Abstract
Despite lacking the adaptive immunity that is found in higher vertebrates, insects are able to defend themselves from a large battery of pathogens by multiple innate immune responses using molecular mechanisms that are strikingly similar to the innate immune responses of other multicellular organisms, including humans. The fruit fly Drosophila melanogaster is therefore an excellent model organism for studying the basic principles of innate immunity using genetic and molecular biology techniques. In Drosophila, invading pathogens that pass through the epithelial barriers (a first line of self-defense) can encounter humoral and cellular responses that utilize pattern-recognition receptors to identify pathogen-associated molecular patterns in the hemolymph or on the immune cell surface. Some pathogens escape recognition and elimination in the hemolymph by invading the host cytoplasm. Some intracellular pathogens such as Listeria monocytogenes are, nevertheless, eliminated by immune reactions such as autophagy through intracellular identification by pattern-recognition receptors.
Keywords: autophagy, Drosophila, innate immunity, pathogen sensor, peptidoglycan
Introduction
The fruit fly Drosophila melanogaster has emerged as a very useful organism for studying the basic principles of innate immunity because of the evolutionary conservation of innate immunity genes, pathways and effector mechanisms and the well-established techniques for manipulating fruit fly genetics and molecular biology. Isolation of Drosophila mutants that had deficient immune responses against bacterial and fungal infections and subsequent genetic screens revealed two conserved signaling cascades—the Toll and imd pathways—both of which lead to the activation of nuclear factor κB (NF-κB) family transcription factors (1). The initial discovery that the Toll pathway is required for immune responses in Drosophila (2) led directly to the demonstration that vertebrate Toll-like receptors (TLRs) are also required for mammalian immunity (3, 4).
The frontline of insect host defense is epithelial tissues such as the epidermis and trachea, which not only act as mechanical barriers but also produce anti-microbial peptides. Pathogens passing through the epithelial barrier encounter cellular and humoral defense reactions in the hemolymph (Fig. 1) (5). The cellular reactions include phagocytosis and encapsulation by host cells. The humoral reactions include the activation of cascades of constitutive proteins present in the hemolymph, such as those in the prophenoloxidase (proPO) cascade, leading to melanization (deposition of melanin pigments onto pathogens and the wounded sites) and the coagulation cascade, and the activation of intracellular signaling pathways that produce defense proteins such as anti-microbial peptides in immune-responsive tissues and cells.
Fig. 1.
Insect immune responses. (A) Epithelial tissues such as the epidermis and trachea act as a first line of self-defense, not only functioning as mechanical barriers but also producing anti-microbial peptides to prevent infections. Black rods indicate pathogens. (B) As insects have open vascular systems, pathogens passing through the epithelial barrier are identified by host proteins called pattern-recognition receptors and activate humoral and cellular responses. The humoral responses depend on primary and secondary responses. The primary response is mediated by the activation of the cascade present in the hemolymph, such as the proPO cascade, leading to melanization, and coagulation. The secondary response requires gene activation of defense proteins such as anti-microbial peptides in immune tissues such as the fat body, which is the functional equivalent of the mammalian liver. The cellular responses include phagocytosis and encapsulation. Black rods indicate pathogens. Open dots indicate pigmented melanin.
Drosophila mutants that fail to produce anti-microbial peptides are susceptible to bacterial and fungal infections, indicating that anti-microbial peptide induction has an important role in insect host defense systems. In response to microbial infections, seven different types of anti-microbial peptides (Attacin, Cecropin, Defensin, Diptericin, Drosocin, Drosomycin and Metchnikowin) with different anti-microbial specificities are secreted into the hemolymph from the fat body in Drosophila; the fat body is the functional equivalent of the mammalian liver. The induction of anti-microbial peptides is mediated by the Toll and imd pathways in Drosophila (Fig. 2), in which Toll and imd are pivotal molecules and which are mechanistically similar to the mammalian TLR/IL-1R signaling pathway and tumor necrosis factor α receptor signaling pathways, respectively (1, 5). This review focuses on the pathogen-recognition systems that mediate Drosophila immune responses.
Fig. 2.
Recognition of microorganisms and induction of innate immune responses in Drosophila. (A) The primary humoral response, i.e. activation of the proPO cascade leading to melanization, is induced by the cleavage of proPO to PO by a serine protease, proPO-activating enzyme (PPAE). (B) A constitutive hemolymph protein, PGRP-LE, binds to monomeric and polymeric DAP-type peptidoglycans (PGNs), which are components of many Gram-negative and some Gram-positive bacteria, and activates the proPO cascade upstream of PPAE. PGRP-LE is also involved in activating the secondary humoral response, the imd pathway-dependent induction of antibacterial peptides. (C) Membrane PGRP-LC is also required for monomeric and polymeric DAP-type peptidoglycan-mediated activation of the imd pathway. (D) PGRP-SA in the hemolymph is involved in the recognition of Gram-positive bacteria with Lys-type peptidoglycans and is required for activation of the Toll pathway in cooperation with GNBP1. PGRP-SD has some redundant functions with PGRP-SA and GNBP1. (E) GNBP3 is involved in yeast-mediated activation of the Toll pathway. (F) The activation of the Toll pathway is mediated by its active ligand, Spz, cleaved from pro-Spz by the serine protease cascade, including the SPE, Spheroide, Spirit, Sphinx, Persephone, ModSP and Grass. (G) In addition to the extracellular functions, PGRP-LE induces anti-microbial peptides through the imd pathway and autophagy in the cytoplasm. Signaling via the Toll or imd pathway activates NF-κB and subsequent transcription of genes for anti-microbial peptides such as Drosomycin and Diptericin.
Microbial recognition in the Toll and imd pathways
The Toll pathway is activated primarily in response to fungal and some Gram-positive bacterial infections, whereas the imd pathway is activated predominantly in response to Gram-negative and other Gram-positive bacterial infections (6). Therefore, Drosophila possesses specific mechanisms to distinguish different pathogens. In innate immunity, conserved molecular patterns of pathogens are thought to be identified by so-called pattern-recognition receptors of the host defense systems (7).
In contrast to mammalian TLRs, which recognize pathogen components such as lipopolysaccharides, the ligand of the Drosophila Toll receptor is Spätzle (Spz), which is produced by proteolysis of an endogenous protein, pro-Spz, in response to infection. Therefore, Drosophila Toll does not act as a pattern-recognition receptor but rather mediates the downstream signaling of such receptors. In Drosophila, some peptidoglycan recognition protein (PGRP) family members and Gram-negative-binding protein (GNBP) family members act as pattern-recognition receptors upstream of either the Toll pathway or the imd pathway.
PGRP and GNBP were first purified from the hemolymph of the silkworm, Bombyx mori, on the basis of their affinity to peptidoglycan (a cell wall component of bacteria) and to Gram-negative bacteria, respectively (8, 9). Later, cDNA cloning of these proteins revealed that they are evolutionarily conserved: there are 13 members in Drosophila; 7 members in the mosquito, Anopheles gambiae and 4 members in both mouse and humans as PGRP families; GNBPs belong to the family of β-glucan-binding proteins (10–14). In Drosophila, there are two classes of PGRP genes: those with long transcripts (e.g. PGRP-LB and -LC) and those with short transcripts (e.g. PGRP-SA and -SD). The PGRP family has a PGRP domain in the C-terminal region, which has some amino acid sequence similarity to peptidoglycan-degrading enzymes with N-acetylmuramyl-alanine amidase activity, such as bacteriophage lysozymes.
Upstream of the Toll pathway, PGRP-SA, PGRP-SD, GNBP1 and GNBP3 act as pattern-recognition receptors. PGRP-SA, a hemolymph protein identified from a loss-of-function mutant screen, is required for activation of the Toll pathway in response to Gram-positive bacterial infections (15). PGRP-SA cooperates with GNBP1 to activate the Toll pathway in response to Gram-positive bacteria (16). PGRP-SD has some redundancy and recognizes Gram-positive bacteria with PGRP-SA and GNBP1 (17). GNBP3 is required for activation of the Toll pathway in response to fungal infections including yeast (18). Downstream of these recognition receptors, pro-Spz is cleaved by a serine protease, the Spz-processing enzyme (SPE) (19). SPE-mediated processing is regulated by several serine proteases such as Persephone, Spirit, Spheroide, Sphinx, Grass and ModSP (20–23).
On the other hand, PGRP-LC and PGRP-LE act as pattern-recognition receptors upstream of the imd pathway. PGRP-LC, a membrane PGRP family member, is required for activation of the imd pathway in response to Gram-negative bacterial infections (24–26).
The susceptibility of the PGRP-LC mutant to Gram-negative bacterial infections is higher than that of other imd pathway mutants, suggesting that there is an activator of the imd pathway in addition to PGRP-LC (24). Consistent with this hypothesis, PGRP-LE, a constitutive hemolymph protein, selectively activates the imd pathway (27). The PGRP-LC/PGRP-LE double-mutant is much more susceptible to Escherichia coli infection than either single mutant alone, indicating that PGRP-LC and PGRP-LE have redundant functions in producing resistance to Gram-negative bacterial infections (27). PGRP-LE activates the proPO cascade, which branches from the PGRP-LE-mediated activation of the imd pathway upstream of a serine protease in the proPO cascade called proPO-activating enzyme (27, 28). In addition to the extracellular functions of PGRP-LE in the hemolymph, PGRP-LE acts as a co-receptor of PGRP-LC on the surface of immune cells and as an intracellular receptor in immune-reactive cells as mentioned below (29).
Drosophila PGRP family members distinguish the structural diversity of peptidoglycans (30). Peptidoglycans are essential cell wall components of almost all bacteria except mycoplasma, which lacks a cell wall. The peptidoglycan layer is thicker in Gram-positive bacteria than Gram negative. Peptidoglycan is a polymer of β-1,4-linked N-acetylglucosamine (GlcNAc) and N-acetylmuramic acid (MurNAc) that can be cross-linked by short stem peptides. The glycan chain is relatively conserved in all bacteria, whereas the amino acid composition of the cross-linking stem peptides and the linkage between stem peptides are diversified depending on the bacterial species (31).
Many Gram-negative bacteria and some Gram-positive bacteria such as the Bacillus species have directly cross-linked diaminopimelic acid (DAP)-containing peptidoglycans and preferentially activate the imd pathway in Drosophila. Many Gram-positive bacteria have peptidoglycans containing lysine instead of DAP with cross-linking peptides between stem peptides and preferentially activate the Toll pathway. Consistent with the finding that PGRP-LE selectively activates the imd pathway in vivo, PGRP-LE selectively binds to DAP-type peptidoglycans, but not to Lys-containing peptidoglycans in vitro (27). PGRP-LC is required for DAP-type peptidoglycan-mediated activation of the imd pathway in vivo and in vitro (32, 33).
The minimum structure of the DAP-type peptidoglycan required for PGRP-LC-mediated activation of the imd pathway is a monomer of GlcNAc and MurNAc with an internal 1,6-anhydro bond attached to a tripeptide containing DAP, known as tracheal cytotoxin (TCT) (34). The heterodimer complex of PGRP-LCx (an isomer of PGRP-LC that has affinity to all types of peptidoglycans) and PGRP-LCa (which has no affinity for peptidoglycans) acts as a TCT receptor, and the PGRP-LCx homodimer complex acts as a receptor for polymeric DAP-type peptidoglycan in vitro (35). The structures of the PGRP domain of PGRP-LE and the TCT complex suggest that the binding of the ligand to PGRP-LE mediates receptor polymerization (36).
These results are consistent with the findings that PGRP-LC and PGRP-LE act synergistically to resist bacteria with DAP-type peptidoglycans such as E. coli and Bacillus megaterium. The minimum structure of the Lys-type peptidoglycan required for PGRP-SA-mediated activation of the Toll pathway comprises two units consisting of GlcNAc–MurNAc attached to Lys-containing tetrapeptides that are covalently dimerized by an interpeptide (37). Drosophila PGRP family members therefore distinguish the structural diversity of peptidoglycans and activate the appropriate immune responses.
In addition to PGRPs acting as pattern-recognition receptors, PGRP family members such as PGRP-SC and PGRP-LB have enzyme activity that hydrolyzes the lactylamide bond between the glycan strand and the stem peptides of peptidoglycans (38–40). The N-acetylmuramyl-alanine amidase activity is consistent with the structural similarity of the PGRP domain to N-acetylmuramyl-alanine amidase that was mentioned earlier. The degraded peptidoglycans lose their ability to stimulate immune responses, suggesting a scavenger function of the enzyme PGRP family members and negative feedback regulation of immune responses by enzymic PGRP family members (41).
Intracellular roles of PGRP-LE
Mosaic analysis (in which mutant cells exist alongside wild-type ones) that was based on clonal expression of a single gene in Drosophila revealed that, in the fat body, PGRP-LE activates the imd pathway in a non-cell-autonomous manner, which is due to the extracellular function of PGRP-LE, as described above (28). In the malphigian tubules (the functional equivalent of the mammalian kidney), however, PGRP-LE activates the imd pathway to induce anti-microbial peptides in a cell-autonomous manner (28). The suggestion of an intracellular function of PGRP-LE is supported by the findings that the delivery of TCT into S2 cells (a Drosophila macrophage-like cell line) induces anti-microbial peptides in a PGRP-LE-dependent manner (29). A conserved motif in PGRP-LE and PGRP-LC is required for their intracellular signal transduction and shows sequence similarity with the receptor-interacting protein (RIP) homotypic interaction motif (RHIM) in mammalian Toll/IL-1R-containing adaptor-inducing IFN-β (TRIF) and RIP1, which are required for TRIF-mediated and TLR3-mediated NF-κB activation, respectively, suggesting an evolutionarily conserved signaling mechanism (29).
The intracellular function of PGRP-LE suggests that PGRP-LE is involved in the recognition of intracellular bacterial pathogens. Intracellular pathogens, a diverse group of organisms that cause such serious diseases as tuberculosis and malaria in humans, invade cells and are thus able to escape humoral and cell surface innate immune receptors (42). Supporting this hypothesis, the PGRP-LE mutant is susceptible to infection by Listeria monocytogenes, an intracellular bacterium with DAP-containing peptidoglycans that infects both mammals and Drosophila (43). In contrast to Gram-negative bacterial infections, the survival rate of the double-mutant PGRP-LE/PGRP-LC to L. monocytogenes infection is similar to that of the PGRP-LE mutant, suggesting that PGRP-LE and PGRP-LC do not have redundant functions in producing resistance to L. monocytogenes (43).
In vitro infection experiments using phagocytic insect blood cells called hemocytes revealed that PGRP-LE is essential for the resistance against intracellular growth of bacteria, but, interestingly, inhibition of the bacterial growth is not dependent on the known innate immune signaling pathways, the Toll and imd pathways (43). In mammals, autophagy—a bulk self-degradation system for the turnover of proteins and organelles that is conserved from yeast to humans (44)—functions as an innate immune response against intracellular bacteria, viruses and parasites in cultured cells (45–51). In Drosophila, autophagy is induced in hemocytes in response to L. monocytogenes infection and is crucial for host survival against Listeria infection as well as for the inhibition of bacterial growth in hemocytes (43); similarly, autophagy is induced in mammalian cells infected with intracellular pathogens.
PGRP-LE is co-localized with infected L. monocytogenes in the cytoplasm and is required for the intracellular infection-dependent induction of autophagy in hemocytes (43). PGRP-LE is also required for the autophagy in hemocytes that is mediated by DAP-type peptidoglycan, but not for autophagy mediated by Lys-type peptidoglycan, suggesting the existence of another intracellular sensor for bacteria having Lys-type peptidoglycans (43). Consistent with these findings, intracellular bacterial growth is not suppressed in S2 cells in which PGRP-LE is not expressed, but forced expression of PGRP-LE suppresses intracellular bacterial growth in S2 cells (43).
The PGRP-LE-dependent suppression of intracellular bacterial growth is thus mediated by autophagy but not by the Toll and imd pathways (43). In this situation, the induction of anti-microbial peptides in response to L. monocytogenes infection in S2 cells is PGRP-LE dependent. The induction of anti-microbial peptides is mediated by the imd pathway, but not by the common autophagy pathway (43). Therefore, PGRP-LE has an essential role in detecting intracellular bacteria through DAP-type peptidoglycans to induce autophagy as an innate immune response, which indicates a direct link between pathogen recognition and the induction of autophagy (Fig. 3). The pathway linking pathogen sensors and autophagy is independent of the known innate immune signaling pathways—the Toll and imd pathways.
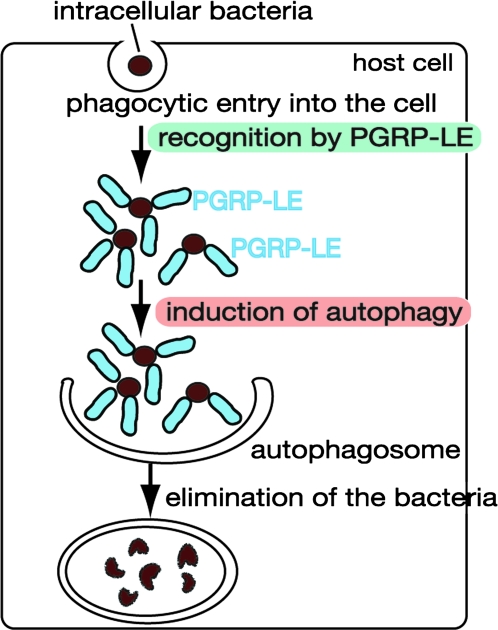
Fig. 3.
Recognition of intracellular bacteria and induction of autophagy by PGRP-LE. Intracellular bacteria such as L. monocytogenes are recognized by PGRP-LE in the cytoplasm through DAP-type peptidoglycans, which induces autophagy. The bacteria are then killed and degraded in the autophagosomes. PGRP-LE-mediated recognition of intracellular bacteria also activate the imd pathway, which induces the expression of anti-microbial peptides (see Fig. 2), but autophagy induction is independent of the imd pathway.
Concluding remarks
In Drosophila, some PGRP family members and GNBP family members act as pathogen sensors. PGRP-LE and PGRP-LC play a crucial role in detecting extracellular and intracellular bacteria with DAP-type peptidoglycans to induce various innate immune responses such as anti-microbial peptide production, melanization and autophagy. As the conserved RHIM-like domain of PGRP-LE and PGRP-LC is required for the induction of anti-microbial peptides, PGRP-LE and PGRP-LC are suggested to share a common intracellular mechanism to activate the imd pathway.
PGRP-LE-mediated induction of autophagy is independent of the imd pathway, suggesting that another intracellular signaling pathway links cytoplasmic receptors and autophagy. There are several cytoplasmic pathogen sensors such as nucleotide-binding oligomerization domain-like receptors in mammals (52). It is important to determine whether mammalian cytoplasmic sensors are involved in the induction of autophagy and, if so, if the mechanism is also conserved in mammals and insects similar to the signaling pathways that lead to the activation of NF-κB family transcription factors. Identification of the pathways and mechanisms of autophagy induced by pathogenic infection and studies involving their manipulation will potentially lead to novel therapeutic treatments of infectious diseases.
Funding
Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan; Japan Society for the Promotion of Science; Program for the Promotion of Basic Research Activities for Innovative Biosciences; National Institutes of Health grant (AI074958); Naito Foundation; Uehara Memorial Foundation.
References
- 1.Lemaitre B, Hoffmann J. The host defense of Drosophila melanogaster. Annu. Rev. Immunol. 2007;25:697. doi: 10.1146/annurev.immunol.25.022106.141615. [DOI] [PubMed] [Google Scholar]
- 2.Lemaitre B, Nicolas E, Michaut L, Reichhart JM, Hoffmann JA. The dorsoventral regulatory gene cassette spatzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell. 1996;86:973. doi: 10.1016/s0092-8674(00)80172-5. [DOI] [PubMed] [Google Scholar]
- 3.Medzhitov R, Preston-Hurlburt P, Janeway CA., Jr. A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature. 1997;388:394. doi: 10.1038/41131. [DOI] [PubMed] [Google Scholar]
- 4.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 5.Hultmark D. Drosophila immunity: paths and patterns. Curr. Opin. Immunol. 2003;15:12. doi: 10.1016/s0952-7915(02)00005-5. [DOI] [PubMed] [Google Scholar]
- 6.Lemaitre B, Reichhart JM, Hoffmann JA. Drosophila host defense: differential induction of antimicrobial peptide genes after infection by various classes of microorganisms. Proc. Natl Acad. Sci. USA. 1997;94:14614. doi: 10.1073/pnas.94.26.14614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Janeway CA., Jr. Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb. Symp. Quant. Biol. 1989;54:1. [Google Scholar]
- 8.Yoshida H, Kinoshita K, Ashida M. Purification of a peptidoglycan recognition protein from hemolymph of the silkworm, Bombyx mori. J. Biol. Chem. 1996;271:13854. doi: 10.1074/jbc.271.23.13854. [DOI] [PubMed] [Google Scholar]
- 9.Lee WJ, Lee JD, Kravchenko VV, Ulevitch RJ, Brey PT. Purification and molecular cloning of an inducible gram-negative bacteria-binding protein from the silkworm, Bombyx mori. Proc. Natl Acad. Sci. USA. 1996;93:7888. doi: 10.1073/pnas.93.15.7888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ochiai M, Ashida M. A pattern recognition protein for peptidoglycan. Cloning the cDNA and the gene of the silkworm, Bombyx mori. J. Biol. Chem. 1999;274:11854. doi: 10.1074/jbc.274.17.11854. [DOI] [PubMed] [Google Scholar]
- 11.Werner T, Liu G, Kang D, Ekengren S, Steiner H, Hultmark D. A family of peptidoglycan recognition proteins in the fruit fly Drosophila melanogaster. Proc. Natl Acad. Sci. USA. 2000;97:13772. doi: 10.1073/pnas.97.25.13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Christophides GK, et al. Immunity-related genes and gene families in Anopheles gambiae. Science. 2002;298:159. doi: 10.1126/science.1077136. [DOI] [PubMed] [Google Scholar]
- 13.Kang D, Liu G, Lundstrom A, Gelius E, Steiner H. A peptidoglycan recognition protein in innate immunity conserved from insects to humans. Proc. Natl Acad. Sci. USA. 1998;95:10078. doi: 10.1073/pnas.95.17.10078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu C, Xu Z, Gupta D, Dziarski R. Peptidoglycan recognition proteins: a novel family of four human innate immunity pattern recognition molecules. J. Biol. Chem. 2001;276:34686. doi: 10.1074/jbc.M105566200. [DOI] [PubMed] [Google Scholar]
- 15.Michel T, Reichhart JM, Hoffmann JA, Royet J. Drosophila Toll is activated by Gram-positive bacteria through a circulating peptidoglycan recognition protein. Nature. 2001;414:756. doi: 10.1038/414756a. [DOI] [PubMed] [Google Scholar]
- 16.Gobert V, Gottar M, Matskevich AA, et al. Dual activation of the Drosophila toll pathway by two pattern recognition receptors. Science. 2003;302:2126. doi: 10.1126/science.1085432. [DOI] [PubMed] [Google Scholar]
- 17.Bischoff V, Vignal C, Boneca IG, Michel T, Hoffmann JA, Royet J. Function of the drosophila pattern-recognition receptor PGRP-SD in the detection of Gram-positive bacteria. Nat. Immunol. 2004;5:1175. doi: 10.1038/ni1123. [DOI] [PubMed] [Google Scholar]
- 18.Gottar M, Gobert V, Matskevich AA, et al. Dual detection of fungal infections in Drosophila via recognition of glucans and sensing of virulence factors. Cell. 2006;127:1425. doi: 10.1016/j.cell.2006.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jang IH, et al. A Spätzle-processing enzyme required for toll signaling activation in Drosophila innate immunity. Dev. Cell. 2006;10:45. doi: 10.1016/j.devcel.2005.11.013. [DOI] [PubMed] [Google Scholar]
- 20.Ligoxygakis P, Pelte N, Hoffmann JA, Reichhart JM. Activation of Drosophila Toll during fungal infection by a blood serine protease. Science. 2002;297:114. doi: 10.1126/science.1072391. [DOI] [PubMed] [Google Scholar]
- 21.Kambris Z, Brun S, Jang IH, et al. Drosophila immunity: a large-scale in vivo RNAi screen identifies five serine proteases required for Toll activation. Curr. Biol. 2006;16:808. doi: 10.1016/j.cub.2006.03.020. [DOI] [PubMed] [Google Scholar]
- 22.El Chamy L, Leclerc V, Caldelari I, Reichhart JM. Sensing of ‘danger signals’ and pathogen-associated molecular patterns defines binary signaling pathways ‘upstream’ of Toll. Nat. Immunol. 2008;9:1165. doi: 10.1038/ni.1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buchon N, Poidevin M, Kwon HM, et al. A single modular serine protease integrates signals from pattern-recognition receptors upstream of the Drosophila Toll pathway. Proc. Natl Acad. Sci. USA. 2009;106:12442. doi: 10.1073/pnas.0901924106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gottar M, Gobert V, Michel T, et al. The Drosophila immune response against Gram-negative bacteria is mediated by a peptidoglycan recognition protein. Nature. 2002;416:640. doi: 10.1038/nature734. [DOI] [PubMed] [Google Scholar]
- 25.Choe KM, Werner T, Stoven S, Hultmark D, Anderson KV. Requirement for a peptidoglycan recognition protein (PGRP) in Relish activation and antibacterial immune responses in Drosophila. Science. 2002;296:359. doi: 10.1126/science.1070216. [DOI] [PubMed] [Google Scholar]
- 26.Rämet M, Manfruelli P, Pearson A, Mathey-Prevot B, Ezekowitz RA. Functional genomic analysis of phagocytosis and identification of a Drosophila receptor for E. coli. Nature. 2002;416:644. doi: 10.1038/nature735. [DOI] [PubMed] [Google Scholar]
- 27.Takehana A, Katsuyama T, Yano T, Oshima Y, Takada H, Aigaki T, Kurata S. Overexpression of a pattern-recognition receptor, peptidoglycan-recognition protein-LE, activates imd/relish-mediated antibacterial defense and the prophenoloxidase cascade in Drosophila larvae. Proc. Natl Acad. Sci. USA. 2002;99:13705. doi: 10.1073/pnas.212301199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takehana A, Yano T, Mita S, Kotani A, Oshima Y, Kurata S. Peptidoglycan recognition protein (PGRP)-LE and PGRP-LC act synergistically in Drosophila immunity. EMBO J. 2004;23:4690. doi: 10.1038/sj.emboj.7600466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaneko T, Yano T, Aggarwal K, et al. PGRP-LC and PGRP-LE have essential yet distinct functions in the drosophila immune response to monomeric DAP-type peptidoglycan. Nat. Immunol. 2006;7:715. doi: 10.1038/ni1356. [DOI] [PubMed] [Google Scholar]
- 30.Kurata S. Recognition of infectious non-self and activation of immune responses by peptidoglycan recognition protein (PGRP)-family members in Drosophila. Dev. Comp. Immunol. 2004;28:89. doi: 10.1016/s0145-305x(03)00121-6. [DOI] [PubMed] [Google Scholar]
- 31.Schleifer KH, Kandler O. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol. Rev. 1972;36:407. doi: 10.1128/br.36.4.407-477.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leulier F, Parquet C, Pili-Floury S, et al. The Drosophila immune system detects bacteria through specific peptidoglycan recognition. Nat. Immunol. 2003;4:478. doi: 10.1038/ni922. [DOI] [PubMed] [Google Scholar]
- 33.Kaneko T, Goldman WE, Mellroth P, et al. Monomeric and polymeric gram-negative peptidoglycan but not purified LPS stimulate the Drosophila IMD pathway. Immunity. 2004;20:637. doi: 10.1016/s1074-7613(04)00104-9. [DOI] [PubMed] [Google Scholar]
- 34.Stenbak CR, Ryu JH, Leulier F, et al. Peptidoglycan molecular requirements allowing detection by the Drosophila immune deficiency pathway. J. Immunol. 2004;173:7339. doi: 10.4049/jimmunol.173.12.7339. [DOI] [PubMed] [Google Scholar]
- 35.Mellroth P, Karlsson J, Hakansson J, Schultz N, Goldman WE, Steiner H. Ligand-induced dimerization of Drosophila peptidoglycan recognition proteins in vitro. Proc. Natl Acad. Sci. USA. 2005;102:6455. doi: 10.1073/pnas.0407559102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lim J-H, Kim M-S, Kim H-E, et al. Structural basis for preferential recognition of diaminopimelic acid-type peptidoglycan by a subset of peptidoglycan recognition proteins. J. Biol. Chem. 2006;281:8286. doi: 10.1074/jbc.M513030200. [DOI] [PubMed] [Google Scholar]
- 37.Filipe SR, Tomasz A, Ligoxygakis P. Requirements of peptidoglycan structure that allow detection by the Drosophila Toll pathway. EMBO Rep. 2005;6:327. doi: 10.1038/sj.embor.7400371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mellroth P, Karlsson J, Steiner H. A scavenger function for a Drosophila peptidoglycan recognition protein. J. Biol. Chem. 2003;278:7059. doi: 10.1074/jbc.M208900200. [DOI] [PubMed] [Google Scholar]
- 39.Zaidman-Remy A, Hervé M, Poidevin M, et al. The Drosophila amidase PGRP-LB modulates the immune response to bacterial infection. Immunity. 2006;24:463. doi: 10.1016/j.immuni.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 40.Bischoff V, Vignal C, Duvic B, Boneca IG, Hoffmann JA, Royet J. Downregulation of the Drosophila immune response by peptidoglycan-recognition proteins SC1 and SC2. PloS Pathog. 2006;2:e14. doi: 10.1371/journal.ppat.0020014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Steiner H. Peptidoglycan recognition proteins: on and off switches for innate immunity. Immunol. Rev. 2004;198:83. doi: 10.1111/j.0105-2896.2004.0120.x. [DOI] [PubMed] [Google Scholar]
- 42.Cossart P, Sansonetti P. Bacterial invasion: the paradigms of enteroinvasive pathogens. Science. 2004;304:242. doi: 10.1126/science.1090124. [DOI] [PubMed] [Google Scholar]
- 43.Yano T, Mita S, Ohmori H, et al. Autophagic control of Listeria through intracellular innate immune recognition in Drosophila. Nat. Immunol. 2008;9:908. doi: 10.1038/ni.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mizushima N, Levine B, Cuervo AM, Klionsky D. Autophagy fights disease though cellular self-digestion. Nature. 2008;451:1069. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Levine B, Deretic V. Unveiling the roles of autophagy in innate and adaptive immunity. Nat. Rev. Immunol. 2007;7:767. doi: 10.1038/nri2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nakagawa I, Amano A, Mizushima N, et al. Autophagy defends cells against invading Group A Streptococcus. Science. 2004;306:1037. doi: 10.1126/science.1103966. [DOI] [PubMed] [Google Scholar]
- 47.Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119:753. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 48.Ogawa M, Yoshimori T, Suzuki T, Sagara H, Mizushima N, Sasakawa C. Escape of intracellular Shigella from autophagy. Science. 2005;307:727. doi: 10.1126/science.1106036. [DOI] [PubMed] [Google Scholar]
- 49.Ling YM, Shaw MH, Ayala C, et al. Vacuolar and plasma membrane stripping and autophagic elimination of Toxoplasma gondii in primed effector macrophages. J. Exp. Med. 2006;203:2063. doi: 10.1084/jem.20061318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Andrade RM, Weaaendarp M, Gubbels M-J, Striepen B, Subauste CS. CD40 induces macrophage anti-Toxoplasma gondii activity by triggering autophagy-dependent fusion of pathogen-containing vacuoles and lysosomes. J. Clin. Invest. 2006;116:2366. doi: 10.1172/JCI28796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Singh SB, Davis AS, Taylor GA, Deretic V. Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science. 2006;313:1438. doi: 10.1126/science.1129577. [DOI] [PubMed] [Google Scholar]
- 52.Meylan E, Tschopp J, Karin M. Intracellular pattern recognition receptors in the host response. Nature. 2006;442:39. doi: 10.1038/nature04946. [DOI] [PubMed] [Google Scholar]