Abstract
A key intermediate in translocation is an ‘unlocked state' of the pre-translocation ribosome in which the P-site tRNA adopts the P/E hybrid state, the L1 stalk domain closes and ribosomal subunits adopt a ratcheted configuration. Here, through two- and three-colour smFRET imaging from multiple structural perspectives, EF-G is shown to accelerate structural and kinetic pathways in the ribosome, leading to this transition. The EF-G-bound ribosome remains highly dynamic in nature, wherein, the unlocked state is transiently and reversibly formed. The P/E hybrid state is energetically favoured, but exchange with the classical P/P configuration persists; the L1 stalk adopts a fast dynamic mode characterized by rapid cycles of closure and opening. These data support a model in which P/E hybrid state formation, L1 stalk closure and subunit ratcheting are loosely coupled, independent processes that must converge to achieve the unlocked state. The highly dynamic nature of these motions, and their sensitivity to conformational and compositional changes in the ribosome, suggests that regulating the formation of this intermediate may present an effective avenue for translational control.
Keywords: EF-G, ribosome, single molecule, translation, translocation
Introduction
The rate of protein synthesis in the cell is controlled, in part, by the mechanism of translation elongation (Mathews et al, 2007). Central to this multi-step process is the precise translocation of mRNA and tRNA substrates through the biochemically and structurally defined aminoacyl (A), peptidyl (P) and exit (E) sites of the two-subunit, ∼3-MDa ribosome (Frank et al, 2007; Shoji et al, 2009). High-resolution structures of the pre-translocation ribosome complex, bearing deacylated P-site and peptidyl-A-site tRNA represent a relatively stable, ‘locked' configuration of the particle in which both tRNAs are bound classically, oriented perpendicular to the interface of the large and small subunits (50S and 30S in bacteria) (Munro et al, 2009b; Figure 1A). During translocation, substrate–ribosome contacts evidenced through these investigations must be remodelled for P- and A-site tRNAs to move to the E- and P-sites, respectively.
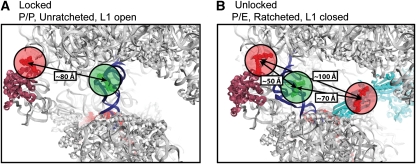
Figure 1.
Structural models of the ribosome with fluorescent components. The distances shown are in agreement with those predicted by smFRET experiments. Structural models were constructed according to published procedures (Tung et al, 2002; Munro et al, 2009a). The P-site tRNA is in blue and the L1 stalk is in purple. (A) The locked ribosome configuration with the tRNA in the classical P site, and the L1 stalk in the open position, consistent with the low-FRET (0.1) state observed in smFRET trajectories acquired from complexes with labelled L1 and P-site tRNA. (B) The unlocked ribosome configuration stabilized by EF-G (shown in cyan) in which the P/E hybrid state is formed, the L1 stalk is closed and the subunits are ratcheted—consistent with the high-FRET (0.65) state observed with labelled L1 and P-site tRNA, and the low-FRET (0.25) state observed with labelled EF-G and P-site tRNA.
Translocation occurs spontaneously in vitro at slow rates (Gavrilova et al, 1976; Bergemann and Nierhaus, 1983; Cukras et al, 2003; Fredrick and Noller, 2003). However, in vivo, rapid substrate movements on the ribosome are catalysed by a conserved GTPase, elongation factor-G (EF-G; Wintermeyer et al, 2004). Pre-steady state biochemical investigations (Savelsbergh et al, 2003; Peske et al, 2004; Pan et al, 2007) suggest that EF-G accelerates translocation by promoting a rate-limiting ‘unlocking' process within the ribosome complex, which immediately precedes substrate movement. Although the mechanism of unlocking remains unknown, cryo-electron microscopy (cryo-EM) studies suggest that the EF-G-bound ribosome adopts an ‘unlocked state' characterized by formation of the P/E tRNA hybrid state, movement of the L1 stalk domain into a closed position and a ratchet-like re-organization at the subunit interface (Valle et al, 2003; Connell et al, 2007; Frank et al, 2007; Taylor et al, 2007) (Figure 1B). Hybrid tRNA configurations arise from the movement of the 3′-CCA termini of A- and P-site tRNAs to adjacent binding sites in the large subunit before codon–anticodon movement in the small subunit (forming A/P and P/E states, respectively; Moazed and Noller, 1989; Sharma et al, 2004; Blanchard et al, 2004b; Dorner et al, 2006; Munro et al, 2007; Walker et al, 2008). Closure of the L1 stalk domain, comprised of helices 76–78 of 23S rRNA and the ribosomal protein L1, describes its movement from an open position distal to the subunit interface towards the E site, where it can make stabilizing contacts with the P/E hybrid tRNA (Valle et al, 2003; Connell et al, 2007; Munro et al, 2007; Taylor et al, 2007; Cornish et al, 2009). Subunit ratcheting involves rotation of the 30S subunit with respect to the 50S subunit, resulting in the remodelling of numerous bridging contacts found at the subunit interface (Valle et al, 2003; Connell et al, 2007; Taylor et al, 2007; Cornish et al, 2009).
Although the order and timing of ‘unlocking' events required for substrate movement (defined through rapid kinetics measurements) and conformational events critical to ‘unlocked state' formation (defined through cryo-EM studies of the EF-G-bound ribosome) remains unclear, conformational processes critical for unlocked state formation occur in the pre-translocation ribosome and have been implicated in the translocation mechanism (Dorner et al, 2006; Horan and Noller, 2007; Pan et al, 2007; Walker et al, 2008). Contemporary translocation models therefore posit that the unlocked state must be achieved before the unlocking events that immediately precede substrate movement (Wintermeyer et al, 2004; Frank et al, 2007; Pan et al, 2007; Shoji et al, 2009; Munro et al, 2009b). Consistent with this idea, single-molecule fluorescence resonance energy transfer (smFRET) studies have demonstrated the spontaneous nature of hybrid state formation (Blanchard et al, 2004b; Munro et al, 2007), L1 stalk closure (Fei et al, 2008; Cornish et al, 2009) and subunit ratcheting (Cornish et al, 2008). Cryo-EM studies have recently shown that the unlocked state can be spontaneously achieved in the pre-translocation ribosome (Agirrezabala et al, 2008; Julian et al, 2008).
Presently, the pre-translocation ribosome complex is thought to spontaneously transition between just two globally distinct conformations: a locked and an unlocked state (Agirrezabala et al, 2008; Fei et al, 2008; Julian et al, 2008). In this view, conformational transitions critical for unlocked state formation occur in a single, concerted event. However, two- and three-colour smFRET studies of the pre-translocation complex demonstrate that the process by which the unlocked state is achieved may be substantially more complex (Munro et al, 2009a). In this study, P/E hybrid state formation and L1 stalk closure are shown to be largely independent conformational events that can occur on distinct time scales. These data suggested that spontaneous unlocked state formation is achieved through a relatively low-probability event, minimally requiring the convergence of P/E hybrid state formation, L1 stalk closure and subunit ratcheting. In this view, a key aspect of EF-G's function in translocation may be to accelerate the rate at which these conformational processes converge.
Here, EF-G's ability to promote unlocked state formation was probed on two distinct ribosome complexes containing deacylated tRNA in the P site using two- and three-colour smFRET imaging. Such complexes, lacking peptidyl-tRNA at the A site and, therefore, unable to translocate, serve as effective model systems for examining EF-G's effect on conformational events in the ribosome and the order and timing of events leading to unlocked state formation (Valle et al, 2003; Connell et al, 2007; Taylor et al, 2007; Fei et al, 2008). As previously reported, formation of the unlocked state was directly monitored through the site-specific labelling of the L1 protein and P-site tRNA (Fei et al, 2008; Munro et al, 2009a). Ribosomal complexes bearing P-site tRNA were shown to spontaneously achieve the unlocked state, a process that was dramatically enhanced in the presence of EF-G. However, contrary to the ribosome transiting between just two states, evidence obtained from multiple structural perspectives, site-directed mutagenesis, structural modelling and kinetic fingerprinting revealed that the EF-G-bound complex was highly dynamic in nature. On this complex the unlocked state can be achieved through multiple structural and kinetic pathways. Within the EF-G-bound ribosome, P-site tRNA favours the P/E hybrid state but remains in dynamic exchange with the classical P/P configuration and the L1 stalk adopts a fast dynamic mode characterized by rapid cycles of closure and opening. These new data reveal that P/E hybrid state formation, L1 stalk closure and subunit ratcheting are not strictly coupled processes, corroborating the notion that the unlocked state is achieved through the convergence of independent structural events (Munro et al, 2009a). These insights suggest a model in which EF-G promotes the formation of the unlocked state by stabilizing a ratcheted conformation of the ribosome, wherein the rate of P/E hybrid state formation and L1 stalk closure become highly accelerated.
Results
The unlocked state of the ribosome is transient in nature
As described elsewhere (Munro et al, 2009a), conformational changes leading to the unlocked state can be monitored on site-specifically labelled L1- and P-site tRNA pre-translocation complexes using prism-based, wide-field smFRET imaging techniques. Here, data were acquired on two distinct ribosome complexes site-specifically labelled at the L1 protein with an acceptor fluorophore and with either a donor labelled P-site initiator tRNAfMet or an elongator tRNAPhe. Conformational dynamics of these complexes were examined at 40-ms time resolution both in the absence and presence of native and site-specifically labelled EF-G. In total, ribosomal complexes containing five distinct sites of L1 labelling and two distinct sites of P-site tRNA labelling were considered (Supplementary Figure S1). All labelling sites yielded qualitatively similar FRET behaviours, strongly suggesting that the present observations are dye independent. Here, only data obtained from complexes labelled at position 55 of the L1 protein and position 8 located at the elbow region of P-site tRNA are described (see Materials and methods section). These labelling positions provided the greatest fluorophore incorporation into the ribosome, the largest dynamic range in FRET values observed, and showed isotropic fluorophore tumbling (Supplementary Table S1). Such complexes were fully active in tRNA selection, peptide bond formation, hybrid state formation and translocation (Munro et al, 2009a). Complexes labelled in this manner displayed FRET transitions indicative of intrinsic, uncoupled motions of the L1 stalk and the P-site tRNA that lead to reductions in their intermolecular distance (Munro et al, 2009a).
Initially, ribosomal complexes containing deacylated P-site tRNAfMet were examined in the absence of EF-G. Similar to the pre-translocation complex, this system was dynamic in nature and smFRET trajectories were best idealized to a four-state Markov chain (Qin, 2004; Munro et al, 2009a; Supplementary Figure S2). Analysis of the occupancy of non-zero FRET states revealed the existence of a predominant low-FRET (∼0.1) state (∼80% occupancy), with infrequent and short-lived transitions to intermediate- (∼0.25, ∼0.4) and high-FRET (∼0.65) configurations (Supplementary Figure S3A, Table 1). Computational models showed that the low-FRET state, corresponding to an inter-dye distance of ∼80 Å (R0=56 Å for the Cy3/5 dye pair), was consistent with a locked ribosome structure in which the P-site tRNA is classically bound, the L1 stalk is in the open position and the subunits are unratcheted (Figure 1A). Structural modelling indicated that the high-FRET state, corresponding to an inter-dye distance of ∼50 Å, could be modelled as an unlocked ribosome configuration in which the P/E hybrid state is formed, the L1 stalk is closed and the subunits are in the ratcheted conformation (Figure 1B). Modelling this distance constraint by L1 closure and P/E hybrid state formation alone was not feasible without invoking substantial distortions of the P-site tRNA body or movements of the L1 stalk to closed configurations, not observed previously on such complexes (data not shown).
Table 1.
EF-G binding modulates the observed dynamics of the P-site tRNA and the L1 stalk
| P-site | A-site | L1-tRNA FRET | EF-G-tRNA FRET | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Occupancies (%) | Rates (s−1) | Occupancies (%) | Rates (s−1) | ||||||||||
| 0.1 | 0.25 | 0.4 | 0.65 | klow → | k→ low | khigh → | k→ high | 0.65 | 0.25 | kP/P | kP/E | ||
| tRNAfMet | Empty | 83±1 | 12±1 | 2.8±0.6 | 1.9±0.6 | 0.54±0.07 | 9.4±0.8 | 25±1 | 0.03±0.01 | ND | ND | ND | ND |
| tRNAfMet | EF-G(GDPNP) | 69±1 | 9.9±0.6 | 6.6±0.4 | 14±1 | 3.5±0.2 | 8.9±0.6 | 17±1 | 1.0±0.1 | 19±1 | 81±1 | 1.8±0.1 | 13±1 |
| tRNAPhe | Empty | 83±1 | 7.0±0.9 | 3.6±0.4 | 5.9±0.7 | 1.2±0.1 | 11±1 | 17±1 | 0.31±0.01 | ND | ND | ND | ND |
| tRNAPhe | EF-G(GDPNP) | 13±1 | 13±1 | 18±1 | 56±2 | 15±1 | 1.5±0.2 | 11±1 | 12±1 | 3.9±0.7 | 96±1 | ∼0.3 | ⩾25 |
| Idealization of smFRET trajectories obtained from complexes bearing labelled L1 and P-site tRNA yielded occupancies in four FRET states. The rates of transition between the observed FRET states were determined by maximum likelihood optimization in QuB (Qin et al, 1996; Qin, 2004). The complete set of transition rates is reported in Supplementary Table S2. klow → is defined as k0.1 → 0.25+k0.1 → 0.4. The method of calculating k→ low and k→ high has been described in detail previously (Munro et al, 2009a). The occupancies in the classical P-site and the P/E hybrid state, and the rates of transition between those states (kP/P and kP/E, respectively) were estimated by fitting smFRET trajectories obtained from complexes bearing labelled P-site tRNA and EF-G to a two-state model. Each rate is reported as the mean calculated from three distinct data sets±s.e. | |||||||||||||
As described elsewhere (Munro et al, 2009a), the rates of interconversion between FRET states were characterized through maximum likelihood optimization (Qin et al, 1996). For simplicity, the complete results of this analysis (Supplementary Table S2) are summarized as the average transition rates into, and out of, low- (k→ low and klow →) and high- (k→ high and khigh →) FRET states, those structurally consistent with locked and unlocked states, respectively (Table 1). The observed rates exiting the low-FRET state were slow (∼0.5 per s) and only modestly dependent on the aminoacylation state of P-site tRNA (data not shown), consistent with previous reports showing that spontaneous subunit ratcheting can occur in complexes containing aminoacyl-tRNA at the P site (Cornish et al, 2009). Fluctuations to the intermediate-FRET states may also include isolated L1 stalk motions into closed positions and/or movements of P-site tRNA towards the E site (Supplementary Figure S4).
The ribosome becomes highly dynamic in the presence of EF-G
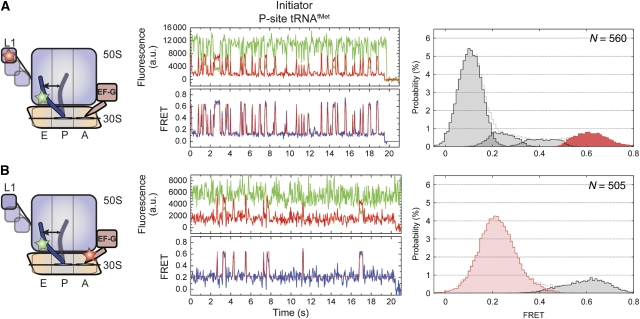
EF-G, in the presence of the non-hydrolysable GTP analogue, GDPNP, binds with high affinity to ribosome complexes programmed with deacylated P-site tRNA and a vacant A site (Zavialov and Ehrenberg, 2003). EF-G(GDPNP) binding at the A site promotes the P/E hybrid state, L1 stalk closure and subunit ratcheting (Valle et al, 2003; Connell et al, 2007; Spiegel et al, 2007; Taylor et al, 2007). To understand how EF-G affects the order and timing of conformational events in the ribosome leading to the unlocked state, experiments were performed under steady-state conditions on L1- and deacylated P-site tRNAfMet-labelled complexes in the presence of saturating EF-G (10 μM) and GDPNP (2 mM) concentrations. Consistent with EF-G's established role in promoting conformational events in the ribosome favouring the unlocked state, a ∼seven-fold increase in the rate exiting the low-FRET state (k→ low) was observed (∼3.6 per s; Table 1). Consequently, occupancy in the high-FRET, unlocked state increased from 1 to 14% (Figure 2A). The presence of EF-G led to an overall increase in dynamics. The increase in unlocked state occupancy resulted from a ∼30-fold acceleration of the rate at which the unlocked state was formed (k→ high) (Table 1). Thus, in complexes containing P-site initiator tRNAfMet the unlocked state was highly unstable in nature, in which the low-FRET state remained ∼68% occupied even under saturating EF-G concentrations (Rodnina et al, 1997; Zavialov and Ehrenberg, 2003).
Figure 2.
EF-G(GDPNP) binding at the A-site stabilizes the P/E hybrid state and lowers the activation barriers for L1 stalk motions. Here the dynamics of the EF-G-bound ribosome complex with P-site initiator tRNAfMet are shown. Two unique labelling schemes were used for smFRET imaging: (A) labelled L1 and P-site tRNA in the presence of 10 μM EF-G and 2 mM GDPNP, and (B) labelled P-site tRNA in the presence of 0.1 μM labelled EF-G and 2 mM GDPNP. (Left panels) Cartoon models of the EF-G-bound complex indicating the dynamic elements and the sites of labelling. (Centre panels) Single-molecule fluorescence (Cy3, green; Cy5, red) and FRET (blue) trajectories. The idealization is overlaid in red on the FRET trace. (Right panels) Histograms indicating the occupancies in the observed FRET states as determined by idealization. Highlighted in red/pink are those states that relate to formation of the unlocked ribosome. ‘N' denotes the number of individual smFRET trajectories included in the analysis.
To examine the extent to which classical (P/P) state occupancy contributes to the prevalence of the low-FRET state, identical experiments were performed on complexes bearing a G2252C mutation in the P-loop of 23S rRNA. This mutation promotes P/E hybrid state formation by destabilizing P-site tRNA interactions with the peptidyltransferase centre (PTC) of the large subunit (Dorner et al, 2006; Munro et al, 2007, 2009a). Destabilization of the classical (P/P) tRNA configuration in this manner only modestly increased dynamics and unlocked state occupancy (Supplementary Figure S5). The low-FRET state remained predominant (∼50%), suggesting additional complexities that may either reflect rapid EF-G binding and dissociation events, or that the low-FRET state is an aggregate of ribosome conformations, consisting of both classical (P/P) and hybrid (P/E) tRNA configurations.
On the EF-G-bound ribosome, P-site tRNA favours the P/E hybrid state but remains dynamic
To directly examine the stability of the EF-G–ribosome complex and the position of P-site tRNA, smFRET experiments were performed on P-site tRNAfMet-labelled ribosome complexes in the presence of site-specifically labelled EF-G. Here, non-natural amino-acid chemistry (Chin et al, 2003) was used to label otherwise native EF-G with Alexafluor647N (A647N) at position 526 within domain IV (see Materials and methods section). Consistent with a long-lived EF-G-ribosome interaction, smFRET trajectories obtained in the presence of 100 nM A647N-labelled EF-G, persisted for ∼4 s on average, limited by A647N photobleaching (Figure 2B and Supplementary Figure S6). Dynamic events, evidenced in the majority of smFRET trajectories, were idealized using a two-state model that showed the system largely occupied a stable, low-FRET (∼0.2) configuration (∼82% occupancy) with short-lived excursions (∼100 ms) to a high-FRET (∼0.65) state (18% occupancy; Table 1).
Structural modelling suggested that the observed dynamics of the EF-G(GDPNP)-bound complex may represent P-site tRNA exchange between hybrid and classical positions. The low-FRET state, corresponding to an estimated inter-dye distance of ∼70 Å, was consistent with cryo-EM models of the ratcheted, EF-G-bound ribosome bearing tRNA in the P/E hybrid state (Figure 1B). Excursions to high FRET, corresponding to an inter-dye distance of ∼50 Å, could be explained by modelling a classical (P/P) tRNA position on the EF-G-bound ribosome (Supplementary Figure S4A). This model was initially tested by preparing similar complexes in which EF-G was stabilized in a post-GTP hydrolysis state, in the presence of GTP and the antibiotic fusidic acid (Rodnina et al, 2001). Consistent with the idea that such complexes should more closely resemble the post-translocation state, in which P-site tRNA occupies a classical position and subunits return to an unratcheted conformation, high-FRET state occupancy increased substantially (32 versus 18%; Supplementary Figure S7; Valle et al, 2003; Gao et al, 2009).
To further test the nature of fluctuations in the EF-G-bound ribosome, point mutations were made in 23S rRNA, predicted to specifically alter the classical-hybrid tRNA equilibrium. Consistent with dynamics stemming from a classical-hybrid exchange of P-site tRNA, a G2252C mutation, which promotes the P/E hybrid state, decreased high-FRET state occupancy in the EF-G(GDPNP) complex by ∼two-fold (Supplementary Figure S8A) (Munro et al, 2007). A C2394A mutation, which disrupts tRNA binding at the large subunit E site to destabilize the P/E hybrid state, increased high-FRET state occupancy by ∼2.5-fold (Supplementary Figure S8C; Walker et al, 2008). These data argue that the dynamics observed between domain IV of EF-G and P-site tRNA represent a persistent hybrid-classical exchange on the EF-G-bound ribosome. Excursions of P-site tRNA to the classical (P/P) state may either reflect transient unratcheting events accompanied by tRNA movement or P-site tRNA movements on the ratcheted ribosome. Although neither model can be excluded in principle, the latter view seems more plausible given that the rates of P-site tRNA motions into, and out of, the classical state (∼2 and ∼13 per s, respectively) are approximately one to two orders of magnitude faster than ratcheting–unratcheting exchange rates previously reported for the EF-G(GDPNP) complex (∼0.02 and ∼1.2 per s for unratcheting and ratcheting, respectively; Cornish et al, 2008).
Two important conclusions about the nature of the EF-G-ribosome interaction can be drawn from these data. First, the lifetime of FRET observed between P-site tRNA and EF-G (⩾4 s), suggests that the EF-G-ribosome interaction is relatively stable. Thus, residual low-FRET state occupancy observed in L1- and P-site tRNA-labelled complexes cannot be explained by cycles of EF-G binding and release. Second, although the P/E hybrid tRNA configuration is >80% occupied while EF-G is bound to the ribosome, the mutagenesis data showing that P-site tRNA remains in dynamic exchange with the classical (P/P) configuration suggests that EF-G motions do not substantially contribute to the dynamic FRET signal. EF-G motions may not be observed either because EF-G's interaction with the ratcheted ribosome is highly stable or because EF-G motions occur much faster than the time scale of imaging.
In the dye-labelled EF-G experiment, P/E hybrid state formation (kP/E≈13 per s) occurs ∼3.5-fold faster than the rate exiting low FRET (klow →) and 13-fold faster that the rate entering the unlocked state (k→ high) observed on equivalent L1- and P-site tRNA-labelled complex (Table 1). In light of EF-G's established propensity to stabilize the P/E hybrid state and subunit ratcheting, these observations suggest that the predominant low-FRET state observed on the later complex corresponds to an aggregate of both locked and P/E hybrid, ratcheted ribosome conformations. Consistent with pre-translocation complex dynamics (Munro et al, 2009a), these data may be explained if L1 stalk motions are largely uncoupled from those of P-site tRNA.
The identity of P-site tRNA influences the stability of the unlocked state
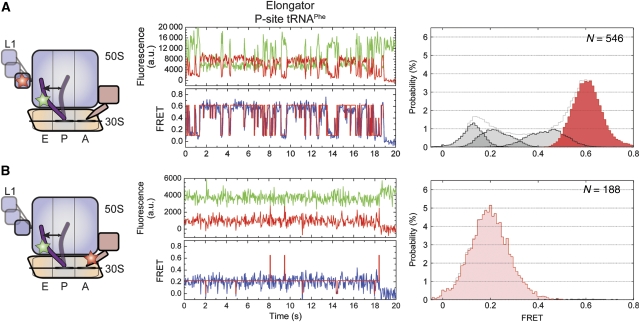
To probe whether the unlocked state is similarly unstable on complexes containing elongator tRNA, analogous experiments were performed on identically labelled L1- and P-site tRNAPhe complexes (see Materials and methods section; Figure 3). Here, both in the presence and absence of EF-G(GDPNP), the FRET states observed had values similar to those identified in complexes containing P-site tRNAfMet (Figure 3A; Supplementary Figure S3B). However, rates of FRET fluctuations and relative occupancies of each FRET state were substantially altered. In the absence of EF-G, these distinctions were relatively modest, in which low- and high-FRET state occupancies were 80 and 7%, respectively, (Supplementary Figure S3B); transition rates out of low- (klow →) and into high-FRET (k→ high) states were ∼2 and ∼0.6 per s, respectively (Table 1 and Supplementary Table S1). However, in the presence of EF-G, the relative occupancies of low- and high-FRET states were inverted from those observed for tRNAfMet to 13 and 57%, respectively (versus 68% low FRET and 14% high FRET; Table 1). Such differences could be attributed to a five-fold increase in the rate out of the low-FRET state (klow →≈17 per s) and an ∼11-fold increase in the rate entering the high-FRET, unlocked state (k→ high≈11 per s; Table 1). Thus, consistent with cryo-EM structures of similar EF-G-bound ribosome complexes (Valle et al, 2003; Connell et al, 2007; Taylor et al, 2007), the unlocked state in the P-site tRNAPhe complex represents the predominant configuration of the system. Yet, similar to the EF-G-bound tRNAfMet complex, this system was also highly dynamic in nature, rapidly and reversibly exiting the unlocked state.
Figure 3.
With tRNAPhe in the P site, the complex has greater occupancy in the P/E hybrid and unlocked states. Here the dynamics of the EF-G-bound ribosome complex with P-site elongator tRNAPhe shown. Two unique labelling schemes were used for smFRET imaging: (A) labelled L1 and P-site tRNA in the presence of 10 μM EF-G and 2 mM GDPNP, and (B) labelled P-site tRNA in the presence of 0.1 μM labelled EF-G and 2 mM GDPNP. (Left panels) Cartoon models of the EF-G-bound complex indicating the dynamic elements and the sites of labelling. (Centre panels) Single-molecule fluorescence (Cy3, green; Cy5, red) and FRET (blue) trajectories. The idealization is overlaid in red on the FRET trace. (Right panels) Histograms indicating the occupancies in the observed FRET states as determined by idealization. Highlighted in red/pink are those states that relate to formation of the unlocked ribosome.
Experiments performed in the presence of site-specifically labelled EF-G showed that distinctions from the tRNAfMet complex could be partially attributed to increased P/E hybrid state stability in the EF-G-bound, P-site tRNAPhe complex (>99% occupied). Transitions to the classical P/P state (∼0.65 FRET) were rarely observed and the rate exiting this state (⩾25 per s) largely exceeded the experimental time resolution (Figure 3B). In line with observations made on the tRNAfMet complex, these data demonstrate that EF-G's propensity to accelerated the dynamics in L1- and P-site tRNA-labelled complexes (Figures 2A and 3A) largely represent dynamics of the L1 stalk on the P/E hybrid, ratcheted ribosome. They also corroborate the idea that the low-FRET state represents an aggregate of ribosome conformations corresponding to both locked (Figure 1A) and partially unlocked (P/E hybrid, ratcheted) ribosome conformations.
EF-G stabilizes a fast dynamic mode of the ribosome
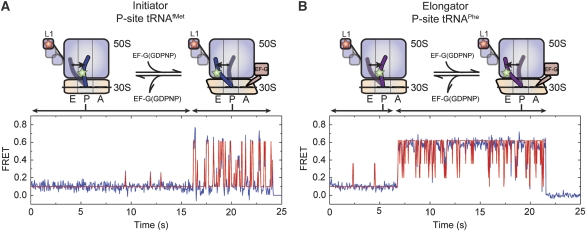
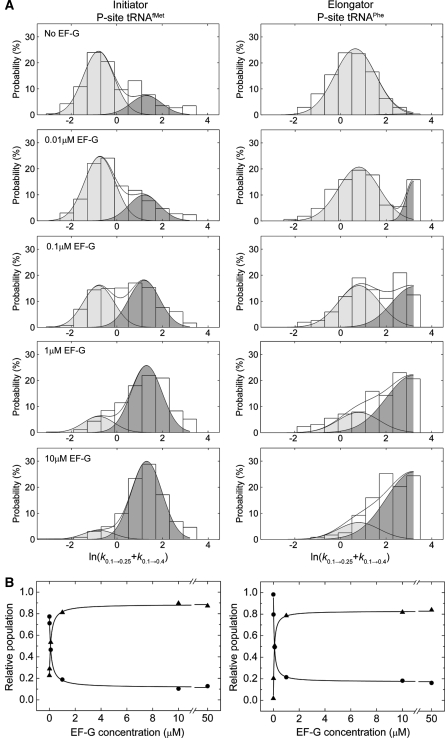
To further explore the nature of the EF-G-ribosome interaction and the EF-G-induced acceleration of ribosome dynamics, the kinetic parameters of motion in L1- and P-site tRNA-labelled ribosome complexes were quantitatively examined in the absence and presence of increasing concentrations of EF-G (0–50 μM; 2 mM GDPNP). Consistent with the intrinsically dynamic nature of both free and EF-G-bound ribosome complexes, as well as a long-lived EF-G-ribosome interaction, a small subset (∼1%) of individual particles were observed to make a clear switch between fast and slow dynamic modes on both P-site tRNAfMet and tRNAPhe complexes (compare Figures 2A, 3A, 4A and B). To quantitatively assess these two distinct dynamic modes, the rate exiting low FRET (klow →) was determined for each individual molecule. In the limit in which mode switching occurs rarely in individual smFRET trajectories, two distinct distributions of transition rates should be observed. Here, the mean rate of each distribution is expected to estimate the rate of L1 stalk closure on the structurally distinct low-FRET state configurations.
Figure 4.
Direct observation of dynamic mode switching observed in smFRET trajectories. Ribosome complexes with labelled L1 and either (A) P-site tRNAfMet or (B) P-site tRNAPhe show dramatic switches in the dynamic mode of the L1 stalk in the presence of intermediate EF-G concentrations (0.1–1 μM). These trajectories represent a minority of molecules (∼1–4%), consistent with slow EF-G binding and release relative to the observation window (∼30sec). FRET traces are shown in blue with idealization, estimated by HMM analysis (Qin, 2004), overlaid in red.
As anticipated by the model, the rates exiting low FRET for both P-site tRNAfMet- and tRNAPhe-bound ribosome complexes exhibited a distinctly bimodal nature at each point in the titration (Figure 5). In the absence of EF-G(GDPNP), transition rates were dominated (∼70–90%) by a slower dynamic mode, in which klow → was ∼0.5 and ∼2 per s for tRNAfMet- and tRNAPhe-bound complexes, respectively. In the presence of saturating EF-G concentrations, transition rates were dominated (∼70–90%) by a faster dynamic mode, in which klow → was ∼4 and ⩾25 per s for tRNAfMet- and tRNAPhe-bound complexes, respectively. The well-defined nature of these distributions is consistent with the occurrence of rare mode-switching events within individual trajectories (limited by the rate of photobleaching), as well as the rates out of low FRET determined in the absence and presence of EF-G (Table 1). Across the EF-G titration, the relative populations of ribosomes exhibiting slow and fast dynamic modes inverted in a concentration-dependent manner, whereas the mean rates of both dynamic modes remained unchanged. For both systems, the subpopulation of ribosome complexes exhibiting a faster dynamic mode saturated at ∼85% at ⩾10 μM EF-G (2 mM GDPNP). Consistent with previous studies (Rodnina et al, 1997; Zavialov and Ehrenberg, 2003), for both systems the midpoint of the EF-G titration was found to be ∼80 nM (Figure 5B).
Figure 5.
EF-G-mediated dynamic mode switching in the ribosome reveals the existence of two distinct low-FRET states. (A) Histograms indicating the distributions of the logarithm of the transition rate out of the low-FRET state (klow →=k0.1 → 0.25+k0.1 → 0.4), calculated for each individual molecule (see Materials and methods section), as a function of EF-G concentration. At each concentration, including in the absence of EF-G, a bimodal distribution of rates is observed that represent slow (light grey) and fast (dark grey) dynamic modes of the ribosome. The populations in each mode were estimated by fitting the distributions to a double Gaussian function (R2>0.9) with mean values μ=−0.8 (s.d., σ=1.0; klow →slow≈0.5 per s) and μ=1.4 (σ=1.0; klow →fast≈4 per s) for the tRNAfMet-bound complex (left panels), and μ=0.8 (σ=1.2; klow →slow≈2 per s) and μ=3.2 (σ=1.2; klow →fast⩾25 per s) for the tRNAPhe-bound complex (right panels). In the case of tRNAPhe, only a portion of the fast dynamic mode distribution was observed due to the short low-FRET state lifetime relative to the experimental time resolution. (B) Plots showing the change in integrated areas of the slow and fast dynamic modes for each system as a function of EF-G concentration. Fitting changes in integrated areas of slow and fast populations to hyperbolic functions provides an estimate of the apparent KD (∼80 nM) for EF-G(GDPNP) binding to the ribosome (R2>0.9).
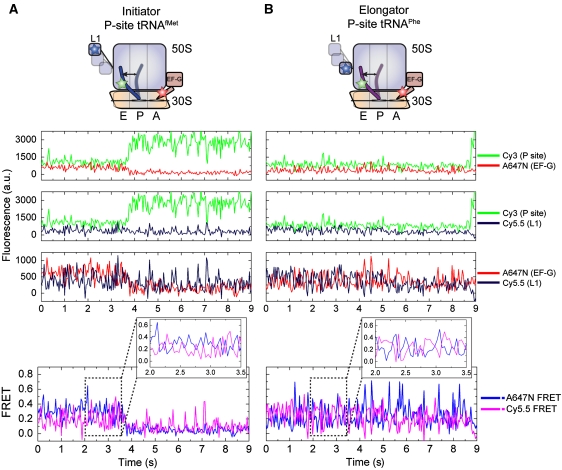
Three-colour FRET experiments display uncoupled motions of P-site tRNA and L1 on the EF-G-bound ribosome
To directly test whether dynamics in the EF-G-bound ribosome are dominated by rapid cycles of L1 stalk closure and opening, three-colour smFRET experiments were performed on complexes bearing Cy3-labelled P-site tRNA and Cy5.5-labelled L1 in the presence of 100 nM A647N-labelled EF-G (2 mM GDPNP; Hohng et al, 2004). Here, the direct illumination of the Cy3 fluorophore on P-site tRNA on the EF-G-bound ribosome is anticipated to yield non-zero FRET efficiencies to both Cy5.5-L1 and A647N-EF-G. In the unlocked state, in which the P/E hybrid state is occupied and the L1 stalk is closed (Figure 1B), FRET efficiency from Cy3-tRNA to Cy5.5-L1 should be substantial, whereas the efficiency from Cy3 to A647N-EF-G should be relatively weak. Non-zero FRET between Cy5.5-L1 and A647N-EF-G is also anticipated in the unlocked state as structural models suggest the closed L1 stalk and domain IV of EF-G are separated by ∼100 Å in this configuration. As shown through previous experiments with fluorescently labelled EF-G, when EF-G is bound to the ribosome, the P/E hybrid state is highly favoured on both P-site tRNAfMet and tRNAPhe complexes and excursions to the classical P/P state are rare. Thus, a relatively stable low-FRET state is expected between Cy3-tRNA and A647N-EF-G. Transitions of the L1 stalk between closed and open configurations on the ratcheted, P/E hybrid state ribosome should be principally characterized by anti-correlated fluctuations in FRET between Cy3-tRNA and Cy5.5-L1, as well as between Cy5.5-L1 and A647N-EF-G. When the L1 stalk is open, FRET between Cy5.5-L1 and A647N-EF-G should be negligible.
The FRET signatures characteristic of rapid L1 stalk opening and closing cycles were observed for both P-site tRNAfMet- and tRNAPhe-bound complexes (Figure 6). Here, the signal-to-noise ratio and the observed FRET values were lower than observed for analogous two-colour FRET experiments owing to changes in optical components required for the experiment (see Materials and methods section). Consistent with two-colour experiments, in both systems fluorescence in line with low-FRET state occupancy was observed between A647N-EF-G and Cy3-tRNA, whereas Cy3-tRNA and Cy5.5-L1 fluorescence signals were more dynamic in nature (Figure 6, fluorescence panels). The existence of FRET between Cy5.5-L1 and A647N-EF-G, revealed by the anti-correlated nature of fluorescence and FRET fluctuations in the in Cy5.5- and A647N-imaging channels (∼−0.7 correlation coefficient versus ∼0.04 after A647N photobleaching; Figure 6, FRET panel), established both the presence of active Cy5.5 and A647N fluorophores, and the highly dynamic nature of both ribosome complexes. Given that the P/E hybrid state was shown to be relatively stable in such complexes (Figures 2B and 3B), such data strongly support the view that the L1 stalk adopts a fast dynamic mode of closure and opening on the EF-G-bound ribosome. Here again, the mean Cy5.5 FRET values suggest that an open L1 stalk configuration is favoured on the tRNAfMet complex, whereas a closed L1 stalk configuration is favoured on the tRNAPhe complex.
Figure 6.
Three-colour smFRET displays uncoupled motions of P-site tRNA the L1 stalk on the EF-G-bound ribosome. smFRET trajectories acquired from ribosome complexes bearing Cy5.5-labelled L1, Cy3-labelled P-site tRNA, in the presence of 100 nM A647N-labelled EF-G(GDPNP). Fluorescence and FRET trajectories are shown for complexes containing (A) P-site tRNAfMet or (B) P-site tRNAPhe. Anti-correlated changes in A647N and Cy5.5 fluorescence, resulting of FRET from A647N to Cy5.5 in the unlocked configuration, verify the presence of both fluorophores.
Discussion
Using an integrated methodological approach, including two- and three-colour smFRET imaging, structural modelling and site-directed mutagenesis, conformational events intrinsic to ribosomes carrying deacylated tRNA in the P site, both in the absence and presence of EF-G, have been shown to occur spontaneously and through largely uncoupled processes. With EF-G stably bound to the A site, the rates of P/E hybrid state formation, L1 stalk closure and formation of the unlocked state were shown to be highly accelerated. Although such systems provide only a model of EF-G's interaction with the pre-translocation complex, these investigations shed important light on the nature and coupling of conformational events underpinning unlocked state formation. This state, minimally requiring P/E hybrid state formation, L1 stalk closure and subunit ratcheting (Valle et al, 2003; Connell et al, 2007; Taylor et al, 2007), serves as a key intermediate in the translocation mechanism formed before the directional movement of paired mRNA–tRNA substrates with respect to the small subunit (Frank et al, 2007; Pan et al, 2007; Walker et al, 2008). An understanding of the mechanism and regulation of events required to achieve this state may, therefore, provide insights relevant to translocation. Such studies may also be important to understanding other translation processes that depend critically on large-scale, and potentially complex, conformational changes in the ribosome, including the mechanisms of translation initiation, termination and recycling (Marshall et al, 2009; Savelsbergh et al, 2009; Sternberg et al, 2009).
As shown for pre-translocation ribosome (Munro et al, 2009a), the complexes investigated revealed that P/E hybrid state formation, L1 stalk closure and subunit ratcheting are interrelated processes, but largely independent in nature. These observations suggest that the process of achieving this state is neither concerted nor defined by a strict sequence of conformational events. Instead, congruent with both induced-fit (Koshland, 1995) and conformational capture mechanisms (Leulliot and Varani, 2001), unlocked state formation may be a low probability event controlled by the relative rates, and reversibility, of distinct conformational degrees of freedom within the ribosome particle and the frequency with which they converge. In this view, although formation of the unlocked state can occur through a concerted process with finite probability, the uncoupled nature of the underlying events instead suggest a multi-step, multi-pathway mechanism. In this light, an important finding of this study, which may be critical to understanding EF-G's role in translocation, is that EF-G(GDPNP) binding to the A site stabilizes a fast dynamic mode of the particle, in which P/E hybrid state formation, L1 stalk closure and unlocked state formation become highly accelerated.
EF-G-induced remodelling of the ribosome's free energy landscape
On the EF-G-bound ribosome P-site tRNA was observed to remain dynamic, sampling both classical (P/P) and hybrid (P/E) states. Here, the relative stabilities of the classical and hybrid P-site tRNA configurations were inverted, and P/E state formation occurred approximately five to seven-fold faster, relative to comparable pre-translocation complexes (Munro et al, 2007, 2009a). These data can be explained by EF-G's known propensity to stabilize a ratcheted ribosome conformation (Valle et al, 2003; Connell et al, 2007; Ermolenko et al, 2007; Taylor et al, 2007; Cornish et al, 2008) and the idea that the organization of elements at the subunit interface and P-site tRNA position are structurally linked (Agirrezabala et al, 2008; Julian et al, 2008). The sensitivity of P-site tRNA motions in the EF-G-bound ribosome to single base-pair mutations in the large subunit (Supplementary Figures S5 & S8), as well as the nucleotide state of EF-G (Supplementary Figure S7), suggests a complex relationship between these two structural processes. However, given that the rates of the hybrid-classical exchange (Table 1) are approximately one to two orders of magnitude faster than subunit ratcheting rates reported for similar systems (Cornish et al, 2008), P-site tRNA excursions between classical and hybrid states can be largely viewed as occurring independently of the subunit ratcheting–unratcheting process.
Although P-site tRNA motions likely contribute meaningfully to the fast dynamics observed in the EF-G-bound ribosome (Figures 2, 3, 4, 5 and 6), investigations in which P/E hybrid state occupancy was highly stabilized (exceeding >98% in complexes containing P-site tRNAPhe) demonstrated that L1 stalk closure and opening dominate the dynamics observed. Such measurements suggest that the rate of L1 stalk closure in the EF-G-bound ribosome may increase by as much as ∼10–20-fold compared with the unbound, vacant A-site complex. Thus, by increasing both the rate of P/E hybrid state formation and L1 stalk closure, EF-G, through either direct or indirect mechanisms, was observed to accelerate the effective rate of unlocked state formation (k→ high) by ∼20–30-fold.
The nature of P-site tRNA alters the metastable energy landscape governing unlocked state formation
As shown for the rates of subunit ratcheting (Cornish et al, 2008) and tRNA dynamics within the A and P sites (Munro et al, 2009a), the observed rates of L1 stalk closure and unlocked state occupancy, both in the absence and presence of EF-G, were strongly dependent on the identity of the P-site tRNA (Table 1). These observations reflect both the exquisite sensitivity of the smFRET measurement and the metastable nature of the ribosome energy landscape, in which even subtle changes in the composition of the ribosome complex can give rise to discernible structural and kinetic signatures (Munro et al, 2009b). Although the molecular origins of these distinctions are presently unclear, one plausible explanation is differences in the stabilities and/or positions of the P/E hybrid state in the tRNAfMet and tRNAPhe complexes. Such distinctions may affect the local environment of the L1 stalk and/or other structural degrees of freedom in the E site. Although the tRNAPhe complex predominantly occupied (57%) the high-FRET, unlocked state, in line with cryo-EM observations (Valle et al, 2003; Connell et al, 2007; Taylor et al, 2007), substantial low-FRET state occupancy was present in the EF-G-bound ribosome for both systems investigated (13–68%). Here, the evidence suggests that the P/E hybrid (Figures 2B and 3B, and Supplementary Figure S5) and ratcheted states (Cornish et al, 2008) predominate (>80–95%). Although the molecular features of this low-FRET state must await structural determination, extended open and/or highly rotated L1 stalk positions on an otherwise unlocked particle may adequately explain these data. Structural modelling showed that the observed FRET value, corresponding to a distance of ∼80 Å, could be satisfied by a simple pivot-like motion of the L1 stalk to an extended open conformation, as seen in a high-resolution structure of the isolated 50S subunit from Dinococcus radiodurans (Harms et al, 2001; Supplementary Figure S4C). Here, modelled L1 stalk motions were hinged near the highly conserved junction of helices H75, H76 and H79 in domain V of 23S rRNA (Cannone et al, 2002; Korostelev et al, 2006; Selmer et al, 2006).
EF-G-independent dynamic mode switching in the ribosome
By directly observing the lifetime of the EF-G-ribosome interaction (⩾4 s) (Supplementary Figure S6), and determining the apparent KD of this interaction (∼80 nM; Figure 5), the on-rate of the EF-G-ribosome interaction, evidenced by a fast dynamic mode of the ribosome, was estimated to be ∼1.5 μM−1 sec−1. Such studies suggest that EF-G forms a high-affinity interaction with the particle at a rate ∼100-times slower than the previously determined bimolecular rate constant of EF-G's initial interaction with the A site (Rodnina et al, 1997). This observation suggests that one or more relatively slow conformational processes limits the rate at which EF-G interacts detectably with the ribosome. Here, observation of a fast dynamic mode in the ribosome in the absence of EF-G suggests that a slow process, intrinsic to the system may principally control the mode-switching event. Given the observation that EF-G stabilizes this state, mode switching may reflect slow, spontaneous subunit ratcheting (Agirrezabala et al, 2008; Cornish et al, 2008; Julian et al, 2008). In this view, rather than explicitly accelerating the rate at which this process occurs, EF-G may instead bind to and stabilize the spontaneously achieved ratcheted conformation. In so doing, binding of EF-G at the A-site increases the probability of unlocked state formation by preventing spontaneous unratcheting (Cornish et al, 2008).
Although the finding that the unlocked state is spontaneously achieved and transient in nature is consistent with the theoretical underpinnings of an unlocked ribosome conformation (Spirin, 2009), understanding how EF-G accelerates unlocked state formation on the pre-translocation ribosome complex remains to be answered. In such complexes, A-site-bound peptidyl-tRNA should prevent domain IV of EF-G from forming stable interactions with the decoding site. Yet, EF-G accelerates the rate of translocation in a manner that is strongly enhanced by GTP hydrolysis (Rodnina et al, 1997; Pan et al, 2007). Pre-steady state measurements of EF-G's initial interactions with the ribosome, in which the order and timing of conformational events are directly measured, will be critical for examining these questions. Such experiments will benefit from distinct labelling strategies that enable the direct monitoring of conformational process in EF-G as well as the ribosome.
Uncoupled conformational events in the ribosome may provide a regulatory mechanism
The spontaneous, multi-step and multi-pathway nature of unlocked state formation has now been shown for both the pre-translocation (Munro et al, 2009a) and EF-G-bound ribosome complex containing both initiator and elongator tRNA substrates in the P site. These findings also hold under both high- and low-magnesium ion concentrations (data not shown). The mechanistic significance of these insights must now be explored. In the model proposed, the convergence of P/E hybrid state formation, subunit ratcheting and L1 stalk closure limit the rate of unlocked state formation. This in turn limits the rate of substrate movement during translocation. Consistent with this model the rates of unlocked state formation observed here (Table 1) and elsewhere (Munro et al, 2009a) correlate with both factor-free translocation rates (Gavrilova et al, 1976; Bergemann and Nierhaus, 1983; Cukras et al, 2003; Fredrick and Noller, 2003) and EF-G-catalysed translocation rates observed for pre-translocation complexes containing either P-site tRNAfMet or tRNAPhe. The mechanistic underpinnings of this correlation must now be further examined. Here, conditions, factors, small-molecules and mutations that stimulate the coupling of these events are predicted to accelerate translocation, whereas those that promote uncoupling are anticipated to inhibit this process.
To this end, it will be critical to examine the role of small-molecule antibiotics known to inhibit the translocation mechanism. Investigations exploring the role of translation factors and mRNA elements that either directly or indirectly interact with the E site to affect the L1 stalk (Blaha and Nierhaus, 2001; Andersen et al, 2006; Schuler et al, 2006; Costantino et al, 2008) may also provide new and unexpected insights. Notably, the stability of the unlocked state seems principally limited by dynamics of the L1 stalk. Factors that regulate L1 stalk motion and/or position may therefore be anticipated to have strong contributions to the translation mechanism (Valle et al, 2003; Schuler et al, 2006; Fei et al, 2008; Cornish et al, 2009). The highly dynamic and loosely coupled nature of L1 stalk motions, and the sensitivity of such motions to conformational and compositional changes in the ribosome, suggests that regulation of the L1 stalk domain may be an effective avenue for achieving translational control. Future investigations exploring how conformational coupling contributes to the cellular regulation of protein synthesis will doubtless be aided by improved strategies for probing conformational changes in the ribosome from multiple structural perspectives simultaneously.
Materials and methods
Preparation of dye-labelled components
Ribosomal protein L1 was cloned into the pET SUMO vector (Invitrogen). On the basis of solvent accessibility and sequence conservation, cysteine residues were introduced through site-directed mutagenesis (Stratagene) at six positions. The mutated His6-SUMO-L1 fusion protein was purified by nickel chromatography and labelled with Cy5–maleimide (GE Healthcare) by incubation for 2 h on ice in a buffer containing 20 mM MES (pH 6.0), 500 mM KCl and 0.1 mM TCEP. Unbound dye was removed by nickel chromatography. The fluorescently labelled protein was further purified by ion exchange chromatography on a MonoS 5/50 GL column (GE Healthcare). The His6-SUMO affinity tag was removed by proteolytic cleavage.
Elongation factor-G was cloned and purified as previously described (Blanchard et al, 2004a). The non-natural amino-acid p-acetyl-L-phenylalanine was introduced at position 526 of EF-G through suppression of an amber stop codon mutated into the EF-G construct by site-directed mutagenesis (Chin et al, 2003). The orthogonal keto group was labelled with A647N hydroxylamine (Invitrogen) by incubation for 2 h on ice in a buffer containing 95 mM succinic acid (pH 4.0), 7 mM KCl and 5 mM BME. The reaction was quenched with 20 mM cyanoborohydride, and subsequently neutralized by the addition of 150 mM Tris (pH 8.5). Unbound dye was removed by nickel chromatography.
All tRNAs were purchased (Sigma), fluorescently labelled at s4U8 with Cy3 maleimide and purified as previously described (Munro et al, 2007).
Purification of ribosome complexes
The 70S ribosomes were isolated from Escherichia coli BL21(DE3) or BL21(DE3) ΔL1 (Munro et al, 2007). ΔL1 ribosomes were reconstituted with Cy5-labelled L1 by incubation with excess protein for 10 min at 37°C, followed by 10 min on ice in Tris Polymix buffer (50 mM Tris–Acetate (pH 7.5), 100 mM KCl, 15 mM Mg(OAc)2, 5 mM NH4OAc, 0.5 mM Ca(OAc)2, 0.1 mM EDTA, 5 mM putrescine, 1 mM spermidine and 5 mM BME). Ribosomes were initiated with fMet-tRNAfMet(Cy3-s4U8) or NAc-Phe-tRNAPhe(Cy3-s4U8) and 5′-biotinylated mRNA as previously described (Munro et al, 2007, 2009a). After surface immobilization of ribosome complexes inside microfluidic reaction chambers (Munro et al, 2007), the P-site tRNA was deacylated by incubation with 2 mM puromycin for 10 min at room temperature.
Five of the six sites of L1 labelling yielded high incorporation efficiency into the ΔL1 ribosomes, and detectable FRET to the P-site tRNA: S55C, S72C, S117C, T173C and T202C. Specific and uniform L1 reconstitution was verified based on structural agreements observed with high-resolution structural models of the ribosome (Korostelev et al, 2006; Selmer et al, 2006), and the similarity in kinetic behaviours observed for each site of labelling. Here, only those results obtained for ribosomes containing L1(Cy5-S55C) are described.
Acquisition and analysis of smFRET data
Equilibrium smFRET experiments were performed in Tris Polymix buffer in the presence of an optimized oxygen-scavenging and triplet state-quenching cocktail (Dave et al, 2009). The Cy3 fluorophore was illuminated under ∼0.5 kW cm−2 intensity at 532 nm (Laser Quantum), in which the signal-to-noise ratio was maximized (>8:1), whereas maintaining acceptable photobleaching rates (∼10–30 s). Movies were acquired in Metamorph (Molecular Imaging) and single-molecule fluorescence trajectories were extracted in Matlab (Mathworks).
From the Cy3 and Cy5 fluorescence trajectories, the efficiency of FRET was calculated according to FRET=ICy5/(ICy3+ICy5). The FRET state occupancies and transition rates were estimated first by idealization of the smFRET trajectories to Markov chain models using the segmental k-means algorithm implemented in QuB (Qin, 2004). The trajectories acquired from complexes with labelled L1 and P-site tRNA were idealized to the four-state model shown in Supplementary Figure S2; data acquired on complexes with labelled EF-G and P-site tRNA were fit to a two-state mode. The distributions of data points assigned to each FRET state were then formed using the idealization (Figures 2 and 3); zero-FRET state distributions are not shown so that the low-FRET state could be more clearly visualized. The details of model selection and kinetic analysis were as described (Munro et al, 2009a). Briefly, the resulting sequence of dwell times was used to optimize the model by maximum likelihood optimization in QuB (Qin et al, 1996). One kinetic model was generated for each trace. The data sets were then split into three groups. For each group, the distributions of log-transformed transition rates were fit to Gaussians. The resulting fits were transformed back to linear space by exponentiation. The mean rates obtain from the three groups are reported in Supplementary Table S2 along with the s.e. values. The complete set of rate constants are summarized by the rates of transition into and out of the low- and high-FRET states, as defined (Table 1; Munro et al, 2009a).
Structural modelling
Modelling of the ribosomal subunits, the L1 stalk, the fluorophores and the tRNAs were previously described (Munro et al, 2009a). The conformation of EF-G was determined by cryo-EM reconstructions of EF-G bound to the ratcheted 70S ribosome, which were obtained from the protein data bank (Valle et al, 2003; PDB accession code ). This was the basis of all-atom modelling, which used the high-resolution structure of EF-G (Hansson et al, 2005; PDB accession code ) to homology model EF-G on the ratcheted ribosome. The final model was docked into the cryo-EM reconstructions.
Supplementary Material
Since the publication of this paper, the authors have noticed that technical issues connected with bulk anisotropy fluorimetry had led to slightly modified values in Supplementary Table 1. These errors have been corrected in this file as of 17 February 2010. The corrections pertain only to control measurements and do not affect the interpretation of the data.
Acknowledgments
We thank Mark Cava (RSP Amino Acids) for his assistance with fluorescently labelling EF-G, and members of the Blanchard laboratory for critical discussions during the writing of this paper. This study was supported by NIH grant 1R01GM079238-01, the Alice Bohmfalk Charitable Trust, and NYSTAR.
Footnotes
The authors declare that they have no conflict of interest.
References
- Agirrezabala X, Lei J, Brunelle JL, Ortiz-Meoz RF, Green R, Frank J (2008) Visualization of the hybrid state of tRNA binding promoted by spontaneous ratcheting of the ribosome. Mol Cell 32: 190–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen CB, Becker T, Blau M, Anand M, Halic M, Balar B, Mielke T, Boesen T, Pedersen JS, Spahn CM, Kinzy TG, Andersen GR, Beckmann R (2006) Structure of eEF3 and the mechanism of transfer RNA release from the E-site. Nature 443: 663–668 [DOI] [PubMed] [Google Scholar]
- Bergemann K, Nierhaus KH (1983) Spontaneous, elongation factor G independent translocation of Escherichia coli ribosomes. J Biol Chem 258: 15105–15113 [PubMed] [Google Scholar]
- Blaha G, Nierhaus KH (2001) Features and functions of the ribosomal E site. Cold Spring Harb Symp Quant Biol 66: 135–146 [DOI] [PubMed] [Google Scholar]
- Blanchard SC, Gonzalez RL, Kim HD, Chu S, Puglisi JD (2004a) tRNA selection and kinetic proofreading in translation. Nat Struct Mol Biol 11: 1008–1014 [DOI] [PubMed] [Google Scholar]
- Blanchard SC, Kim HD, Gonzalez RL Jr, Puglisi JD, Chu S (2004b) tRNA dynamics on the ribosome during translation. Proc Natl Acad Sci USA 101: 12893–12898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannone JJ, Subramanian S, Schnare MN, Collett JR, D'Souza LM, Du Y, Feng B, Lin N, Madabusi LV, Muller KM, Pande N, Shang Z, Yu N, Gutell RR (2002) The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics 3: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin JW, Cropp TA, Anderson JC, Mukherji M, Zhang Z, Schultz PG (2003) An expanded eukaryotic genetic code. Science 301: 964–967 [DOI] [PubMed] [Google Scholar]
- Connell SR, Takemoto C, Wilson DN, Wang H, Murayama K, Terada T, Shirouzu M, Rost M, Schuler M, Giesebrecht J, Dabrowski M, Mielke T, Fucini P, Yokoyama S, Spahn CM (2007) Structural basis for interaction of the ribosome with the switch regions of GTP-bound elongation factors. Mol Cell 25: 751–764 [DOI] [PubMed] [Google Scholar]
- Cornish PV, Ermolenko DN, Noller HF, Ha T (2008) Spontaneous intersubunit rotation in single ribosomes. Mol Cell 30: 578–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornish PV, Ermolenko DN, Staple DW, Hoang L, Hickerson RP, Noller HF, Ha T (2009) Following movement of the L1 stalk between three functional states in single ribosomes. Proc Natl Acad Sci USA 106: 2571–2576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantino DA, Pfingsten JS, Rambo RP, Kieft JS (2008) tRNA–mRNA mimicry drives translation initiation from a viral IRES. Nat Struct Mol Biol 15: 57–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cukras AR, Southworth DR, Brunelle JL, Culver GM, Green R (2003) Ribosomal proteins S12 and S13 function as control elements for translocation of the mRNA–tRNA complex. Mol Cell 12: 321–328 [DOI] [PubMed] [Google Scholar]
- Dave R, Terry DS, Munro JB, Blanchard SC (2009) Mitigating unwanted photophysical processes for improved single-molecule fluorescence imaging. Biophys J 96: 2371–2381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorner S, Brunelle JL, Sharma D, Green R (2006) The hybrid state of tRNA binding is an authentic translation elongation intermediate. Nat Struct Mol Biol 13: 234–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermolenko DN, Majumdar ZK, Hickerson RP, Spiegel PC, Clegg RM, Noller HF (2007) Observation of intersubunit movement of the ribosome in solution using FRET. J Mol Biol 370: 530–540 [DOI] [PubMed] [Google Scholar]
- Fei J, Kosuri P, MacDougall DD, Gonzalez RL Jr (2008) Coupling of ribosomal L1 stalk and tRNA dynamics during translation elongation. Mol Cell 30: 348–359 [DOI] [PubMed] [Google Scholar]
- Frank J, Gao H, Sengupta J, Gao N, Taylor DJ (2007) The process of mRNA–tRNA translocation. Proc Natl Acad Sci USA 104: 19671–19678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredrick K, Noller H (2003) Catalysis of ribosomal translocation by sparsomycin. Science 300: 1159–1162 [DOI] [PubMed] [Google Scholar]
- Gao YG, Selmer M, Dunham CM, Weixlbaumer A, Kelley AC, Ramakrishnan V (2009) The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science 326: 694–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrilova LP, Kostiashkina OE, Koteliansky VE, Rutkevitch NM, Spirin AS (1976) Factor-free (‘non-enzymic') and factor-dependent systems of translation of polyuridylic acid by Escherichia coli ribosomes. J Mol Biol 101: 537–552 [DOI] [PubMed] [Google Scholar]
- Hansson S, Singh R, Gudkov AT, Liljas A, Logan DT (2005) Crystal structure of a mutant elongation factor G trapped with a GTP analogue. FEBS Lett 579: 4492–4497 [DOI] [PubMed] [Google Scholar]
- Harms J, Schluenzen F, Zarivach R, Bashan A, Gat S, Agmon I, Bartels H, Franceschi F, Yonath A (2001) High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell 107: 679–688 [DOI] [PubMed] [Google Scholar]
- Hohng S, Joo C, Ha T (2004) Single-molecule three-color FRET. Biophys J 87: 1328–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horan LH, Noller HF (2007) Intersubunit movement is required for ribosomal translocation. Proc Natl Acad Sci USA 104: 4881–4885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julian P, Konevega AL, Scheres SH, Lazaro M, Gil D, Wintermeyer W, Rodnina MV, Valle M (2008) Structure of ratcheted ribosomes with tRNAs in hybrid states. Proc Natl Acad Sci USA 105: 16924–16927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korostelev A, Trakhanov S, Laurberg M, Noller HF (2006) Crystal structure of a 70S ribosome–tRNA complex reveals functional interactions and rearrangements. Cell 126: 1065–1077 [DOI] [PubMed] [Google Scholar]
- Koshland DE Jr (1995) The key-lock theory and the induced fit theory. Angew Chem Int Ed Engl 33: 2375–2378 [Google Scholar]
- Leulliot N, Varani G (2001) Current topics in RNA–protein recognition: control of specificity and biological function through induced fit and conformational capture. Biochemistry 40: 7947–7956 [DOI] [PubMed] [Google Scholar]
- Marshall RA, Aitken CE, Puglisi JD (2009) GTP hydrolysis by IF2 guides progression of the ribosome into elongation. Mol Cell 35: 37–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews MB, Sonenberg N, Hershey JW (2007) Origins and principles of translational control. In Translational Control in Biology and Medicine, Mathews MB, Sonenberg N, Hershey JW (eds). pp 1–40. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Moazed D, Noller HF (1989) Intermediate states in the movement of transfer RNA in the ribosome. Nature 342: 142–148 [DOI] [PubMed] [Google Scholar]
- Munro JB, Altman RB, O'Connor N, Blanchard SC (2007) Identification of two distinct hybrid state intermediates on the ribosome. Mol Cell 25: 505–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro JB, Altman RB, Tung CS, Cate JHD, Sanbonmatsu KY, Blanchard SC (2009a) Spontaneous formaton of the unlocked state of the ribosome is a multistep process. Proc Natl Acad Sci USA (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro JB, Sanbonmatsu KY, Spahn CM, Blanchard SC (2009b) Navigating the ribosome's metastable energy landscape. Trends Biochem Sci 34: 390–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D, Kirillov S, Cooperman BS (2007) Kinetically competent intermediates in the translocation step of protein synthesis. Mol Cell 25: 519–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peske F, Savelsbergh A, Katunin VI, Rodnina MV, Wintermeyer W (2004) Conformational changes of the small ribosomal subunit during elongation factor G-dependent tRNA–mRNA translocation. J Mol Biol 343: 1183–1194 [DOI] [PubMed] [Google Scholar]
- Qin F (2004) Restoration of single-channel currents using the segmental k-means method based on hidden Markov modeling. Biophys J 86: 1488–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin F, Auerbach A, Sachs F (1996) Estimating single-channel kinetic parameters from idealized patch-clamp data containing missed events. Biophys J 70: 264–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodnina MV, Savelsbergh A, Katunin VI, Wintermeyer W (1997) Hydrolysis of GTP by elongation factor G drives tRNA movement on the ribosome. Nature 385: 37–41 [DOI] [PubMed] [Google Scholar]
- Rodnina MV, Semenkov YP, Savelsbergh A, Katunin VI, Peske F, Wilden B, Wintermeyer W (2001) Mechanism of tRNA translocation on the ribosome. Mol Biol 35: 559–568 [PubMed] [Google Scholar]
- Savelsbergh A, Katunin VI, Mohr D, Peske F, Rodnina MV, Wintermeyer W (2003) An elongation factor G-induced ribosome rearrangement precedes tRNA–mRNA translocation. Mol Cell 11: 1517–1523 [DOI] [PubMed] [Google Scholar]
- Savelsbergh A, Rodnina MV, Wintermeyer W (2009) Distinct functions of elongation factor G in ribosome recycling and translocation. RNA 15: 772–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler M, Connell SR, Lescoute A, Giesebrecht J, Dabrowski M, Schroeer B, Mielke T, Penczek PA, Westhof E, Spahn CM (2006) Structure of the ribosome-bound cricket paralysis virus IRES RNA. Nat Struct Mol Biol 13: 1092–1096 [DOI] [PubMed] [Google Scholar]
- Selmer M, Dunham CM, Murphy FV, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V (2006) Structure of the 70S ribosome complexed with mRNA and tRNA. Science 313: 1935–1942 [DOI] [PubMed] [Google Scholar]
- Sharma D, Southworth DR, Green R (2004) EF-G-independent reactivity of a pre-translocation-state ribosome complex with the aminoacyl tRNA substrate puromycin supports an intermediate (hybrid) state of tRNA binding. RNA 10: 102–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji S, Walker SE, Fredrick K (2009) Ribosomal translocation: one step closer to the molecular mechanism. ACS Chem Biol 4: 93–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegel PC, Ermolenko DN, Noller HF (2007) Elongation factor G stabilizes the hybrid-state conformation of the 70S ribosome. RNA 13: 1473–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spirin AS (2009) The ribosome as a conveying thermal ratchet machine. J Biol Chem 284: 21103–21119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg SH, Fei J, Prywes N, McGrath KA, Gonzalez RL Jr (2009) Translation factors direct intrinsic ribosome dynamics during translation termination and ribosome recycling. Nat Struct Mol Biol 16: 861–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor DJ, Nilsson J, Merrill AR, Andersen GR, Nissen P, Frank J (2007) Structures of modified eEF2.80S ribosome complexes reveal the role of GTP hydrolysis in translocation. EMBO J 26: 2421–2431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung C, Joseph S, Sanbonmatsu K (2002) All-atom homology model of the Escherichia coli 30S ribosomal subunit. Nat Struct Biol 9: 750–755 [DOI] [PubMed] [Google Scholar]
- Valle M, Zavialov AV, Sengupta J, Rawat U, Ehrenberg M, Frank J (2003) Locking and unlocking of ribosomal motions. Cell 114: 123–134 [DOI] [PubMed] [Google Scholar]
- Walker SE, Shoji S, Pan D, Cooperman BS, Fredrick K (2008) Role of hybrid tRNA-binding states in ribosomal translocation. Proc Natl Acad Sci USA 105: 9192–9197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wintermeyer W, Peske F, Beringer M, Gromadski KB, Savelsbergh A, Rodnina MV (2004) Mechanisms of elongation on the ribosome: dynamics of a macromolecular machine. Biochem Soc Trans 32(Pt 5): 733–737 [DOI] [PubMed] [Google Scholar]
- Zavialov AV, Ehrenberg M (2003) Peptidyl-tRNA regulates the GTPase activity of translation factors. Cell 114: 113–122 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Since the publication of this paper, the authors have noticed that technical issues connected with bulk anisotropy fluorimetry had led to slightly modified values in Supplementary Table 1. These errors have been corrected in this file as of 17 February 2010. The corrections pertain only to control measurements and do not affect the interpretation of the data.