investigate further the importance of nuclear β-actin in the transactivation of the target genes. We selected the β-actin–regulated gene SLC11A1 that is important in macrophage-mediated natural resistance to a variety of intracellular pathogens. We also selected the nontarget gene SCG2 as a control. We compared the SLC11A1 or SCG2 promoter-driven luciferase activity between HL-60 cells transfected with siRNA specific for β-actin and those with control siRNA. As shown in Figure 8A, β-actin knockdown had no effect on both SLC11A1 and SCG2 promoter-driven transactivation of luciferase activity in untreated HL-60 cells; however, in PMA-treated cells, β-actin knockdown significantly decreased the luciferase activity driven by the SLC11A1 promoter but not the SCG2 promoter. As shown in Figure 8B, transfection of β-actin siRNA significantly decreased the nuclear β-actin level in PMA-treated HL-60 cells but had no effect on nuclear marker HDAC1 expression. Similarly, blocking the PMA-induced accumulation of nuclear β-actin by p38 inhibitor III inhibits the luciferase activity driven by SLC11A1 promoter but not SCG2 promoter (Figure 8C). To further assess the role of nuclear β-actin in SLC11A1 mRNA expression, HL-60 cells were treated with PMA for 30 h to induce cell adherence and SLC11A1 mRNA expression. The adherent cells were then microinjected into the nuclei with an antibody against β-actin or with mouse nonspecific IgG as a control together with TRITC-labeled dextran to identify the injected cells. Eighteen hours after microinjection, the cells were fixed and mRNA in situ hybridization was performed to detect the level of SLC11A11 mRNA expression. As shown in Figure 8D, PMA-induced SLC11A1 mRNA expression in HL-60 cells were significantly inhibited when cells were injected with β-actin antibody but not mouse nonspecific IgG. These data indicate that β-actin is involved in the transcription of SLC11A1 gene. Our results demonstrate that nuclear accumulation of β-actin is associated with the transactivation of the SLC11A1 gene.
Figure 8.
Nuclear β-actin is required for PMA-induced transactivation of SLC11A1. HL-60 cells were respectively transfected with siRNA control or siRNAs specific for β-actin. Twenty-four hours after transfection, cells were transiently cotransfected again with the luciferase reporter vector pREP4-SLC11A1-Luc or pREP4-SCG2-Luc with pRL-CMV. Transfected cells were cultured for 48 h with or without PMA (10 ng/ml). (A) Total protein extracts were prepared and the luciferase activities driven by SLC11A1 or SCG promoter were analyzed by the dual-luciferase reporter assay system. (B) Nuclear extracts were prepared and β-actin and HDAC1 expression was detected by Western blot analysis. (C) HL-60 cells were transiently cotransfected with the luciferase reporter vector pREP4-SLC11A1-Luc or pREP4-SCG2-Luc with pRL-CMV. The cells were treated with the p38 MAPK inhibitor (p38 inhibitor III; 20 μM) at 24 h posttransfection and then treated with PMA (10 ng/ml) 1 h later. The luciferase activities were analyzed at 48 h after PMA treatment. The data shown (mean ± SE) are the averages of three independent experiments performed in triplicate. **p < 0.001, compared with the respective group of untreated cells. (D) Microinjection of antibody to β-actin inhibits SLC11A1 transcription in vivo. HL-60 cells were treated with PMA (10 ng/ml) for 30 h and were then microinjected into the nuclei with an antibody against β-actin or with mouse IgG as a control, together with TRITC-labeled dextran to identify the injected cells. Eighteen hours after microinjection, the cells were fixed with 3.7% formaldehyde and hybridized in situ with biotinylated anti-sense RNA probe to SLC11A11 mRNA. The biotinylated probe was developed for visualization with alkaline phosphatase-mediated techniques using the ELF97 mRNA in situ hybridization kit. Cells containing the biotinylated probes were then detected using streptavidin-alkaline phosphatase conjugate and the ELF97 phosphatase substrate.
DISCUSSION
Evidence has gradually been accumulating to prove that actin is present in the nucleus and plays key roles in nuclear functions, although it is still not clear whether its occurrence in nuclei is universal. The human promyelocytic leukemia cell line HL-60 has been used as a unique model to study the cellular and molecular events involved in the proliferation and differentiation of myeloid cells. The cell line can be induced to differentiate into monocytic cells by treatment with 1α, 25-dihydroxyvitamin D3 or into macrophage-like cells by treatment with phorbol esters, such as PMA, or into granulocytic cells by treatment with retinoic acid (Trayner et al., 1998). In this study, we found that although HL-60 cells and human monocytes express substantial levels of actin, it is predominantly cytoplasmic and is scarcely detectable in the nuclei in both types of untreated cells; however, PMA treatment induces actin accumulation in the nucleus. We also observed an increase in β-actin synthesis after PMA treatment. Total β-actin content and cytoplasmic β-actin content increased marginally during the course of macrophage differentiation. In contrast, there was a dramatic increase of β-actin content in the nucleus. These results suggest that increased β-actin in the nucleus most likely arises from the cytoplasmic pool of actin. The mechanism of β-actin translocation into the nucleus remains to be elucidated.
Given the numerous data demonstrating the regulation of nuclear import and export of proteins such as transcription factors, it seems reasonable to propose that actin might appear in the nucleus at high concentrations only when signaling cascades either increase the rate of import or inhibit export. Several cell signaling pathways can be activated by PMA treatment, but the signaling pathway(s) whereby PMA regulates the translocation of actin from the cytoplasm to nuclei remains unknown. Previous studies have established an essential role of PKC signaling in PMA-induced myeloid differentiation (Tonetti et al., 1994; Schultz et al., 1997). It has also been demonstrated that the MEK/ERK MAPK signaling pathway is activated and plays a critical role, during PMA-induced macrophage-like differentiation of HL-60 cells (Miranda et al., 2002). In this study, we found that PKC and MEK/ERK inhibitors potently inhibit PMA-induced differentiation and at the same time block the translocation of cytoplasmic β-actin into the nuclei. In addition, we demonstrated that although the p38 MAPK signaling pathway is not essential for differentiation (Supplemental Figure S1), as reported previously, it is necessary for β-actin translocation.
The impact of actin and actin-binding protein on chromatin remodeling and gene transcription has long been an interesting issue for cell biologists. Egly et al. (1984) showed that a purified protein with many of the characteristics of actin was able to stimulate transcription by RNAP II in vitro and that this activity was positively correlated with the concentration of G-actin in the in vitro system. Several recent studies have implicated actin in the regulation of RNAP II-mediated transcription (Percipalle et al., 2003; Hofmann et al., 2004; Kukalev et al., 2005). Our present studies demonstrate that nuclear β-actin plays an important role in transcription regulation during PMA-induced differentiation of HL-60 cells toward macrophages. Interestingly, although nuclear actin was barely detectable in untreated HL-60 cells by Western blot and immunofluorescence analysis, 25 gene promoters bound by β-actin were identified by ChIP-on-chip analysis. The majority of these genes (18 of 25 genes) are involved in cellular response to stimuli and chemotaxis. After treatment with PMA for 48 h, 827 gene promoters displayed enriched binding by actin. These genes are involved in multiple functions, including signaling pathway, cell growth and differentiation, apoptosis, ion transport, and immune response. Notably, β-actin not only directly functions in chromatin remodeling, transcription, RNA splicing, and nucleocytoplasmic trafficking but also regulates the genes involved in the biological processes mentioned above (Supplemental Table S4). Our data also belie the notion that β-actin nonspecifically interacts with the promoter region; if this were the case, β-actin would have been found at the promoter in all the genes and in the absence of PMA treatment. The complex nature of β-actin activity is observed not only in its diverse involvement in many different biological processes but also in the ways it controls transcription of its target genes: 1) Ou et al. demonstrated that β-actin specifically binds to a 27-nt repeat element in intron 4 of the endothelial nitric-oxide synthase gene and regulates the expression of this gene (Wang et al., 2002; Ou et al., 2005); 2) β-actin participates in chromatin remodeling as a component of the human chromatin remodeling complex (BAF), which interacts with chromatin during gene activation (Zhao et al., 1998; Rando et al., 2002; Song et al., 2007); and 3) β-actin plays a direct role in RNA transcription as a part of the preinitiation complex with RNAP II (Hofmann et al., 2004). Recently, we have found that β-actin is recruited to SLC11A1 gene promoter via interaction with transcription factor ATF-3, which binds to a activator protein-1–like element of this promoter (unpublished data). Therefore, β-actin might also adopt different ways to regulate the identified target genes in differentiating HL-60 cells.
Here, by using coimmunoprecipitation, we demonstrated a specific interaction between nuclear actin and RNAP II in PMA-treated HL-60 cells and human monocytes; this interaction is not seen in either type of untreated cells. Through ChIP-on-chip assay, we demonstrated that β-actin was associated with few gene promoters of PMA-induced genes; however, after PMA treatment, β-actin was recruited to the promoter regions of many genes. These results suggest that the association of β-actin and RNAP II depends on the differentiation status of HL-60 cells, and the RNAP II complex containing β-actin could regulate the RNAP II-mediated transcription of the PMA-induced genes. Several possible mechanisms could be responsible for this association. PMA treatment could 1) induce enough concentration of monomeric β-actin and/or β-actin-binding proteins to accumulate in the nuclei and affect the formation of β-actin-containing RNAP II complexes; 2) cause covalent modification of β-actin and/or β-actin-binding protein (phosphorylation or methylation) and make the interaction between β-actin and RNAP II possible; and 3) cause covalent modification of RNAP II, as Sjolinder et al. (2005) reported that β-actin binds to the hyperphosphorylated C-terminal domain of the largest subunit of Pol II. Our results also revealed a strong positive correlation between PMA-induced recruitment of β-actin and the recruitment of RNAP II at the promoters.
Previous studies have demonstrated that the recruitment of a transcription factor or modulator to its target genes does not mean that the transcriptional status of the target genes must be activated or repressed (Martone et al., 2003; Blais et al., 2005). In this study, regulation of the six target genes by β-actin was analyzed by real-time RT-PCR in response to PMA treatment. The results indicated that all except for one (TCF23), of the target genes were dependent on β-actin for their transcriptional regulation after PMA treatment. Such an observation could be explained by combinatorial regulation by recruitment of additional transcriptional factors or modulators. Therefore, knowledge of the location of a given transcriptional modulator does not provide information about whether the modulator actually regulates a nearby gene under the prevailing conditions. Therefore additional functional analyses are always required to complement ChIP-on-chip data and thereby achieve a more accurate depiction of regulatory networks.
The SLC11A1 gene encodes the natural resistant-associated macrophage protein 1. It has been shown to be associated with susceptibility to infectious diseases, such as tuberculosis, leprosy, leishmaniasis, and HIV infection (Blackwell et al., 2001; Donninger et al., 2004). It has been proposed as a candidate gene for autoimmune diseases such as rheumatoid arthritis, juvenile rheumatoid arthritis, sarcoidosis, and Crohn's diseases (Blackwell et al., 2001; Dubaniewicz et al., 2005). In humans, SLC11A1 is expressed in monocytes/macrophages and polymorphonuclear neutrophils (Cellier et al., 1997; Roig et al., 2002). SLC11A1 gene expression is low or undetectable in normal and quiescent cells. These include transformed human cell lines from either erythroid or lymphoid T or B lineages to the progenitors of the monocyte/macrophage lineage (KG1, U937, and THP), the promyelocytic leukemia cell line HL-60, and human monocytes (Cellier et al., 1997; Xu et al., 2005). It can however be strongly induced in these cells differentiated toward either the macrophage or the granulocyte pathway. In this study, we found that β-actin accumulation in nucleus is required for the PMA-induced SLC11A1 gene expression. To confirm the function of nuclear β-actin on PMA-induced expression of the SLC11A1 gene, we either knocked down the expression of β-actin or used the p38 MAPK inhibitor to block the nuclear accumulation of β-actin. We found that SLC11A1 promoter-driven luciferase activity was reduced significantly or was even completely inhibited. Furthermore, microinjection of specific β-actin antibodies into nuclei could inhibit PMA-induced SLC11A1 mRNA expression. These results indicate that nuclear β-actin is involved in the PMA-induced transcriptional activation of the SLC11A1 gene.
To our knowledge, this study provides the first direct proof that translocation of β-actin into the nucleus is associated with transcriptional regulation of gene expression during macrophage differentiation. The study of nuclear translocation of β-actin is likely to contribute to better understanding cellular gene expression in response to extracellular stimuli.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Maryse Thivierge for monocyte preparation, to Claude Lachance for data processing, and to Gabriella Wojewodka for a critical review of the manuscript. This work was supported by Canadian Institute of Health Research (CIHR) grant FRN 36337 (to D. R.), a National Sciences and Engineering Research Council of Canada grant 6844 (to D. R.), a Strategic Training Centre in Infectious Diseases and Autoimmunity CIHR grant (to Y. Z.), and Canadian Institute of Health Research grant MOP 6822 (to M.R.-P.).
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-06-0534) on January 6, 2010.
REFERENCES
- Ascough K. R. Endocytosis: actin in the driving seat. Curr. Biol. 2004;14:R124–R126. [PubMed] [Google Scholar]
- Bettinger B. T., Gilbert D. M., Amberg D. C. Actin up in the nucleus. Nat. Rev. Mol. Cell Biol. 2004;5:410–415. doi: 10.1038/nrm1370. [DOI] [PubMed] [Google Scholar]
- Blackwell J. M., et al. SLC11A1 (formerly NRAMP1) and disease resistance. Cell Microbiol. 2001;3:773–784. doi: 10.1046/j.1462-5822.2001.00150.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blais A., Tsikitis M., Costa-Alvear D., Sharan R., Kluger Y., Dynlacht B. D. An initial blueprint for myogenic differentiation. Genes Dev. 2005;19:553–569. doi: 10.1101/gad.1281105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blessing C. A., Ugrinova G. T., Goodson H. V. Actin and ARPs: action in the nucleus. Trends Cell Biol. 2004;14:435–442. doi: 10.1016/j.tcb.2004.07.009. [DOI] [PubMed] [Google Scholar]
- Brakebusch C., Fassler R. The integrin-actin connection, an eternal love affair. EMBO J. 2003;22:2324–2333. doi: 10.1093/emboj/cdg245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cellier M., Shustik C., Dalton W., Rich E., Hu J., Malo D., Schurr E., Gros P. Expression of the human NRAMP1 gene in professional primary phagocytes: studies in blood cells and in HL-60 promyelocytic leukemia. J. Leukoc. Biol. 1997;61:96–105. doi: 10.1002/jlb.61.1.96. [DOI] [PubMed] [Google Scholar]
- Chen M., Shen X. Nuclear actin and actin-related proteins in chromatin dynamics. Curr. Opin. Cell Biol. 2007;19:326–330. doi: 10.1016/j.ceb.2007.04.009. [DOI] [PubMed] [Google Scholar]
- Courgeon A. M., Maingourd M., Maisonhaute C., Montmory C., Rollet E., Tanguay R. M., Best-Belpomme M. Effect of hydrogen peroxide on cytoskeletal proteins of Drosophila cells: comparison with heat shock and other stresses. Exp. Cell Res. 1993;204:30–37. doi: 10.1006/excr.1993.1005. [DOI] [PubMed] [Google Scholar]
- Dennis G., Jr., B.T., Sherman B. T., Hosak D. A., Yang J., Gao W., Lane H. C., Lempicki R. A. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:R60.1–R60.10. [PubMed] [Google Scholar]
- Donninger H., Cashmore T. J., Scriba T., Petersen D. C., Janse van R. E., Hayes V. M. Functional analysis of novel SLC11A1 (NRAMP1) promoter variants in susceptibility to HIV-1. J. Med. Genet. 2004;41:e49. doi: 10.1136/jmg.2003.013318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubaniewicz A., Jamieson S. E., Dubaniewicz-Wybieralska M., Fakiola M., Nancy M. E., Blackwell J. M. Association between SLC11A1 (formerly NRAMP1) and the risk of sarcoidosis in Poland. Eur. J. Hum. Genet. 2005;13:829–834. doi: 10.1038/sj.ejhg.5201370. [DOI] [PubMed] [Google Scholar]
- Egly J. M., Miyamoto N. G., Moncollin V., Chambon P. Is actin a transcription initiation factor for RNA polymerase B? EMBO J. 1984;3:2363–2371. doi: 10.1002/j.1460-2075.1984.tb02141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fomproix N., Percipalle P. An actin-myosin complex on actively transcribing genes. Exp. Cell Res. 2004;294:140–148. doi: 10.1016/j.yexcr.2003.10.028. [DOI] [PubMed] [Google Scholar]
- Gonsior S. M., Platz S., Buchmeier S., Scheer U., Jockusch B. M., Hinssen H. Conformational difference between nuclear and cytoplasmic actin as detected by a monoclonal antibody. J. Cell Sci. 1999;112:797–809. doi: 10.1242/jcs.112.6.797. [DOI] [PubMed] [Google Scholar]
- Hofmann W., et al. Cofactor requirements for nuclear export of Rev response element (RRE)- and constitutive transport element (CTE)-containing retroviral RNAs. An unexpected role for actin. J. Cell Biol. 2001;152:895–910. doi: 10.1083/jcb.152.5.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann W. A., et al. Actin is part of pre-initiation complexes and is necessary for transcription by RNA polymerase II. Nat. Cell Biol. 2004;6:1094–1101. doi: 10.1038/ncb1182. [DOI] [PubMed] [Google Scholar]
- Hu P., Wu S., Hernandez N. A role for beta-actin in RNA polymerase III transcription. Genes Dev. 2004;18:3010–3015. doi: 10.1101/gad.1250804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauss S. W., Chen C., Penman S., Heald R. Nuclear actin and protein 4. 1, essential interactions during nuclear assembly in vitro. Proc. Natl. Acad. Sci. USA. 2003;100:10752–10757. doi: 10.1073/pnas.1934680100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauss S. W., Heald R., Lee G., Nunomura W., Gimm J. A., Mohandas N., Chasis J. A. Two distinct domains of protein 4.1 critical for assembly of functional nuclei in vitro. J. Biol. Chem. 2002;277:44339–44346. doi: 10.1074/jbc.M204135200. [DOI] [PubMed] [Google Scholar]
- Kukalev A., Nord Y., Palmberg C., Bergman T., Percipalle P. Actin and hnRNP U cooperate for productive transcription by RNA polymerase II. Nat. Struct. Mol. Biol. 2005;12:238–244. doi: 10.1038/nsmb904. [DOI] [PubMed] [Google Scholar]
- Liu R., Liu H., Chen X., Kirby M., Brown P. O., Zhao K. Regulation of CSF1 promoter by the SWI/SNF-like BAF complex. Cell. 2001;106:309–318. doi: 10.1016/s0092-8674(01)00446-9. [DOI] [PubMed] [Google Scholar]
- Martone R., et al. Distribution of NF-kappaB-binding sites across human chromosome 22. Proc. Natl. Acad. Sci. USA. 2003;100:12247–12252. doi: 10.1073/pnas.2135255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda M. B., McGuire T. F., Johnson D. E. Importance of MEK-1/-2 signaling in monocytic and granulocytic differentiation of myeloid cell lines. Leukemia. 2002;16:683–692. doi: 10.1038/sj.leu.2402400. [DOI] [PubMed] [Google Scholar]
- Olave I. A., Reck-Peterson S. L., Crabtree G. R. Nuclear actin and actin-related proteins in chromatin remodeling. Annu. Rev. Biochem. 2002;71:755–781. doi: 10.1146/annurev.biochem.71.110601.135507. [DOI] [PubMed] [Google Scholar]
- Ou H., Shen Y. H., Utama B., Wang J., Wang X., Coselli J., Wang X. L. Effect of nuclear actin on endothelial nitric oxide synthase expression. Arterioscler. Thromb. Vasc. Biol. 2005;25:2509–2514. doi: 10.1161/01.ATV.0000189306.99112.4c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendleton A., Pope B., Weeds A., Koffer A. Latrunculin B or ATP depletion induces cofilin-dependent translocation of actin into nuclei of mast cells. J. Biol. Chem. 2003;278:14394–14400. doi: 10.1074/jbc.M206393200. [DOI] [PubMed] [Google Scholar]
- Percipalle P., Fomproix N., Kylberg K., Miralles F., Bjorkroth B., Daneholt B., Visa N. An actin-ribonucleoprotein interaction is involved in transcription by RNA polymerase II. Proc. Natl. Acad. Sci. USA. 2003;100:6475–6480. doi: 10.1073/pnas.1131933100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percipalle P., Zhao J., Pope B., Weeds A., Lindberg U., Daneholt B. Actin bound to the heterogeneous nuclear ribonucleoprotein hrp36 is associated with Balbiani ring mRNA from the gene to polysomes. J. Cell Biol. 2001;153:229–236. doi: 10.1083/jcb.153.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philimonenko V. V., et al. Nuclear actin and myosin I are required for RNA polymerase I transcription. Nat. Cell Biol. 2004;6:1165–1172. doi: 10.1038/ncb1190. [DOI] [PubMed] [Google Scholar]
- Rando O. J., Zhao K., Janmey P., Crabtree G. R. Phosphatidylinositol-dependent actin filament binding by the SWI/SNF-like BAF chromatin remodeling complex. Proc. Natl. Acad. Sci. USA. 2002;99:2824–2829. doi: 10.1073/pnas.032662899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roig E. A., Richer E., Canonne-Hergaux F., Gros P., Cellier M. F. Regulation of NRAMP1 gene expression by 1alpha,25-dihydroxy-vitamin D(3) in HL-60 phagocytes. J. Leukoc. Biol. 2002;71:890–904. [PubMed] [Google Scholar]
- Sanger J. W., Gwinn J., Sanger J. M. Dissolution of cytoplasmic actin bundles and the induction of nuclear actin bundles by dimethyl sulfoxide. J. Exp. Zool. 1980a;213:227–230. doi: 10.1002/jez.1402130210. [DOI] [PubMed] [Google Scholar]
- Sanger J. W., Sanger J. M., Kreis T. E., Jockusch B. M. Reversible translocation of cytoplasmic actin into the nucleus caused by dimethyl sulfoxide. Proc. Natl. Acad. Sci. USA. 1980b;77:5268–5272. doi: 10.1073/pnas.77.9.5268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer U., Hinssen H., Franke W. W., Jockusch B. M. Microinjection of actin-binding proteins and actin antibodies demonstrates involvement of nuclear actin in transcription of lampbrush chromosomes. Cell. 1984;39:111–122. doi: 10.1016/0092-8674(84)90196-x. [DOI] [PubMed] [Google Scholar]
- Schultz H., Engel K., Gaestel M. PMA-induced activation of the p42/44ERK- and p38RK-MAP kinase cascades in HL-60 cells is PKC dependent but not essential for differentiation to the macrophage-like phenotype. J. Cell. Physiol. 1997;173:310–318. doi: 10.1002/(SICI)1097-4652(199712)173:3<310::AID-JCP2>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Shen X., Ranallo R., Choi E., Wu C. Involvement of actin-related proteins in ATP-dependent chromatin remodeling. Mol. Cell. 2003;12:147–155. doi: 10.1016/s1097-2765(03)00264-8. [DOI] [PubMed] [Google Scholar]
- Sjolinder M., Bjork P., Soderberg E., Sabri N., Farrants A. K., Visa N. The growing pre-mRNA recruits actin and chromatin-modifying factors to transcriptionally active genes. Genes Dev. 2005;19:1871–1884. doi: 10.1101/gad.339405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S. S., Kelly K. H., Jockusch B. M. Actin co-purifies with RNA polymerase II. Biochem. Biophys. Res. Commun. 1979;86:161–166. doi: 10.1016/0006-291x(79)90395-4. [DOI] [PubMed] [Google Scholar]
- Song Z., Wang M., Wang X., Pan X., Liu W., Hao S., Zeng X. Nuclear actin is involved in the regulation of CSF1 gene transcription in a chromatin required, BRG1 independent manner. J. Cell. Biochem. 2007;102:403–411. doi: 10.1002/jcb.21300. [DOI] [PubMed] [Google Scholar]
- Suetsugu S., Takenawa T. Regulation of cortical actin networks in cell migration. Int. Rev. Cytol. 2003;229:245–286. 245–286. doi: 10.1016/s0074-7696(03)29006-9. [DOI] [PubMed] [Google Scholar]
- Thuraisingam T., Xu Y. Z., Moisan J., Lachance C., Garnon J., Di M. S., Gaestel M., Radzioch D. Distinct role of MAPKAPK-2 in the regulation of TNF gene expression by Toll-like receptor 7 and 9 ligands. Mol. Immunol. 2007;44:3482–3491. doi: 10.1016/j.molimm.2007.03.019. [DOI] [PubMed] [Google Scholar]
- Tonetti D. A., Henning-Chubb C., Yamanishi D. T., Huberman E. Protein kinase C-beta is required for macrophage differentiation of human HL-60 leukemia cells. J. Biol. Chem. 1994;269:23230–23235. [PubMed] [Google Scholar]
- Trayner I. D., Bustorff T., Etches A. E., Mufti G. J., Foss Y., Farzaneh F. Changes in antigen expression on differentiating HL60 cells treated with dimethylsulphoxide, all-trans retinoic acid, alpha1,25-dihydroxyvitamin D3 or 12-O-tetradecanoyl phorbol-13-acetate. Leukoc. Res. 1998;22:537–547. doi: 10.1016/s0145-2126(98)00041-1. [DOI] [PubMed] [Google Scholar]
- Wang J., Dudley D., Wang X. L. Haplotype-specific effects on endothelial NO synthase promoter efficiency: modifiable by cigarette smoking. Arterioscler. Thromb. Vasc. Biol. 2002;22:e1–e4. doi: 10.1161/01.ATV.0000016248.51577.1F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y. Z., Di M. S., Gallouzi I., Rola-Pleszczynski M., Radzioch D. RNA-binding protein HuR is required for stabilization of SLC11A1 mRNA and SLC11A1 protein expression. Mol. Cell. Biol. 2005;25:8139–8149. doi: 10.1128/MCB.25.18.8139-8149.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K., Wang W., Rando O. J., Xue Y., Swiderek K., Kuo A., Crabtree G. R. Rapid and phosphoinositol-dependent binding of the SWI/SNF-like BAF complex to chromatin after T lymphocyte receptor signaling. Cell. 1998;95:625–636. doi: 10.1016/s0092-8674(00)81633-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.