Summary
Programmed cell death (or apoptosis) is an evolutionarily conserved, genetically controlled suicide mechanism for cells that, when deregulated, can lead to developmental defects, cancers, and degenerative diseases [1, 2]. In C. elegans, DNA damage induces germ cell death by signaling through cep-1/p53, ultimately leading to the activation of CED-3/caspase [3–13]. It has been hypothesized that the major regulatory events controlling cell death occur by cell-autonomous mechanisms, that is, within the dying cell. In support of this, genetic studies in C. elegans have shown that the core apoptosis pathway genes ced-4/APAF-1 and ced-3/caspase are required in cells fated to die [9]. However, it is not known whether the upstream signals that activate apoptosis function in a cell-autonomous manner. Here we show that kri-1, an ortholog of KRIT1/CCM1, which is mutated in the human neurovascular disease cerebral cavernous malformation [14, 15], is required to activate DNA damage-dependent cell death independently of cep-1/p53. Interestingly, we find that kri-1 regulates cell death in a cell-nonautonomous manner, revealing a novel regulatory role for nondying cells in eliciting cell death in response to DNA damage.
Keywords: DEVBIO, CELLCYCLE
Highlights
► kri-1 is a novel positive regulator of C. elegans germ cell apoptosis ► kri-1 regulates cell death independently of cep-1/p53 ► kri-1 regulates cell death cell nonautonomously, possibly by cross-tissue signaling
Results and Discussion
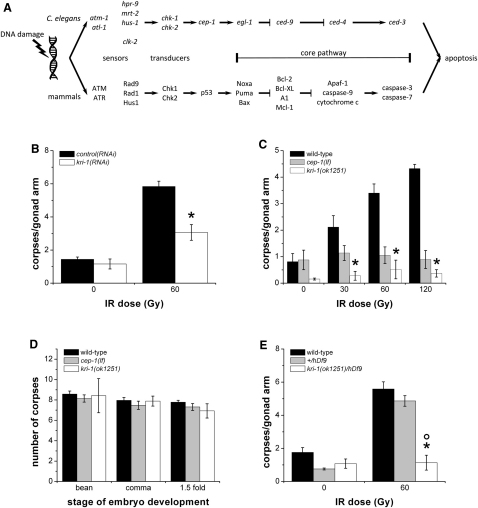
In an RNA interference (RNAi) screen unrelated to apoptosis, we serendipitously uncovered a cep-1/p53-interacting gene, kri-1, the ortholog of human KRIT1/CCM1, which is frequently mutated in the neurovascular disease cerebral cavernous malformation [14, 15]. Because this gene had been previously shown to integrate signals from reproductive tissues (germ cells) to elicit longevity effects in nonreproductive (somatic) tissues [16] and interacts with cep-1, an important mediator of germ cell death (Figure 1A) [7, 8], we asked whether kri-1 is involved in a novel, cell-nonautonomous mechanism to regulate germ cell death. To test this, we first investigated whether kri-1 regulates cell death like cep-1, by quantifying the number of germ cell corpses in wild-type animals fed bacteria producing double-stranded RNA against a control gene or kri-1 exposed to ionizing radiation (IR) (Figure 1B). We found that knockdown of kri-1 by RNAi significantly reduced the number of germ cell corpses after DNA damage (IR) compared to animals fed control RNAi (p = 0.01), suggesting that kri-1 is required for germ cell death. We verified this initial observation by performing a dose-response analysis of the kri-1(ok1251) deletion mutant. In contrast to wild-type animals, kri-1(ok1251) deletion mutants did not exhibit an increase in germ cell apoptosis after exposure to increasing doses of IR (Figure 1C; see also Figure S1 available online). This was reminiscent of cep-1 loss-of-function (lf) mutants that are also resistant to IR-induced apoptosis. Therefore, we examined whether kri-1 regulates germ cell death specifically, like cep-1, or whether it regulates cell death in all cells, like ced-3, by quantifying apoptosis in developing embryos of wild-type animals and cep-1(lf) and kri-1(ok1251) mutants. We found that developmental cell death was unaffected in kri-1(ok1251) mutants, suggesting that the regulation of cell death by kri-1 is specific to germ cells, like cep-1 (Figure 1D). Finally, to determine whether the ok1251 allele is a null, we performed a deficiency analysis by crossing ok1251 into a strain containing the hDf9 deficiency that removes the kri-1 locus and quantified the number of germ cell corpses after DNA damage (Figure 1E). Strains containing the ok1251 allele in trans to hDf9 were as resistant to damage-induced germ cell apoptosis as ok1251 homozygotes, suggesting that ok1251 is a null allele. Collectively, these and further observations (see below) indicate that kri-1 is specifically required for germ cell death in response to DNA damage.
Figure 1.
kri-1 Is Required for DNA Damage-Induced Germ Cell Death Specifically
(A) The conserved genetic pathway regulating cell death in mammals and C. elegans. Sensors and transducers relay DNA damage signals to CEP-1/p53, which transcriptionally activates one or more BH3-only genes to promote apoptosis.
(B) Wild-type animals fed control(RNAi) (black) or kri-1(RNAi) (white) were subjected to ionizing radiation (IR) at 20°C, and germ cell apoptosis was quantified 24 hr later. Data represent mean ± standard error of the mean (SEM) of three independent experiments and at least 55 germlines in total per strain per condition. ∗p < 0.05 versus wild-type. See Table S1 for full list of p values.
(C) Synchronized wild-type (black), cep-1(lf) (gray), and kri-1(ok1251) (white) young adult animals were treated with increasing doses of IR, and germ cell apoptosis was scored as above. Data represent mean ± SEM of at least three independent experiments and at least 50 germlines in total per strain per condition. ∗p < 0.05 versus wild-type.
(D) Embryos at the indicated developmental stages were scored for apoptosis with Nomarski optics in wild-type (black), cep-1(lf) (gray bars), and kri-1(ok1251) (white) animals. Data represent mean ± SEM of three independent experiments and at least 45 embryos in total per strain per stage.
(E) Apoptosis was scored in wild-type animals (black), a strain with a wild-type copy of kri-1 in trans to the hDf9 deficiency (gray), and ok1251 in trans to hDf9 (white) treated with IR as above. Data represent mean ± SEM of at least four independent experiments and at least 25 germlines in total per strain per condition. ∗p < 0.05 versus wild-type; °p < 0.01 between kri-1(ok1251)/hDf9 and +/hDf9. See also Figure S1.
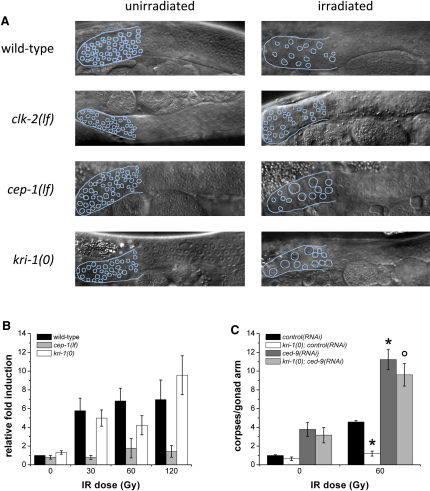
Given that kri-1 is required to promote germ cell death in response to DNA damage, we were interested to know at which step in the pathway it might be functioning (Figure 1A). In the C. elegans germline, the DNA damage checkpoint genes (hpr-9, mrt-2, hus-1, and clk-2) are required to both transiently arrest mitotic proliferation and activate cep-1-dependent apoptosis of damaged germ cells [5, 6]. To ascertain whether kri-1 is functioning in an analogous manner (i.e., upstream of cep-1), we tested whether kri-1 null (0) mutants mimic the germline phenotypes of checkpoint gene mutants. In contrast to clk-2 mutants that are defective in cell-cycle arrest, we found that kri-1 was not required for IR-induced arrest of mitotically proliferating cells (Figure 2A; Figure S2A), implying that kri-1 acts downstream or independently of the DNA damage checkpoint. To delineate whether kri-1 is required to transduce signals to the CEP-1 protein and therefore allow apoptosis to occur, we examined the activity of CEP-1 by quantifying the transcript levels egl-1, a proapoptotic target gene of CEP-1 [17, 18]. Consistent with previous work, egl-1 transcript levels as assessed by real-time quantitative PCR (qPCR) increased in response to DNA damage in wild-type animals, but not in cep-1(lf) mutants (Figure 2B). Interestingly, egl-1 induction in kri-1(0) mutants was similar to that seen in wild-type animals, indicating that the transcriptional activity of CEP-1 is induced normally in the absence of kri-1. This is consistent with kri-1 promoting damage-induced apoptosis downstream or independently of cep-1. Such a model raised the possibility that cep-1 might regulate kri-1 transcription or KRI-1 protein localization in response to DNA damage and that this was required to promote germ cell death. However, neither kri-1 transcript levels nor GFP::KRI-1 localization was significantly affected by IR or cep-1 status (Figures S2B–S2D).
Figure 2.
kri-1 Functions Downstream of the Checkpoint Genes but Upstream of ced-9
(A) Synchronized hermaphrodites at the fourth larval stage (L4) were treated with IR, and the number of nuclei per unit area in the mitotic region of the germline was quantified 24 hr later at 20°C. The mitotic region and nuclei have been outlined for clarity. Representative images from three independent experiments are shown.
(B) RNA was isolated by TRIzol from synchronized wild-type (black), cep-1(lf) (gray), and kri-1(0) (white) mutants, and egl-1 transcript levels were measured by quantitative real-time PCR. Data represent mean ± SEM of three independent experiments.
(C) Synchronized wild-type and kri-1(0) L4 animals fed control(RNAi) (Y95B8A_84.g, a nonexpressed gene) (black and white, respectively) or ced-9(RNAi) (dark gray and light gray, respectively) were subjected to IR, and germ cell death was quantified as described above. Data represent mean ± SEM of three independent experiments and at least 25 germlines in total per strain per condition. ∗p < 0.01 versus wild-type; °p < 0.05 versus kri-1(0); control(RNAi). See also Figure S2.
The data above suggest a model wherein kri-1 functions downstream of or in parallel to the key decision-making step in the cell death pathway and likely regulates components of the core death pathway (i.e., egl-1, ced-9, ced-4, and ced-3). To investigate this further, we examined the epistatic relationship between kri-1 and ced-9. Healthy cells require functional CED-9/BCL2 to prevent ectopic activation of CED-3/caspase by CED-4 (Figure 1A). We reasoned that if kri-1 functions downstream of ced-9, ablation of kri-1 would suppress the increased cell death caused by ced-9(lf); on the other hand, the converse would be true if kri-1 acts upstream of ced-9. Knockdown of ced-9 by RNAi (>50% knockdown; Figure S2E) caused a significant increase in apoptosis both before and after DNA damage, but this was unaffected by loss of kri-1 (Figure 2C), which we confirmed in kri-1(0); ced-9(lf) double mutants (data not shown). This indicates that kri-1 is not functioning strictly downstream of ced-9 (i.e., in a manner similar to ced-4 or ced-3). To be sure, we quantified the mRNA of both ced-4 and ced-3 by qPCR and found that their levels were not affected in kri-1(0) mutants in response to IR (Figures S2F–S2H); in addition, CED-4 protein expression and localization was not affected in kri-1(0) mutants (data not shown). Therefore, we infer from these results that kri-1 acts upstream of, or parallel to, ced-9.
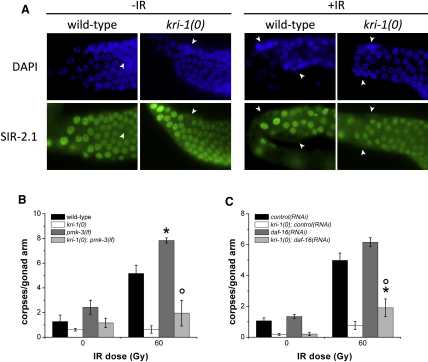
Because kri-1 functions independently of cep-1 and impinges on the core death pathway, we were interested to know whether kri-1 is cooperating with other genes known to regulate germ cell death independently or downstream of cep-1. In particular, the histone deacetylase sir-2.1 [19], the MAP kinase pmk-3 [20], and the retinoblastoma (RB) ortholog lin-35 [21] have all been shown to regulate germ cell death independently of cep-1. In addition to activating cell death independently of cep-1 in a manner similar to kri-1, the SIR-2.1 protein exits the nuclei of germ cells after DNA damage [19]. To determine whether the relocalization of SIR-2.1 is required for kri-1-mediated germ cell death, we immunostained kri-1(0) animals with SIR-2.1 antibodies to ascertain whether SIR-2.1 protein levels or localization was altered. Although we found that kri-1 did not affect the SIR-2.1 protein staining pattern (Figure 3A), it still remained possible that kri-1 and sir-2.1 function in the same pathway. To address this, we created a double heterozygous mutant containing both the kri-1(0) and sir-2.1(lf) mutations (kri-1(0)/+; sir-2.1(lf)/+), and we observed wild-type levels of germ cell apoptosis in response to DNA damage (data not shown), suggesting that these genes operate in different pathways. In contrast to sir-2.1 and kri-1, which positively regulate germline apoptosis, the MAP kinase gene pmk-3 inhibits germline apoptosis independently of cep-1 [20]. Therefore, we tested whether kri-1 was required for germ cell death caused by loss of function of pmk-3. We created kri-1(0); pmk-3(lf) double mutants and found that germ cell death was suppressed to the same degree as kri-1(0) single mutants (Figure 3B), suggesting that kri-1 is epistatic to pmk-3 and does not regulate cell death through pmk-3. We do not believe that pmk-3 regulates kri-1 because kri-1 transcript levels and protein localization remained unchanged in pmk-3(lf) mutants (Figure S3). Finally, because lin-35 positively regulates germ cell apoptosis by controlling the levels of the CED-9 protein (i.e., loss of lin-35 lead to an increase in CED-9 protein levels) [21], we tested whether kri-1 functions through lin-35/RB by quantifying CED-9 protein levels in kri-1(0) animals by western blot. We found that CED-9 protein levels were unaffected in kri-1(0) (A. Ross, personal communication; data not shown), suggesting that kri-1 does not regulate germline apoptosis through this pathway.
Figure 3.
kri-1 Functions Independently of Known cep-1-Independent Pathways
(A) Wild-type and kri-1(0) animals were immunostained with DAPI or SIR-2.1 antibodies before and after IR. The images show the pachytene region of the germline. Arrowheads indicate nuclei positive for DAPI staining but negative for SIR-2.1 protein expression. Representative images of at least three independent experiments are shown.
(B) Wild-type (black), kri-1(0) (white), pmk-3(lf) (dark gray), and kri-1(0); pmk-3(lf) (light gray) animals were synchronized and scored for apoptosis as described in Figure 1B. Data represent mean ± SEM of three independent experiments and at least 50 germlines in total per strain per condition. ∗p < 0.05 versus wild-type; °p < 0.05 versus pmk-3(lf).
(C) Germline apoptosis was quantified in synchronized wild-type and kri-1(0) mutants fed control(RNAi) (black and white, respectively) or daf-16(RNAi) (dark and light gray, respectively) and treated with IR as described above. Data represent mean ± SEM of four independent experiments and at least 25 germlines in total per strain per condition. ∗p < 0.01 versus wild-type; °p < 0.01 versus daf-16(RNAi). See also Figure S3.
Additionally, it has been shown that kri-1 influences the localization of the forkhead transcription factor DAF-16 in the intestine by responding to signals from the germline and regulating worm life span [16] and that DAF-16 may negatively regulate IR-induced germ cell apoptosis [17]. These two pieces of evidence suggested that kri-1 might function through daf-16 to regulate germ cell death. Animals fed daf-16(RNAi) exhibited wild-type levels of germ cell death in response to IR (Figure 3C), consistent with published results reporting that DAF-16 has a weak effect on germ cell death [17, 19]. We tested whether kri-1(0) could suppress apoptosis in animals fed daf-16(RNAi) and found that it did (Figure 3C), which we confirmed by creating kri-1(0) daf-16(lf) double mutants (data not shown). This suggests that kri-1 does not require daf-16 to mediate its apoptotic function. Alternatively, it was possible that daf-16 regulates kri-1 to mediate germ cell death; however, neither kri-1 transcript nor protein levels were significantly affected in daf-16(lf) mutants (Figure S3). These observations reveal that kri-1 is involved in a novel pathway that regulates germ cell death in response to DNA damage.
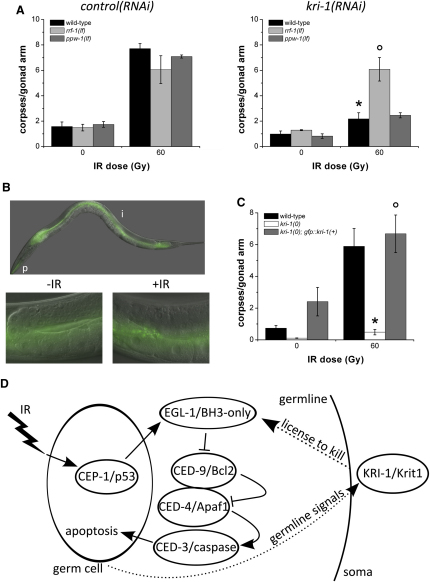
There are two possible mechanisms by which kri-1 may promote germ cell death. The first is a cell-autonomous mechanism, in which kri-1 regulates the core death pathway (EGL-1 or CED-9) in germ cells to initiate cell death. Alternatively, it is possible that kri-1 regulates cell death outside of germ cells (i.e., from somatic cells) via a novel pathway. In support of the latter hypothesis, kri-1 is required to extend the life span of worms through its effects on DAF-16 in the intestine, possibly by receiving signals from germ cells [16]; in addition, microarray data suggest that kri-1 is not expressed in the germline [22]. To distinguish between these possibilities, we took advantage of tissue-specific RNAi in C. elegans and selectively knocked down kri-1 in germ cells and the soma in rrf-1(lf) [23] and ppw-1(lf) [24] mutants, respectively, and quantified IR-induced germ cell apoptosis [21] (Figure 4A). Wild-type, rrf-1(lf), and ppw-1(lf) mutants fed bacteria producing control(RNAi) had similar numbers of germ cell corpses after DNA damage. Ablation of kri-1 by RNAi in wild-type animals inhibited DNA damage-induced germ cell apoptosis to the same extent as kri-1(0) mutants. However, selective knockdown of kri-1 in germ cells in rrf-1(lf) mutants caused an increase in IR-induced apoptosis, suggesting that kri-1 expression in germ cells is not required to promote apoptosis. Conversely, specific knockdown of kri-1 in the soma in ppw-1(lf) mutants prevented germ cell death, suggesting that kri-1 is required in somatic tissue to regulate germ cell death. In support of this contention, we were able to rescue damage-induced germ cell apoptosis to wild-type levels by expressing GFP::KRI-1 from a somatic extrachromosomal array (Figures 4B and 4C). Although it is possible that low-level expression of GFP::KRI-1 in the germline may account for this observation, the fact that extrachromosomal arrays are generally silenced in the C. elegans germline [25] strongly supports a model in which kri-1 is required in nondying somatic cells to promote germ cell death.
Figure 4.
kri-1 Regulates Germ Cell Death from Somatic Tissues by a Cell-Nonautonomous Mechanism
(A) Germ cell death was quantified after treatment with IR in wild-type (black), rrf-1(lf) (light gray), and ppw-1(lf) (dark gray) L4 worms fed bacteria producing double-stranded RNA against a control gene (left panel) or kri-1 (right panel). Data represent mean ± SEM of three independent experiments and at least 35 germlines in total per strain per condition. ∗p < 0.01 versus control(RNAi); °p < 0.01 versus kri-1(RNAi).
(B) GFP::KRI-1 expressed under the control of the endogenous kri-1 promoter is detectable in the pharynx (p) and intestine (i) of transgenic animals (top panel). GFP::KRI-1 is excluded from the germline in unirradiated animals (bottom left panel) and does not change localization after irradiation (bottom right panel). Representative images of at least three independent experiments are shown.
(C) Apoptotic germ cells were quantified in wild-type (black), kri-1(0) (white), and a kri-1(0) strain expressing a wild-type copy of GFP::KRI-1 in the soma (dark gray). Data represent mean ± SEM of three independent experiments and at least 40 germlines in total per strain per condition. ∗p < 0.01 versus wild-type; °p < 0.01 versus kri-1(0).
(D) Model depicting somatic requirement for kri-1 in promoting germ cell death in response to DNA damage. We hypothesize that there are “license to kill” factors secreted from the soma into the germline to mediate cell death. Solid lines represent known regulatory interactions; dotted lines represent hypothetical interactions.
Collectively, these data imply a novel mechanism whereby somatic cells communicate with germ cells to promote their death in response to DNA damage (Figure 4D). Indeed, other genes have been shown to participate in germline-soma signaling during proliferation and differentiation of the germline [26, 27], dauer formation, and life-span control [28, 29], confirming that these two tissues can signal to each other in response to certain stimuli. The fact that kri-1/KRIT1 regulates germ cell death from cells outside of the germline independently of cep-1/p53 implies that cells not fated to die (somatic cells) in C. elegans can regulate the core death pathway in germ cells by novel, cell-nonautonomous mechanisms. In the case of kri-1, there are several ways in which it may be performing this function. First, we examined the possibility that kri-1 may be acting through daf-9 and daf-12 to promote apoptosis, in a manner analogous to its proposed role in life-span control [16], by feeding kri-1(RNAi) to daf-9(lf) and daf-12(lf) mutants. We found that both mutants exhibited a resistance to apoptosis when fed kri-1(RNAi), suggesting that kri-1 does not act through these two genes (Figure S4). Second, although we have shown that kri-1 does not affect the transcript levels of the BH3-only gene egl-1, it is possible that the kri-1 is required to activate the EGL-1 protein. Similar to mammalian BH3-only proteins, EGL-1 may require other coactivating proteins or modifications in order to induce cell death in germ cells. For example, BID, BAD, and BIM/BMF are proapoptotic BH3-only proteins regulated at the posttranslational level through cleavage, phosphorylation, and sequestration by interacting proteins, respectively [30]. Therefore, it is formally possible that KRI-1 may facilitate the activation of EGL-1 by similar transcription-independent mechanisms. Alternatively, KRI-1 may be involved in receiving signals from the germ cells, which results in the subsequent release of death-inducing factors. Although further studies are required to resolve the biochemical mechanism by which KRI-1 dictates germ cell death from the soma, our observations reveal a novel cross-tissue signaling mechanism whereby somatic tissue can promote germ cell death in response to DNA damage in C. elegans, which may have broader applicability to cell death in general.
Acknowledgments
We thank the Caenorhabditis Genetics Center, which is supported by the National Institutes of Health National Center for Research Resources, for worm strains. We are grateful to C. Kenyon (University of California, San Francisco) for providing the muEx353 strain and B. Conradt (Dartmouth University) for CED-9 antibody. This work was supported by a Canadian Institutes of Health Research award to W.B.D.; a Research Training Competition (Restracomp) award from the SickKids Foundation to S.I.; and Wellcome Trust, Cancer Research UK, and American Institute for Cancer Research awards to A.G. We thank M. Irwin, M. Zhen, A. Perrin, A. Ross, I. Watson, and J. Lau for critical input on the manuscript and fruitful discussions.
Published online: February 4, 2010
Footnotes
Supplemental Information includes four figures, Supplemental Experimental Procedures, and one table and can be found with this article online at doi:10.1016/j.cub.2009.12.032.
Supplemental Information
References
- 1.Ellis H.M., Horvitz H.R. Genetic control of programmed cell death in the nematode C. elegans. Cell. 1986;44:817–829. doi: 10.1016/0092-8674(86)90004-8. [DOI] [PubMed] [Google Scholar]
- 2.Hipfner D.R., Cohen S.M. Connecting proliferation and apoptosis in development and disease. Nat. Rev. Mol. Cell Biol. 2004;5:805–815. doi: 10.1038/nrm1491. [DOI] [PubMed] [Google Scholar]
- 3.Ahmed S., Hodgkin J. MRT-2 checkpoint protein is required for germline immortality and telomere replication in C. elegans. Nature. 2000;403:159–164. doi: 10.1038/35003120. [DOI] [PubMed] [Google Scholar]
- 4.Gartner A., Milstein S., Ahmed S., Hodgkin J., Hengartner M.O. A conserved checkpoint pathway mediates DNA damage-induced apoptosis and cell cycle arrest in C. elegans. Mol. Cell. 2000;5:435–443. doi: 10.1016/s1097-2765(00)80438-4. [DOI] [PubMed] [Google Scholar]
- 5.Hofmann E.R., Milstein S., Boulton S.J., Ye M., Hofmann J.J., Stergiou L., Gartner A., Vidal M., Hengartner M.O. Caenorhabditis elegans HUS-1 is a DNA damage checkpoint protein required for genome stability and EGL-1-mediated apoptosis. Curr. Biol. 2002;12:1908–1918. doi: 10.1016/s0960-9822(02)01262-9. [DOI] [PubMed] [Google Scholar]
- 6.Ahmed S., Alpi A., Hengartner M.O., Gartner A. C. elegans RAD-5/CLK-2 defines a new DNA damage checkpoint protein. Curr. Biol. 2001;11:1934–1944. doi: 10.1016/s0960-9822(01)00604-2. [DOI] [PubMed] [Google Scholar]
- 7.Derry W.B., Putzke A.P., Rothman J.H. Caenorhabditis elegans p53: Role in apoptosis, meiosis, and stress resistance. Science. 2001;294:591–595. doi: 10.1126/science.1065486. [DOI] [PubMed] [Google Scholar]
- 8.Schumacher B., Hofmann K., Boulton S., Gartner A. The C. elegans homolog of the p53 tumor suppressor is required for DNA damage-induced apoptosis. Curr. Biol. 2001;11:1722–1727. doi: 10.1016/s0960-9822(01)00534-6. [DOI] [PubMed] [Google Scholar]
- 9.Yuan J.Y., Horvitz H.R. The Caenorhabditis elegans genes ced-3 and ced-4 act cell autonomously to cause programmed cell death. Dev. Biol. 1990;138:33–41. doi: 10.1016/0012-1606(90)90174-h. [DOI] [PubMed] [Google Scholar]
- 10.Conradt B., Horvitz H.R. The C. elegans protein EGL-1 is required for programmed cell death and interacts with the Bcl-2-like protein CED-9. Cell. 1998;93:519–529. doi: 10.1016/s0092-8674(00)81182-4. [DOI] [PubMed] [Google Scholar]
- 11.del Peso L., González V.M., Núñez G. Caenorhabditis elegans EGL-1 disrupts the interaction of CED-9 with CED-4 and promotes CED-3 activation. J. Biol. Chem. 1998;273:33495–33500. doi: 10.1074/jbc.273.50.33495. [DOI] [PubMed] [Google Scholar]
- 12.Chen F., Hersh B.M., Conradt B., Zhou Z., Riemer D., Gruenbaum Y., Horvitz H.R. Translocation of C. elegans CED-4 to nuclear membranes during programmed cell death. Science. 2000;287:1485–1489. doi: 10.1126/science.287.5457.1485. [DOI] [PubMed] [Google Scholar]
- 13.Schumacher B., Hanazawa M., Lee M.H., Nayak S., Volkmann K., Hofmann E.R., Hengartner M., Schedl T., Gartner A. Translational repression of C. elegans p53 by GLD-1 regulates DNA damage-induced apoptosis. Cell. 2005;120:357–368. doi: 10.1016/j.cell.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 14.Sahoo T., Johnson E.W., Thomas J.W., Kuehl P.M., Jones T.L., Dokken C.G., Touchman J.W., Gallione C.J., Lee-Lin S.Q., Kosofsky B. Mutations in the gene encoding KRIT1, a Krev-1/rap1a binding protein, cause cerebral cavernous malformations (CCM1) Hum. Mol. Genet. 1999;8:2325–2333. doi: 10.1093/hmg/8.12.2325. [DOI] [PubMed] [Google Scholar]
- 15.Laberge-le Couteulx S., Jung H.H., Labauge P., Houtteville J.P., Lescoat C., Cecillon M., Marechal E., Joutel A., Bach J.F., Tournier-Lasserve E. Truncating mutations in CCM1, encoding KRIT1, cause hereditary cavernous angiomas. Nat. Genet. 1999;23:189–193. doi: 10.1038/13815. [DOI] [PubMed] [Google Scholar]
- 16.Berman J.R., Kenyon C. Germ-cell loss extends C. elegans life span through regulation of DAF-16 by kri-1 and lipophilic-hormone signaling. Cell. 2006;124:1055–1068. doi: 10.1016/j.cell.2006.01.039. [DOI] [PubMed] [Google Scholar]
- 17.Quevedo C., Kaplan D.R., Derry W.B. AKT-1 regulates DNA-damage-induced germline apoptosis in C. elegans. Curr. Biol. 2007;17:286–292. doi: 10.1016/j.cub.2006.12.038. [DOI] [PubMed] [Google Scholar]
- 18.Schumacher B., Schertel C., Wittenburg N., Tuck S., Mitani S., Gartner A., Conradt B., Shaham S. C. elegans ced-13 can promote apoptosis and is induced in response to DNA damage. Cell Death Differ. 2005;12:153–161. doi: 10.1038/sj.cdd.4401539. [DOI] [PubMed] [Google Scholar]
- 19.Greiss S., Hall J., Ahmed S., Gartner A. C. elegans SIR-2.1 translocation is linked to a proapoptotic pathway parallel to cep-1/p53 during DNA damage-induced apoptosis. Genes Dev. 2008;22:2831–2842. doi: 10.1101/gad.482608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lettre G., Kritikou E.A., Jaeggi M., Calixto A., Fraser A.G., Kamath R.S., Ahringer J., Hengartner M.O. Genome-wide RNAi identifies p53-dependent and -independent regulators of germ cell apoptosis in C. elegans. Cell Death Differ. 2004;11:1198–1203. doi: 10.1038/sj.cdd.4401488. [DOI] [PubMed] [Google Scholar]
- 21.Schertel C., Conradt B. C. elegans orthologs of components of the RB tumor suppressor complex have distinct pro-apoptotic functions. Development. 2007;134:3691–3701. doi: 10.1242/dev.004606. [DOI] [PubMed] [Google Scholar]
- 22.Reinke V., Smith H.E., Nance J., Wang J., Van Doren C., Begley R., Jones S.J., Davis E.B., Scherer S., Ward S., Kim S.K. A global profile of germline gene expression in C. elegans. Mol. Cell. 2000;6:605–616. doi: 10.1016/s1097-2765(00)00059-9. [DOI] [PubMed] [Google Scholar]
- 23.Sijen T., Fleenor J., Simmer F., Thijssen K.L., Parrish S., Timmons L., Plasterk R.H., Fire A. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell. 2001;107:465–476. doi: 10.1016/s0092-8674(01)00576-1. [DOI] [PubMed] [Google Scholar]
- 24.Tijsterman M., Okihara K.L., Thijssen K., Plasterk R.H.A. PPW-1, a PAZ/PIWI protein required for efficient germline RNAi, is defective in a natural isolate of C. elegans. Curr. Biol. 2002;12:1535–1540. doi: 10.1016/s0960-9822(02)01110-7. [DOI] [PubMed] [Google Scholar]
- 25.Kelly W.G., Xu S., Montgomery M.K., Fire A. Distinct requirements for somatic and germline expression of a generally expressed Caernorhabditis elegans gene. Genetics. 1997;146:227–238. doi: 10.1093/genetics/146.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Killian D.J., Hubbard E.J.A. C. elegans pro-1 activity is required for soma/germline interactions that influence proliferation and differentiation in the germ line. Development. 2004;131:1267–1278. doi: 10.1242/dev.01002. [DOI] [PubMed] [Google Scholar]
- 27.Kimble J.E., White J.G. On the control of germ cell development in Caenorhabditis elegans. Dev. Biol. 1981;81:208–219. doi: 10.1016/0012-1606(81)90284-0. [DOI] [PubMed] [Google Scholar]
- 28.Apfeld J., Kenyon C. Cell nonautonomy of C. elegans daf-2 function in the regulation of diapause and life span. Cell. 1998;95:199–210. doi: 10.1016/s0092-8674(00)81751-1. [DOI] [PubMed] [Google Scholar]
- 29.Wolkow C.A., Kimura K.D., Lee M.S., Ruvkun G. Regulation of C. elegans life-span by insulinlike signaling in the nervous system. Science. 2000;290:147–150. doi: 10.1126/science.290.5489.147. [DOI] [PubMed] [Google Scholar]
- 30.Puthalakath H., Strasser A. Keeping killers on a tight leash: Transcriptional and post-translational control of the pro-apoptotic activity of BH3-only proteins. Cell Death Differ. 2002;9:505–512. doi: 10.1038/sj.cdd.4400998. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.