Abstract
Yersinia pestis, the agent of plague, is transmitted to mammals by infected fleas. Y. pestis exhibits a distinct life stage in the flea, where it grows in the form of a cohesive biofilm that promotes transmission. After transmission, the temperature shift to 37°C induces many known virulence factors of Y. pestis that confer resistance to innate immunity. These factors are not produced in the low-temperature environment of the flea, however, suggesting that Y. pestis is vulnerable to the initial encounter with innate immune cells at the flea bite site. In this study, we used whole-genome microarrays to compare the Y. pestis in vivo transcriptome in infective fleas to in vitro transcriptomes in temperature-matched biofilm and planktonic cultures, and to the previously characterized in vivo gene expression profile in the rat bubo. In addition to genes involved in metabolic adaptation to the flea gut and biofilm formation, several genes with known or predicted roles in resistance to innate immunity and pathogenicity in the mammal were upregulated in the flea. Y. pestis from infected fleas were more resistant to phagocytosis by macrophages than in vitro-grown bacteria, in part attributable to a cluster of insecticidal-like toxin genes that were highly expressed only in the flea. Our results suggest that transit through the flea vector induces a phenotype that enhances survival and dissemination of Y. pestis after transmission to the mammalian host.
Author Summary
Bubonic plague cycles depend on the ability of Yersinia pestis to alternately infect two very different hosts—a mammal and a flea. Like any arthropod-borne pathogen, Y. pestis must sense host-specific environmental cues and regulate gene expression accordingly to produce a transmissible infection in the flea after being taken up in a blood meal, and again when it exits the flea and enters the mammal. We examined the Y. pestis phenotype at the point of transmission by in vivo gene expression analyses, the first description of the transcriptome of an arthropod-borne bacterium in its vector. In addition to genes associated with physiological adaptation to the flea gut, several Y. pestis virulence factors required for resistance to innate immunity and dissemination in the mammal were induced in the flea, suggesting that the arthropod life stage primes Y. pestis for successful infection of the mammal.
Introduction
Arthropod-borne transmission of bacterial pathogens is somewhat rare but has evolved in a phylogenetically diverse group that includes the rickettsiae, Borrelia spirochetes, and the gram-negative bacteria Francisella tularensis and Yersinia pestis, the plague bacillus. Y. pestis circulates among many species of wild rodents, its primary reservoir hosts, via flea bite. As it alternates between fleas and mammals, it is postulated that Y. pestis regulates gene expression appropriately to adapt to the two disparate host environments, and that different sets of genes are required to produce a transmissible infection in the flea and disease in the mammal.
Many important Y. pestis virulence factors that are required for plague in mammals have been identified, and most of them are induced by a temperature shift from <26°C to 37°C, which mimics the transition from a flea to the warm-blooded host [1]. To date, only three transmission factors (genes specifically required to produce a transmissible infection in the flea) have been characterized. One, the yersinia murine toxin (ymt) gene, encodes a phospholipase D that is required for survival in the flea midgut [2]. The other two, (hmsHFRS and gmhA), are responsible for an extracellular polysaccharide and a lipopolysaccharide (LPS) core modification that are required for normal biofilm formation and blockage in the flea [3],[4]. Biofilm development in the flea digestive tract is important for biological transmission [5],[6],[7]. After being taken up in a blood meal, Y. pestis proliferates in the lumen of the flea midgut to form cohesive multicellular biofilm aggregates. In some infected fleas, the proventricular valve between the midgut and esophagus is colonized. The subsequent growth and consolidation of the adherent Y. pestis biofilm amongst the rows of cuticle-covered spines that line the proventriculus interferes with normal blood feeding, resulting in regurgitation of bacteria and transmission. Fleas with a completely blocked proventriculus make prolonged, repeated attempts to feed, increasing the opportunities for transmission.
Formation of a Y. pestis biofilm in vitro and in the flea proventriculus depends on synthesis of an extracellular polysaccharide matrix (ECM) that is synthesized only at temperatures below 26°C [3],[7]. In common with many other bacteria, ECM synthesis in Y. pestis is controlled by intracellular levels of cyclic di-GMP, which are determined by competing activities of the hmsT diguanylate cyclase and hmsP phosphodiesterase gene products [8],[9]. Bacterial adhesins are typically required for initial adherence and autoaggregation in biofilm development [10], but such factors have yet to be identified in Y. pestis.
In a previous study, we reported the in vivo gene expression profile of Y. pestis during bubonic plague in rats [11]. In this study, we characterized the Y. pestis transcriptome in blocked Xenopsylla cheopis rat fleas, an important vector of plague to humans. Comparing the Y. pestis gene expression profile in the flea to those of in vitro biofilm and planktonic cells cultured at the low temperature typical of the flea implicated several genes in a flea-specific adaptive response and in proventricular blockage. In addition, comparing the gene expression patterns in the flea and in the rat bubo confirmed that distinct subsets of genes are differentially expressed during the Y. pestis life cycle. Notably, several genes with known or predicted roles in protection against the mammalian innate immune system and in pathogenesis were upregulated in the flea, suggesting that transit through the insect vector preinduces a phenotype that enhances Y. pestis survival and dissemination in the mammal after flea-borne transmission.
Results/Discussion
Transcriptional profile of Y. pestis in the flea
Little is known about the environmental conditions in the flea digestive tract, how Y. pestis adapts to them, or the physiological state of the bacteria at transmission when they exit the flea and enter the mammal. Adult fleas are obligate blood feeders and take frequent blood meals, consisting primarily of protein and lipid with relatively little carbohydrate. Flea proteases, lipases, and other digestive enzymes begin to process the blood meal in the midgut immediately after feeding, yielding amino acids and peptides, glycerol, fatty acids, and simple carbohydrates [12]. This provides the “medium” for Y. pestis growth, but these and other factors such as pH, oxygen tension, osmolarity, and flea antibacterial immune components are poorly defined. During the first week after being ingested in an infectious blood meal, Y. pestis grows rapidly in the flea midgut to form large bacterial aggregates. Bacterial load peaks at about 106 cells per flea as the Y. pestis biofilm accumulates in the proventriculus to cause blockage, and then plateaus [2],[3].
In this study, we determined the Y. pestis gene expression profile in infective, blocked fleas, in which the proventriculus was occluded with a mature bacterial biofilm. Y. pestis KIM6+, which lacks the 70-kb virulence plasmid that is not required for flea infection or blockage [3] was used for this analysis. Blockage occurred between 1.5 and 3.5 weeks after the initial infectious blood meal, during which time the fleas fed on uninfected mice twice weekly. The Y. pestis in vivo biofilm transcriptome was compared to the transcriptomes of in vitro biofilm and planktonic cultures grown at 21°C, the same temperature at which the fleas were maintained.
Expression of 55% of Y. pestis ORFs was detected in the flea samples; and 74 to 79% in the in vitro biofilm, exponential phase planktonic and stationary phase planktonic cultures. Principal component analysis to visualize overall clustering of the microarray data showed that the transcriptional profiles were reproducible and discrete for the in vitro and in vivo conditions (Fig. 1A). Profiles of the exponential and stationary phase planktonic cultures clustered most closely, whereas the profiles from in vitro and in vivo biofilm growth were more distinct from each other and from the planktonic culture profiles. There were 214 Y. pestis genes whose expression was significantly upregulated and 56 genes downregulated in the flea compared to all in vitro growth conditions (Fig. 1B; Tables S1 and S2). Quantitative RT-PCR analysis of a subset of Y. pestis genes differentially expressed in the flea was confirmatory of the microarray results (Fig. S2).
Figure 1. Distinct transcriptional profile of Y. pestis in infected fleas.
(A) Principal Component Analysis (PCA) representation of replicate microarray gene expression profiles of Y. pestis KIM6+ from blocked fleas (blue symbols) and from in vitro flowcells, exponential phase planktonic cultures, and stationary phase planktonic cultures (red, green, and purple symbols, respectively). (B) Venn diagrams representing the number of Y. pestis genes upregulated or downregulated ≥2-fold in the flea relative to in vitro culture conditions.
Y. pestis metabolic adaptation to the flea gut environment
Of the 214 genes upregulated in the flea gut compared to all in vitro conditions, 78 are metabolic genes, 60 of which are involved in uptake and catabolism of amino acids and carbohydrates (Table S1). In particular, genes involved in transport and catabolism of the L-glutamate group of amino acids (Gln, His, Arg, and Pro) were specifically upregulated in the flea (Fig. 2). The degradation of these amino acids gives rise to L-glutamate and the TCA cycle intermediates succinate, formate, and α-ketoglutarate. The gabD and gabT genes involved in the production of succinate from γ-aminobutyrate (GABA), another member of the L-glutamate group, were also highly induced in the flea. The gabD gene functions to produce succinate from both GABA and hydroxyphenylacetate (HPA), an aromatic degradation product of Tyr and Phe; and the HPA transport (hpaX) and catabolism (hpaCBIFHDE) genes of Y. pestis were also highly upregulated in the flea gut (Table S1, Fig. 2). As Y. pestis does not have homologs of genes required to produce GABA or HPA, these metabolites may be taken up from the flea digestive tract. Alternatively, the gabD and gabT gene products might act in the reverse direction to synthesize GABA, which has osmoprotective properties [13]. The central role of the L-glutamate family of amino acids may also confer this advantage in the flea gut, because Glu and Pro are osmoprotectants. Interestingly, both glutamate and GABA are important neurotransmitters at the neuromuscular junction of insects, and the concentration of glutamate is very low in insect hemolymph, suggesting that it is converted to glutamine before it is absorbed [14]. Insect midgut epithelium is typified by multiple amino acid transporters with specific substrates and rapid absorption kinetics, but different amino acids enter the hemocoel at different rates and amounts [14],[15]. Thus, Y. pestis metabolism in the flea may reflect the available pool of amino acids in the midgut.
Figure 2. Y. pestis amino acid uptake and catabolism pathways upregulated in the flea.
Periplasmic and inner membrane uptake proteins for proline (PutP), histidine (HisJMPQ), glutamine (GlnHPQ), arginine (ArtIMPQ), spermidine (PotABCD), and hydroxyphenylacetate (HpaX) are indicated from left to right. Genes encoding catabolic enzymes leading to glutamate and TCA cycle intermediates are also shown. Symbols labeled in blue indicate genes upregulated ≥2-fold in the flea compared to all in vitro conditions (Table S1).
In contrast to the amino acids, hexoses do not appear to be an important energy source during infection of the flea. Only the genes encoding for chitobiose phosphotransferase (PTS) uptake and utilization systems (chbBC; chbF), and for a PTS system of unknown specificity (frwBCD) were significantly upregulated in the flea [16],[17]. Chitobiose could be present in the flea gut due to turnover of the chitin layer on the proventricular spines. Expression of the glucose PTS system was only slightly increased relative to LB cultures, and other PTS systems were downregulated (Table S2). Glycolytic pathways were not upregulated in the flea; instead, available hexoses and the gluconeogenesis pathway may be used to synthesize polysaccharide components required for cell growth. Upregulation of the actP and acs genes in the flea, which direct the uptake of acetate and its conversion to acetyl-CoA, also suggests that insufficient acetyl-CoA is produced by glycolysis to potentiate the TCA cycle. The switch from acetate secretion to acetate uptake is typical of growth in a glucose-limited, amino acid rich environment [18]. In contrast to hexose uptake systems, Y. pestis genes that encode permeases for the pentoses ribose, xylose, and arabinose were induced in the flea gut. Acquisition of pentoses from the environment may be important because Y. pestis does not possess glucose 6-phosphate dehydrogenase activity, the first step of the pentose phosphate pathway [19].
Although the flea gut contains lipid derived from the blood meal, Y. pestis does not appear to use it as a major energy source. None of the fatty acid uptake or catabolism genes were upregulated in the flea compared to growth in LB. However, genes for glycerol and glycerol-3-phosphate uptake and utilization were upregulated, suggesting that flea digestion products derived from blood glycerolipids may be used by Y. pestis. In summary, Y. pestis appears to use amino acids, particularly the L-glutamate family, as primary carbon, nitrogen, and energy sources in the flea. Amino acid carbon is presumably funneled into the TCA cycle, the genes for which are highly expressed in the flea (Table S3).
Y. pestis genes involved in infection and biofilm formation in the flea
Because blockage of the flea vector is essentially a biofilm phenomenon, Y. pestis genes whose expression patterns are significantly upregulated in the flea and flowcell biofilms relative to planktonic cultures (Table S4) might indicate that they are transmission factors. Several studies comparing the transcriptional profiles of Escherichia coli and other gram negative bacteria during biofilm and planktonic growth in vitro have been published [20],[21],[22],[23]. Certain genes whose mutational loss resulted in an altered biofilm phenotype have been identified in these studies; but in general a consistent, distinct biofilm gene expression profile has not emerged. This is probably because different media and experimental systems have been employed and the fact that a biofilm consists of a physiologically heterogeneous community [24],[25]. Nevertheless, common biofilm-related adaptations include the repression of motility and the induction of specific adhesins, an extracellular polysaccharide matrix (ECM), and an envelope stress response (ESR) [10],[23]. However, Y. pestis is constitutively nonmotile, and synthesis of the Hms-dependent biofilm ECM is regulated post-translationally [26]. The ymt gene was among the most highly expressed genes in the flea (Table S3), but neither it nor the known transmission factors (hmsHFRS, hmsT, hmsP, and gmhA) showed significantly higher expression in the flea than in vitro at 21°C, indicating that they are induced primarily by low temperature, and not by environmental factors specific to the flea gut. Y. pestis homologs of two genes with previously identified roles in biofilm, yidE, which encodes a hyperadherence factor in E. coli [27], and cpxP, a member of the cpxPAR ESR system, were upregulated in the flowcell; but predicted adhesin genes were not upregulated.
The transcriptional profile of Y. pestis in blocked fleas showed greater similarity to the transcriptional profile reported for E. coli in mature, four-day-old in vitro biofilms [23]. In addition to yidE and cpxP, other Y. pestis predicted adhesins and components of an ESR were upregulated in the flea. The Y. pestis homologs of Pseudomonas aeruginosa cupA1 and cupA3 in a predicted fimbrial biosynthesis operon and yapL, a predicted autotransporter adhesin similar to E. coli tibA, were specifically upregulated in the flea (Table S1). The cupA fimbrial locus and tibA are important for surface adherence and for biofilm formation in P. aeruginosa and E. coli, respectively [28],[29]. Evidence for induction of an ESR in the flea included the high expression levels of rpoE, the gene for the alternate transcription factor σE (as well as the anti-σE negative regulator genes rseA and rseB), cpxP; and pspA and pspG, components of the phage-shock protein (Psp) response (Tables S1 and S3). These genes were also found to be upregulated in mature E. coli biofilms [23], suggesting that the three prominent ESR systems are important for integrating signals required for survival in a biofilm.
Because homologs of the yidE, cpxP, tibA (yapL), cupA fimbriae, and pspABC genes were upregulated in the flea and have been shown to be involved in biofilm formation in other bacteria [23],[27],[28],[29], we made a series of Y. pestis strains containing deletions of these loci. However, the single loss of any of these genes did not result in a noticeable defect in biofilm formation in vitro, or in flea infection or blockage (data not shown). These genes may contribute to biofilm formation, but are not individually essential for this phenotype. Although genes in the polyamine transport gabTpotDBC locus are among the most highly induced genes in the flea (Table S1) and polyamines are essential for Y. pestis biofilm formation [30], we have previously reported that a Y. pestis Δpot mutant has no defect in flea infection or blockage [31]. This is likely due to the fact that Y. pestis is able to synthesize polyamines de novo.
Differential gene expression during the Y. pestis life cycle
With this study, the in vivo transcriptome of Y. pestis in blocked fleas and in the rat bubo [11] have now both been characterized. A comparison of normalized gene expression levels from the two data sets provides insight into the biology of the flea-mammal life cycle. About 15% of Y. pestis genes showed significantly higher relative expression levels or expression only in the flea than in the bubo; 24% were more highly expressed in the bubo than in the flea; and 61% were not differentially expressed in the two hosts (Fig. 3).
Figure 3. Distinct Y. pestis gene expression profiles in flea and rat hosts.
(A) Hierarchichal clustering of normalized microarray data sets of Y. pestis gene expression in the rat bubo and the flea. The scale indicates relative transcript levels (blue = low; red = high) for all 4,638 Y. pestis genes on the microarray. (B) Percentages of Y. pestis genes that are differentially regulated (or not) in the flea and in the rat bubo.
Several virulence factors were differentially regulated in the two hosts, but others were not (Table 1). In addition to the known temperature-induced virulence factors, iron acquisition systems, including the ybt and yfe operons that are required for virulence; and oxidative and nitrosative stress response genes, including the hmp virulence factor, are highly upregulated in the rat bubo, but not the flea. The analysis also reinforces the model that Y. pestis produces a hexaacylated lipid A in the flea, and that the change to the less immunostimulatory tetraacylated form occurs only after transmission [32]. Other virulence and transmission factors were not differentially regulated, including the hms genes; and the Y. pestis plasminogen activator (pla), critical for dissemination from extravascular tissue at the fleabite site [33], and ymt were highly expressed in both hosts (Table S3 and [11]). The Y. pestis outer surface protein gene yadB, recently shown to be required for dissemination and bubonic plague pathogenesis from a subcutaneous inoculation site [34], was significantly upregulated in both the flea and the bubo compared to in vitro conditions (Tables 1, S1).
Table 1. Differential expression of Y. pestis pathogenesis-related genes in the flea.
| Gene or system | Description/function | Expression level in flea relative to*: | |||
| Flow cell | Exp. phase | Stat. phase | Bubo | ||
| Outer surface components | |||||
| caf1R | F1 capsule gene regulator | f | f | f | −2.4 |
| caf1M | F1 capsule periplasmic chaperone | f | f | f | −20 |
| caf1A | F1 capsule outer membrane usher | 7.1 | 5.3 | 4.0 | −36 |
| caf1 | F1 capsule subunit protein | 6.8 | 5.0 | 4.5 | −42 |
| pla | plasminogen activator | 0.9 | 1.0 | 0.8 | −1.5 |
| yadB | YadA-like protein | f | f | f | 1.1 |
| yadC | YadA-like protein | 2.7 | 3.3 | 1.9 | f |
| psaE | pH 6 antigen regulator | −4.4 | −7.9 | −20 | −12.6 |
| psaF | pH 6 antigen hypothetical protein | 1.2 | 1.0 | −2.7 | −0.8 |
| psaA | pH 6 antigen fimbrial subunit | ns | ns | ns | b |
| psaB | pH 6 antigen chaperone | ns | ns | ns | ns |
| psaC | pH 6 antigen usher | 3.4 | 3.4 | 2.5 | −0.6 |
| lpxP | hexaacylated lipid A synthesis | 2.2 | 3.4 | 3.2 | 1.8 |
| msbB | hexaacylated lipid A synthesis | 1.5 | 1.6 | 1.4 | 2.2 |
| Iron acquisition systems | |||||
| Ybt | yersiniabactin siderophore, 10 genes | 1.5–4.2 | −1.4–1.8 | −4.6–1.0 | −15–−84 |
| Yfe | ABC iron transporter, 5 genes | −1.4–1.8 | −1.6–1.3 | −1.4–1.6 | −1.2–−14 |
| Toxins | |||||
| yitR | positive regulator of yitABC | 10.3 | 47 | 23 | f |
| yitABC | insecticidal-like toxin complex | 5.6–11 | 21–27 | 8.8–10 | 17–25 |
| yipB | insecticidal-like toxin complex | 4.0 | 10 | 5.6 | 2.2 |
| Global regulatory systems | |||||
| rovA | regulator of virulence factors | −7.3 | −6.6 | −5.9 | −1.4 |
| rovM | repressor of rovA | 9.1 | 7.1 | 5.9 | 11.1 |
| phoP | regulator of PhoPQ regulon | 2.1 | 2.2 | 2.2 | 2.3 |
| Other | |||||
| mgtC | survival in macrophages | 3.0 | 5.9 | 4.3 | 10.0 |
| mviM | Salmonella mouse virulence factor | 2.4 | 2.9 | 1.5 | 2.9 |
*: f, expression detected in flea but not comparison condition; b, expression detected in bubo but not flea; ns, no expression detected in flea or comparison condition.
Expression of genes in the pH 6 antigen locus (psaEFABC), responsible for the synthesis and transport of the PsaA fimbriae that enhance resistance to phagocytosis by macrophages [35],[36], were higher in the bubo than the flea, although the usher protein gene psaC was upregulated in the flea compared to in vitro growth (Tables 1, S1). The psa locus is regulated by RovA [36]. Consistent with these findings, rovA expression was downregulated in the flea; whereas expression of rovM, a negative regulator of rovA [37], was upregulated.
The transcriptional regulator gene phoP of the PhoPQ two-component regulatory system and the PhoP-regulated mgtC gene were expressed at levels >2-fold higher in fleas than in any other condition (Tables 1, S1, S3). PhoP and MgtC are established virulence factors known to be important for survival of Y. pestis and other gram-negative bacteria in macrophages and for resistance to cationic antimicrobial peptides (CAMPs) of the mammalian innate immune response [38],[39],[40]. The PhoPQ system is induced in low Mg2+ or low pH environments, or by exposure to CAMPs [41],[42],[43]. The Mg2+ concentration and pH of the flea digestive tract have not been defined, so the inducing stimulus is unknown, but CAMPs are induced and secreted into the gut by blood feeding insects when they take a blood meal containing bacteria [44],[45]. X. cheopis fleas encode homologs of the insect CAMPs cecropin and defensin, and mount an inducible antibacterial response to infection (unpublished data). Thus, the PhoPQ regulatory system may be induced by the flea's immune system in response to Y. pestis in the midgut. Despite the upregulation of phoP in the flea, with the notable exception of mgtC there was little correlation between predicted PhoP-regulated genes in vitro and genes upregulated in the flea [39],[46],[47]. Differential regulation of members of the PhoP regulon may occur depending on the inducing stimulus, however [48].
Induction of a phagocytosis-resistant phenotype in the flea
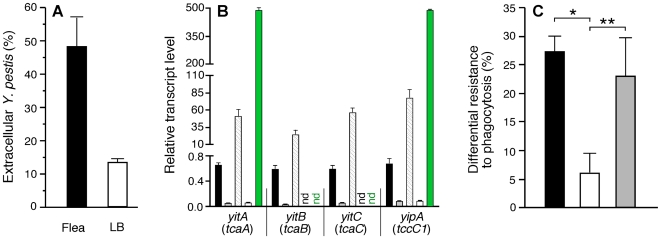
Soon after transmission, Y. pestis would be expected to encounter rapidly-responding phagocytic cells in the dermis. To assess the overall effect of the flea-specific phenotype on this encounter, we compared the interaction of Y. pestis recovered from infected fleas and from in vitro cultures with murine bone marrow macrophages. Bacteria from fleas showed significantly lower levels of phagocytosis (Fig. 4A). We have previously reported analogous findings using human polymorphonuclear leukocytes (PMNs) [7].
Figure 4. Phagocytosis-resistant phenotype of Y. pestis isolated from fleas correlates with expression level of the yit-yip insecticidal-like toxin genes.
(A) The percentage of extracellular Y. pestis KIM6+ 1 hour after addition to murine bone marrow macrophages are shown for bacteria from in vitro cultures (LB) or from infected fleas. The mean and SEM of five independent experiments done in duplicate are shown; P<0.0001. (B) Relative transcript levels of insecticidal-like toxin genes in Y. pestis KIM6+ wt grown in LB (black bars), ΔyitR mutant grown in LB (grey bars), ΔyitR mutant from fleas (white bars), and the complemented ΔyitR mutant from LB (hatched bars) and from fleas (green bars); nd = not done. The mean and SEM of three independent experiments done in triplicate are shown. Values corresponding to separate segments of the y-axis are significantly different (P<0.001); values for LB-grown wt bacteria (black bars) are also significantly different from values represented by the grey and white bars (P<0.05). (C) Differential resistance to phagocytosis by murine macrophages (% extracellular flea-derived bacteria minus % extracellular in vitro-grown bacteria) of Y. pestis KIM6+ wt (black bar, n = 3), ΔyitR mutant (white bar, n = 3), and complemented ΔyitR mutant (grey bar, n = 2). The mean and standard error of the n experiments done in duplicate are indicated; *P<0.01; ** P = 0.06.
The yit and yip genes in a Y. pestis locus (y0181–0191) that encode predicted insecticidal-like toxins of the toxin complex (Tc) family and three linked phage-related genes were upregulated 4- to 50-fold in the flea midgut (Tables 1 and S1). We previously reported that the genes for these Tc-like proteins are highly expressed in fleas, but that their products are nontoxic to fleas [49]. yitR, a LysR-type regulator that activates the Tc-like yit genes [50], was upregulated >10-fold in the flea, but its expression was not detected in the rat bubo (Table 1). The specific induction in the flea of yitR and genes in the adjacent Tc-like yit and yip loci suggests that they are involved in adaptation to and colonization of the flea. However, deletion of yitR or yitA-yipB (y0183–y0191) does not affect the ability of Y. pestis KIM6+ to infect or block fleas (data not shown). These observations, and the fact that the Yersinia Tc proteins have toxicity to certain eukaryotic cell lines in vitro [50],[51], prompted us to investigate a possible post-transmission antiphagocytic role for these proteins in the mammalian host.
To determine if the insecticidal-like toxins were involved in resistance to phagocytosis, we repeated the macrophage experiments with a Y. pestis ΔyitR mutant, which as expected showed greatly reduced expression of the yit and yip genes in vitro and in the flea (Fig. 4B). Loss of yitR significantly reduced the increased resistance to phagocytosis of Y. pestis isolated from infected fleas (Fig. 4C).
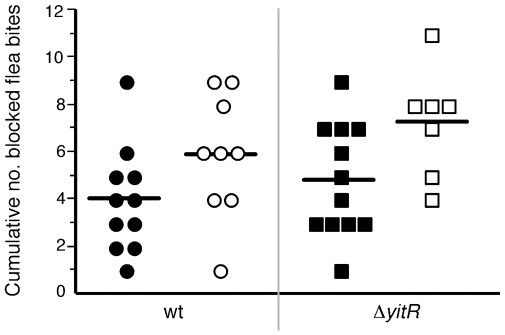
Since the yit and yip genes are not required for Y. pestis to produce a transmissible infection in fleas, it was possible to compare the virulence of wild-type and ΔyitR Y. pestis following transmission by fleabite. The incidence rate and time to disease onset were identical for both Y. pestis strains, demonstrating that expression of yit and yip is not essential for flea-borne transmission or disease (data not shown). On average, the mice challenged with Y. pestis ΔyitR-infected fleas, both those that developed disease and those that did not, received a higher cumulative number of bites from blocked fleas than the mice challenged with Y. pestis-infected fleas, but this difference was not statistically significant (Fig. 5). However, it was not possible to detect any relatively minor difference in LD50 because the number of bacteria transmitted by a blocked flea varies widely [1],[52]. Even a small decrease in LD50 provided by the Yit-Yip proteins would be significant at the ecological level in the maintenance of plague transmission cycles, because the transmission efficiency of blocked fleas is very low– often only a few or no bacterial cells are transmitted in an individual fleabite [52]. Because phoP is required by Y. pestis to produce a transmissible infection in fleas (unpublished data), it was not possible to similarly assess the effect on disease transmission of phoP induction in the flea.
Figure 5. Mean and range of the cumulative number of blocked flea bites received by mice.
Circles and squares indicate individual mice challenged by fleas infected with wt or ΔyitR Y. pestis 195/P, respectively. Filled symbols indicate mice that developed terminal plague; open symbols indicate mice that did not develop disease.
Does transit through the flea vector preadapt Y. pestis to resist mammalian innate immunity?
When Y. pestis is transmitted into the dermis by an infected flea, it is immediately exposed to the mammalian innate immune system. The most important antiphagocytic virulence factors, the cytotoxic Yersinia outer proteins (Yops), part of the T3SS encoded by the Y. pestis virulence plasmid and the F1 capsule encoded by the pMT1 plasmid, are not present at this initial stage of infection. Their expression is strictly temperature-regulated and are not produced in vivo until 3–5 hours after the temperature shift to 37°C that accompanies transmission [1],[3],[53],[54]. Consequently, Y. pestis grown at <28°C in vitro are initially susceptible to in vivo uptake and killing by phagocytes until the Yop and F1 virulence factors are produced, effectively preventing further phagocytosis [53],[54]. Our results indicate that Y. pestis entering the mammal from an infective flea is relatively resistant to macrophages, as well as PMNs [7]; a vector-specific phenotype that is not related to the T3SS or capsule.
Coming from the flea, Y. pestis is also associated with the biofilm ECM, identical or closely related to the poly-β-1,6-N-acetyl glucosamine ECM of staphylococcal biofilms, which has been shown to provide protection from innate immune components [55],[56]. In addition, although the antiphagocytic F1 capsule and Psa fimbriae do not appear to be produced in the flea, upregulation in the flea of most F1 genes in the cafRcaf1M1A1 locus and the Psa usher protein gene psaC (Tables 1, S1) suggests that components of the F1 and Psa translocation system are made, which may prime Y. pestis for rapid secretion of these extracellular virulence factors after transmission. The upregulation of the innate immunity resistance genes phoP and mgtC suggest that those Y. pestis that are phagocytized may be prepared for resistance to CAMPs and intracellular survival while still in the flea vector. Finally, the major essential virulence factors yadBC and pla, essential for Y. pestis dissemination from the dermis, were maximally or very highly expressed in the flea (Tables 1, S3). Besides degrading plasminogen, the Pla protease may also inactivate CAMPs, particularly when the F1 capsule is not present [57], which matches the phenotype of Y. pestis in the flea.
In summary, Y. pestis appears to be prepared for pathogenesis in the mammal while still in the flea vector. The biofilm phenotype of Y. pestis and the virulence factors upregulated or highly expressed in the flea may enhance the earliest stages of plague pathogenesis while the full complement of temperature-shift-regulated virulence factors is still being induced. Increased resistance to innate immunity that is preinduced in the flea vector may be critical to productive transmission because blocked fleas transmit relatively few bacteria, often below the LD50 of Y. pestis grown in vitro at <28°C [1],[52].
Materials and Methods
Ethics statement
All animals were handled in strict accordance with good animal practice as defined by NIH animal care and use policies and the Animal Welfare Act, USPHS; and all animal work was approved by the Rocky Mountain Laboratories Animal Care and Use Committee.
Bacterial strains and growth conditions for in vitro transcriptome analyses
Y. pestis KIM6+, which lacks the 70-kb virulence plasmid that is not required for flea infection or blockage, was used for gene expression analyses. A KIM6+ strain with an in-frame deletion that eliminated amino acids 28–281 of the predicted 291 amino acid residue yitR (y0181) gene product was produced by allelic exchange, using the pCVD442 suicide vector system [11]. This mutant was complemented by electroporation with a recombinant pWKS130 plasmid containing the wild-type yitR promoter and orf. The ΔyitR mutant was also transformed with pWKS130 alone to generate an empty vector control strain. For in vitro planktonic samples, bacteria were grown from frozen stocks in brain heart infusion (BHI) medium at 28°C, followed by two successive transfers in Luria Bertani broth supplemented with 100 mM MOPS, pH 7.4 (LB/MOPS) at 21°C. An inoculum of 104 cells/ml was added to 50 ml of LB/MOPS and incubated at 21°C with shaking at 250 rpm until exponential (OD600 = 2.5) or stationary phase (OD600 = 4.5). Approximately 0.5 ml of the exponential phase culture and 0.25 ml of the stationary phase culture was resuspended in 1 ml and 0.5 ml, respectively, of RNAprotect bacterial reagent (Qiagen; Valencia, CA), incubated for 5 min at room temperature, and centrifuged at 21°C for 5 min prior to RNA extraction.
For in vitro biofilms, 400 µl of a 107/ml bacterial suspension was injected into a flowcell (Stovall; Greensboro, NC) that was connected to a reservoir of LB/MOPS at 21°C. Following a 30 min incubation period to allow the bacteria to adhere to the glass surface of the flow cell, LB/MOPS was pumped through the flow cell at a rate of 0.3 ml/min. After 48 hours, the flowcell was disconnected and the thick Y. pestis biofilm was harvested and treated with 0.5ml of RNAprotect similarly to the planktonic cultures.
Flea infections and collection of samples for in vivo transcriptome analyses
X. cheopis fleas were infected with Y. pestis KIM6+ by using a previously described artificial feeding system [3]. The infectious blood meal was prepared by growing Y. pestis KIM6+ overnight at 37°C in BHI medium, without aeration. A cell pellet containing 109 bacterial cells was resuspended in 1 ml PBS and added to 5 ml heparinized mouse blood. Fleas that took a blood meal were maintained at 21°C and 75% relative humidity, fed twice weekly on uninfected mice, and monitored for proventricular blockage as previously described [3]. On the day blockage was diagnosed, the digestive tract was dissected out and macerated in RNAprotect, a process that required about 1 min. Thirty midguts from blocked fleas were pooled for each of the two biological replicates. Midguts from 60 uninfected fleas were also collected as controls to assess background hybridization of flea RNA to the microarray.
A flea-borne transmission model [58] was used to determine Y. pestis infectivity after challenge by flea bite. Fleas were infected with Y. pestis 195/P, a fully virulent wild-type strain, or with a Y. pestis 195/P ΔyitR mutant constructed as described above. Between 2–3 weeks after infection, the time required for Y. pestis to block fleas with a proventricular biofilm, groups of 20–40 fleas were applied to a restrained mouse and allowed to feed for 60 min. The fleas were then recovered and examined under a dissecting microscope to determine how many had taken a normal blood meal (unblocked or non-infective fleas) and how many were blocked (infective fleas). After challenge, mice were monitored and euthanized upon the appearance of signs of terminal illness. Mice that did not develop any symptoms after one week following a challenge were re-challenged. A total of 9–10 BALB/cAnN and 10 RML Swiss-Webster mice were challenged with each strain.
RNA isolation, amplification, and microarray
RNA was isolated from six independent samples from in vitro and flow cell cultures and two independent samples from pooled blocked fleas (Fig. S1) using the RNeasy Mini Kit (Qiagen). Flea-derived RNA samples were secondarily split into three technical replicates each. RNA integrity was verified on a Bioanalyzer 2100 (Agilent Technologies; Santa Clara, CA). Total RNA (100 ng) was amplified and labeled with modified biotin-11-CTP (Perkin Elmer; Waltham, MA) and biotin-16-UTP (Roche Molecular Biochemicals, Pleasanton, CA) by using the Message-Amp II-Bacteria amplified antisense RNA (aRNA) kit (Ambion; Austin, TX). Amplified RNA was then fragmented using Ambion's Fragmentation reagent (Applied Biosystems), hybridized to the RML custom Affymetrix GeneChip that contains sequences for all Y. pestis predicted ORFs, and scanned. The amplification step did not affect the relative transcript signals obtained by microarray (data not shown).
Microarray data analysis
Affymetrix GeneChip Operating Software (GCOS v1.4, GEO platform GPL2129, http://www.affymetrix.com) was used for initial analysis of the microarray data at the probe-set level. All *.cel files, representing individual biological replicates, were scaled to a trimmed mean of 500 using a scale mask consisting of only the Yersinia pestis KIM6+ probe-sets to produce the *.chp files. A pivot table with all samples was created including calls, call p-value and signal intensities for each gene. The pivot table was then imported into GeneSpring GX 7.3 (http://www.chem.agilent.com), where hierarchical clustering (condition tree) using a Pearson correlation similarity measure with average linkage was used to produce the dendrogram indicating that biological replicates grouped together. The pivot table was also imported into Partek Genomics Suite software (Partek Inc.; St. Louis, MO) to produce a principal components analysis (PCA) plot as a second statistical test for the grouping of biological replicates. ANOVA was run from this data set to produce a false discovery rate report producing false positive reduced p-values for each comparison of interest.
The correlated replicates of all test conditions and controls were combined, and quality filters based upon combined calls and signal intensities were used to further evaluate individual gene comparisons. Present and marginal calls were treated as the same whereas absent calls were negatively weighted and eliminated from calculations. Ratios of test/control values and associated t-test and ANOVA p-values values of all individual genes passing the above filters were determined using GeneSpring, SAM, and Partek software. The microarray data have been deposited in the NCBI GEO public database (accession number GSE16493).
To compare differential in vivo gene expression patterns in the flea and the rat, the average hybridization signal for each individual Y. pestis gene was divided by the average signal of all 4,683 genes on the microarray for both the flea microarray (this study) and the rat bubo microarray [11] data sets. Gene by gene comparisons of these normalized expression data sets were used for Fig. 3 and Tables 1, S5, and S6).
Macrophage phagocytosis assay
Murine bone marrow-derived macrophages were prepared as described [59],[60] and cultured in Dulbecco's Modified Eagles medium (DMEM) supplemented with 5 mM L-glutamine, 25 mM HEPES, 10% heat-inactivated fetal bovine serum, 5 mM non-essential amino acids, and 10 ng/ml CSF-1 (PeproTech; Rocky Hills, NJ). 1-ml suspensions of Y. pestis KIM6+ containing pAcGFP1 (Clontech; Mountain View, CA) from 21°C stationary phase LB/MOPS cultures, or from triturated midguts dissected from fleas 2 to 3 weeks after infection were treated for 15 sec in a FastPrep FP120 using lysing matrix H (Qbiogene; Carlsbad, CA) to disrupt bacterial aggregates, quantified by Petroff-Hausser direct count, and diluted in DMEM to ∼1×106 bacteria/ml. 0.1 ml of bacterial suspension was added to tissue culture plate wells containing ∼1×105 differentiated primary macrophages cultured on 12 mm glass coverslips in 1 ml DMEM. The plates were not centrifuged after addition of the bacteria, and midgut triturate from an equivalent number of uninfected fleas was added to the in vitro-derived bacterial suspensions used for these experiments. After 1 h incubation at 37°C and 5% CO2, the medium was removed and the cells washed, fixed in 2.5% paraformaldehyde for 10 min at 37°C, and then rewashed. Extracellular bacteria were labelled by indirect immunofluorescence as described [60] using a 1∶50,000 dilution of hyperimmune rabbit anti-Y. pestis polyclonal antibody [7] and a 1∶400 dilution of AlexaFluor 568-conjugated goat anti-rabbit antibody (Invitrogen; Carlsbad, CA). The percentage of extracellular bacteria was determined by dividing the number of red-fluorescent bacteria by the total number (red- and green only-fluorescent) bacteria associated with individual macrophages. To calculate differential resistance to phagocytosis for a given strain, the average percent extracellular LB-grown bacteria was subtracted from the average percent extracellular flea-derived bacteria. Results from 2–3 independent experiments performed in triplicate were analyzed by unpaired two-tailed t-test.
Quantitative RT-PCR
Independent RNA samples were prepared from blocked fleas and in vitro biofilm and planktonic cultures as described for the microarray experiments, except that the RNA was not amplified. Samples were treated with rDnase I (Ambion) and confirmed by PCR to be free of genomic DNA contamination. cDNA was synthesized from the RNA and used for quantitative PCR on an ABI Prism 7900 sequence detection system (Taqman, Applied Biosystems). The reactions contained oligonucleotide primers and probes designed using Primer Express version 2.0 software (Applied Biosystems) and the Taqman Universal PCR Master Mix (Applied Biosystems). For each primer-probe set assay, a standard curve was prepared using known concentrations of Y. pestis KIM6+ genomic DNA and used to transform CT values into relative DNA quantity. The quantity of cDNA for each experimental gene was normalized relative to the quantity of the reference gene crr (y1485), and the ratio of the normalized quantity of each gene in the flea samples to the normalized quantity in the in vitro samples was calculated (Fig. S2). Primer and probe sets used are listed in Table S7.
Supporting Information
Representative electrophoretograms of total RNA extracted from dissected flea digestive tracts. Electrophoretograms derived from uninfected (A) and blocked (B) flea digestive tracts are shown, with prokaryotic and eukaryotic rRNA peaks indicated.
(0.97 MB TIF)
Quantitative reverse transcription (QRT) PCR confirmation of microarray results. The quantity of each mRNA was determined relative to that of the reference gene crr (y1485). Fold-differences in transcript levels of the 12 Y. pestis genes in the flea compared to (A) in vitro biofilm, (B) exponential phase planktonic cultures, and (C) stationary phase planktonic cultures are shown as determined by microarray (grey bars) and QRT-PCR (black bars). *gabT transcript was detected by microarray in the flea samples only.
(1.92 MB TIF)
Y. pestis genes upregulated ≥2-fold in the flea relative to all in vitro conditions.
(0.32 MB DOC)
Y. pestis genes downregulated ≥2-fold in the flea relative to all in vitro conditions.
(0.14 MB DOC)
The 100 most highly expressed Y. pestis genes in the flea.
(0.22 MB DOC)
Y. pestis genes upregulated ≥2-fold in the flea and flowcell biofilms relative to planktonic culture conditions.
(0.12 MB DOC)
Y. pestis genes with significantly higher relative expression levels in the flea gut than in the rat bubo.
(0.28 MB DOC)
Y. pestis genes with significantly higher relative expression levels in the rat bubo than in the flea.
(0.51 MB DOC)
Primers and probes used for quantitative RT-PCR.
(0.06 MB DOC)
Acknowledgments
We thank Mary Burtnick, Leigh Knodler, Robin Ireland, and Jeffrey Shannon for help with the macrophage experiments. We also thank Patti Rosa, Frank Gherardini, Mollie Jewett, and Catherine Bosio for critical reading of the manuscript.
Footnotes
The authors have declared that no competing interests exist.
This work was supported by the Division of Intramural Research, NIAID, NIH. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Perry RD, Fetherston JD. Yersinia pestis– etiologic agent of plague. Clin Microbiol Rev. 1997;10:35–66. doi: 10.1128/cmr.10.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hinnebusch BJ, Rudolph AE, Cherepanov P, Dixon JE, Schwan TG, et al. Role of Yersinia murine toxin in survival of Yersinia pestis in the midgut of the flea vector. Science. 2002;296:733–735. doi: 10.1126/science.1069972. [DOI] [PubMed] [Google Scholar]
- 3.Hinnebusch BJ, Perry RD, Schwan TG. Role of the Yersinia pestis hemin storage (hms) locus in the transmission of plague by fleas. Science. 1996;273:367–370. doi: 10.1126/science.273.5273.367. [DOI] [PubMed] [Google Scholar]
- 4.Darby C, Ananth SL, Tan L, Hinnebusch BJ. Identification of gmhA, a Yersinia pestis gene required for flea blockage, by using a Caenorhabditis elegans biofilm system. Infect Immun. 2005;73:7236–7242. doi: 10.1128/IAI.73.11.7236-7242.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bacot AW, Martin CJ. Observations on the mechanism on the transmission of plague by fleas. J Hyg Plague. 1914;13(Suppl 3):423–439. [PMC free article] [PubMed] [Google Scholar]
- 6.Darby C, Hsu JW, Ghori N, Falkow S. Caenorhabditis elegans: plague bacteria biofilm blocks food intake. Nature. 2002;417:243–244. doi: 10.1038/417243a. [DOI] [PubMed] [Google Scholar]
- 7.Jarrett CO, Deak E, Isherwood KE, Oyston PC, Fischer ER, et al. Transmission of Yersinia pestis from an infectious biofilm in the flea vector. J Infect Dis. 2004;190:783–792. doi: 10.1086/422695. [DOI] [PubMed] [Google Scholar]
- 8.Kirillina O, Fetherston JD, Bobrov AG, Abney J, Perry RD. HmsP, a putative phosphodiesterase, and HmsT, a putative diguanylate cyclase, control Hms-dependent biofilm formation in Yersinia pestis. Mol Microbiol. 2004;54:75–88. doi: 10.1111/j.1365-2958.2004.04253.x. [DOI] [PubMed] [Google Scholar]
- 9.Bobrov AG, Kirillina O, Perry RD. The phosphodiesterase activity of the HmsP EAL domain is required for negative regulation of biofilm formation in Yersinia pestis. FEMS Microbiol Lett. 2005;247:123–130. doi: 10.1016/j.femsle.2005.04.036. [DOI] [PubMed] [Google Scholar]
- 10.Beloin C, Da Re S, Ghigo J-M. Colonization of abiotic surfaces. In: Böck A, Curtis R III, Kaper JB, Neidhardt FC, Nyström K, et al., editors. EcoSal—Escherichia coli and Salmonella: cellular and molecular biology. Washington, D.C.: ASM Press; 2005. [Google Scholar]
- 11.Sebbane F, Lemaitre N, Sturdevant DE, Rebeil R, Virtaneva K, et al. Adaptive response of Yersinia pestis to extracellular effectors of innate immunity during bubonic plague. Proc Natl Acad Sci U S A. 2006;103:11766–11771. doi: 10.1073/pnas.0601182103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Terra WR, Ferreira C, Jordao BP, Dillon RJ. Digestive enzymes. In: Lehane MJ, Billingsley PF, editors. Biology of the insect midgut. London: Chapman & Hall; 1996. pp. 153–194. [Google Scholar]
- 13.Ogahara T, Ohno M, Takayama M, Igarashi K, Kobayashi H. Accumulation of glutamate by osmotically stressed Escherichia coli is dependent on pH. J Bacteriol. 1995;177:5987–5990. doi: 10.1128/jb.177.20.5987-5990.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chapman RF. The insects. Structure and function. Cambridge, UK: Cambridge University Press; 1998. [Google Scholar]
- 15.Boudko DY, Kohn AB, Meleshkevitch EA, Dasher MK, Seron TJ, et al. Ancestry and progeny of nutrient amino acid transporters. Proc Natl Acad Sci U S A. 2005;102:1360–1365. doi: 10.1073/pnas.0405183101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keyhani NO, Roseman S. Wild-type Escherichia coli grows on the chitin disaccharide, N,N′-diacetylchitobiose, by expressing the cel operon. Proc Natl Acad Sci U S A. 1997;94:14367–14371. doi: 10.1073/pnas.94.26.14367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reizer J, Reizer A, Saier MH., Jr Novel phosphotransferase system genes revealed by bacterial genome analysis-a gene cluster encoding a unique Enzyme I and the proteins of a fructose-like permease system. Microbiology. 1995;141:961–971. doi: 10.1099/13500872-141-4-961. [DOI] [PubMed] [Google Scholar]
- 18.Wolfe AJ. The acetate switch. Microbiol Mol Biol Rev. 2005;69:12–50. doi: 10.1128/MMBR.69.1.12-50.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mortlock RP, Brubaker RR. Glucose-6-phosphate dehydrogenase and 6-phosphogluconate dehydrogenase activities of Pasteurella pestis and Pasteurella pseudotuberculosis. J Bacteriol. 1962;84:1122–1123. doi: 10.1128/jb.84.5.1122-1123.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Whiteley M, Bagera MG, Bumgarner RE, Parsek MR, Teitzel GM, et al. Gene expression in Pseudomonas aeruginosa biofilms. Nature. 2001;413:860–864. doi: 10.1038/35101627. [DOI] [PubMed] [Google Scholar]
- 21.Waite RD, Paccanaro A, Papakonstantinopoulou A, Hurst JM, Saqi M, et al. Clustering of Pseudomonas aeruginosa transcriptomes from planktonic cultures, developing and mature biofilms reveals distinct expression profiles. BMC Genomics. 2006;7:162. doi: 10.1186/1471-2164-7-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schembri MA, Kjaergaard K, Klemm P. Global gene expression in Escherichia coli biofilms. Mol Microbiol. 2003;48:253–267. doi: 10.1046/j.1365-2958.2003.03432.x. [DOI] [PubMed] [Google Scholar]
- 23.Beloin C, Valle J, Latour-Lambert P, Faure P, Kzreminski M, et al. Global impact of mature biofilm lifestyle on Escherichia coli K-12 gene expression. Mol Microbiol. 2004;51:659–674. doi: 10.1046/j.1365-2958.2003.03865.x. [DOI] [PubMed] [Google Scholar]
- 24.Lazazzera BA. Lessons from DNA microarray analysis: the gene expression profile of biofilms. Curr Opin Microbiol. 2005;8:222–227. doi: 10.1016/j.mib.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 25.Stewart PS, Franklin MJ. Physiological heterogeneity in biofilms. Nat Rev Microbiol. 2008;6:199–210. doi: 10.1038/nrmicro1838. [DOI] [PubMed] [Google Scholar]
- 26.Perry RD, Bobrov AG, Kirillina O, Jones HA, Pedersen L, et al. Temperature regulation of the hemin storage (Hms+) phenotype of Yersinia pestis is posttranscriptional. J Bacteriol. 2004;186:1638–1647. doi: 10.1128/JB.186.6.1638-1647.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Torres AG, Jeter C, Langley W, Matthysse AG. Differential binding of Escherichia coli O157:H7 to alfalfa, human epithelial cells, and plastic is mediated by a variety of surface structures. Appl Environ Microbiol. 2005;71:8008–8015. doi: 10.1128/AEM.71.12.8008-8015.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vallet I, Olson JW, Lory S, Lazdunski A, Filloux A. The chaperone/usher pathways of Pseudomonas aeruginosa: identification of fimbrial gene clusters (cup) and their involvement in biofilm formation. Proc Natl Acad Sci U S A. 2001;98:6911–6916. doi: 10.1073/pnas.111551898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sherlock O, Vejborg RM, Klemm P. The TibA adhesin/invasin from enterotoxigenic Escherichia coli is self recognizing and induces bacterial aggregation and biofilm formation. Infect Immun. 2005;73:1954–1963. doi: 10.1128/IAI.73.4.1954-1963.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patel CN, Wortham BW, Lines JL, Fetherston JD, Perry RD, et al. Polyamines are essential for the formation of plague biofilm. J Bacteriol. 2006;188:2355–2363. doi: 10.1128/JB.188.7.2355-2363.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vadyvaloo V, Jarrett C, Sturdevant D, Sebbane F, Hinnebusch BJ. Analysis of Yersinia pestis gene expression in the flea vector. Adv Exp Med Biol. 2007;603:192–200. doi: 10.1007/978-0-387-72124-8_16. [DOI] [PubMed] [Google Scholar]
- 32.Rebeil R, Ernst RK, Jarrett CO, Adams KN, Miller SI, et al. Characterization of late acyltransferase genes of Yersinia pestis and their role in temperature-dependent lipid A variation. J Bacteriol. 2006;188:1381–1388. doi: 10.1128/JB.188.4.1381-1388.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sebbane F, Jarrett CO, Gardner D, Long D, Hinnebusch BJ. Role of the Yersinia pestis plasminogen activator in the incidence of distinct septicemic and bubonic forms of flea-borne plague. Proc Natl Acad Sci U S A. 2006;103:5526–5530. doi: 10.1073/pnas.0509544103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Forman S, Wulff CR, Myers-Morales T, Cowan C, Perry RD, et al. yadBC of Yersinia pestis, a new virulence determinant for bubonic plague. Infect Immun. 2008;76:578–587. doi: 10.1128/IAI.00219-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang X, Lindler LE. The pH 6 antigen is an antiphagocytic factor produced by Yersinia pestis independent of Yersinia outer proteins and capsule antigen. Infect Immun. 2004;72:7212–7219. doi: 10.1128/IAI.72.12.7212-7219.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cathelyn JS, Crosby SD, Lathem WW, Goldman WE, Miller VL. RovA, a global regulator of Yersinia pestis, specifically required for bubonic plague. Proc Natl Acad Sci U S A. 2006;103:13514–13519. doi: 10.1073/pnas.0603456103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heroven AK, Dersch P. RovM, a novel LysR-type regulator of the virulence activator gene rovA, controls cell invasion, virulence and motility of Yersinia pseudotuberculosis. Mol Microbiol. 2006;62:1469–1483. doi: 10.1111/j.1365-2958.2006.05458.x. [DOI] [PubMed] [Google Scholar]
- 38.Oyston PC, Dorrell N, Williams K, Li SR, Green M, et al. The response regulator PhoP is important for survival under conditions of macrophage-induced stress and virulence in Yersinia pestis. Infect Immun. 2000;68:3419–3425. doi: 10.1128/iai.68.6.3419-3425.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grabenstein JP, Fukuto HS, Palmer LE, Bliska JB. Characterization of phagosome trafficking and identification of PhoP-regulated genes important for survival of Yersinia pestis in macrophages. Infect Immun. 2006;74:3727–3741. doi: 10.1128/IAI.00255-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blanc-Potard AB, Groisman EA. The Salmonella selC locus contains a pathogenicity island mediating intramacrophage survival. Embo J. 1997;16:5376–5385. doi: 10.1093/emboj/16.17.5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Groisman EA, Kayser J, Soncini FC. Regulation of polymixin resistance and adaptation to low-Mg2+ environments. J Bacteriol. 1997;179:7040–7045. doi: 10.1128/jb.179.22.7040-7045.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bader MW, Sanowar S, Daley ME, Schneider AR, Cho U, et al. Recognition of antimicrobial peptides by a bacterial sensor kinase. Cell. 2005;122:461–472. doi: 10.1016/j.cell.2005.05.030. [DOI] [PubMed] [Google Scholar]
- 43.Prost LR, Miller SI. The Salmonellae PhoQ sensor: mechanisms of detection of phagosome signals. Cell Microbiol. 2008;10:576–582. doi: 10.1111/j.1462-5822.2007.01111.x. [DOI] [PubMed] [Google Scholar]
- 44.Dimopoulus G, Richman A, Müller H-M, Kafatos FC. Molecular immune responses of the mosquito Anopheles gambiae to bacteria and malaria parasites. Proc Natl Acad Sci U S A. 1997;94:11508–11513. doi: 10.1073/pnas.94.21.11508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lehane MJ, Wu D, Lehane SM. Midgut-specific immune molecules are produced by the blood-sucking insect Stomoxys calcitrans. Proc Natl Acad Sci U S A. 1997;94:11502–11507. doi: 10.1073/pnas.94.21.11502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou D, Han Y, Qin L, Chen Z, Qiu J, et al. Transcriptome analysis of the Mg2+-responsive PhoP regulator in Yersinia pestis. FEMS Microbiol Lett. 2005;250:85–95. doi: 10.1016/j.femsle.2005.06.053. [DOI] [PubMed] [Google Scholar]
- 47.Perez JC, Groisman EA. Transcription factor function and promoter architecture govern the evolution of bacterial regulons. Proc Natl Acad Sci U S A. 2009;106:4319–4324. doi: 10.1073/pnas.0810343106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miyashiro T, Goulian M. Stimulus-dependent differential regulation in the Escherichia coli PhoQ-PhoP system. Proc Natl Acad Sci U S A. 2007;104:16305–16310. doi: 10.1073/pnas.0700025104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Erickson DL, Waterfield NR, Vadyvaloo V, Long D, Fischer ER, et al. Acute oral toxicity of Yersinia pseudotuberculosis to fleas: implications for the evolution of vector-borne transmission of plague. Cell Microbiol. 2007;9:2658–2666. doi: 10.1111/j.1462-5822.2007.00986.x. [DOI] [PubMed] [Google Scholar]
- 50.Gendlina I, Held KG, Bartra SS, Gallis BM, Doneanu CE, et al. Identification and type III-dependent secretion of the Yersinia pestis insecticidal-like proteins. Mol Microbiol. 2007;64:1214–1227. doi: 10.1111/j.1365-2958.2007.05729.x. [DOI] [PubMed] [Google Scholar]
- 51.Hares MC, Hinchliffe SJ, Strong PC, Eleftherianos I, Dowling AJ, et al. The Yersinia pseudotuberculosis and Yersinia pestis toxin complex is active against cultured mammalian cells. Microbiology. 2008;154:3503–3517. doi: 10.1099/mic.0.2008/018440-0. [DOI] [PubMed] [Google Scholar]
- 52.Lorange EA, Race BL, Sebbane F, Hinnebusch BJ. Poor vector competence of fleas and the evolution of hypervirulence in Yersinia pestis. J Infect Dis. 2005;191:1907–1912. doi: 10.1086/429931. [DOI] [PubMed] [Google Scholar]
- 53.Burrows TW, Bacon GA. The basis of virulence in Pasteurella pestis: the development of resistance to phagocytosis in vitro. Br J Exp Pathol. 1956;37:286–299. [PMC free article] [PubMed] [Google Scholar]
- 54.Cavanaugh DC, Randall R. The role of multiplication of Pasteurella pestis in mononuclear phagocytes in the pathogenesis of flea-borne plague. J Immunol. 1959;83:348–363. [PubMed] [Google Scholar]
- 55.Erickson DL, Jarrett CO, Callison JA, Fischer ER, Hinnebusch BJ. Loss of a biofilm-inhibiting glycosyl hydrolase during the emergence of Yersinia pestis. J Bacteriol. 2008;190:8163–8170. doi: 10.1128/JB.01181-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vuong C, Voyich JM, Fischer ER, Braughton KR, Whitney AR, et al. Polysaccharide intercellular adhesin (PIA) protects Staphylococcus epidermidis against major components of the human innate immune system. Cell Microbiol. 2004;6:269–275. doi: 10.1046/j.1462-5822.2004.00367.x. [DOI] [PubMed] [Google Scholar]
- 57.Galván EM, Lasaro MAS, Schifferli DM. Capsular antigen Fraction 1 and Pla modulate the susceptibility of Yersinia pestis to pulmonary antimicrobial peptides such as cathelicidin. Infect Immun. 2008;76:1456–1464. doi: 10.1128/IAI.01197-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sebbane F, Jarrett C, Gardner D, Long D, Hinnebusch BJ. The Yersinia pestis caf1M1A1 fimbrial capsule operon promotes transmission by flea bite in a mouse model of bubonic plague. Infect Immun. 2009;77:1222–1229. doi: 10.1128/IAI.00950-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bosio CM, Elkins KL. Susceptibility to secondary Francisella tularensis live vaccine strain infection in B-cell-deficient mice is associated with neutrophilia but not with defects in specific T-cell-mediated immunity. Infect Immun. 2001;69:194–203. doi: 10.1128/IAI.69.1.194-203.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Celli J. Intracellular localization of Brucella abortus and Francisella tularensis in primary murine macrophages. Methods Mol Biol. 2008;431:133–145. doi: 10.1007/978-1-60327-032-8_11. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Representative electrophoretograms of total RNA extracted from dissected flea digestive tracts. Electrophoretograms derived from uninfected (A) and blocked (B) flea digestive tracts are shown, with prokaryotic and eukaryotic rRNA peaks indicated.
(0.97 MB TIF)
Quantitative reverse transcription (QRT) PCR confirmation of microarray results. The quantity of each mRNA was determined relative to that of the reference gene crr (y1485). Fold-differences in transcript levels of the 12 Y. pestis genes in the flea compared to (A) in vitro biofilm, (B) exponential phase planktonic cultures, and (C) stationary phase planktonic cultures are shown as determined by microarray (grey bars) and QRT-PCR (black bars). *gabT transcript was detected by microarray in the flea samples only.
(1.92 MB TIF)
Y. pestis genes upregulated ≥2-fold in the flea relative to all in vitro conditions.
(0.32 MB DOC)
Y. pestis genes downregulated ≥2-fold in the flea relative to all in vitro conditions.
(0.14 MB DOC)
The 100 most highly expressed Y. pestis genes in the flea.
(0.22 MB DOC)
Y. pestis genes upregulated ≥2-fold in the flea and flowcell biofilms relative to planktonic culture conditions.
(0.12 MB DOC)
Y. pestis genes with significantly higher relative expression levels in the flea gut than in the rat bubo.
(0.28 MB DOC)
Y. pestis genes with significantly higher relative expression levels in the rat bubo than in the flea.
(0.51 MB DOC)
Primers and probes used for quantitative RT-PCR.
(0.06 MB DOC)