Abstract
FANCM binds and remodels replication fork structures in vitro. We report that in vivo, FANCM controls DNA chain elongation in an ATPase-dependent manner. In the presence of replication inhibitors that do not damage DNA, FANCM counteracts fork movement, possibly by remodelling fork structures. Conversely, through damaged DNA, FANCM promotes replication and recovers stalled forks. Hence, the impact of FANCM on fork progression depends on the underlying hindrance. We further report that signalling through the checkpoint effector kinase Chk1 prevents FANCM from degradation by the proteasome after exposure to DNA damage. FANCM also acts in a feedback loop to stabilize Chk1. We propose that FANCM is a ringmaster in the response to replication stress by physically altering replication fork structures and by providing a tight link to S-phase checkpoint signalling.
Keywords: DNA replication, FANCM, Fanconi anemia, fork regression
Introduction
Replication forks frequently stall at genomic sequences that are difficult to replicate as well as at endogenous DNA lesions. Persistent blocks to DNA replication can provoke the loss of replisome components, thereby rendering stalled forks vulnerable to nucleolytic degradation (Cobb et al, 2003; Tourriere and Pasero, 2007). Aberrant cleaving of stalled forks yields double-strand breaks, chromosome instability and ultimately cell death. DNA interstrand crosslinking agents, which are widely used in cancer chemotherapy, cause one of the most toxic lesions for DNA replication. In higher eukaryotes, Fanconi anemia (FA) proteins are implicated in tolerating interstrand DNA crosslinks.
FA is a genetic disease associated with bone marrow failure, developmental anomalies and early onset cancer (Tischkowitz and Hodgson, 2003). Cellular hallmarks of FA include radial chromosomes and hypersensitivity to DNA interstrand crosslinks (Schroeder et al, 1964; Auerbach and Wolman, 1976). Thirteen FA complementation groups have been assigned and the corresponding genes cloned. The FA group M protein contains an amino-terminal superfamily 2 helicase motif, and a carboxyl-terminal ERCC4-like domain (Meetei et al, 2005). In response to DNA replication stress, FANCM promotes the loading of the FA core complex onto chromatin (Kim et al, 2008). Subsequently, the FA core complex monoubiquitinates FANCD2 and FANCI (Garcia-Higuera et al, 2001; Smogorzewska et al, 2007). Monoubiquitination of FANCD2 and FANCI presumably targets FANCD2 and FANCI to DNA repair sites (Garcia-Higuera et al, 2001; Smogorzewska et al, 2007). Additionally, and independently of the FA ubiquitin ligase complex, FANCM associates with the checkpoint protein HCLK2 and facilitates the ATR/Chk1-mediated checkpoint response at stalled replication forks (Collis et al, 2008). The integrity of the ATPase domain of FANCM is necessary for its function in ATR/Chk1 signalling and in tolerance to the DNA crosslinker, mitomycin C, but dispensable for the monoubiquitination of FANCD2 and FANCI (Collis et al, 2008; Xue et al, 2008). This underlines the observation that FANCM has at least two independent functions in a cell and that these are either ATP dependent or independent.
Biochemical studies have shown that FANCM, and its yeast ortholog Fml1, can bind and translocate the branch point of model replication forks and Holliday junctions (Sun et al, 2008; Xue et al, 2008; Gari et al, 2008b). Recombinant FANCM and Fml1 can promote the reversal of replication fork structures in vitro (Sun et al, 2008; Gari et al, 2008a). On the basis of these biochemical findings, it has been proposed that FANCM may sense and remodel stalled replication forks (Collis et al, 2008; Sun et al, 2008; Gari et al, 2008a). There is, however, no direct evidence that FANCM facilitates the progression of replication forks in vivo.
Fork regression has been implicated in protecting and resuming stalled forks (Heller and Marians, 2006). In vitro, spontaneous fork regression relieves topological constraints, that is positive supercoiling ahead of the fork (Postow et al, 2001b). In bacteria, fork regression was observed after UV damage and is thought to facilitate the access of DNA repair enzymes to the blocking lesion (Courcelle et al, 2003). Recently, it has been shown that in bacteriophage T4, fork regression driven by the T4 helicase is the primary mechanism to reactivate stalled replication forks in vivo (Long and Kreuzer, 2009). Regressed forks accumulate in checkpoint-deficient rad53 yeast cells treated with hydroxyurea (HU), but form very rarely in UV-treated rad14rad53 cells, or in wild-type cells (Sogo et al, 2002; Lopes et al, 2006). Although we cannot rule out the possibility that fork regression may be too transient to be detected in wild-type cells, it has been proposed that regressed forks represent pathological structures in eukaryotes (Sogo et al, 2002; Lopes et al, 2006).
This work monitors the progression of replication forks by visualizing replicons at the single molecule level. We find that FANCM ensures steady progression of replication forks in vivo. FANCM promotes replication through damaged DNA and the integrity of its ATPase domain is necessary for the reactivation of stalled forks after DNA damage. This implies that remodelling of stalled replication forks by FANCM might be a mechanism to resume DNA synthesis in human cells. We also investigated FANCM's function in replication in the presence of replication inhibitors that do not physically block replication. During HU and aphidicolin treatment, FANCM seems to counteract fork movement. Furthermore, we provide evidence that FANCM and Chk1 are reciprocally required to stabilize each other. We propose that FANCM restores coupling of the leading and lagging strand synthesis after replication stress. Therefore, FANCM is essential for proper replication dynamics, whereby its effect on replication depends on the nature of the lesion encountered.
Results
FANCM is essential for constant progression of replication forks
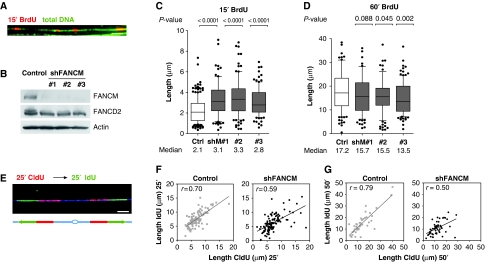
To follow dynamics of replication forks in vivo, we pulse labelled DNA of human cells with nucleotide analogs (BrdU, CldU or IdU) and spread the DNA fibres on glass slides (Jackson and Pombo, 1998). After revealing the newly synthesized DNA through immunofluorescence with antibodies directed against the indicated nucleotides, the track length was measured. Whole fibres were also stained with antibodies against guanosine (Figure 1A), to distinguish intact fibres from broken ends. FANCM was efficiently knocked down by a short-hairpin (sh) RNA approach in HeLa cells with three different oligonucleotides against the FANCM sequence (Figure 1B). Replication tracks were pulse labelled with BrdU for a short time (15 min), to reduce the number of replication tracks that result from the fusion of converging replication forks (Figure 1A) and thereby increasing the probability to visualize single replication forks. For all three shFANCM constructs used, we observed a statistically significant (P<0.0001) increase in track length of newly synthesized DNA (Figure 1C, n>100 for each condition) compared with control cells transfected with the empty pSUPER vector. Consistently with what was observed in cancerous HeLa cells, the knockdown of FANCM in the fetal lung fibroblast cell line MRC5 led to a 23% increase in the median length of BrdU tracks (Supplementary Figure 1A and B). Thus, during a short labelling period, the apparent rate of BrdU incorporation is higher in the absence of FANCM, suggesting that FANCM opposes fork movement.
Figure 1.
FANCM controls DNA chain elongation. (A) HeLa cells were labelled for 15 min with BrdU, DNA was stretched out on glass slides and newly synthesized DNA was revealed by immunofluorescence (red). Total DNA was visualized with an antibody against guanosine (green). The bar represents 10 μm. (B) The knockdown of FANCM in HeLa cells was verified 3 days after transfection by western blot with an antibody against FANCM. The depletion of FANCM was confirmed by reduced monoubiquitination of FANCD2 (slower migrating band is absent in lanes 2–4). To induce the monoubiquitination of FANCD2, cells were exposed to 1 μM aphidicolin for 1 h. Actin was used as a loading control. (C) Graphic representation of the 15-min BrdU track lengths measured in μm (y axis, n>100 for each condition). The bar dissecting the box represents the median of the data points, the whiskers span the 10- and 90%-percentile and the dots represent data points that lay outside these percentiles. The P-values relative to the pSUPER-puro control cells were determined by Mann–Whitney test and are depicted above the graph. Three shFANCM plasmids (shM#1, 2 and 3) were used to deplete FANCM. (D) HeLa cells were pulse labelled with BrdU for 60 min and track length was plotted on the y axis. Three different oligonucleotides against the FANCM sequence were used to knock down FANCM protein levels. The median of track length for all shFANCM constructs used was shorter than for control cells. For plasmids shFANCM #2 and #3, the distribution of track length was significantly different as determined by Mann–Whitney test (P-values above graph). (E) Replication forks were pulse labelled with two pulses of equal length of 25 min or 50 min; The first pulse (CldU) was revealed in red and the second (IdU) in green. The integrity of the fibres was assessed with an antibody against guanosine (blue). The bar represents 5 μm. (F) DNA of control and FANCM shRNA HeLa cells were labelled with a 25-min CldU pulse, followed by an IdU pulse of equal length. Note that the data points are more dispersed for FANCM shRNA cells compared with control cells. (G) The length of the first 50 min (CldU) pulse was plotted on the x axis and the value for the second 50 min pulse (IdU) on the y axis (n>50). The linear regression is represented with a bold line. The correlation coefficient r is represented in the top left corner.
Next, we tested whether the increased rate of nucleotide incorporation could be maintained in FANCM shRNA cells over a longer labelling period. We observed that the replication tracks in HeLa cells were statistically shorter with two out of three constructs used (Figure 1D) when pulse labelled for 60 min. A similar reduction of the median track length (−32%) was observed in MRC5 cells depleted for FANCM (Supplementary Figure 1C). This suggests that the elevated rate of DNA chain elongation cannot be maintained over 60 min in FANCM shRNA cells. During 60 min pulses, fusion and termination events occur, and the probability that replication forks stall at endogenous impediments is increased. Conversely, during the short 15 min pulse, forks that pause are probably not detected, due to limitations in image resolution. Taken together, the short pulse labelling provides information about the speed of the replication forks that is increased in FANCM shRNA cells, whereas the longer labelling gives a more global picture of fork progression over longer DNA stretches.
To further underline our conclusion that replication does not progress constantly in FANCM shRNA cells over time, we pursued replication forks by labelling with two different nucleotide analogs that can be distinguished with specific antibodies. We first gave a pulse with the first nucleotide (CldU) followed by a pulse of equal length with a second nucleotide (IdU) (Figure 1E). The length of the first pulse was plotted on the x axis and the corresponding track length of the second pulse on the y axis. If all the replication forks were to move at a perfectly constant speed during both pulses, all the data points would lie on a diagonal of a 45° angle. Furthermore, if there were neither initiation, nor pausing, nor termination events occurring, the correlation coefficient r should be equal to 1. The linear correlation of the data points collected from the HeLa cells transfected with the control plasmid and pulse labelled with pulses of equal length with CldU and IdU, almost forms an angle of 45° (n>50 for each condition, Figure 1F and G). For cells lacking FANCM, a linear regression with a flatter angle was observed. This means that the track length measured for the first pulse was on average longer than for the second pulse. For FANCM shRNA cells, the correlation coefficient dropped from 0.70 (control) to 0.59 (for 25 min pulses) and from 0.79 to 0.50 (for 50 min pulses), suggesting that replication forks progressed unsteadily and paused more frequently in the absence of FANCM. The same phenotype was observed when MRC5 cells were labelled with two 25 min or two 50 min pulses (Supplementary Figure 2).
Taken together, our results suggest that FANCM opposes the rate of DNA chain elongation and at the same time assures constant progression of replication forks over time.
FANCM protects the genome from formation of single-stranded DNA
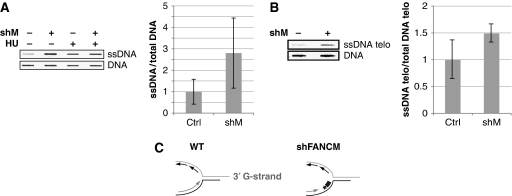
We predicted that uncontrolled DNA chain elongation in the absence of FANCM would lead to an accumulation of aberrant DNA structures, such as single-stranded (ss) DNA. As FANCM has a function in DNA damage signalling (Collis et al, 2008), classical protein markers of the DNA damage response (e.g. phospho-Chk1 or its downstream targets) may underestimate the level of abnormal DNA structures in FANCM-depleted cells. To probe ssDNA directly, we extracted DNA under non-denaturing conditions and immobilized it on a nitrocellulose membrane. An antibody that recognizes guanine only in its ssDNA form was used to reveal total ssDNA. We observed an increase in the amount of ssDNA in FANCM-depleted cells (Figure 2A).
Figure 2.
FANCM prevents the accumulation of ssDNA. (A) DNA from shFANCM and control HeLa cells was extracted under non-denaturing conditions and slot blotted. ssDNA was revealed by western blotting with an antibody that recognizes guanosine only in its ssDNA state. To control DNA loading, the membrane was denatured and blotted with the same antibody against guanosine. HU treatment was used as positive control for ssDNA formation. (B) Native DNA extracts were blotted as before, and telomeric ssDNA was revealed by Southern blotting with a radiolabelled G-rich probe (TTAGGG). The membrane was denatured and blotted with the same probe as loading control. (C) Cartoon representing a replication fork progressing towards the telomeric end with the 3′G-rich overhang. About 97% of replication origins used to replicate telomeres were reported to lie within the subtelomeric region. The radioactively (*) labelled, G-rich probe (thick grey line) can detect ssDNA on the C-strand arising from a stalled fork. The accumulation of ssDNA in shFANCM RNA cells (right panel) could be due to increased stalling of replication forks in the telomeric repeats.
An accumulation of ssDNA may reflect the formation of pathological structures during DNA replication through endogenous DNA lesions or secondary DNA structures. To test whether FANCM also has a function in the maintenance of sequences that are intrinsically difficult to replicate, we focused on telomeres, which consist of a highly conserved repetitive (TTAGGG) DNA sequence (Sfeir et al, 2009). The high content in guanosine renders telomeres prone to form secondary structures such as G-quartets (Rhodes and Giraldo, 1995). Model replication forks frequently regress in telomeric repeats (Fouche et al, 2006) forming a four-stranded structure that resembles a Holliday Junction and presumably block replication. We used the same approach as in Figure 2A and performed a native Southern blot with a radioactively labelled TTAGGG probe. We observed an increase of ssDNA in our samples stemming from shFANCM-treated cells compared with control cells (Figure 2B). Quantification of three independent experiments showed that on average there was 49% more ssDNA in FANCM shRNA cells compared with control cells at telomeres. Note that the G-probe can only anneal on the C-strand and therefore changes in signal are not due to differences in the telomeric 3′ G-overhangs (Figure 2C). Mostly, replication of telomeres initiates in the subtelomeric region, and only about 3% of telomeric DNA shows an internal origin of replication (Sfeir et al, 2009). A stalled replication fork moving through the telomeric repeats towards the end of the chromosome would leave a gap of ssDNA at the fork. This gap on the C-strand could be detected by our G-probe. Therefore, the ssDNA observed could stem from a blocked replication fork that is going towards the telomere end and uses the C-strand as a template (Figure 2C).
FANCM's function in opposing replication fork progression requires its ATPase activity and is independent of FANCD2
We next assessed whether the slowing of replication movement depends on the ATPase activity of FANCM. We used HEK293 cells that stably knockdown FANCM by expressing an siRNAmir and that are either complemented with siRNA-resistant WT FANCM or with the ATPase mutant K117R FANCM (Collis et al, 2008) and labelled these cells for 15 min with BrdU. The median length of replication tracks in cells complemented with K117R FANCM was 69% longer than in cells complemented with WT FANCM (Figure 3A, n>100 for each cell line). Cells expressing the ATPase-dead mutant K117R also mimicked the phenotype of shFANCM cells after 60 min pulse labelling with BrdU (Figure 3B), and increased levels of ssDNA were detected in HEK293 cells complemented with siRNA-resistant K117R FANCM (Figure 3C).
Figure 3.
FANCM's ability to slow down replication forks relies on its ATPase activity (A) HEK293 cells stably depleted for FANCM and complemented by either WT FANCM or the ATPase mutant (K117R) FANCM were pulse labelled for 15 min with BrdU and track length was analysed. Cells expressing the ATPase mutant displayed increased track length (median=2.3 μm, n=195) compared with control cells (median=1.4 μm, n=219). The P-value (<0.0001) is displayed above the graph. (B) Analysis of replication tracks after 60 min pulse labelling with BrdU: WT FANCM, median=18.5 μm, n=104; K117R FANCM, median=10.1 μm, n=138, P<0.0001. (C) DNA from WT FANCM and K117R FANCM cells was extracted under non-denaturing conditions, dot blotted, and ssDNA was revealed, as described in Figure 2. On the right, a histogram shows the result of the quantification of ssDNA levels in two independent experiments (# 1 and # 2). FANCM's ability to slow down replication forks is independent of FANCD2. (D) Two different oligonucleotides against the FANCD2 sequence were cloned in the pSUPER-puro plasmid (shD2#1 and 2) to deplete FANCD2 in HeLa cells. (E) shFANCD2 and control cells were pulse labelled 15 min with BrdU and track length of newly synthesized DNA was determined. The distribution of BrdU tracks in shFANCD2 cells was significantly (P<0.05) inferior compared with control cells. (F) Same experiment as in (E), but this time DNA was pulse labelled for 60 min. The median of the track length of FANCD2 shRNA cells is significantly shorter than the one of control cells (P<0.0001).
We wanted to confirm that the phenotype observed when FANCM is impaired is not due to a defect in ubiquitination of FANCD2. The knockdown of FANCD2 in HeLa cells by two different hairpins (Figure 3D) caused a reduction in the length of replication tracks labelled for 15 min with BrdU (−21 and −39% for the respective construct used, Figure 3E, n>100). This is in sharp contrast to FANCM shRNA cells, which showed an increased track length after 15′BrdU labelling. FANCD2 shRNA cells also showed a reduction in the median track length when labelled for 60 min with BrdU (Figure 3F). Taken together, these results suggest that the slowing of replication fork progression by FANCM is dependent on its ATPase activity, however, is independent of FANCD2. This is consistent with the observation that the ATPase domain of FANCM is not essential for the activation by monoubiquitination of FANCD2 (Xue et al, 2008; Singh et al, 2009).
FANCM opposes progression of replication forks in the presence of replication inhibitors that do not damage DNA
To determine whether FANCM is essential for replication in the presence of drugs that inhibit DNA chain elongation, we measured replication fork progression in cells treated with aphidicolin. Aphidicolin (aph) inhibits the primase and DNA polymerases directly (Arabshahi et al, 1988), without damaging DNA.
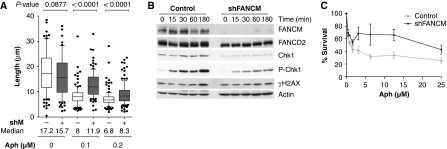
After exposure of HeLa cells to aphidicolin in the presence of BrdU (60 min), we observed that overall replication tracks of control and FANCM shRNA cells decreased with increasing doses of aphidicolin (Figure 4A). The median length of replication tracks of FANCM shRNA cells was longer than the median length of tracks from control cells exposed to the same dose of aphidicolin. This suggests that FANCM opposes the progression of replication forks in the presence of aphidicolin.
Figure 4.
FANCM counteracts replication in the presence of aphidicolin. (A) HeLa cells were exposed to different concentrations of aphidicolin for 60 min in the presence of BrdU. DNA track length of >100 tracks was plotted on the y axis for each concentration of aphidicolin. The median of track length was 9% shorter for untreated shFANCM (shM#1) cells. This value was higher in the presence of aphidicolin and absence of FANCM (+48% for 0.1 μM, +23% for 0.2 μM aph). (B) Control and FANCM shRNA cells were exposed for 3 h to 4 μM aphidicolin. In FANCM-depleted cells, phosphorylation of Chk1 and monoubiquitination of FANCD2 is diminished. (C) HeLa cells depleted for FANCM survived better during chronic exposure to aphidicolin than control cells (mean and s.e.m. plotted).
Recently, it has been reported that FANCM has a function in S-phase checkpoint signalling (Collis et al, 2008). To verify whether under our experimental conditions FANCM is also involved in the activation of the S-phase checkpoint, FANCM shRNA and control cells were exposed to 4 μM aphidicolin for up to 3 h and decreased levels of Chk1 and phospho Ser345 Chk1 were observed in cells depleted for FANCM (Figure 4B).
We also quantified the survival of shFANCM and control cells after chronic exposure to aphidicolin by measuring DNA content (Figure 4C). Surprisingly, after 2 days of incubation in medium-containing aphidicolin, the density of FANCM-depleted cells was higher than the density of control cells. This observation may reflect that the increased rate of DNA chain elongation in FANCM-deficient cells partially compensates the slowing of replication forks induced by aphidicolin.
To verify that the increased track length is not exclusive for aphidicolin, we repeated these experiments with another non-damaging replication inhibitor, that is HU, which lowers the dNTP pool in a cell. FANCM shRNA cells exposed to HU showed increased replication track length, decreased checkpoint signalling and augmented cell survival when compared with pSUPER control cells (Supplementary Figure 3). We did not observe, however, a dramatic change in apoptotic activity and cell cycle profile of control and shFANCM cells exposed to HU (Supplementary Figure 4).
In conclusion, FANCM facilitates Chk1 phosphorylation and restrains DNA chain elongation in cells treated with HU or aphidicolin.
FANCM promotes DNA replication through damaged DNA
To assess whether FANCM affects replication through damaged DNA, FANCM shRNA and control cells were exposed to two types of DNA-damaging agents, that is camptothecin (CPT) and UV light. CPT and UV were chosen, as FANCM, but not FA core complex-deficient cells, has been shown to be sensitive to those agents (Rosado et al, 2009; Singh et al, 2009).
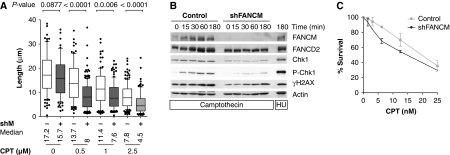
CPT covalently traps topoisomerase I onto DNA and ultimately leads to the formation of DSBs. CPT inhibited replication as manifested by an overall decrease in track lengths in control and FANCM shRNA cells (Figure 5A). We observed significantly shorter replication tracks for FANCM shRNA cells treated with CPT compared with control cells. Although in untreated FANCM shRNA cells the median of 60 min BrdU track lengths is only 9% shorter than the median length of control cells, the median in the presence of CPT is decreased by 41% for 0.5 μM CPT, by 33% for 1 μM CPT and by 44% for 2.5 μM CPT.
Figure 5.
FANCM promotes DNA chain elongation through CPT-damaged DNA. (A) The reduction in replication track length was greater in shFANCM than in control cells (−41% for 0.5 μM CPT, −33% for 1 μM and −44% for 2.5 μM). (B) DNA damage checkpoint signalling is defective in shFANCM cells. Cells were exposed to 2.5 μM CPT for up to 3 h. FANCM shRNA cells showed diminished phosphorylation of Chk1 and H2AX on western blots. (C) In the presence of CPT, the survival of FANCM shRNA cells is more severely impaired than the survival of control cells (mean and s.e.m. plotted).
After CPT treatment, we observed a defect in DNA damage checkpoint activation manifested by a decrease in phospho Ser345 Chk1 and γH2AX (Figure 5B). In agreement with FANCM being required for DNA chain elongation in the presence of CPT, FANCM is important for cell survival after chronic exposure to CPT (Figure 5C). However, we were not able to observe an important difference in the apoptotically induced cleavage of poly ADP ribose polymerase 1 (PARP-1) or the cell cycle profile between shFANCM and control cells exposed to CPT (Supplementary Figure 5).
To test whether FANCM promotes DNA chain elongation specifically in DNA damaged after exposure to CPT, we exposed FANCM shRNA and control cells to UV light. UV induces formation of pyrimidine dimers and less frequently pyrimidine-pyrimidone (6-4) photoproducts (Glickman et al, 1986). Similarly to what has been observed after CPT treatment, cell survival, Chk1 phosphorylation and replication fork progression were impaired in FANCM shRNA cells exposed to UV (Supplementary Figure 6).
Thus, FANCM has a function in replication through UV- and CPT-damaged DNA and is required for a complete DNA damage response.
FANCM is essential for resumption of DNA replication after DNA damage
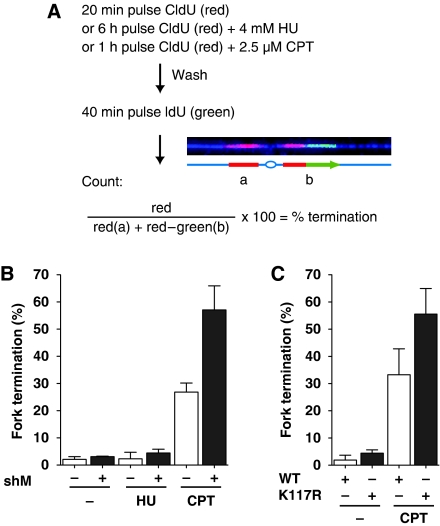
Fork remodelling has been proposed to allow the resumption of DNA synthesis after a replication block (Heller and Marians, 2006). To distinguish ongoing replication from termination, we used a two colour pulse labelling approach, consisting of a first pulse revealed in red (CldU) and a second pulse in green (IdU). Red only tracks stand for forks that terminated during the first pulse, whereas ongoing forks are labelled with red and green (Figure 6A). The percentage of forks that terminated was calculated as the number of red tracks divided by the total number of forks (red plus red–green) and multiplied by 100. For untreated FANCM shRNA HeLa cells, the amount of terminated forks was only slightly higher than for control cells (average of three independent experiments, n>500 total for FANCM shRNA and control). The same observation held true for forks that were released after 6 h exposure to 4 mM HU. This implies that FANCM is not necessary to maintain replisomes at HU-stalled forks, a prerequisite for fork reactivation. Strikingly, the percentage of forks that terminated after exposure to 2.5 μM CPT for 1 h was higher (57% on average) in the absence of FANCM compared with control cells (27%). A similar increase in fork termination after CPT treatment was observed in FANCM siRNA cells expressing the FANCM ATPase mutant (K117R) compared with cells complemented with the WT FANCM (Figure 6C). This suggests that remodelling of the replication fork by FANCM is required for carrying on DNA replication after a physical block to replication induced by CPT.
Figure 6.
FANCM promotes fork reactivation after CPT treatment. (A) Outline of the protocol used to quantify replication fork reactivation after treatment with replication-blocking agents. Cells were pulse labelled with CldU (red) during exposure to HU or CPT and released in the presence of IdU (green). Forks that terminate during the CldU pulse are red, ongoing forks are red and green. (B) Cells were labelled according to the protocol depicted in (A). The percentage of terminated forks was determined by counting red tracks and dividing them by the total number of forks (red and red–green tracks) and multiplied by 100. The mean and standard deviation of three independent experiments are represented in the graph (n>500). In the presence and absence of HU FANCM shRNA cells reactivated almost as many forks as wild-type cells. Whereas CPT prevented 26.8% of forks in control cells from resuming replication, this number reached 57.1% in cells depleted for FANCM. (C) HEK293 FANCM siRNA cells expressing either the ATPase mutant K117R or WT FANCM were labelled with CldU and IdU in the absence or presence of CPT as described in (A). Whereas in WT FANCM expressing cells 33.3% of the forks were not able to resume replication after CPT treatment, 55.6% of the forks terminated in the ATPase mutant K117R cells (average of three independent experiments, n>500 each).
FANCM and Chk1 mutually stabilize each other
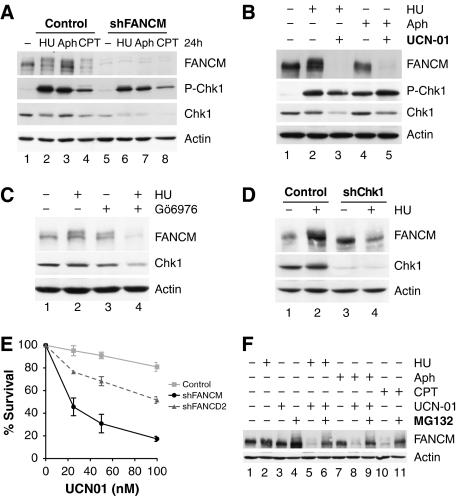
To examine the S-phase checkpoint response in FANCM-knockdown cells after sustained damage, we treated FANCM shRNA and control cells for 24 h with a DNA-damaging agent (CPT) and replication inhibitors (HU and aphidicolin). We observed that Chk1 was almost as strongly phosphorylated in FANCM shRNA as in the control cells (Figure 7A). Surprisingly, we noticed that FANCM levels decreased after treatment with CPT (Figure 7A, lane 4). As the Chk1 kinase is degraded after CPT treatment (Zhang et al, 2005), we hypothesized that Chk1 could be involved in the stabilization of FANCM. To further investigate this notion, we treated cells with either HU or aphidicolin for 8 h both in the presence and absence of the Chk1 inhibitor, UCN01, and assayed FANCM protein levels by western blotting. FANCM was strikingly destabilized in the presence of replication inhibitors and UCN01 (Figure 7B, lanes 3 and 5). As UCN01 also inhibits kinases other than Chk1 (Bain et al, 2007), we aimed to limit off-target effects by using another Chk1 inhibitor Gö6976 (Kohn et al, 2003), which has a different spectrum for non-Chk1 kinases. FANCM levels decreased in the presence of Gö6976 and HU in a similar manner to UCN01 (Figure 7C). Furthermore, FANCM was destabilized in the presence of HU when Chk1 was knocked down using specific shRNA directed against Chk1 (Figure 7D, lane 4) as compared with control cells treated with HU (Figure 7D, lane 2).
Figure 7.
FANCM and Chk1 stabilize each other after exposure to DNA-damaging agents. (A) HeLa cells depleted for FANCM phosphorylate Chk1 almost as robustly as control cells when exposed to drugs for 24 h (1 mM HU, 1 μM aphidicolin and 50 nM camptothecin). FANCM is destabilized after CPT treatment for 24 h (upper panel). Chk1 is destabilized in the presence of CPT, but more importantly in the absence of FANCM and presence of DNA-damaging agents (lower panel). (B) FANCM is degraded in the presence of the replication inhibitors aphidicolin (1 μM) and HU (1 mM) on addition of the Chk1 inhibitor UCN01 (0.3 μM, 24 h; lanes 3 and 5). (C) Less FANCM is present in HeLa cells exposed to 1 mM HU for 8 h in the presence of 1.5 μM Gö6976, a Chk1 inhibitor. (D) FANCM protein levels decrease in Chk1 shRNA HeLa cells in the presence of HU (1 mM, 8 h). Compare lanes 2 and 4. (E) FANCM shRNA HeLa cells survive less well in the presence of UCN01, whereas FANCD2 shRNA cells show an intermediate phenotype. (F) FANCM destabilization by 1 μM aphidicolin or 1 mM HU in the presence of 0.1 μM UCN01 can be reversed on inhibition of the proteasome by MG132 (25 μM, 16 h; lanes 6, 9 and 11).
Interestingly, we also found a decrease of Chk1 levels in FANCM shRNA cells (Figures 4A, 5A and 7A). This destabilization of Chk1 was exacerbated in the presence of DNA replication inhibitors (HU and aphidicolin) as well as after CPT treatment. Thus, after prolonged exposure to DNA-damaging agents, cells lacking FANCM are proficient in phosphorylating Chk1, but overall Chk1 protein levels are decreased. Moreover, cells lacking adequate FANCM levels were more sensitive to UCN01 than FANCD2 shRNA or control cells (Figure 7E), likely as a result of the decreased Chk1 levels. Apoptotic activity and cell cycle profiles of control and shFANCM cells exposed to UCN01 are shown in Supplementary Figure 7.
We observed that FANCM was destabilized after CPT treatment in control cells, but not after HU and aphidicolin (Figure 7A). Consistently, a difference between aphidicolin and HU versus CPT is that the latter does not induce strong Chk1 activation. Taken together, these data suggest that FANCM stabilizes Chk1 and Chk1 activity is required for FANCM stability in the presence of replication inhibitors and DNA-damaging agents.
Earlier reports indicate that FANCM is degraded by ubiquitin-dependent degradation through the proteasome in mitosis (Yen and Elledge, 2008; Kee et al, 2009). We investigated whether FANCM is also degraded through the proteasome after exposure to DNA-damaging agents. Strikingly, FANCM was no longer degraded in the presence of replication inhibitors (HU and aphidicolin) and under suppression of Chk1 activity (by UCN01) when the proteasome was inhibited by MG132 (Figure 7F, lanes 6 and 9). Similarly, FANCM is stabilized in the presence of CPT and MG132 (Figure 7F, lane 11).
Together, these results suggest that Chk1 and FANCM mutually stabilize each other and both contribute to promote cell survival. We have shown that FANCM regulates DNA chain elongation and provide evidence for FANCM providing a link between Chk1 signalling and DNA replication.
Discussion
This study shows that FANCM is required for normal DNA replication, and that this requirement is exacerbated when cells are exposed to inhibitors of DNA replication. Further, we find that FANCM and Chk1 positively regulate each other during DNA replication stress. This work provides evidence that FANCM's activity at replication forks is coupled to Chk1 signalling and ensures optimal replisome progression through replication obstacles.
Accumulating evidence suggests that FANCM has a general function in DNA replication, which is unique in regards to other FA core complex proteins. Unlike mice deficient for other FA core complex proteins, Fancm-deficient mice exhibit increased cancer incidence, and increased spontaneous sister chromatid exchanges (SCEs) that may reflect increased stress during DNA replication (Bakker et al, 2009). An elevated basal level of SCEs is also observed in ΔFANCM DT40 cells (Rosado et al, 2009). In addition to MMC sensitivity, cells lacking FANCM are uniquely sensitive to UV and CPT (Collis et al, 2008; Bakker et al, 2009; Rosado et al, 2009; Singh et al, 2009). The C-terminus of FANCM is necessary for resistance to CPT, but not essential for cellular tolerance of DNA crosslinks (Singh et al, 2009). Altogether, these observations support the notion that FANCM has additional functions in the tolerance of DNA replication stress, independently of the FA core complex (Collis et al, 2008).
In the absence of FANCM, the association of FANCA in chromatin and the monoubiquitination of FANCD2 induced by DNA crosslinks are still detectable, albeit at strongly reduced levels (Bakker et al, 2009; Rosado et al, 2009; Singh et al, 2009). Furthermore, the ATPase activity of FANCM is dispensable for FANCD2 monoubiquitination (Xue et al, 2008; Rosado et al, 2009; Singh et al, 2009). Here, we show that FANCM promotes the recovery of replication forks stalled by CPT-stabilized topoisomerase I cleavage complexes, in an ATPase-dependent manner. In vitro, FANCM binds avidly to branched DNA structures and can promote replication fork reversal (Xue et al, 2008; Gari et al, 2008a, 2008b). Thus, we believe that the function of FANCM in fork recovery reflects its ability to remodel replication forks and is independent of FANCM's function in FANCD2 monoubiquitination. We observe that FANCM's activity also controls DNA chain elongation in the absence of exogenous sources of DNA replication stress. In cells lacking FANCM, the progression of replication forks is accelerated, but unsteady. Faster fork movement in FANCM-depleted cells is also observed when replication forks are slowed down after treatment of cells with HU or aphidicolin. FANCM presumably controls the progression of replication through natural impediments by ensuring constant fork movement and limiting the formation of pathological replication structures.
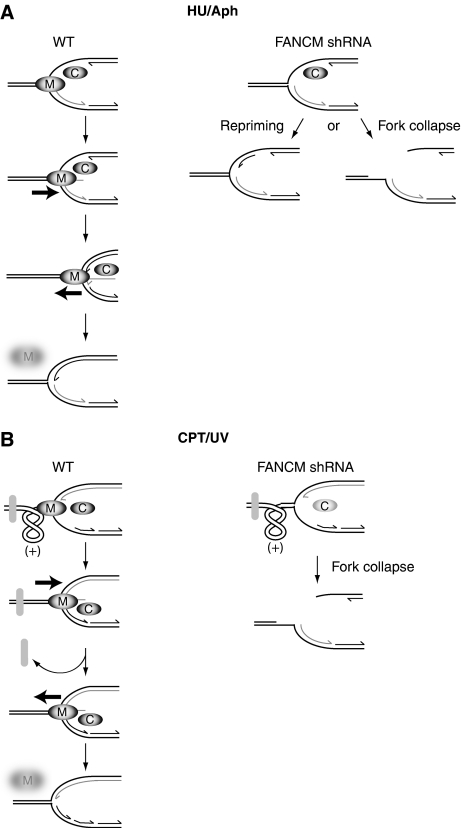
Our systematic analysis of replication dynamics in cells exposed to different types of replication inhibitors lead us to suggest the following model as to how fork remodelling by FANCM may facilitate the progression of replication forks. We propose that FANCM prevents the accumulation of ssDNA through fork reversal and thereafter promotes coupling of leading and lagging strand DNA synthesis (Figure 8). In the presence of HU and aphidicolin, long stretches of ssDNA are exposed. We find that in such conditions, Chk1 is activated and in turn its activation prevents FANCM from proteasome-mediated degradation (Figures 7 and 8A). Surprisingly, FANCM also promotes a positive feedback loop to confer Chk1 stability (Figure 7A). We suggest that when either the leading or lagging strand DNA polymerase is blocked, FANCM counteracts DNA chain elongation by fork reversal. The ongoing fork is set back to where it was blocked and leading and lagging strand DNA polymerases can be re-coupled. FANCM-mediated fork regression is expected to expose ssDNA on the newly synthesized strand. The concerted action of DNA synthesis combined with reversal of the branch point by FANCM (or some other branch point translocase such as BLM) in the opposite direction may lead to the re-annealing of the newly synthesized DNA that was previously extruded during fork reversal. Thereby, the ssDNA disappears and the replication fork is re-established. The vanishing of ssDNA (and subsequent checkpoint down-regulation) provides a stop signal for FANCM's fork reversal activity by targeting FANCM for degradation. According to our model, in FANCM shRNA cells, the accumulation of ssDNA is expected to promote unscheduled re-priming of DNA synthesis. This ‘runaway' DNA chain elongation may account for the longer replication track lengths observed in FANCM shRNA cells in the absence (Figure 1C) and the presence of replication inhibitors that do not damage DNA (HU, Supplementary Figure 3A and aphidicolin Figure 4A). Furthermore, this model explains how FANCM counteracts fork movement and thereby in the long run ensures that replication forks move at a constant speed (Figure 1F and G).
Figure 8.
(A) FANCM could prevent the accumulation of ssDNA by coupling leading and lagging strand synthesis. In wild-type cells, if leading strand synthesis is halted and lagging strand synthesis continues (left panel), long stretches of ssDNA are exposed. Chk1 (C) is activated at ssDNA and FANCM (M) is stabilized. FANCM could then regress the lagging strand by extruding ssDNA (bold arrow represents direction of fork reversal). FANCM continues fork reversal until the gap of ssDNA at the fork disappears. FANCM, or another branch point translocase (e.g. BLM), helps reversing the fork in the opposite direction (bold arrow), DNA chain elongation of the leading strand proceeds and the extruded ssDNA of the last synthesized Okazaki fragment is reannealed with the template strand. FANCM is not stabilized by Chk1 anymore and degraded by the proteasome and the newly coupled leading and lagging strand forks can resume replication. In FANCM shRNA cells, the occurrence of ssDNA could lead to unscheduled repriming events and thereby account for the longer replication tracks and the accumulation of ssDNA observed when FANCM is depleted. Alternatively, in the absence of FANCM, stalled forks may be subjected to nucleolytic cleavage and collapse. (B) Replication-blocking lesions (grey oval), for example, caused by CPT or UV, lead to the accumulation of positive supercoiling ahead of the replication fork. CPT brings about low levels of ssDNA and weak Chk1 activity. In wild-type cells, FANCM is stabilized locally and fork reversal by FANCM (bold arrow indicates direction) could relieve the topological strain and facilitate the access of repair proteins. FANCM, or another branch point translocase, would then regress the fork in the reverse direction (bold arrow), a new Okazaki fragment would be synthesized and the forks of the leading and lagging strand recoupled. In FANCM shRNA cells (right panel), the repriming of DNA replication is probably inhibited by the physical block and the positive supercoiling that occurs between the hindrance and the fork.
Agents like CPT and UV physically hinder the replication fork. It is important to note that besides topoisomerase inhibitors, a variety of DNA lesions, such as UV-induced DNA lesions, can lead to an accumulation of topoisomerase 1 cleavage complexes (Lanza et al, 1996; Pommier, 2006). A fork that advances towards a block gradually forces the DNA ahead of it to form positive supercoiling (Courcelle et al, 2003; Koster et al, 2007). CPT causes only small amounts of ssDNA (Davies et al, 2008) and hence Chk1 is only slightly activated (Figure 5B). We propose that ssDNA occurs only locally restricted at the blocked fork (Figure 8B). FANCM alleviates the torsional strain by regressing the fork and allowing the intertwines to spread over a larger DNA region, (Postow et al, 2001a). Thereby, FANCM would provide space for DNA repair enzymes to gain access to the lesion. After the obstacle has been removed, the replication fork is restored by FANCM or another branch point translocase (e.g. BLM) by migrating the branch point in the opposite direction. Leading and lagging strand are coupled and DNA chain elongation proceeds. The disappearance of ssDNA triggers the proteasomal degradation of FANCM. In FANCM shRNA cells exposed to agents that physically block replication, the persistent supercoils ahead of the fork could eventually lead to fork collapse (Figure 6B). By removing the supercoiling ahead of the fork through fork reversal, we explain how FANCM may be required to promote progression of replication forks through DNA-containing blocks to replication induced by CPT or UV. This could account for the observed slower progression of replication forks and increased frequency of fork termination in CPT or UV-treated cells lacking FANCM's activity (Figures 5A and 6; Supplementary Figure 6A).
DNA lesions promote uncoupling of leading and lagging strand replication (Sogo et al, 2002; Lopes et al, 2006). To sum up, in our model following uncoupling of leading and lagging strand synthesis, the fork remodelling activity of FANCM opposes the movement of the ongoing fork to limit the accumulation of abnormal DNA structures. FANCM's fork reversal activity is only shut off when coupled leading and lagging strand synthesis is restored and the ssDNA signal disappears. We propose that coordination between leading and lagging strand synthesis by FANCM accounts for constant replication fork progression.
We found that FANCM is stabilized by Chk1 activity after DNA replication stress (Figure 7). After the induction of DNA replication stress, Chk1 is protected by HCLK2 from rapid proteasome-dependent degradation (Zhang et al, 2005; Collis et al, 2007). FANCM interacts with HCLK2 (Collis et al, 2008), and we find that FANCM has also an impact on the stability of Chk1. Altogether, these studies suggest that the stability of proteins associated in DNA damage signalling complexes is highly regulated during DNA replication stress. Interestingly, a non-degradable form of FANCM leads to more radial chromosomes after MMC treatment (Kee et al, 2009). Kee et al suggested that FANCM has to be cleared from chromosomes at the G2/M transition to prevent chromosomal instability. Similarly, the regulated degradation of FANCM in S phase may be necessary to prevent chromosomal instability after DNA damage. Polo-like kinase Plk1 is required for the degradation of FANCM in mitosis (Kee et al, 2009), but not after DNA damage (our unpublished data). Thus, the kinase and E3-ubiquitin ligase responsible for degrading FANCM after DNA damage and in the absence of a strong Chk1 response remain to be discovered.
A number of proteins involved in tolerance of DNA replication stress are known to modulate replication dynamics. In bacteria, translesion DNA polymerases hold up replication fork progression possibly to allow more time and space for repairing damaged DNA (Indiani et al, 2009). PARP-1 decelerates replication in chicken DT40 and HeLa cells after damage (Sugimura et al, 2008). The Rad51 paralog XRCC3 slows down replication after DNA damage induced by cisplatin and UV in chicken DT40 cells (Henry-Mowatt et al, 2003). FANCM counteracts fork movement in the absence of exogenous sources of DNA damage, and controls DNA replication after exposure to replication inhibitors that do not damage DNA and following contact with damaging agents. FANCM's response is coordinated with S-phase checkpoint signalling. Therefore, we propose that FANCM orchestrates the response to replication stress in mammalian cells by both directly affecting DNA chain elongation and through stimulating S-phase checkpoint signalling.
Materials and methods
Cell culture, transfection and drugs
HeLa and MRC5 cells were grown under standard conditions in Dulbecco's modified Eagle medium supplemented with 10% FCS and 1% penicillin-streptomycin. HEK293 FANCM siRNAmir cells expressing either the WT or the K117R mutant, a gift from Stephen West (Collis et al, 2008), were maintained in the presence of 50 μg/ml hygromycin (PAA laboratories) and 1 μg/ml puromycin (Sigma). Transfections were carried out by lipofectamine (Invitrogen) and following the manufacturer's instructions. The following 19 nt target sequences were cloned in pSUPER carrying a puromycin resistance for selection (Azzalin and Lingner, 2006): for FANCM 5′-CAACAGTGGTGAATAGTAA-3′ (#1), 5′-TAGTGTCAATAAGAACAAG-3′ (#2), 5′-AGGCTGTGCAACAAGTTAT-3′ (#3), for FANCD2 5′-CTAGCACCGTATTCAAGTA-3′ (#1), 5′-GGATTGTCTAACACCATCA-3′ (#2) and for Chk1 5′-CTGAAGAAGCAGTCGCAGT-3′ (Xiao et al, 2003). At 24 h after transfection, 1 μg/ml of puromycin was added and the cells were selected for another 48 h (except for shChk1 24 h). HU, aphidicolin, CPT, Gö6976, UCN01 and MG132 were purchased from Sigma. UV-C irradiation was carried out with a Stratalinker at a wavelength of 254 nm (Stratagene).
DNA fibre analysis
Asynchronously growing HeLa, MRC5 and HEK293 FANCM siRNAmir cells were labelled with 30 μM BrdU. For double labelling, cells were labelled with 30 μM CldU, washed with PBS and exposed to 250 μM IdU. Cells were lysed and DNA fibres were stretched onto glass slides as reported earlier (Jackson and Pombo, 1998). The DNA fibres were denatured with 2.5 M HCl for 1 h, washed with PBS and blocked with 2% BSA in PBST for 30 min. The newly replicated BrdU and CldU tracks were revealed with and antibody against BrdU (Abcam) and IdU was visualized by another anti-BrdU (Beckton Dickinson). All the DNA fibres were also stained with an antibody against guanosine (Argene) to stain all DNA. The following secondary antibodies were used: anti-mouse Alexa 488 (Molecular Probes), anti-rat Cy3 (Jackson Immunoresearch), anti-mouse biotin (Jackson Immunoresearch). Streptavidin APC (Biolegend) was used against biotinylated anti-mouse. Microscopy was carried out with a Zeiss Axioplan equipped with an Axiocam camera.
Statistical analysis
The probability that two data sets stem from the same distribution was assayed by a non-parametrical Mann–Whitney test (Prism software).
Immunoblotting
Cells were directly lysed in 1 × sample buffer, sonicated and boiled. Samples were resolved by SDS–PAGE (6–14%) and blotted onto nitrocellulose membrane. Antibodies against the following proteins and peptides were used in this study: FANCD2 (Novus Biologicals), FANCM (a gift from Weidong Wang), β-actin (Abcam), γH2AX (upstate), two different phospho-Chk1(Ser345) (Cell Signaling), Chk1 (Santa Cruz), PARP1 (Cell Signaling Technology), α-tubulin (Sigma) and histone H3 (Abcam). The following secondary antibodies were used: anti-rabbit-HRP and anti-mouse-HRP (both Jackson Immunoresearch).
Slot blotting
DNA was extracted under non-denaturing conditions with a genome wizard genomic DNA purification kit (Promega). DNA was sonicated to determine its concentration by nanodrop (Thermo Scientific). Equal amounts of DNA were bound to a nitrocellulose membrane through slot blotting (Biorad). DNA was crosslinked to the membrane by UV light (Stratalinker from Stratgene). Then the membrane was processed by standard western blotting and incubated with an antibody against guanosine (Argene) for detection of ssDNA. The DNA was denatured (0.4 M NaOH, 0.6 M NaCl) and neutralized (1.5 M NaCl, 0.5 M Tris pH 7.4). After washing with PBS, the membrane was immunoblotted again with the antibody against guanine to detect total DNA. To specifically detect telomeric signal, DNA was processed as above; however, membranes were pre-hybridized for 1 h in CHURCH buffer at 50°C after incubation overnight at 50°C in CHURCH buffer with a 32P radiolabelled heterogeneous telomeric probe that was labelled in a strand-specific manner with the Rad prime random labelling kit (Invitrogen). Following hybridization, washing steps included 2X with 2XSSC 0.1% SDS and once with 0.2XSSC 0.1% SDS at 50°C. Signals were detected with a phosphoimager (Fuji) and quantified using the AIDA software package (raytest).
Cell survival assay
Hela cells were transfected with episomal pSUPER plasmids carrying a puromycin resistance (a gift from Joachim Lingner). Three days after transfection, 1500 cells/well were treated with the respective drugs for another 2 days (aphidicolin and CPT), 3 days (HU) or 5 days (UCN01). Each assay was done in triplicate. Cell survival was determined by staining nucleic acids with CyQUANT GR dye (Invitrogen). Fluorescence readings were normalized to those obtained from untreated cells, assumed to yield 100% cell survival.
FACS
106 HeLa cells were harvested, washed with PBS and fixed in 70% cold ethanol. After a washing step with PBS, cells were RNAseA (0.2 μg/ml) treated for 15 min at 37°C. DNA was stained with propidium iodide (final concentration: 40 μg/ml) and analysed by FacsScan (Becton Dickinson). Cell cycle profiles were analysed by the FlowJo Software.
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Figure 5
Supplementary Figure 6
Supplementary Figure 7
Supplementary Figure Legends
Review Process File
Acknowledgments
We thank Peter Beard, Joachim Lingner, Weidong Wang, Stephen West and Simon Boulton for sharing reagents. We are grateful to Massimo Lopes for helpful and thorough comments on the manuscript and Chantal Décaillet for technical support. SL-G is financially supported by a Marie Heim-Vögtlin fellowship (PMPDA3-118607) of the Swiss National Science Foundation (SNSF) and AC by an SNSF professorship (PP00A-118991).
Footnotes
The authors declare that they have no conflict of interest.
References
- Arabshahi L, Brown N, Khan N, Wright G (1988) Inhibition of DNA polymerase alpha by aphidicolin derivatives. Nucleic Acids Res 16: 5107–5113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auerbach AD, Wolman SR (1976) Susceptibility of Fanconi's anaemia fibroblasts to chromosome damage by carcinogens. Nature 261: 494–496 [DOI] [PubMed] [Google Scholar]
- Azzalin CM, Lingner J (2006) The human RNA surveillance factor UPF1 is required for S phase progression and genome stability. Curr Biol 16: 433–439 [DOI] [PubMed] [Google Scholar]
- Bain J, Plater L, Elliott M, Shpiro N, Hastie CJ, McLauchlan H, Klevernic I, Arthur JS, Alessi DR, Cohen P (2007) The selectivity of protein kinase inhibitors: a further update. Biochem J 408: 297–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker ST, van de Vrugt HJ, Rooimans MA, Oostra AB, Steltenpool J, Delzenne-Goette E, van der Wal A, van der Valk M, Joenje H, te Riele H, de Winter JP (2009) Fancm-deficient mice reveal unique features of Fanconi anemia complementation group M. Hum Mol Genet 18: 3484–3495 [DOI] [PubMed] [Google Scholar]
- Cobb JA, Bjergbaek L, Shimada K, Frei C, Gasser SM (2003) DNA polymerase stabilization at stalled replication forks requires Mec1 and the RecQ helicase Sgs1. EMBO J 22: 4325–4336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collis SJ, Barber LJ, Clark AJ, Martin JS, Ward JD, Boulton SJ (2007) HCLK2 is essential for the mammalian S-phase checkpoint and impacts on Chk1 stability. Nat Cell Biol 9: 391–401 [DOI] [PubMed] [Google Scholar]
- Collis SJ, Ciccia A, Deans AJ, Horejsi Z, Martin JS, Maslen SL, Skehel JM, Elledge SJ, West SC, Boulton SJ (2008) FANCM and FAAP24 function in ATR-mediated checkpoint signaling independently of the fanconi anemia core complex. Mol Cell 32: 313–324 [DOI] [PubMed] [Google Scholar]
- Courcelle J, Donaldson JR, Chow KH, Courcelle CT (2003) DNA damage-induced replication fork regression and processing in Escherichia coli. Science 299: 1064–1067 [DOI] [PubMed] [Google Scholar]
- Davies AA, Huttner D, Daigaku Y, Chen S, Ulrich HD (2008) Activation of ubiquitin-dependent DNA damage bypass is mediated by replication protein a. Mol Cell 29: 625–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouche N, Ozgur S, Roy D, Griffith JD (2006) Replication fork regression in repetitive DNAs. Nucleic Acids Res 34: 6044–6050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Higuera I, Taniguchi T, Ganesan S, Meyn MS, Timmers C, Hejna J, Grompe M, D'Andrea AD (2001) Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol Cell 7: 249–262 [DOI] [PubMed] [Google Scholar]
- Gari K, Decaillet C, Delannoy M, Wu L, Constantinou A (2008a) Remodeling of DNA replication structures by the branch point translocase FANCM. Proc Natl Acad Sci USA 105: 16107–16112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gari K, Decaillet C, Stasiak AZ, Stasiak A, Constantinou A (2008b) The Fanconi anemia protein FANCM can promote branch migration of Holliday junctions and replication forks. Mol Cell 29: 141–148 [DOI] [PubMed] [Google Scholar]
- Glickman BW, Schaaper RM, Haseltine WA, Dunn RL, Brash DE (1986) The C-C (6-4) UV photoproduct is mutagenic in Escherichia coli. Proc Natl Acad Sci USA 83: 6945–6949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller RC, Marians KJ (2006) Replisome assembly and the direct restart of stalled replication forks. Nat Rev Mol Cell Biol 7: 932–943 [DOI] [PubMed] [Google Scholar]
- Henry-Mowatt J, Jackson D, Masson JY, Johnson PA, Clements PM, Benson FE, Thompson LH, Takeda S, West SC, Caldecott KW (2003) XRCC3 and Rad51 modulate replication fork progression on damaged vertebrate chromosomes. Mol Cell 11: 1109–1117 [DOI] [PubMed] [Google Scholar]
- Indiani C, Langston LD, Yurieva O, Goodman MF, O'Donnell M (2009) Translesion DNA polymerases remodel the replisome and alter the speed of the replicative helicase. Proc Natl Acad Sci USA 106: 6031–6038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson DA, Pombo A (1998) Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J Cell Biol 140: 1285–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee Y, Kim JM, D'Andrea AD (2009) Regulated degradation of FANCM in the Fanconi anemia pathway during mitosis. Genes Dev 23: 555–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JM, Kee Y, Gurtan A, D'Andrea AD (2008) Cell cycle-dependent chromatin loading of the Fanconi anemia core complex by FANCM/FAAP24. Blood 111: 5215–5222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohn EA, Yoo CJ, Eastman A (2003) The protein kinase C inhibitor Go6976 is a potent inhibitor of DNA damage-induced S and G2 cell cycle checkpoints. Cancer Res 63: 31–35 [PubMed] [Google Scholar]
- Koster DA, Palle K, Bot ES, Bjornsti MA, Dekker NH (2007) Antitumour drugs impede DNA uncoiling by topoisomerase I. Nature 448: 213–217 [DOI] [PubMed] [Google Scholar]
- Lanza A, Tornaletti S, Rodolfo C, Scanavini MC, Pedrini AM (1996) Human DNA topoisomerase I-mediated cleavages stimulated by ultraviolet light-induced DNA damage. J Biol Chem 271: 6978–6986 [DOI] [PubMed] [Google Scholar]
- Long DT, Kreuzer KN (2009) Fork regression is an active helicase-driven pathway in bacteriophage T4. EMBO Rep 10: 394–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes M, Foiani M, Sogo JM (2006) Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol Cell 21: 15–27 [DOI] [PubMed] [Google Scholar]
- Meetei AR, Medhurst AL, Ling C, Xue Y, Singh TR, Bier P, Steltenpool J, Stone S, Dokal I, Mathew CG, Hoatlin M, Joenje H, de Winter JP, Wang W (2005) A human ortholog of archaeal DNA repair protein Hef is defective in Fanconi anemia complementation group M. Nat Genet 37: 958–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pommier Y (2006) Topoisomerase I inhibitors: camptothecins and beyond. Nat Rev Cancer 6: 789–802 [DOI] [PubMed] [Google Scholar]
- Postow L, Crisona NJ, Peter BJ, Hardy CD, Cozzarelli NR (2001a) Topological challenges to DNA replication: conformations at the fork. Proc Natl Acad Sci USA 98: 8219–8226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postow L, Ullsperger C, Keller RW, Bustamante C, Vologodskii AV, Cozzarelli NR (2001b) Positive torsional strain causes the formation of a four-way junction at replication forks. J Biol Chem 276: 2790–2796 [DOI] [PubMed] [Google Scholar]
- Rhodes D, Giraldo R (1995) Telomere structure and function. Curr Opin Struct Biol 5: 311–322 [DOI] [PubMed] [Google Scholar]
- Rosado IV, Niedzwiedz W, Alpi AF, Patel KJ (2009) The Walker B motif in avian FANCM is required to limit sister chromatid exchanges but is dispensable for DNA crosslink repair. Nucleic Acids Res 37: 4360–4370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder TM, Anschutz F, Knopp A (1964) Spontaneous chromosome aberrations in familial panmyelopathy. Humangenetik 1: 194–196 [DOI] [PubMed] [Google Scholar]
- Sfeir A, Kosiyatrakul ST, Hockemeyer D, MacRae SL, Karlseder J, Schildkraut CL, de Lange T (2009) Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell 138: 90–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh TR, Bakker ST, Agarwal S, Jansen M, Grassman E, Godthelp BC, Ali AM, Du CH, Rooimans MA, Fan Q, Wahengbam K, Steltenpool J, Andreassen PR, Williams DA, Joenje H, de Winter JP, Meetei AR (2009) Impaired FANCD2 monoubiquitination and hypersensitivity to camptothecin uniquely characterize Fanconi anemia complementation group M. Blood 114: 174–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smogorzewska A, Matsuoka S, Vinciguerra P, McDonald ER III, Hurov KE, Luo J, Ballif BA, Gygi SP, Hofmann K, D'Andrea AD, Elledge SJ (2007) Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell 129: 289–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogo JM, Lopes M, Foiani M (2002) Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science 297: 599–602 [DOI] [PubMed] [Google Scholar]
- Sugimura K, Takebayashi S, Taguchi H, Takeda S, Okumura K (2008) PARP-1 ensures regulation of replication fork progression by homologous recombination on damaged DNA. J Cell Biol 183: 1203–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Nandi S, Osman F, Ahn JS, Jakovleska J, Lorenz A, Whitby MC (2008) The FANCM ortholog Fml1 promotes recombination at stalled replication forks and limits crossing over during DNA double-strand break repair. Mol Cell 32: 118–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischkowitz MD, Hodgson SV (2003) Fanconi anaemia. J Med Genet 40: 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourriere H, Pasero P (2007) Maintenance of fork integrity at damaged DNA and natural pause sites. DNA Repair (Amst) 6: 900–913 [DOI] [PubMed] [Google Scholar]
- Xiao Z, Chen Z, Gunasekera AH, Sowin TJ, Rosenberg SH, Fesik S, Zhang H (2003) Chk1 mediates S and G2 arrests through Cdc25A degradation in response to DNA-damaging agents. J Biol Chem 278: 21767–21773 [DOI] [PubMed] [Google Scholar]
- Xue Y, Li Y, Guo R, Ling C, Wang W (2008) FANCM of the Fanconi anemia core complex is required for both monoubiquitination and DNA repair. Hum Mol Genet 17: 1641–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen HC, Elledge SJ (2008) Identification of SCF ubiquitin ligase substrates by global protein stability profiling. Science 322: 923–929 [DOI] [PubMed] [Google Scholar]
- Zhang YW, Otterness DM, Chiang GG, Xie W, Liu YC, Mercurio F, Abraham RT (2005) Genotoxic stress targets human Chk1 for degradation by the ubiquitin-proteasome pathway. Mol Cell 19: 607–618 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Figure 5
Supplementary Figure 6
Supplementary Figure 7
Supplementary Figure Legends
Review Process File