Abstract
Tooth development is a highly heritable process which relates to other growth and developmental processes, and which interacts with the development of the entire craniofacial complex. Abnormalities of tooth development are common, with tooth agenesis being the most common developmental anomaly in humans. We performed a genome-wide association study of time to first tooth eruption and number of teeth at one year in 4,564 individuals from the 1966 Northern Finland Birth Cohort (NFBC1966) and 1,518 individuals from the Avon Longitudinal Study of Parents and Children (ALSPAC). We identified 5 loci at P<5×10−8, and 5 with suggestive association (P<5×10−6). The loci included several genes with links to tooth and other organ development (KCNJ2, EDA, HOXB2, RAD51L1, IGF2BP1, HMGA2, MSRB3). Genes at four of the identified loci are implicated in the development of cancer. A variant within the HOXB gene cluster associated with occlusion defects requiring orthodontic treatment by age 31 years.
Author Summary
Genome-wide association studies have been used to identify genetic variants conferring susceptibility to diseases, intermediate phenotypes, and physiological traits such as height, hair color, and age at menarche. Here we analyze the NFBC1966 and ALSPAC birth cohorts to investigate the genetic determinants of a key developmental process: primary tooth development. The prospective nature of our studies allows us to exploit accurate measurements of age at first tooth eruption and number of teeth at one year, and also provides the opportunity to assess whether genetic variants affecting these traits are associated with dental problems later in the life course. Of the genes that we find to be associated with primary tooth development, several have established roles in tooth development and growth, and almost half have proposed links with the development of cancer. We find that one of the variants is also associated with occlusion defects requiring orthodontic treatment later in life. Our findings should provide a strong foundation for the study of the genetic architecture of tooth development, which as well as its relevance to medicine and dentistry, may have implications in evolutionary biology since teeth represent important markers of evolution.
Introduction
Heritability of primary tooth emergence is estimated to be over 70% [1]. Abnormalities in tooth development are common with tooth agenesis alone affecting up to 10% of the population, ranking it as the most common developmental anomaly in humans [2]. Such abnormalities contribute to a variety of challenging and expensive orthodontic, prosthetic and surgical treatments and account for approximately 6% of all dental health care attendances [3]. Many genes implicated in primary dentition have regulatory functions important to several developmental processes in the embryo [4], and the developing tooth is a useful model for the study of organogenesis [5]. However, despite substantial research into tooth development in mice and human malformation syndromes [5], the genetic determinants of the normal variation in human tooth development have not been established.
To identify genetic loci regulating primary dentition we performed a general population based genome-wide association (GWA) study of tooth development in infancy among individuals from the 1966 Northern Finland Birth Cohort (NFBC1966) and the Avon Longitudinal Study of Parents and Children (ALSPAC). Specifically, we tested for associations with time to first tooth eruption and number of teeth by one year of age. These phenotypes are relevant to later tooth development because teeth largely acquire their final form at a very early age [6]. The availability of longitudinal birth cohort data allowed us to investigate life-course associations with dental occlusion defects.
Results
We tested 300,766 SNPs common to both studies (each used the Illumina platform). The analyses were adjusted for sex, gestational age and population structure (Materials and Methods). Results for the two cohorts were combined using fixed effects inverse variance meta-analysis. Five genetic loci were identified at genome-wide significance (P<5×10−8). Table 1 shows the top-ranking SNPs at each locus (see also Figure 1 and Figure 2, Figures S1, S2, S3). For all SNPs the allele associated with a delay in tooth eruption was associated with fewer teeth at the end of infancy. Table S1 shows details of the functions of genes linked to the identified loci.
Table 1. The top GWA signals at each locus from a meta-analysis of NFBC1966 and ALSPAC.
| In (closest) gene/locus | SNP | Chromosome (Position) | Effect/Other allele | Frequency of effect allele NFBC ALSPAC | Time to first tooth eruption | Number of teeth | ||||||||
| P value NFBC | P value ALSPAC | % var NFBC/ALSPAC | Overall P | P value NFBC | P value ALSPAC | % var NFBC/ALSPAC | Overall P | |||||||
| SNPs at genome-wide significance in meta-analysis ( P <5×10−8) | ||||||||||||||
| (KCNJ2) | rs8079702 | 17 (65,702,421) | G/A | 0.391 | 0.424 | 1.89×10−17 | 1.62×10−6 | 1.62/1.48 | 3.77×10−22 | 7.78×10−12 | 3.22×10−4 | 1.15/1.05 | 1.24×10−14 | |
| EDA | rs4844096 | X (68,722,043) | G/A | 0.423 | 0.466 | 6.21×10−6 | 3.56×10−4 | 0.42/0.77 | 2.61×10−8 | 3.79×10−9 | 2.65×10−3 | 0.73/0.52 | 4.57×10−11 | |
| “ | rs5936487 | X (68,809,641) | G/A | 0.390 | 0.462 | 6.65×10−8 | 6.90×10−5 | 0.50/0.99 | 6.18×10−11 | 3.89×10−8 | 2.10×10−3 | 0.52/0.57 | 3.36×10−10 | |
| MSRB3 | rs10506525 | 12 (64,069,645) | C/T | 0.266 | 0.375 | 1.29×10−6 | 5.80×10−4 | 0.46/0.71 | 6.17×10−9 | 2.56×10−4 | 5.60×10−4 | 0.18/0.74 | 8.67×10−7 | |
| IGF2BP1 | rs9674544 | 17 (44,439,710) | G/A | 0.462 | 0.484 | 2.97×10−5 | 5.44×10−3 | 0.25/0.40 | 8.33×10−7 | 2.29×10−4 | 9.86×10−7 | 0.27/1.61 | 1.55×10−8 | |
| RAD51L1 | rs1956529 | 14 (67,858,677) | T/C | 0.383 | 0.364 | 1.07×10−3 | 5.07×10−4 | 0.16/0.73 | 1.32×10−5 | 4.95×10−6 | 1.23×10−3 | 0.51/0.60 | 2.59×10−8 | |
| SNPs with suggestive evidence (5×10−6> P >5×10−8) in meta-analysis | ||||||||||||||
| 2q35 | rs6435957 | 2 (217,586,454) | T/C | 0.371 | 0.309 | 0.021 | 0.250 | 0.21/0.00 | 9.99×10−3 | 6.85×10−6 | 0.017 | 0.37/1.10 | 3.64×10−7 | |
| 6q21 | rs9386463 | 6 (106,200,750) | G/A | 0.446 | 0.483 | 4.55×10−6 | 0.047 | 0.38/0.14 | 5.99×10−7 | 8.29×10−3 | 0.586 | 0.08/0.00 | 0.011 | |
| (HOXB1, HOXB2) | rs6504340 | 17 (43,972,018) | G/A | 0.224 | 0.209 | 3.00×10−3 | 0.136 | 0.12/0.01 | 9.40×10−4 | 2.86×10−5 | 6.00×10−3 | 0.44/0.34 | 6.06×10−7 | |
| 6q22 | rs2817937 | 6 (121,140,156) | C/T | 0.124 | 0.097 | 0.110 | 0.400 | 0.10/0.00 | 0.070 | 6.23×10−6 | 0.152 | 0.25/0.01 | 3.00×10−6 | |
| (HMGA2) | rs12424086 | 12 (64,650,776) | C/T | 0.228 | 0.304 | 7.78×10−5 | 0.027 | 0.29/0.22 | 7.57×10−6 | 9.77×10−5 | 0.011 | 0.22/0.41 | 3.64×10−6 | |
SNPs at genome wide significance P<5×10−8 and SNPS with suggestive evidence at 5×10−6>P>5×10−8. The P value of each cohort is corrected for sex, gestational age and population structure using principal components and genomic control. The combined P value is calculated using a fixed effects inverse variance meta-analysis. When no gene is within 50 kb of the SNP the chromosome band is given. Positions of SNPs are reported in NCBI build 36 coordinates. The alleles all refer to the forward strand. The effect allele is defined as the allele associated with later tooth eruption and a smaller number of teeth. % var is the percentage of variance explained by each SNP. P values attaining overall GWA significance are in bold.
Figure 1. Linkage disequilibrium and association at loci reaching genome-wide significance for primary tooth development in meta-analysis of NFBC1966 and ALSPAC.
(A) KCNJ2 gene region for time to first tooth eruption. (B) EDA gene region for time to first tooth. (C) MSRB3 gene region for time to first tooth. (D) IGF2BP1 gene region for number of teeth at 12 months (SNP with high P at 44000 kb is that near HOXB2, rs6504340). Note: This is a gene-rich region, so most genes are omitted to simplify the plot. (E) RAD51L1 gene region for number of teeth at 12 months. -log10 p-value is plotted against genomic position (NCBI build 36). Most significant SNP in each region is plotted in blue, r2 with top SNP is colour coded red (0.8 – 1.0), orange (0.5 – 0.8), yellow (0.2 – 0.5), and white <0.2. Gene annotations are based on Genome Browser (RefSeq Genes) and arrows represent direction of transcription. Recombination rate is estimated by LDhat using HapMap CEU sample. All r2 values are calculated in NFBC1966.
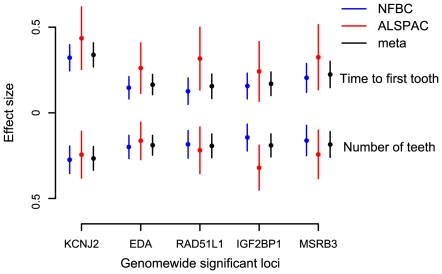
Figure 2. Meta-analysis for primary tooth development by genotype for the five SNPs attaining genome-wide significance.
Estimates and 95% confidence intervals for regression coefficients are given for the effect of delayed teething allele in Gaussian regression on time to first tooth and an ordinal regression on number of teeth.
The strongest association with both phenotypes was for SNP rs8079702, located 15 kb downstream of KCNJ2 (inward rectifier potassium channel 2) (P = 3.77×10−22 for time of first tooth, P = 1.24×10−14 for number of teeth; Table 1). There are no SNPs in KCNJ2 in our data, but rs8079702 had highest correlation with SNP rs4328485 which was the closest available SNP to KCNJ2 (r2 = 0.17; 1 kb away). KCNJ2 has been implicated in Pierre Robin sequence [7] and Andersen-Tawil syndrome [8], which show abnormalities in tooth development (missing teeth, delays in eruption) and are characterized by craniofacial anomalies such as narrowing of the jaw and cleft palate [8]. The second strongest association was for SNP rs5936487, located within the EDA (ectodermal dysplasia protein) gene (P = 6.18×10−11 for time of first tooth, P = 3.36×10−10 for number of teeth). EDA was fundamental in forming the first teeth in organisms [9], and mutations cause hypohidrotic ectodermal dysplasia (HED) and non-syndromic disorders of tooth agenesis [10].
The three remaining loci at genome-wide significance (P<5×10−8) have SNPs located within the genes RAD51L1 (RAD51-like1), IGF2BP1 (insulin-like growth factor 2 mRNA binding protein 1) and MSRB3 (methionine sulfoxide reductase B3). RAD51L1 is involved in DNA repair and a variant in the gene has been found to confer susceptibility to breast cancer [11]. It is responsible for protein kinase activity, and the injection of activators of protein kinase C (PKC) in rats causes delays in tooth eruption [12]. IGF2BP1 regulates the growth factor IGF2, and knockouts of the gene in mice suggest a role in organ development [13], while its expression is associated with ovarian cancer [14]. A microarray study in the developing mouse molar tooth found MSRB3 to be in the top 100 most expressed genes of 34,000 examined [15].
Each of the associated SNPs explain a small fraction of the residual phenotypic variation in time to first tooth (0.2%–1.6%, NFBC1966; 0.4%–1.5%, ALSPAC) and number of teeth by one year (0.2%–1.2%, NFBC1966; 0.5%–1.6%, ALSPAC), after controlling for sex and gestational age. Selecting the SNP with the most extreme signal for either phenotype to represent each locus (“top SNPs”), and analysing them together, the additive effects of these five top SNPs explain 2.9% of the variance of both tooth eruption time and number of teeth in the NFBC1966, and 4.2% and 4.0% of the variance in tooth eruption and number of teeth in ALSPAC. Without a suitable external replication cohort these estimates were derived in the two discovery cohorts and therefore may overestimate the true values due to the “winner's curse”. GWA studies have thus far explained only a small proportion of heritability [16], and our estimates are comparable with the variance explained in human height by a GWA study [17]. In order to identify variants with lower effect sizes or rarer variants larger sample sizes would be required.
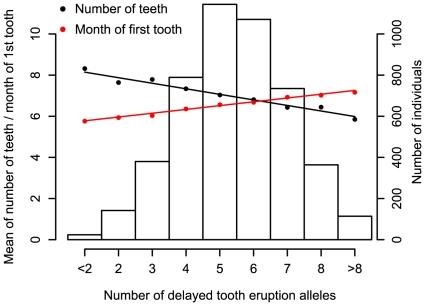
We also summarized the predictive power of the five top SNPs by defining a ‘delayed tooth eruption’ measure as the number of alleles across the SNPs that delay tooth eruption. Figure 3 shows the number of delayed tooth eruption alleles against the mean of both time to first tooth eruption and number of teeth by one year in NFBC1966. Individuals with 8 or more delayed eruption alleles (10% of NFBC1966) have an average of 1.5 fewer teeth at 12 months, and later tooth eruption by 1.1 months, compared to individuals with 3 or fewer such alleles (11% of NFBC1966). Figure S4 shows the same plot for time to first tooth in ALSPAC.
Figure 3. The combined impact of the delayed tooth eruption alleles in 5 identified loci at P<5×10−8 in the NFBC1966.
Subjects are classified by the number of delayed tooth eruption alleles. SNPs are chosen so that they had the strongest signal for number of teeth at each locus. Mean time of first tooth eruption is plotted in red and number of teeth by the age of one year in black. The bars represent the number of individuals for each count of ‘delayed tooth eruption’ alleles. The line through points is a linear regression fit.
In addition to the five loci attaining genome wide significance, there were 5 loci with SNPs that had P-values between 5×10−6 and 5×10−8 (Table 1). We investigated the biological functions of nearby genes to see if any of these loci were related to tooth development. These signals included SNP rs6504340, which is located between the developmental regulatory genes HOXB1 (homeobox B1) and HOXB2 (homeobox B2). Although previous studies have indicated that tooth development is independent of a Hox patterning program [18], Homeobox genes have recently been shown to be expressed in the dental mesenchyme in the pharyngeal teeth of bony fishes [9]. SNP rs6504340 lies 500 kb upstream of rs9674544 in IGF2BP1, but the two SNPs show almost no linkage disequilibrium with each other (r2 = 0.002 in NFBC1966 and r2 = 0.006 in ALSPAC). Furthermore, a test for association of rs6504340 conditional on rs9674544 was significant (P = 6.3×10−5 in NFBC1966 and P = 0.01 in ALSPAC; Materials and Methods), indicating that these represent two independent signals (Figure 1). We also identified three SNPs at 2q35, the most significant of which had r2 = 0.48 with a variant associated with breast cancer [11],[19], and SNP rs12424086 located close to the HMGA2 gene and 6 kb away from rs1042725, the SNP identified by a GWA study for adult and childhood height [20].
Given the influence of tooth development on dental occlusion, we hypothesized that genetic determinants of early tooth eruption may associate with dental occlusion later in life. We tested for associations between the SNP with the most extreme signal for either phenotype at each of the 10 identified loci and defects in occlusion requiring orthodontic treatment by the age of 31 years in the NFBC1966 (data not available in ALSPAC). A total of 611 individuals (13.5%) reported a defect in occlusion that had required orthodontic treatment. Of the 10 SNPs tested, SNP rs6504340 (HOXB gene cluster) gave a significant association, where each G allele (associated with delayed tooth eruption and lower number of teeth in infancy, Table 1), increased the odds of having an occlusal defect requiring orthodontic treatment by 35%, after adjusting for sex (odds ratio (OR) = 1.35, 95% CI = 1.16–1.57; P = 1.13×10−4; further adjustment for gestational age did not change the result). A smaller number of teeth at 1 year also predicted higher risk of orthodontic treatment (OR = 1.05, 95% CI = 1.01–1.09; P = 0.009). However, when number of teeth or time to first tooth were included in the model with dental occlusion as outcome, the associations with the G allele remained (P = 0.001, P = 1.71×10−4), suggesting an independent association between rs6504340 and dental occlusion.
Discussion
Teeth and several other organs have common growth and developmental pathways during early life [21]. The genes at the loci identified in our study have roles in organogenesis, growth and developmental processes, and cancer. Mutations in three of the genes lead to altered organogenesis and development; KCNJ2 (teeth, jaws, palates, ears, fingers, toes), EDA (teeth, hair, sweat glands, salivary glands) and IGF2BP1 (intestines) [8],[13],[22]. Of the loci at suggestive levels of significance, the HOXB gene cluster is an established regulator of development, and the HMGA2 gene has previously been associated with adult height [20]. Normal development and cancer both involve shifts between cell proliferation and differentiation [23] and genes regulating organ-specific growth are known to be involved in oncogenesis [24]. A previous study identified a common genetic link between an abnormal tooth development and cancer [25]. From our identified loci, IGF2BP1 and RAD51L1 have been implicated in cancer [11],[14] as have HOXB2, 2q35, and HMGA2 [19],[26],[27].
We provide the first detailed insight into the genetic architecture of primary dentition and our findings could have implications for the study of other developmental and organogenic processes. Exploiting the availability of longitudinal cohort data [28] we found an association between a variant within the HOXB gene cluster and the requirement for orthodontic treatment due to defective occlusion by the age of 31 years. Further GWA studies of developmental processes during infancy may establish whether the genetic determinants of infant development can contribute to the study of chronic diseases, such as cancer, that occur later in life.
Materials and Methods
Study population and phenotype description
The data was derived from two genome-wide scans of the geographically defined prospective birth cohorts; the NFBC1966 and ALSPAC. The NFBC1966 followed pregnancies in the two northernmost provinces of Finland with expected delivery dates in 1966. ALSPAC recruited mothers during pregnancy with expected dates of delivery between April 1991 and December 1992 from Bristol and the surrounding area in the South West of England. A total of 4,564 samples were available from the NFBC1966 and 1,518 from ALSPAC. In both cohorts, two separate measures of primary tooth development were collected: i) date of first tooth eruption (in months), and ii) number of teeth (measured at 12 months in NFBC1966 and 15 months in ALSPAC). In the NFBC1966 date of first tooth eruption and number of teeth was gathered by public health professionals during children's monthly visits to child welfare centers (parents carried a booklet where they had recorded the developmental milestones reached). In ALSPAC, parents reported the date of first tooth eruption and number of teeth at 15 months on a questionnaire. In order to ascertain the accuracy of the parental responses, a subsample were examined and validated by a dentist. Information on date of first tooth eruption was available for 4,523 individuals in the NFBC1966 (99% of available GWA samples) and 1396 (92%) in ALSPAC and for number of teeth, 4,326 (95%) in the NFBC1966 and 1,426 (94%) in ALSPAC. All aspects of the study were reviewed and approved by the Ethics Committee of the University of Oulu and the ALSPAC Law and Ethics Committee and by the respective local research committees. Participants (in NFBC1966) and parents (in ALSPAC) gave written informed consent.
Genotyping
The Illumina HumanCNV370-Duo DNA Analysis BeadChip was used for genotyping the NFBC1966, and Illumina HumanHap317K BeadChip for ALSPAC. The genotyping and quality control procedures have been described elsewhere [29],[30]. SNPs were excluded from the analysis if the call rate in the final sample was <95%, if there was a lack of Hardy-Weinberg Equilibrium (HWE) (P<10−4 in NFBC1966, P<5×10−7 in ALSPAC), or if the MAF was <1%. After quality control, 329,091 SNPs in NFBC1966 and 310,611 in ALSPAC were available. We report here the results from the 300,766 genotyped SNPs common to both studies.
Statistical analyses
Age of first tooth eruption in the NFBC1966 was recorded in months, such that the first tooth could have erupted at any time between the end of previous month and the end of the recorded month. In ALSPAC it was recorded to the nearest month and 3 individuals were recorded as having no teeth after 15 months. To account for the censoring in the two cohorts the outcome was analyzed using parametric survival analysis in the R software package 2.7.1.The Gaussian distribution gave a good fit to the data in both cohorts and was used to model the underlying event time. Number of teeth in the NFBC1966 was recorded at 12 months. In ALSPAC, measurements were taken at around 15 months but there was variability in the exact time of measurement, therefore the ALSPAC analysis was adjusted for age of measurement. Teeth typically erupt in pairs from the upper and lower jaw (75% of children had an even number of teeth in the NFBC1966), making the Poisson distribution inappropriate for modeling the number of teeth. Therefore ordinal logistic regression was used as implemented by the polr function in the R package. Analyses of the X chromosome treated males as homozygous females. The allele frequencies of the identified SNPs on the X chromosome did not differ significantly between the sexes. GWA analyses were adjusted for sex, gestational age and population stratification using principal components (PC). Each analysis was corrected for population stratification separately by including those of the top 10 PCs that were associated with the phenotype at P<0.05 [31]. For number of teeth, PCs 3, 6 and 9 were included in ALSPAC and none in the NFBC1966. For time to first tooth eruption no PCs were included in ALSPAC and PC 2 was included in the NFBC1966. After correction by PCs, the estimated variance inflation factors [32] for date of first tooth eruption were 1.039 and 1.047 in ALSPAC and NFBC1966 respectively, and 1.011 and 1.039 for number of teeth. Genomic control [32] was then used to correct the residual population stratification. The variance inflation factors from the meta-analyses were 1.012 for number of teeth and 1.015 date of first tooth eruption.
Results from the two studies were combined using fixed effects inverse variance meta-analysis [33]. Analyses were performed using the statistical package R and metaMapper (a meta-analysis software developed in-house). Conditional analyses were calculated using the likelihood ratio test comparing ordinal regression models, one including rs9674544 and the other including rs9674544 and rs6504340. Variance explained by each SNP was computed as 1 minus the ratio of variance of residuals of the model with age, gestational age and SNP to variance of residuals of the model with just age and gestational age. To correct for overfitting, each individual's phenotype was estimated from a model that did not include that individual. The total variance explained by the five loci reaching genome-wide significance was calculated similarly using the most associated SNPs for each phenotype at each locus. Additional tests for association with orthodontic treatment used the SNPs most associated with number of teeth at the 10 loci at P<5×10−6. Table 1 reports the top GWA signals at each of the ten loci (i.e. the SNP with the strongest association with either time to first tooth eruption or number of teeth at age 1 year).
URLs
Jackson Laboratory website, http://www.jax.org; NCBI, http://www.ncbi.nlm.nih.gov; R project, www.r-project.org; Uniprot, http://www.uniprot.org.
Supporting Information
Manhattan plots for the 300,766 SNPs from the genome-wide association meta-analysis for (A) time to first tooth eruption, and (B) number of teeth at 12 months. The (blue) line indicates the genome-wide significance threshold (P<5×10−8).
(0.17 MB PDF)
Manhattan plots and linkage disequilibrium (LD) diagrams for five identified loci (P<5×10−8). (A) Locus 17q24 (KCNJ2), (B) Locus Xq13 (EDA), (C) Locus 14q24 (RAD51L1), (D) Locus 17q21.4 (IGF2BP1), and (E) Locus 12q14 (MSRB3). The (blue) line indicates the genome-wide significance threshold (P<5×10−8).
(0.89 MB PDF)
Quantile-quantile plots of observed -log10 P values versus the expectation under the null for (A) time to first tooth eruption and (B) number of teeth at 12 months. The most associated 10,000 SNPs from the meta-analysis are shown.
(0.07 MB PDF)
Additive effect of delayed tooth eruption alleles in identified loci in ALSPAC. Subject classified by the number of delayed tooth eruption alleles. SNPs chosen had the strongest signal for time to first tooth eruption at each locus. Mean time of first tooth eruption is plotted in black. The bars represent the number of individuals for each count of “delayed tooth eruption” alleles. Lines through points are linear regression fits.
(0.05 MB PDF)
Summary of the candidate genes located within the top loci.
(0.08 MB PDF)
Acknowledgments
We are grateful to all the families who took part in this study, the midwives for their help in recruiting them, Professors Paula Rantakallio and Jean Golding, founders of these cohort studies, and the whole NFBC 1966 and ALSPAC teams. We thank the Sample Logistics and Genotyping Facilities both at the Wellcome Trust Sanger Institute and the Broad Genotyping Center for generating the ALSPAC and NFBC1966 genome wide genetic data.
This publication is the work of the authors and they will serve as guarantors for the contents of this paper.
Footnotes
The authors have declared that no competing interests exist.
The NFBC1966 received financial support from the Academy of Finland (project grants 104781, 120315, 132797, and Center of Excellence in Complex Disease Genetics); University Hospital Oulu, Biocenter, University of Oulu, Finland; the European Community's Fifth/Seventh Framework Programme (EURO-BLCS, QLG1-CT-2000-01643, FP7/2007-2013); NHLBI grant 5R01HL087679-02 through the STAMPEED program (1RL1MH083268-01); ENGAGE project (HEALTH-F4-2007-201413); the Medical Research Council (studentship grant G0500539, centre grant G0600705); the Wellcome Trust (project grant GR069224), UK; the Research Council UK fellowship; the National Institute of Health Research (NIHR) Biomedical Research Centre Programme at Imperial College; and the Division of Epidemiology, Public Health and Primary Care (studentship grant DFHM G24038). The DNA extractions, sample quality controls, biobank up-keeping, and aliquotting were performed in the National Public Health Institute, Biomedicum Helsinki, Finland, and supported financially by the Academy of Finland and Biocentrum Helsinki. The UK Medical Research Council, the Wellcome Trust, and the University of Bristol provide core support for ALSPAC. CJH is funded by a European Union grant HEALTH-2007-201550 HyperGenes. DME is supported by a Medical Research Council New Investigator Award (MRC G0800582). The ICLS (International Centre for Life Course Studies in Society and Health) is funded by an ESRC award: RES-596-28-0001. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Hughes TE, Bockmann MR, Seow K, Gotjamanos T, Gully N, et al. Strong genetic control of emergence of human primary incisors. J Dent Res. 2007;86:1160–1165. doi: 10.1177/154405910708601204. [DOI] [PubMed] [Google Scholar]
- 2.Nanci A. St Louis: Mosby Elsevier, pg; 2008. Ten Cate's oral histology. Development, structure, and function, 7th ed.96 [Google Scholar]
- 3.Anderson R, Richmond S, Thomas DW. Patient presentation at medical practices with dental problems: an analysis of the 1996 General Practice Morbidity Database for Wales. Br Dent J. 1999;186:297–300. doi: 10.1038/sj.bdj.4800091. [DOI] [PubMed] [Google Scholar]
- 4.Thesleff I. Genetic basis of tooth development and dental defects. Acta Odontol Scand. 2000;58:191–194. doi: 10.1080/000163500750051728. [DOI] [PubMed] [Google Scholar]
- 5.Tucker A, Sharpe P. The cutting-edge of mammalian development; how the embryo makes teeth. Nat Rev Genet. 2004;5:499–508. doi: 10.1038/nrg1380. [DOI] [PubMed] [Google Scholar]
- 6.Koussoulakou DS, Margaritis LH, Koussoulakos SL. A curriculum vitae of teeth: evolution, generation, regeneration. Int J Biol Sci. 2009;5:226–243. doi: 10.7150/ijbs.5.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benko S, Fantes JA, Amiel J, Kleinjan DJ, Thomas S, et al. Highly conserved non-coding elements on either side of SOX9 associated with Pierre Robin sequence. Nat Genet. 2009;41:359–364. doi: 10.1038/ng.329. [DOI] [PubMed] [Google Scholar]
- 8.Yoon G, Oberoi S, Tristani-Firouzi M, Etheridge SP, Quitania L, et al. Andersen-Tawil syndrome: prospective cohort analysis and expansion of the phenotype. Am J Med Genet A. 2006;140:312–321. doi: 10.1002/ajmg.a.31092. [DOI] [PubMed] [Google Scholar]
- 9.Fraser GJ, Hulsey CD, Bloomquist RF, Uyesugi K, Manley NR, et al. An ancient gene network is co-opted for teeth on old and new jaws. PLoS Biol. 2009;7:e31. doi: 10.1371/journal.pbio.1000031. doi: 10.1371/journal.pbio.1000031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mues GI, Griggs R, Hartung AJ, Whelan G, Best LG, et al. From ectodermal dysplasia to selective tooth agenesis. Am J Med Genet A. 2009;149A:2037–2041. doi: 10.1002/ajmg.a.32801. [DOI] [PubMed] [Google Scholar]
- 11.Thomas G, Jacobs KB, Kraft P, Yeager M, Wacholder S, et al. A multistage genome-wide association study in breast cancer identifies two new risk alleles at 1p11.2 and 14q24.1 (RAD51L1). Nat Genet. 2009;41:579–584. doi: 10.1038/ng.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wise GE, Yao S, Liu D. Injections of osteoprotegerin and PMA delay tooth eruption. Clin Anat. 2006;19:19–24. doi: 10.1002/ca.20144. [DOI] [PubMed] [Google Scholar]
- 13.Hansen TV, Hammer NA, Nielsen J, Madsen M, Dalbaeck C, et al. Dwarfism and impaired gut development in insulin-like growth factor II mRNA-binding protein 1-deficient mice. Mol Cell Biol. 2004;24:4448–4464. doi: 10.1128/MCB.24.10.4448-4464.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gu L, Shigemasa K, Ohama K. Increased expression of IGF II mRNA-binding protein 1 mRNA is associated with an advanced clinical stage and poor prognosis in patients with ovarian cancer. Int J Oncol. 2004;24:671–678. [PubMed] [Google Scholar]
- 15.Pemberton TJ, Li F-Y, Oka S, Mendoza-Fandino GA, Hsu Y-H, et al. Identification of novel genes expressed during mouse tooth development by microarray gene expression analysis. Dev Dyn. 2007;236:2245–2257. doi: 10.1002/dvdy.21226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weedon MN, Lango H, Lindgren CM, Wallace C, Evans DM, et al. Genome-wide association analysis identifies 20 loci that influence adult height. Nat Genet. 2008;40:575–583. doi: 10.1038/ng.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.James CT, Ohazama A, Tucker AS, Sharpe PT. Tooth development is independent of a Hox patterning programme. Dev Dyn. 2002;225:332–335. doi: 10.1002/dvdy.10168. [DOI] [PubMed] [Google Scholar]
- 19.Stacey SN, Manolescu A, Sulem P, Rafnar T, Gudmundsson J, et al. Common variants on chromosomes 2q35 and 16q2 confer susceptibility to estrogen receptor-positive breast cancer. Nat Genet. 2007;39:865–869. doi: 10.1038/ng2064. [DOI] [PubMed] [Google Scholar]
- 20.Weedon MN, Lettre G, Freathy RM, Lindgren CM, Voight BF, et al. A common variant of HMGA2 is associated with adult and childhood height in the general population. Nat Genet. 2007;39:1245–1250. doi: 10.1038/ng2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thesleff I, Vaahtokari A, Partanen AM. Common molecular mechanisms regulating the development of teeth and other organs. Int J Dev Biol. 1995;39:35–50. [PubMed] [Google Scholar]
- 22.Mikkola ML, Theleff I. Ectodysplasin signalling in development. Cytokine Growth Factor Rev. 2003;14:211–224. doi: 10.1016/s1359-6101(03)00020-0. [DOI] [PubMed] [Google Scholar]
- 23.Nunes FD, de Almeida FC, Tucci R, de Sousa SC. Homeobox genes: a molecular link between development and cancer. Pesqui Odontol Bras. 2003;17:94–98. doi: 10.1590/s1517-74912003000100018. [DOI] [PubMed] [Google Scholar]
- 24.Hallikas O, Palin K, Sinjashina N, Rautiainen R, Partanen J, et al. Genome-wide prediction of mammalian enhancers based on analysis of transcription-factor binding affinity. Cell. 2006;124:47–59. doi: 10.1016/j.cell.2005.10.042. [DOI] [PubMed] [Google Scholar]
- 25.Lammi L, Arte S, Somer M, Jarvinen H, Lahermo P, et al. Mutations in AXIN2 cause familial tooth agenesis and predispose to colorectal cancer. Am J Hum Genet. 2004;74:1043–1050. doi: 10.1086/386293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Collins Y, Tan DF, Pejovic T, Mor G, Qian F, et al. Identification of differentially expressed genes in clinically distinct groups of serous ovarian carcinomas using cDNA microarray. Int J Mol Med. 2005;14:43–53. [PubMed] [Google Scholar]
- 27.Grier DG, Thompson A, Kwasniewska A, McGonigle GJ, Halliday HL, et al. The pathophysiology of HOX genes and their role in cancer. J Pathol. 2005;205:154–171. doi: 10.1002/path.1710. [DOI] [PubMed] [Google Scholar]
- 28.Manolio TA. Cohort studies and the genetics of complex disease. Nat Genet. 2009;41:5–6. doi: 10.1038/ng0109-5. [DOI] [PubMed] [Google Scholar]
- 29.Sabatti C, Service SK, Hartikainen AL, Pouta A, Ripatti S, et al. Genome-wide association analysis of metabolic traits in a birth cohort from a founder population. Nat Genet. 2009;41:35–46. doi: 10.1038/ng.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Timpson NJ, Tobias JH, Richards JB, Soranzo N, Duncan EL, et al. Common variants in the region around Osterix are associated with bone mineral density and growth in childhood. Hum Mol Genet. 2009;18:1510–1517. doi: 10.1093/hmg/ddp052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Novembre J, Stephens M. Interpreting principal component analyses of spatial population genetic variation. Nat Genet. 2008;40:646–649. doi: 10.1038/ng.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 33.Sutton AJ, Abrams KR, Jones DR, Sheldon TA Song F. Chichester, UK: John Wiley & Sons, pg; 2000. Methods for meta-analysis in medical research.58 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Manhattan plots for the 300,766 SNPs from the genome-wide association meta-analysis for (A) time to first tooth eruption, and (B) number of teeth at 12 months. The (blue) line indicates the genome-wide significance threshold (P<5×10−8).
(0.17 MB PDF)
Manhattan plots and linkage disequilibrium (LD) diagrams for five identified loci (P<5×10−8). (A) Locus 17q24 (KCNJ2), (B) Locus Xq13 (EDA), (C) Locus 14q24 (RAD51L1), (D) Locus 17q21.4 (IGF2BP1), and (E) Locus 12q14 (MSRB3). The (blue) line indicates the genome-wide significance threshold (P<5×10−8).
(0.89 MB PDF)
Quantile-quantile plots of observed -log10 P values versus the expectation under the null for (A) time to first tooth eruption and (B) number of teeth at 12 months. The most associated 10,000 SNPs from the meta-analysis are shown.
(0.07 MB PDF)
Additive effect of delayed tooth eruption alleles in identified loci in ALSPAC. Subject classified by the number of delayed tooth eruption alleles. SNPs chosen had the strongest signal for time to first tooth eruption at each locus. Mean time of first tooth eruption is plotted in black. The bars represent the number of individuals for each count of “delayed tooth eruption” alleles. Lines through points are linear regression fits.
(0.05 MB PDF)
Summary of the candidate genes located within the top loci.
(0.08 MB PDF)