Abstract
Fanconi anaemia is a chromosomal instability disorder associated with cancer predisposition and bone marrow failure. Among the 13 identified FA gene products only one, the DNA translocase FANCM, has homologues in lower organisms, suggesting a conserved function in DNA metabolism. However, a precise role for FANCM in DNA repair remains elusive. Here, we show a novel function for FANCM that is distinct from its role in the FA pathway: promoting replication fork restart and simultaneously limiting the accumulation of RPA-ssDNA. We show that in DT40 cells this process is controlled by ATR and PLK1, and that in the absence of FANCM, stalled replication forks are unable to resume DNA synthesis and genome duplication is ensured by excess origin firing. Unexpectedly, we also uncover an early role for FANCM in ATR-mediated checkpoint signalling by promoting chromatin retention of TopBP1. Failure to retain TopBP1 on chromatin impacts on the ability of ATR to phosphorylate downstream molecular targets, including Chk1 and SMC1. Our data therefore indicate a fundamental role for FANCM in the maintenance of genome integrity during S phase.
Keywords: ATR, DNA repair, DNA replication, FANCM, Fanconi anaemia
Introduction
Strict maintenance of replication fork integrity is vitally important for dividing cells in the face of genotoxic stress caused by endogenous and exogenous agents, which can lead to stalled or collapsed replication forks. In metazoans, such DNA damage results in the activation of the phosphoinositide 3-kinase-like kinases (PIKKs) ataxia-telangiectasia mutated (ATM) and ATM and Rad3-related (ATR) (Abraham, 2001; Zou and Elledge, 2003). On activation, ATR and ATM phosphorylate downstream targets such as the checkpoint effector kinases, Chk1 and Chk2, respectively, which allows cells to recover from replicative stress. This not only ensures accurate DNA replication but also prevents genomic instability, a recognized causative factor in tumour development. This is highlighted by several genome instability and cancer-predisposing human diseases, for example the Bloom and Werner syndromes, Nijmegen breakage syndrome, and Fanconi anaemia (FA).
FA is a rare, recessive, genetically heterogeneous syndrome characterized by progressive bone marrow failure, spontaneous chromosomal instability, and predisposition to cancer. At the molecular level, cells from FA patients are hypersensitive to DNA damage-inducing agents that block progression of replication forks (Patel and Joenje, 2007; Wang, 2007). The cloning of 13 FA genes so far has given us an outline of an evolutionarily conserved DNA damage response pathway. Unfortunately, hardly any of the FA proteins contain known sequence motifs implicated in DNA repair and as a consequence we have no clues to their function in this process. Recent identification of a highly conserved gene in the FA pathway, FANCM, could shed some light on this important issue. This is because the FANCM protein contains a highly conserved amino (N) terminal helicase domain and a carboxy (C) terminal nuclease-like domain, which imply that it might participate directly in DNA repair (Meetei et al, 2005; Mosedale et al, 2005). This domain arrangement is identical to that of the archaeabacterial FANCM orthologue Hef, in which the activities of both domains are coordinated to unwind and cleave a replication fork (Komori et al, 2004).
In line with this, biochemical characterization of human FANCM and its orthologues in Schizosaccharomyces pombe (Fml1) and Saccharomyces cerevisiae (Mph1) revealed a range of activities for these proteins that suggests involvement in processing replication forks, including replication fork regression, Holliday junction branch migration, and D-loop dissolution (Prakash et al, 2005; Gari et al, 2008; Sun et al, 2008). Furthermore, genetic analysis in S. pombe indicates that Fml1 promotes replication fork restart and, similarly to avian FANCM, suppresses sister chromatid recombination (Sun et al, 2008; Rosado et al, 2009). In addition, the C-terminal part of FANCM binds to a homologue of the XPF family of endonucleases, FAAP24, and this interaction promotes FANCM localization to chromatin in vivo and its recognition of replication fork-like DNA structures in vitro (Ciccia et al, 2007; Kim et al, 2008). These observations have led to the proposal that FANCM moves along DNA, sensing, signalling, and then restarting stalled replication forks. The discovery that human and avian cells deficient for FANCM are sensitive to camptothecin (CPT) and UV, both known to interfere with replication, further supports this line of thinking (Bakker et al, 2009; Rosado et al, 2009; Singh et al, 2009). Although this is an attractive hypothesis, the in vivo evidence in support of such a model in vertebrates is still limited.
Recently, it has been shown that FANCM plays a role in establishing the intra-S-phase checkpoint through interactions with ATR and hCLK2, independently of the FA core complex (Collis et al, 2008). ATR is the principal kinase required to activate the cellular response to replication stress (Cortez et al, 2001), and hCLK2 has been shown to be essential for ATR stability and that of its downstream target Chk1 (Collis et al, 2007; Takai et al, 2007). These findings, together with the biochemical activities of FANCM and the observation that FANCM co-purifies with RPA (Ciccia et al, 2007), raise the intriguing possibility that the protein may promote replication fork stability and establishment of the intra-S-phase checkpoint by bringing together the components required for ATR activation and efficient stimulation of ATR-mediated checkpoint signalling.
The work presented here identifies a novel function for FANCM in promoting replication fork restart and ATR-mediated signalling. Interestingly, these functions of FANCM can be dissociated from its role within the FA pathway.
Results
Global replication fork dynamics are altered in FANCM-deficient cells
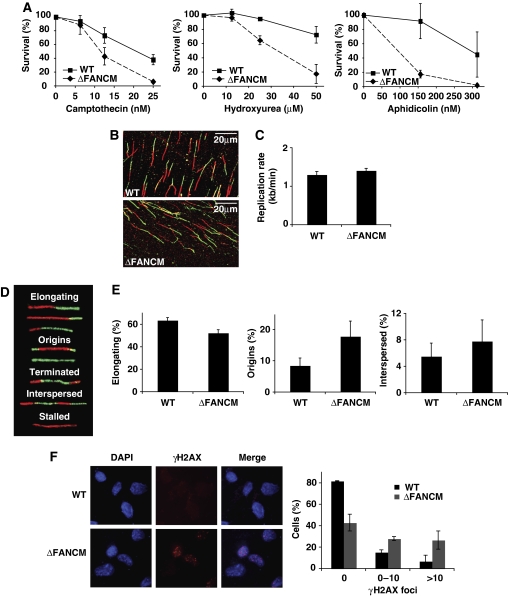
Human, mouse, and chicken cells defective for FANCM show a high level of genomic instability and are hypersensitive to agents that interfere with DNA replication (Figure 1A) (Bakker et al, 2009; Rosado et al, 2009; Singh et al, 2009). In addition, mouse and chicken cells also show high rates of spontaneous sister chromatid exchange (SCE) events suggesting that the protein must, at least in these organisms, somehow act to prevent crossover formation (Sun et al, 2008; Rosado et al, 2009). It is thought that some forms of HR-mediated repair of collapsed replication forks result in such crossovers, giving rise to spontaneous SCEs (Saleh-Gohari et al, 2005). As both spontaneous SCEs and hypersensitivity to replication inhibitors could be attributed to events at blocked replication forks (Lambert et al, 2005), we set out to determine the role of FANCM during DNA replication. The firing of replication origins, replication fork progression, and recovery from stalled forks can be visualized at the single molecule level by the DNA fibre technique.
Figure 1.
FANCM-deficient cells are hypersensitive to agents interfering with replication, display altered global DNA replication, and accumulate spontaneous DNA damage. (A) Cellular sensitivity of WT and ΔFANCM cells to camptothecin (CPT), hydroxyurea (HU), and aphidicolin (APH) as measured by MTS survival assay. Mean values of three independent experiments are shown ±s.e.m. (B) Representative images of actual fibres from WT and ΔFANCM cells. (C) Replication fork velocity in the WT DT40 and ΔFANCM cells used in this study. A minimum of 200 tracts were measured per experiment. Mean values for three independent experiments are shown ±s.e.m. (D) Five classes of replication structures and (E) the relative frequency of occurrence of these different classes. A minimum of 200 fibres were scored per experiment. Mean values of three independent experiments are shown ±s.e.m. (F) γ-H2AX foci in untreated WT and ΔFANCM cells. Left panel shows representative images of γ-H2AX foci. Right panel shows quantification of percentage cells exhibiting γ-H2AX foci out of more than 200 cells per experiment. Bars represent mean values from three independent experiments ±s.e.m.
We first monitored replication in WT DT40 and FANCM-deficient cells in unperturbed S phase using asynchronous log-phase cultures pulse-labelled with iododeoxyuridine (IdU) followed by chlorodeoxyuridine (CldU) (Figure 1B; Supplementary Figure 1A) (Jackson and Pombo, 1998; Zachos et al, 2005; Petermann et al, 2006). The use of an asynchronous cell population avoids any artefacts introduced by synchronization procedures. Under these conditions, WT and FANCM−/− cells showed almost identical fork velocity with an average fork progression rate of 1.3 kb per minute, in agreement with earlier work (Figure 1C; Supplementary Figure 1A) (Maya-Mendoza et al, 2007). These data suggest that, on a global scale, FANCM deficiency does not impact on the processivity of replication polymerases. The double labelling protocol used in this experiment allows one also to define five major patterns of replication structures, namely, elongating forks—category 1; new initiation events (origin firing)—category 2; termination events—category 3; interspersed origins—category 4; and stalled/collapsed forks—category 5 (Figure 1D) (Conti et al, 2007; Maya-Mendoza et al, 2007). Using this classification, we noticed that FANCM mutant cells showed a small but reproducible difference in categories 1, 2, and 4 (Figure 1E). As we observed a difference in structures that are associated with replisome instability, it is conceivable that FANCM−/− cells accumulate increased levels of spontaneous DNA breaks during replication. Phosphorylation of H2AX (γ-H2AX) on S139 is a well-characterized early marker of DNA damage and after exposure to genotoxic stress, γ-H2AX foci are detectable at sites of DNA damage (Foster and Downs, 2005). Log-phase untreated WT and FANCM−/− cells were stained with anti-γ-H2AX antibody and the number of γ-H2AX foci was analysed. We found that the number of foci was significantly higher in FANCM−/− cells when compared with WT cells (Figure 1F); not surprisingly, these γ-H2AX foci colocalized with sites of ongoing DNA replication, indicating that they most likely denote stalled/collapsed forks (Supplementary Figure 1B). This is in line with a recent report by Collis and colleagues who also showed increased γ-H2AX staining in HeLa cells treated with siRNAs targeting FANCM (Collis et al, 2008).
Taken together, these data support a role for FANCM during DNA replication and suggest that in FANCM-deficient cells replication forks are more prone to stalling/collapse leading to spontaneous DNA damage as shown by γ-H2AX staining. This is more than likely reflected in the decreased number of elongating forks and the simultaneous increased frequency of new initiation events and closely spaced origins (Figure 1E; categories 1, 2, and 4).
FANCM is required to prevent activation of latent origins under conditions of replicative stress
Given that the replication pattern was altered in untreated FANCM−/− cells, a logical expectation from this result would be that replication stress would exacerbate these effects. Therefore, to obtain a deeper insight into FANCM's role in replication, we characterized how replication stress influences the global dynamics of replication in FANCM−/− cells.
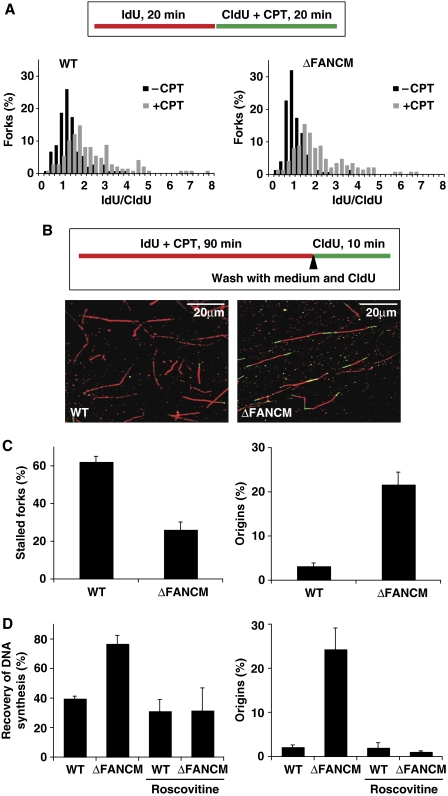
First, we analysed the length of newly replicated DNA tracts in cells exposed to CPT, a well-characterized agent that blocks replication. CPT-induced DNA damage occurs during DNA replication when replication forks collide with trapped Top1 cleavage complexes (Seiler et al, 2007). Log-phase cells were pulse-labelled for 20 min with nucleotide analogues, and CPT was added for 20 min during the second pulse (Figure 2A). CPT treatment caused a two-fold reduction in average fork velocity in both WT and FANCM−/− cells (Figure 2A), indicating that the replication elongation checkpoint in response to replication stress is intact (Conti et al, 2007). This is in marked contrast to Chk1-deficient DT40 cells (Zachos et al, 2005; Petermann et al, 2006; Maya-Mendoza et al, 2007). Interestingly, this was accompanied by an increase in the number of new initiation events in FANCM−/− cells (data not shown). As FANCM−/− cells could properly respond to CPT treatment by downregulating DNA synthesis and FANCM has been reported to function in ATR-mediated checkpoint signalling (Collis et al, 2008), we entertained the idea that increased replication fork instability in this cell line might be due to a problem in facilitating restart of replication forks. To test this hypothesis, we used a modified double labelling protocol that allows study of fork recovery after removal of replication stress (Figure 2B). CPT was added with the first pulse and cells were incubated for 90 min (this treatment did not result in significant cell death as measured by the level of annexin V-positive cells (an early marker of apoptosis) (Supplementary Figure 2A)). After incubation, the first label and CPT were washed out and fork recovery was assessed by labelling replicons with the second label for 5, 10, and 20 min. Under such conditions, the forks that are able to resume DNA synthesis incorporate the second label and appear as red-green tracts (Figure 2B). In WT cells, a majority of forks were unable to resume DNA synthesis after these release times (Figure 2C; Supplementary Figure 2B). In striking contrast to this, we found that FANCM-deficient cells showed that only 25% of forks were unable to resume DNA synthesis after 10 min (P<0.00004; Figure 2C; Supplementary Figure 2B). Interestingly, this very efficient fork recovery was accompanied by a more than six-fold increase in new initiation events (P<0.0008; Figure 2C; Supplementary Figure 2C). A similar pattern of response was also seen on exposure to hydroxyurea (data not shown), suggesting a more general role for FANCM in response to various types of replicative stress. This was an unexpected result and we could envisage at least two possible explanations for this: (i) FANCM−/− cells efficiently restart forks through HR-mediated repair, or (ii) these cells rescue terminally collapsed replication forks by a fork arriving from an adjacent origin. Given that we did not observe a significant increase in Rad51 foci (a marker of HR) after CPT treatment in FANCM-deficient cells (Supplementary Figure 3), and that the processivity of DNA polymerases implicated in HR is relatively slow, we favoured the latter hypothesis. In this scenario, DNA tracts emanating from a neighbouring origin(s) activated in the vicinity of a stalled/collapsed fork should produce detectable DNA tracts. In support of this, it has been reported earlier that dormant licensed origins can be used under conditions of replicative stress and there are at least 10 times more MCM2–7 molecules loaded onto DNA in G1 than there are active replication origins (Ge et al, 2007; Ibarra et al, 2008).
Figure 2.
Analysis of replication fork dynamics and restart in CPT-treated WT and ΔFANCM cells. (A) Top—schematic of the fibre labelling experiment, below—replication fork rates shown as frequency distribution in WT DT40 and ΔFANCM cells treated with CPT for 20 min during the second labelling period. The x axis represents the ratio between the tract length of the first label (IdU) divided by the tract length of the second label (CldU). (B) Top—schematic of the fibre labelling experiment for analysis of replication fork recovery and representative images of actual fibres from WT and ΔFANCM cells, below—effect of 90 min exposure to CPT on replication fork activity in the absence (C) and presence of roscovitine (D). The bars represent mean values from three independent experiments ±s.e.m.
To test whether FANCM−/− cells are able to fire new origins to rescue DNA replication, we carried out an experiment in which the recovery of DNA synthesis (‘fork recovery') after replicative stress was assessed in the presence and absence of an inhibitor of origin firing. For this we used roscovitine, a Cdk inhibitor that prevents origin activation. Roscovitine had no significant effect on the efficiency of origin firing or fork recovery in WT DT40 cells (Figure 2D; Supplementary Figure 4), but in striking contrast to our previous observations, FANCM−/− cells showed significantly reduced recovery of DNA synthesis accompanied by a significant drop in new initiation events (P<0.04; Figure 2D; Supplementary Figure 4). Therefore, we conclude that the significantly increased replication fork recovery seen in this mutant is due to increased firing of neighbouring dormant origins.
Recovery from replication stress in FANCM-deficient cells is caffeine sensitive but ATM- and Chk1 independent
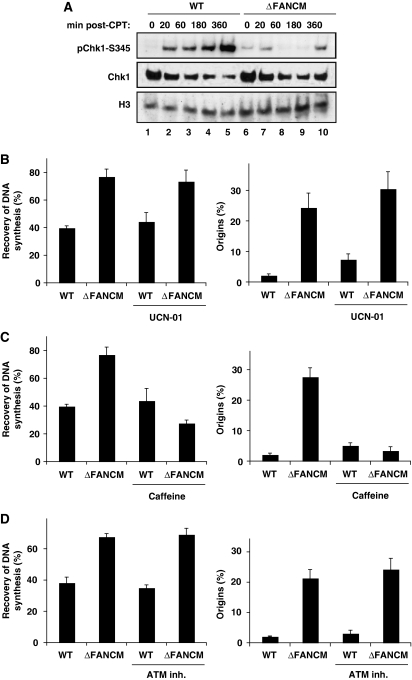
Human and chicken cells defective for Chk1 show multiple intra-S-phase checkpoint defects including a prolonged S phase, increased origin usage, and an inability to resume DNA synthesis on release from replication blocks (Feijoo et al, 2001; Zachos et al, 2005). Recently, human cells depleted of FANCM by siRNAs were shown to be defective in Chk1 phosphorylation in response to replication stress (Collis et al, 2008), and this is also the case in our FANCM-deficient DT40 cell line (Figure 3A). Intriguingly, however, in DT40 Chk1 phosphorylation is compromised but not abolished altogether (compare lines 5 and 10). This is a consistent feature of FANCM−/− cells as the delay, but not loss, of Chk1 phosphorylation is also seen after hydroxyurea treatment (data not shown). Of note is the observation that unperturbed FANCM−/− cells show weak phosphorylation of Chk1 compared to WT cells (Figure 3A, lines 1 and 6), implying that the spontaneous DNA damage present in FANCM−/− cells might be sufficient to provoke low levels of Chk1 activation. Nevertheless, FANCM-deficient cells were unable to effectively stimulate Chk1 phosphorylation in response to replication stress.
Figure 3.
Recovery of DNA synthesis and suppression of excess origin firing in ΔFANCM cells is caffeine sensitive but ATM- and Chk1 independent. (A) Western blot showing levels of Chk1 phosphorylation using a phospho-specific antibody to phosphorylated serine 345 (Chk1-S345) on extracts from WT DT40 and ΔFANCM cells treated with CPT for the indicated periods of time. Histone H3 was used as a loading control. Analysis of recovery of DNA synthesis and activation of latent origins in WT DT40 and ΔFANCM cells treated for 90 min with CPT in the presence and absence of indicated inhibitors (B) UCN-01; (C) caffeine and (D) ATM inhibitor. The bars represent mean values from three independent experiments ±s.e.m.
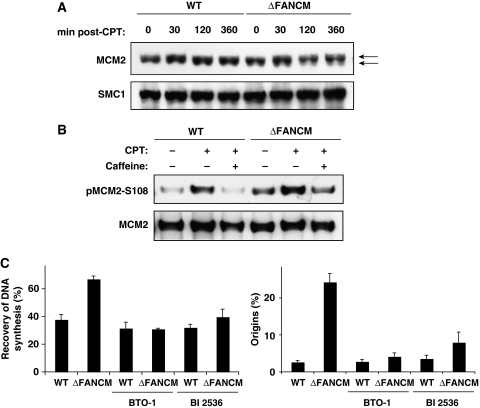
A simple explanation for our results would be that the defect in Chk1 activation in FANCM−/− cells would relieve inhibition of latent origin firing and thus lead to the phenotype described above. To verify whether this was indeed the case, we analysed replication fork recovery in the presence of UCN-01—a potent and selective Chk1 inhibitor widely used in checkpoint studies (Graves et al, 2000). As expected, UCN-01 increased latent origin firing in WT cells, thus confirming that the dose used overrides Chk1-dependent origin control. In FANCM−/− cells the inhibitor slightly potentiated the level of new initiation events in addition to the increase described above (Figure 3B), indicating that the rescue of DNA synthesis in this cell line is most likely mediated by origins that are either fired stochastically or are under the control of Chk1-independent pathway(s).
Although the mechanisms by which dormant origins are activated are still elusive, the existence of an ATR-dependent but Chk1-independent pathway for origin control has been described earlier in Xenopus (Luciani et al, 2004). Considering the limitations of the fibre technique used in this study we could exclude the ‘stochastic' model if the firing of new origins in FANCM mutant cells was, in fact, inhibited in the absence of ATR/ATM kinase activity. As complete ablation of ATR activity is lethal in vertebrate cells and an ATR-specific inhibitor is not yet available (Abraham, 2001; Cortez et al, 2001), we used caffeine, a broad-spectrum inhibitor of PIKKs, including ATR and ATM, to investigate this. In WT cells, caffeine increased origin firing (Figure 3C), in line with its role as an ATR/ATM inhibitor. In striking contrast to this, the presence of caffeine completely abolished excess origin firing in FANCM-deficient cells leading to an increase in replication forks unable to resume DNA synthesis (P<0.007; Figure 3C). Since both ATR and ATM have been shown to regulate origin usage (Shechter et al, 2004), we wished to exclude the possibility of ATM involvement in this process. To do this, we carried out experiments as above but this time using the ATM-specific inhibitor 2-morpholin-4-yl-6-thianthren-1-yl-pyran-4-one. Inhibition of ATM had no effect on origin firing or fork recovery in either WT or FANCM−/− cells (Figure 3D). Therefore, we conclude that the rescue of DNA synthesis in FANCM-deficient cells is caffeine sensitive and most likely carried out by ATR-dependent but Chk1-independent firing of new origins.
In summary, our data show the existence of a novel intrinsic pathway in vertebrates that actively targets origins of replication in the presence of irreparable replication forks. This mechanism promotes initiation of dormant origins in the vicinity of stalled/collapsed forks to ensure completion of DNA replication.
FANCM is required for efficient activation of ATR by promoting TopBP1 loading on chromatin
An unexpected observation reported above is the ability of FANCM-deficient cells to support a low level of Chk1 phosphorylation in unperturbed cells and the defect (but not abrogation) of this response under replicative stress. The current model for Chk1 activation is that in response to DNA damage or replication fork stalling, Chk1 is phosphorylated by ATR on Ser317 and Ser345 in human cells (Zhao and Piwnica-Worms, 2001). ATR is the main kinase to be activated as a result of replicative stress, however it also becomes activated in response to DSBs, albeit with delayed kinetics because of the time required for resection of DSBs and generation of stretches of single-stranded DNA (ssDNA) (Abraham, 2001; Shiotani and Zou, 2009). On the basis of the above, we entertained the idea that in FANCM−/− cells replication forks may collapse more easily, generating DSBs. These then have to be first processed in an ATM-dependent manner before providing an ATR-activating signal. If this was indeed the case, it would provide a plausible explanation for the delayed Chk1 phosphorylation in our FANCM mutant. To test this hypothesis, we examined the kinetics of DSB formation in CPT-treated WT and FANCM−/− cells by pulse field gel electrophoresis (PFGE). In both cell lines increasing doses of CPT induced DSBs; however, we were unable to detect any excess of DSB formation in FANCM−/− cells compared with WT DT40 (Supplementary Figure 5). Consistent with this, FANCM−/− cells also showed a similar number of Rad51 foci on CPT treatment (Supplementary Figure 3) and ATM inhibitor had no effect on recovery of DNA synthesis or firing of new origins (Figure 3D). Therefore, we conclude that the delayed Chk1 phosphorylation in FANCM−/− cells is unlikely to be related to the DSB processing required for generation of ssDNA and thus activation of ATR.
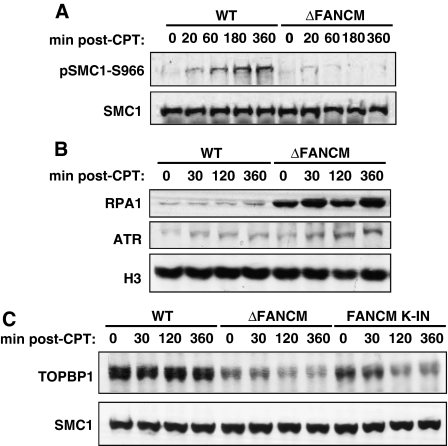
To obtain a deeper insight into the role of FANCM in ATR activation we set out to determine the impact of FANCM disruption on phosphorylation of its downstream targets in addition to Chk1. Claspin selectively abrogates phosphorylation of Chk1 with no effect on other ATR targets; therefore, checking these other targets would allow us to position FANCM in the pathway for Chk1 activation upstream or downstream of claspin. The prediction was that if FANCM functioned downstream of claspin, phosphorylation of other ATR targets in ΔFANCM cells should not be affected. In response to replication stress, SMC1 phosphorylation has been shown to be ATR dependent but Chk1 independent (Liu et al, 2006). Strikingly, the phosphorylation of SMC1 was significantly attenuated in FANCM−/− cells compared to WT DT40 (Figure 4A). SMC1 is also a target for ATM-dependent phosphorylation in response to DSBs (Shiloh, 2003). As mentioned above, we could not detect any excess of DSBs in FANCM-deficient cells, but to exclude a role for ATM in this process we also assessed SMC1 phosphorylation in WT and FANCM−/− cells in the presence of ATM inhibitor. As expected, addition of the inhibitor had little effect on SMC1 phosphorylation (data not shown).
Figure 4.
FANCM is required for efficient activation of ATR in response to CPT by promoting TopBP1 loading on chromatin. (A) Western blot showing phosphorylation status of SMC1 in whole cell extracts from WT and ΔFANCM DT40 cells treated for the indicated times with 1 μM CPT. Phosphorylated SMC1 was detected using a phospho-specific antibody raised against human SMC1 phosphorylated at serine 966 (SMC1-S966). Total SMC1 was used as a loading control. (B) Western blot showing RPA and ATR loading onto chromatin in response to 1 μM CPT treatment in WT and ΔFANCM DT40 cells. Histone H3 was used as a loading control. (C) Western blot for chromatin loading of TopBP1 in WT, ΔFANCM, and FANCM K-IN (knockin) cells after treatment with CPT with SMC1 as the loading control.
In summary, our results suggest that in response to CPT, SMC1 is targeted for ATR-dependent phosphorylation and this event is significantly attenuated in the absence of FANCM. Intriguingly, they also point to a possible role for FANCM in the direct regulation of ATR activity.
Full ATR activation requires a number of cofactors. These include its partner protein ATRIP, the Rad9-Hus1-Rad1 (9-1-1) clamp complex, and TopBP1. All these factors are recruited to sites of DNA damage or replication stress through interactions with RPA-coated ssDNA. As we observed defects in phosphorylation of ATR targets in FANCM-deficient cells, we wished to investigate at what point in the ATR activation process FANCM acts.
FANCM has been shown to bind and promote remodelling of DNA structures resembling stalled replication forks in vitro and it is predicted that in vivo such activities may contribute to generation of RPA-ssDNA (Sogo et al, 2002). On the basis of this, we speculated that perhaps the fork remodelling activity of FANCM is required to generate the RPA-ssDNA that is the initial ATR-activating signal. To test this hypothesis, we probed for the presence of RPA in chromatin fractions from CPT-treated WT and FANCM-deficient cells. WT DT40 cells showed time-dependent accumulation of RPA on chromatin (Figure 4B). The same experiment carried out with FANCM−/− cells showed a marked difference: there was significantly more RPA present on chromatin. We therefore conclude that FANCM limits the accumulation of RPA-ssDNA in response to replication stress.
Although these data are inconsistent with our hypothesis, they are in line with an earlier study reporting an elevated level of RPA-ssDNA in FANCM-depleted HeLa cells (Collis et al, 2008). We speculated therefore that FANCM might be required to stabilize the interaction of ATR-ATRIP with RPA-ssDNA. To test this hypothesis, we looked at the impact of FANCM loss on levels of ATR loading on chromatin in cells exposed to CPT. In both cell lines we could detect ATR on chromatin but with slightly higher levels of ATR present on chromatin in FANCM-deficient cells (Figure 4B). This correlated with the increase in chromatin-bound RPA observed in these cells. We therefore conclude that FANCM ablation does not impact on the recruitment of ATR to damaged chromatin.
Another explanation of the ATR signalling defects we observed would be that FANCM-deficient cells are unable to stimulate recruitment and/or retention of ATR co-activators on chromatin. Depletion of TopBP1 inhibits the damage-inducible phosphorylation of ATR substrates (Liu et al, 2006) because TopBP1 directly stimulates the kinase activity of ATR (Kumagai et al, 2006). The mechanisms that regulate TopBP1 access to ATR are not very well understood but may include the recruitment of TopBP1 to sites of replication stress through its interaction with the 9-1-1 clamp, or with the leading strand DNA polymerase α (Delacroix et al, 2007; Yoo et al, 2007; Yan and Michael, 2009). This could also be tested by analysing the kinetics of chromatin accumulation for the 9-1-1 complex or TopBP1 upon replication stress. Unfortunately, after an extensive screen for antibodies raised against human Rad9, Rad1, and Hus1 we failed to find one that cross-reacts with the chicken proteins. We did, however, succeed in obtaining an antibody that recognizes chicken TopBP1.
Probing for the presence of TopBP1 in chromatin fractions allowed us to test the above-mentioned hypothesis. In WT cells, we observed a significant amount of TopBP1 on chromatin (Figure 4C). The same experiment carried out with FANCM−/− cells showed a marked defect: there was much less TopBP1 present on chromatin, and moreover, cells lacking FANCM could not retain TopBP1 on chromatin after CPT treatment (Figure 4C). Interestingly, chromatin retention of TopBP1 required the ATPase activity of FANCM, as cells expressing ATPase dead FANCM (Rosado et al, 2009) are also deficient in TopBP1 retention on chromatin (Figure 4C). This indicates that FANCM most likely contributes to Chk1 and SMC1 phosphorylation by stimulating ATR activity towards its targets through promoting TopBP1 retention on chromatin.
Polo-like kinase-1 activity is required to activate dormant origins in FANCM mutant cells during exposure to CPT
An interesting observation reported in this study is that FANCM-deficient cells cannot restart stalled replication forks but they instead fire dormant origins to finish replication. Our experiments with caffeine and the ATM inhibitor strongly argue that this is an active process that is caffeine sensitive and most likely requires ATR activity. On the other hand, we showed that FANCM disruption impacts on ATR activity. How could we reconcile this apparently contradictory data? In search for clues regarding the molecular requirements for this process we surveyed the recent literature on the role of ATR in replication checkpoint and controlling origin firing.
Intriguingly, recent work in the Xenopus system has shown that Plx1 (the Xenopus orthologue of human and chicken polo-like kinase-1 (PLK1)) has an important function in the modulation of the checkpoint that controls origin firing under replicative stress (Trenz et al, 2008). In this system, Plx1 binds to chromatin in the presence of stalled forks and alleviates the suppression of the intra-S-phase checkpoint that regulates origin firing. This in turn allows for replication from origins that otherwise would be kept suppressed. Essential for this function is the ATR-dependent phosphorylation of serine 92 of MCM2 to create a docking site for PLK1 (Trenz et al, 2008). In light of this and the data presented above, we speculated that a similar ATR/PLK1-dependent mechanism might function in avian cells. As this process is dependent on ATR-mediated phosphorylation of MCM2 we first checked whether MCM2 was also being phosphorylated in our FANCM−/− strain on replicative stress. In FANCM−/− cells, we could detect a shift in MCM2 mobility by SDS–PAGE in response to CPT treatment, suggesting that under these conditions the protein was modified by phosphorylation, but this modification was not present in WT DT40 cells (Figure 5A). To confirm this, we used a phospho-specific antibody raised against human MCM2 phosphorylated on serine 108 (the equivalent of Xenopus serine 92 and chicken serine 95) in WT and FANCM−/− cells treated with CPT (Figure 5B). As expected, FANCM−/− cells showed increased levels of phosphorylated MCM2, and in addition this phosphorylation was significantly reduced in the presence of caffeine (Figure 5B), indicating that this modification was ATR dependent. This was strongly suggestive that in DT40 a mechanism similar to that described in Xenopus might be in place.
Figure 5.
ATR and PLK kinases control recovery from replicative stress in ΔFANCM cells. (A) Western blot showing MCM2 levels in chromatin fractions derived from WT DT40 and ΔFANCM cells treated with CPT for the indicated periods of time. The arrows indicate the band shift resulting from MCM2 phosphorylation after CPT treatment. Histone H3 was used as a loading control. (B) MCM2 immunoprecipitated from WT and ΔFANCM was blotted for phosphorylation at serine 108 (pMCM2-S108) in cells treated with or without CPT and caffeine. (C) Effect of PLK1 inhibitors on recovery of DNA synthesis and activation of latent origins in WT DT40 and ΔFANCM cells treated for 90 min with CPT in the presence and absence of indicated inhibitors. The bars represent mean values from three independent experiments ±s.e.m.
A simple prediction of this model would be that PLK1 inhibition in FANCM-deficient cells will reduce the origin firing and, consequently, increase the level of stalled forks to the level observed in WT cells. As PLK1 is an essential gene and mouse knockouts are embryonic lethal we decided to inhibit PLK1 by using two well-characterized small molecule inhibitors: BI2536 and benzothiazole-N-oxide (Lansing et al, 2007). Importantly, the use of two independent inhibitors should allow us to rule out any off-target effects of PLK1 inhibition.
To establish a role for PLK1 in controlling excess origin firing in FANCM mutant cells, we analysed replication fork recovery in the presence and absence of PLK1 inhibitors. Under these conditions, the inhibitors had little or no effect on origin firing and fork recovery in WT DT40 cells (Figure 5C). The same experiment carried out with FANCM−/− cells showed a marked difference: in this case both inhibitors blocked the excess origin firing (P<0.006; Figure 5C), reducing the level of fork recovery to the level observed in the WT cells. These data suggest that inhibition of PLK1 activity in FANCM-deficient cells would increase their sensitivity to CPT, and indeed, this is what we observed when we measured the sensitivity of WT and FANCM−/− cells to CPT in the presence and absence of PLK1 inhibitors (Supplementary Figure 6). We therefore conclude that PLK1 is required to activate dormant origins of replication in FANCM-deficient cells to ensure completion of DNA replication.
Taken together, these data suggest that in the absence of FANCM, stalled replication forks cannot be restarted by conventional mechanisms. Instead, to ensure completion of replication, cells use a novel ATR- and PLK1-dependent pathway. As yet, we do not know the precise molecular mechanisms involved, but it is tempting to speculate that, similar to the Xenopus system, in DT40 PLK1 recruitment to chromatin surrounding stalled replication forks would promote phosphorylation-dependent recruitment of CDC45 to neighbouring dormant origins. Such association is a prerequisite for origin activation (Trenz et al, 2008).
FANCM function in replication fork restart is independent of the FA pathway
FANCM is a component of the FA core complex and one of the most highly conserved molecules within the pathway (Meetei et al, 2005; Mosedale et al, 2005). The FA core complex has been shown to have a crucial role in response to DNA damage by promoting monoubiquitination of the FANCD2/FANCI complex. Interestingly, a recent study has also established a novel, FA core complex-independent function for FANCM in suppressing cross-over recombination (Rosado et al, 2009) and in ATR/Chk1 signalling (Collis et al, 2008). Taking all of the above into consideration we wondered whether FANCM's role in promoting replication fork restart, under conditions of replication stress, would be FA dependent or independent.
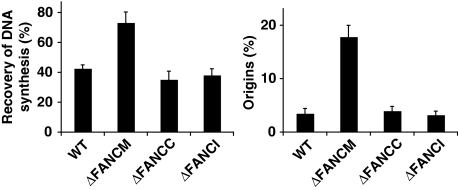
To test this, we analysed replication fork recovery in FA mutant cells deficient for either one of the FA core complex components (FANCC) or a component of the FA ID complex (FANCI). We used the same experimental conditions as described above. Under these conditions, both FANCC and FANCI mutant cells showed similar levels of recovery of DNA synthesis and origin firing to the level observed in WT DT40 cells (Figure 6). In summary, these data suggest that the role for FANCM described here is distinct from its function within the FA pathway.
Figure 6.
FANCM promotes replication fork recovery and inhibition of latent origins by a mechanism independent from its role in the FA pathway. Analysis of recovery of DNA synthesis and activation of latent origins in WT, ΔFANCM, ΔFANCC, and ΔFANCI DT40 cells treated with CPT for 90 min. The bars represent mean values from three independent experiments ±s.e.m.
Discussion
There are a variety of mechanisms involved in restoration of DNA replication after genotoxic stress, and many are imperfectly understood (Kolodner et al, 2002). Here, we identify a novel role for FANCM in this process. We show that upon CPT treatment, in FANCM mutant cells replication forks fail to resume DNA synthesis after removal of the drug. Instead, successful completion of DNA replication is ensured by ATR/PLK1-dependent activation of neighbouring dormant origins. In addition to this, we dissect the ATR/Chk1 signalling pathway and show that in FANCM mutant cells this pathway is compromised resulting in reduced phosphorylation of ATR downstream targets, including Chk1 and SMC1. Surprisingly, though, the same cells progressively accumulate RPA-ssDNA, a prerequisite for ATR activation (Zou and Elledge, 2003; Shiotani and Zou, 2009) suggesting that in this case RPA by itself is not sufficient to efficiently activate ATR and the S-phase checkpoint. Consistent with this, in Xenopus extracts RPA-ssDNA alone does not activate ATR (Stokes et al, 2002). The loading of the 9-1-1 trimer and TopBP1 at sites of DNA damage is required for ATR to phosphorylate at least some of its substrates including Chk1 (Zou et al, 2002). In line with this, we show that FANCM mutant cells are impaired in the stable association of TopBP1 on chromatin after replication stress. Given that efficient ATR-mediated Chk1 phosphorylation requires coordinated action of TopBP1 and claspin, and the fact that claspin is not required for phosphorylation of other ATR targets but exclusively links ATR to Chk1 (Liu et al, 2006), our data support a role for FANCM upstream of TopBP1, probably at the DNA level, in a common pathway leading to Chk1 activation. In support of this, FANCM deficiency also impacts on the ability of ATR to phosphorylate one of its other downstream targets, SMC1. We suspect that the list of ATR-mediated phosphorylation targets that require FANCM is in fact much longer, though at this time we are unable to investigate a larger array of substrates because of the limited availability of suitable antibodies in the DT40 system. The role of TopBP1 in the ATR activation process is not yet clear; certainly, TopBP1 is able to stimulate ATR kinase activity in vitro and is required for optimal ATR-dependent phosphorylation events and checkpoint establishment in vivo (Kumagai et al, 2006; Liu et al, 2006). However, TopBP1 is not required for the basal activity of ATR (Mordes et al, 2008), and may therefore only stimulate ATR activity towards certain substrates. By modulating the localization of TopBP1 in response to replication stress, FANCM may also be involved in ATR substrate selection by controlling the switch from local to global activation of ATR in response to high levels of genotoxic stress.
An unanswered question is how FANCM and its enzymatic activity may promote ATR phosphorylation events. Recently, it was proposed that fork-remodelling activities of FANCM might be required to stabilize stalled replication forks to facilitate access or retention of downstream checkpoint mediators and subsequent fork restart (Collis et al, 2008; Gari et al, 2008). Our data support and further corroborate this concept showing a role for FANCM in promoting TopBP1 loading on chromatin. How TopBP1 is recruited to chromatin, the interdependence of TopBP1 and the 9-1-1 complex in chromatin localization and the impact of FANCM on this process await further investigation, however. The resolution of this important issue will likely require in vitro reconstitution of the signalling cascade using, for example, the Xenopus system.
Checkpoint controls are evolutionarily conserved mechanisms that inhibit cell-cycle progression when DNA is damaged and protect genomic stability (Abraham, 2001; Cortez et al, 2001; Zou and Elledge, 2003). In vertebrate systems, human and chicken cells deprived of Chk1 show multiple S-phase checkpoint defects including a prolonged S-phase, increased origin usage, and inability to resume DNA synthesis upon release from replication block (Feijoo et al, 2001; Zachos et al, 2005). An interesting observation presented here is the low level of Chk1 phosphorylation detected in log-phase FANCM−/− cells suggesting that, to some extent, in unperturbed cells the basal level of ATR activity is not affected. This then, may be sufficient to support global DNA replication in the unperturbed FANCM mutant cells. In line with this, Chk1 and Claspin mutants show strikingly different phenotypes to FANCM with respect to global replication patterns, including replication fork velocity and suppression of Chk1-dependent latent origin firing (Zachos et al, 2003; Petermann et al, 2006, 2008). Thus, our data provide further support to the idea that TopBP1 is required for ATR activation but not for its basal kinase activity (Medhurst et al, 2008; Mordes et al, 2008), and perhaps, this basal activity is enough to support a low level of Chk1 phosphorylation to ensure completion of DNA synthesis.
Intriguingly, however, FANCM−/− cells are unable to efficiently upregulate this response upon replication stress. What could be the reason for this? It is tempting to speculate that a ‘threshold' mechanism exists that regulates the intra-S-phase checkpoint and that this mechanism could involve modulation of the signal (DNA) itself. Indeed, the existence of such a mechanism was suggested in yeast (Shimada et al, 2002). In such a scenario, a certain number of intermediates at stalled forks might be necessary to provoke clamp complex and TopBP1 accumulation and subsequent checkpoint activation. This would ensure that a low level of fork collapses and/or transient pausing/stalling of some forks does not halt the entire replication program, which then would need to be reset (switched off) to accomplish genome duplication. Another possibility, not mutually exclusive, is that ATR activation is regulated by multiple inputs, including DNA structures and assembly of ‘activating' complex(es). FANCM could then stabilize the replisome providing access to specific DNA structures and at the same time function as a loading platform assisting the assembly of the ATR-activating complex(es). Consistent with this, hCLK2 was shown to interact with ATR-ATRIP, Chk1, and TopBP1 (Collis et al, 2007; Danielsen et al, 2009); FANCM, on the other hand, is constitutively bound to chromatin during the S phase (Kim et al, 2008) and interacts with hCLK2 (Collis et al, 2008).
An unexpected finding reported here is that the rescue of DNA synthesis after replicative stress in FANCM mutant cells depends on ATR- and PLK1-regulated origin firing. What would be the physiological relevance of this? One possibility is that immovable replication blocks, dissociation of the replisome, extensive fork remodelling, and/or fork collapse could lead to abnormal four-branched recombination intermediates (reversed forks) and DNA breaks. Such lesions have to be processed to allow replication restart, however in some instances, they may act as ‘fork terminators' leading to irreversible replisome dissociation. In such a situation, the ATR/PLK1-dependent mechanism would ensure stalled fork bypass by recruitment of supplementary origins to complete genome duplication under the stress. Overall, our data and earlier studies clearly show that this indeed could be the case, at least in the DT40 and Xenopus systems (Trenz et al, 2008). The increased level of ATR on chromatin in FANCM−/− cells may explain its ability to promote local phosphorylation of MCM2. An open question is, however, why would ATR signalling promote both inhibition of late origin firing through phosphorylation of Chk1 and origin activation through MCM2 phosphorylation? The most plausible explanation, in our opinion, is that the ATR/PLK1 mechanism acts locally, near the stalled fork, ensuring completion of replication within active replicons. The Chk1 signalling would, on the other hand, enforce a global control of DNA synthesis. In such a scenario, ATR signalling would, at the same time, provide a mechanism for spatial and temporal regulation/control of DNA replication (Cimprich and Cortez, 2008).
Recent studies suggested an involvement of the FA pathway in the activation of dormant origins when replication forks stall (Phelps et al, 2007; Wang et al, 2008). These studies reported that replication stress in FANCC and FANCD2-deficient cells increases the frequency of new origins firing. Consistent with the current understanding of the mechanisms involved in origin control the authors proposed that the FA pathway stabilizes replication forks during genotoxic stress. In line with this, the FA pathway was also shown to have an important function in regulating fragile site stability (Howlett et al, 2005). Our data, on the other hand, argue that FANCM, a component of the FA core complex, promotes replication fork restart by a mechanism independent of the functional FA pathway. This would suggest that FANCM functions upstream of the FA pathway in promoting replication fork restart or, alternatively, FANCM and the FA pathway function in two distinct processes required to support stabilize/restart replication forks. The experiments to clarify this are in progress.
We currently do not know the molecular basis of the putative role for FANCM in replication. However, FANCM clearly has an important function in response to replicative stress and it may now be possible to reconcile the genetic and biochemical work. Our study clearly shows the requirement for FANCM in global DNA replication, in response to replication stress and in activation of the ATR signalling cascade. The biochemical activities of FANCM, such as binding to DNA structures resembling stalled replication forks, branch migration, translocase, and fork regression activities all may participate in stabilizing replication forks and promoting fork restart (Sun et al, 2008). We propose that FANCM recognizes, binds, and possibly remodels stalled replication forks, thus initiating a complex network of responses to stabilize, repair, and/or restart the fork to preserve genome integrity.
Finally, our work identifies yet more additional FA-independent functions for FANCM. Interestingly, the recently reported FANCM knockout mouse shows unique features atypical of other FA mice, including decreased overall survival, increased cancer incidence, and an underrepresentation of female mice (Bakker et al, 2009). As the original human FANCM patient cell line, EUFA867, has recently been shown to harbour additional mutations in FANCA, a true FANCM patient has yet to be identified (Singh et al, 2009). We believe that this provides an ideal opportunity to look back and reconsider whether FANCM can be truly classified as a bona fide FA gene, although perhaps this question will only be answered if a true human FANCM patient can be identified.
Materials and methods
Cells
Chicken DT40 cells were cultured in RPMI 1640 medium (Invitrogen) supplemented with 8% foetal calf serum, 2% chicken serum, and 10 μM β-mercaptoethanol and kept at 38°C in a humidified incubator containing 5% CO2.
Antibodies, mutagens, and chemical inhibitors
Primary antibodies used in this study were purchased from Abcam (BrdU, H3, MCM2), BD Biosciences (BrdU), Bethyl Laboratories (SMC1, SMC1-S966, TopBP1), Cell Signaling Technology (Chk1-S345), Millipore (γ-H2AX), Santa Cruz Biotechnology (ATR, Chk1, Rad51, RPA1), and AbD Serotec (MCM2-S108). Secondary antibodies used for western blotting were purchased from GE Healthcare. Secondary antibodies used for immunofluorescence were from Sigma-Aldrich (Cy3-labelled anti-mouse IgG) and Invitrogen (Alexa Fluor 488-labelled anti-rabbit and anti-rat IgG). CPT, hydroxyurea, and aphidicolin were purchased from Sigma-Aldrich and used at the indicated concentrations. Chemical inhibitors used were from Axon Medchem (BI 2536), Calbiochem (2-morpholin-4-yl-6-thianthren-1-yl-pyran-4-one), and Sigma-Aldrich (BTO-1, caffeine, roscovitine, UCN-01).
Cell fractionation
DT40 cell fractions enriched for chromatin were obtained as follows. Approximately 50 million cells were washed with phosphate-buffered saline (PBS) and lysed on ice in five packed cell volumes of hypotonic buffer (2 mM MgCl2, 3 mM CaCl2, 320 mM sucrose, 1 mM DTT, 10 mM Tris–HCl, pH 7.5) containing 0.3% Nonidet P-40 (NP40). Cells were centrifuged for 2 min at 2800 g and the supernatant containing cytoplasmic proteins was removed. Crude nuclei were washed in hypotonic buffer before the addition of two packed cell volumes of nuclear extraction buffer (1.5 mM MgCl2, 420 mM NaCl, 200 μM EDTA, 25% glycerol, 1 mM DTT, 20 mM HEPES, pH 7.7). Nuclei were extracted for 30 min at 4°C (in buffer containing a final NaCl concentration of 280 mM (Guo et al, 2009)), before being spun down at 8000 g for 15 min. The supernatant containing soluble nuclear proteins was removed and the chromatin pellet was resuspended in denaturing buffer (9 M urea, 150 mM β-mercaptoethanol, 50 mM Tris–HCl, pH 7.4) before sonication, followed by western blotting.
Mutagen sensitivity assays and PFGE
Cells were grown in 96-well plates at a density of 3 × 103 cells per well. CPT, hydroxyurea, or aphidicolin were added at a range of concentrations and incubated for five cell doublings. For survival assays with Plk inhibitors, cells were incubated with 50 μM BTO-1 or 5 nM BI 2536 for 1 h. Subsequently, cells were treated with the indicated doses of CPT for 90 min and then washed and processed as above. MTS (Promega) was added to each well and after incubation at 37°C for 4 h, cell viability was measured by luminometry at 492 nm. Each dose point was measured in quadruplicate and values were plotted relative to untreated controls. PFGE was performed as described earlier (Hanada et al, 2007).
Immunofluorescence
After treatment, DT40 cells were washed in PBS and spun onto poly-L-lysine-coated coverslips. Cells were fixed in 4% (w/v) paraformaldehyde in PBS for 10 min, permeabilized with 0.2% Triton X-100 in PBS for 10 min, and blocked with 10% (w/v) bovine serum albumin (BSA) in PBS for 30 min. Cells were incubated with the indicated antibodies for 1 h in 10% BSA in PBS. After washing in PBS, secondary antibodies were applied for 30 min. Cells were then stained with 4′,6′-diamidino-2-phenylindole (DAPI), and images were captured by fluorescence microscopy using a Zeiss Axioskop 2 Plus microscope. For co-localization with replication foci, cells were labelled with 25 μM CldU for 30 min with or without 100 nM CPT and then processed for γ-H2AX staining as described above. Subsequently, cells were fixed with 2% (w/v) paraformaldehyde in PBS for 10 min and DNA was denatured with 2 M HCl at 37°C for 30 min. Blocking with 10% (w/v) BSA in PBS was for 30 min followed by incubation with an antibody recognizing BrdU for 1 h in 10% BSA in PBS. Coverslips were washed for 10 min with high stringency buffer (0.5 M NaCl, 36 mM Tris pH 8.0, 0.5% Tween-20) and the secondary antibody was applied for 30 min.
DNA fibre analysis
Exponentially, growing DT40 cells were pulse-labelled with 25 μM IdU after incubation with 250 μM CldU for the times specified. Where indicated, cells were pretreated with the following inhibitors: UCN-01 at 300 nM, roscovitine at 100 μM, caffeine at 5 mM, ATM inhibitor at 10 μM, BTO-1 at 50 μM, and BI 2536 at 1 μM for 60 min before IdU labelling and then throughout, including the washing steps. CPT was used at 2.5 μM in all fibre experiments. Fibre spreads were prepared from 1 × 106 cells/ml as described earlier (Jackson and Pombo, 1998). Slides were incubated in 2.5 M HCl for 80 min and then washed several times in PBS, followed by incubation in blocking buffer (2% BSA in PBS) for 20 min. Primary antibodies were diluted in blocking buffer and incubated for 1 h followed by extensive washes in PBS. Secondary antibodies were applied for 30 min, and slides were mounted in Vectashield Mounting Medium (Vector Laboratories). Fibre tracts were examined using a Zeiss LSM 510 Meta confocal microscope. Pictures were taken from randomly selected fields with untangled fibres and analysed using the ImageJ software package. A minimum of 200 individual fibres were analysed for each experiment and the mean of at least three independent experiments presented. The relative frequency of the different classes of replication structures was scored as a percentage of all the different structures counted (Maya-Mendoza et al, 2007) and for fork velocity analysis a conversion factor for the length of a labelled track of 1 μm=2.59 kb was used (Jackson and Pombo, 1998; Petermann et al, 2006).
Statistical analysis
In all fibre experiments, a minimum of 200 individual fibres were analysed in at least three independent experiments and the Student's two-tailed t-test was used to determine statistical significance.
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Figure 5
Supplementary Figure 6
Supplementary Information
Review Process File
Acknowledgments
We thank KJ Patel for FANCM and FANCC and M Takata for FANCI knockout cell lines and I Hickson, P McHugh, F Esashi, and L Wu for invaluable advice. We also thank E Petermann for help with the fibre technique as well as all the members of the DNA repair groups for the technical advice. WN is supported by a Senior International Research Fellowship from the Association for International Cancer Research and by the Polish State Committee for Scientific Research grant (N301 165935).
Footnotes
The authors declare that they have no conflict of interest.
References
- Abraham RT (2001) Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev 15: 2177–2196 [DOI] [PubMed] [Google Scholar]
- Bakker ST, van de Vrugt HJ, Rooimans MA, Oostra AB, Steltenpool J, Delzenne-Goette E, van der Wal A, van der Valk M, Joenje H, Te Riele H, de Winter JP (2009) Fancm-deficient mice reveal unique features of Fanconi anemia complementation group M. Hum Mol Genet 18: 3484–3495 [DOI] [PubMed] [Google Scholar]
- Ciccia A, Ling C, Coulthard R, Yan Z, Xue Y, Meetei AR, Laghmani el H, Joenje H, McDonald N, de Winter JP, Wang W, West SC (2007) Identification of FAAP24, a Fanconi anemia core complex protein that interacts with FANCM. Mol Cell 25: 331–343 [DOI] [PubMed] [Google Scholar]
- Cimprich KA, Cortez D (2008) ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol 9: 616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collis SJ, Barber LJ, Clark AJ, Martin JS, Ward JD, Boulton SJ (2007) HCLK2 is essential for the mammalian S-phase checkpoint and impacts on Chk1 stability. Nat Cell Biol 9: 391–401 [DOI] [PubMed] [Google Scholar]
- Collis SJ, Ciccia A, Deans AJ, Horejsi Z, Martin JS, Maslen SL, Skehel JM, Elledge SJ, West SC, Boulton SJ (2008) FANCM and FAAP24 function in ATR-mediated checkpoint signaling independently of the Fanconi anemia core complex. Mol Cell 32: 313–324 [DOI] [PubMed] [Google Scholar]
- Conti C, Seiler JA, Pommier Y (2007) The mammalian DNA replication elongation checkpoint: implication of Chk1 and relationship with origin firing as determined by single DNA molecule and single cell analyses. Cell Cycle 6: 2760–2767 [DOI] [PubMed] [Google Scholar]
- Cortez D, Guntuku S, Qin J, Elledge SJ (2001) ATR and ATRIP: partners in checkpoint signaling. Science 294: 1713–1716 [DOI] [PubMed] [Google Scholar]
- Danielsen JM, Larsen DH, Schou KB, Freire R, Falck J, Bartek J, Lukas J (2009) HCLK2 is required for activity of the DNA damage response kinase ATR. J Biol Chem 284: 4140–4147 [DOI] [PubMed] [Google Scholar]
- Delacroix S, Wagner JM, Kobayashi M, Yamamoto K, Karnitz LM (2007) The Rad9-Hus1-Rad1 (9-1-1) clamp activates checkpoint signaling via TopBP1. Genes Dev 21: 1472–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feijoo C, Hall-Jackson C, Wu R, Jenkins D, Leitch J, Gilbert DM, Smythe C (2001) Activation of mammalian Chk1 during DNA replication arrest: a role for Chk1 in the intra-S phase checkpoint monitoring replication origin firing. J Cell Biol 154: 913–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster ER, Downs JA (2005) Histone H2A phosphorylation in DNA double-strand break repair. FEBS J 272: 3231–3240 [DOI] [PubMed] [Google Scholar]
- Gari K, Decaillet C, Stasiak AZ, Stasiak A, Constantinou A (2008) The Fanconi anemia protein FANCM can promote branch migration of Holliday junctions and replication forks. Mol Cell 29: 141–148 [DOI] [PubMed] [Google Scholar]
- Ge XQ, Jackson DA, Blow JJ (2007) Dormant origins licensed by excess Mcm2-7 are required for human cells to survive replicative stress. Genes Dev 21: 3331–3341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves PR, Yu L, Schwarz JK, Gales J, Sausville EA, O'Connor PM, Piwnica-Worms H (2000) The Chk1 protein kinase and the Cdc25C regulatory pathways are targets of the anticancer agent UCN-01. J Biol Chem 275: 5600–5605 [DOI] [PubMed] [Google Scholar]
- Guo R, Xu D, Wang W (2009) Identification and analysis of new proteins involved in the DNA damage response network of Fanconi anemia and Bloom syndrome. Methods 48: 72–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada K, Budzowska M, Davies SL, van Drunen E, Onizawa H, Beverloo HB, Maas A, Essers J, Hickson ID, Kanaar R (2007) The structure-specific endonuclease Mus81 contributes to replication restart by generating double-strand DNA breaks. Nat Struct Mol Biol 14: 1096–1104 [DOI] [PubMed] [Google Scholar]
- Howlett NG, Taniguchi T, Durkin SG, D'Andrea AD, Glover TW (2005) The Fanconi anemia pathway is required for the DNA replication stress response and for the regulation of common fragile site stability. Hum Mol Genet 14: 693–701 [DOI] [PubMed] [Google Scholar]
- Ibarra A, Schwob E, Mendez J (2008) Excess MCM proteins protect human cells from replicative stress by licensing backup origins of replication. Proc Natl Acad Sci USA 105: 8956–8961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson DA, Pombo A (1998) Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J Cell Biol 140: 1285–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JM, Kee Y, Gurtan A, D'Andrea AD (2008) Cell cycle-dependent chromatin loading of the Fanconi anemia core complex by FANCM/FAAP24. Blood 111: 5215–5222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodner RD, Putnam CD, Myung K (2002) Maintenance of genome stability in Saccharomyces cerevisiae. Science 297: 552–557 [DOI] [PubMed] [Google Scholar]
- Komori K, Hidaka M, Horiuchi T, Fujikane R, Shinagawa H, Ishino Y (2004) Cooperation of the N-terminal Helicase and C-terminal endonuclease activities of Archaeal Hef protein in processing stalled replication forks. J Biol Chem 279: 53175–53185 [DOI] [PubMed] [Google Scholar]
- Kumagai A, Lee J, Yoo HY, Dunphy WG (2006) TopBP1 activates the ATR-ATRIP complex. Cell 124: 943–955 [DOI] [PubMed] [Google Scholar]
- Lambert S, Watson A, Sheedy DM, Martin B, Carr AM (2005) Gross chromosomal rearrangements and elevated recombination at an inducible site-specific replication fork barrier. Cell 121: 689–702 [DOI] [PubMed] [Google Scholar]
- Lansing TJ, McConnell RT, Duckett DR, Spehar GM, Knick VB, Hassler DF, Noro N, Furuta M, Emmitte KA, Gilmer TM, Mook RA Jr, Cheung M (2007) In vitro biological activity of a novel small-molecule inhibitor of polo-like kinase 1. Mol Cancer Ther 6: 450–459 [DOI] [PubMed] [Google Scholar]
- Liu S, Bekker-Jensen S, Mailand N, Lukas C, Bartek J, Lukas J (2006) Claspin operates downstream of TopBP1 to direct ATR signaling towards Chk1 activation. Mol Cell Biol 26: 6056–6064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luciani MG, Oehlmann M, Blow JJ (2004) Characterization of a novel ATR-dependent, Chk1-independent, intra-S-phase checkpoint that suppresses initiation of replication in Xenopus. J Cell Sci 117: 6019–6030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maya-Mendoza A, Petermann E, Gillespie DA, Caldecott KW, Jackson DA (2007) Chk1 regulates the density of active replication origins during the vertebrate S phase. EMBO J 26: 2719–2731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medhurst AL, Warmerdam DO, Akerman I, Verwayen EH, Kanaar R, Smits VA, Lakin ND (2008) ATR and Rad17 collaborate in modulating Rad9 localisation at sites of DNA damage. J Cell Sci 121: 3933–3940 [DOI] [PubMed] [Google Scholar]
- Meetei AR, Medhurst AL, Ling C, Xue Y, Singh TR, Bier P, Steltenpool J, Stone S, Dokal I, Mathew CG, Hoatlin M, Joenje H, de Winter JP, Wang W (2005) A human ortholog of archaeal DNA repair protein Hef is defective in Fanconi anemia complementation group M. Nat Genet 37: 958–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mordes DA, Glick GG, Zhao R, Cortez D (2008) TopBP1 activates ATR through ATRIP and a PIKK regulatory domain. Genes Dev 22: 1478–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosedale G, Niedzwiedz W, Alpi A, Perrina F, Pereira-Leal JB, Johnson M, Langevin F, Pace P, Patel KJ (2005) The vertebrate Hef ortholog is a component of the Fanconi anemia tumor-suppressor pathway. Nat Struct Mol Biol 12: 763–771 [DOI] [PubMed] [Google Scholar]
- Patel KJ, Joenje H (2007) Fanconi anemia and DNA replication repair. DNA Repair (Amst) 6: 885–890 [DOI] [PubMed] [Google Scholar]
- Petermann E, Helleday T, Caldecott KW (2008) Claspin promotes normal replication fork rates in human cells. Mol Biol Cell 19: 2373–2378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petermann E, Maya-Mendoza A, Zachos G, Gillespie DA, Jackson DA, Caldecott KW (2006) Chk1 requirement for high global rates of replication fork progression during normal vertebrate S phase. Mol Cell Biol 26: 3319–3326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelps RA, Gingras H, Hockenbery DM (2007) Loss of FANCC function is associated with failure to inhibit late firing replication origins after DNA cross-linking. Exp Cell Res 313: 2283–2292 [DOI] [PubMed] [Google Scholar]
- Prakash R, Krejci L, Van Komen S, Anke Schurer K, Kramer W, Sung P (2005) Saccharomyces cerevisiae MPH1 gene, required for homologous recombination-mediated mutation avoidance, encodes a 3′ to 5′ DNA helicase. J Biol Chem 280: 7854–7860 [DOI] [PubMed] [Google Scholar]
- Rosado IV, Niedzwiedz W, Alpi AF, Patel KJ (2009) The Walker B motif in avian FANCM is required to limit sister chromatid exchanges but is dispensable for DNA crosslink repair. Nucleic Acids Res 37: 4360–4370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh-Gohari N, Bryant HE, Schultz N, Parker KM, Cassel TN, Helleday T (2005) Spontaneous homologous recombination is induced by collapsed replication forks that are caused by endogenous DNA single-strand breaks. Mol Cell Biol 25: 7158–7169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler JA, Conti C, Syed A, Aladjem MI, Pommier Y (2007) The intra-S-phase checkpoint affects both DNA replication initiation and elongation: single-cell and -DNA fiber analyses. Mol Cell Biol 27: 5806–5818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shechter D, Costanzo V, Gautier J (2004) ATR and ATM regulate the timing of DNA replication origin firing. Nat Cell Biol 6: 648–655 [DOI] [PubMed] [Google Scholar]
- Shiloh Y (2003) ATM and related protein kinases: safeguarding genome integrity. Nat Rev Cancer 3: 155–168 [DOI] [PubMed] [Google Scholar]
- Shimada K, Pasero P, Gasser SM (2002) ORC and the intra-S-phase checkpoint: a threshold regulates Rad53p activation in S phase. Genes Dev 16: 3236–3252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiotani B, Zou L (2009) Single-stranded DNA orchestrates an ATM-to-ATR switch at DNA breaks. Mol Cell 33: 547–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh TR, Bakker ST, Agarwal S, Jansen M, Grassman E, Godthelp BC, Ali AM, Du CH, Rooimans MA, Fan Q, Wahengbam K, Steltenpool J, Andreassen PR, Williams DA, Joenje H, de Winter JP, Meetei AR (2009) Impaired FANCD2 monoubiquitination and hypersensitivity to camptothecin uniquely characterize Fanconi anemia complementation group M. Blood 114: 174–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogo JM, Lopes M, Foiani M (2002) Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science 297: 599–602 [DOI] [PubMed] [Google Scholar]
- Stokes MP, Van Hatten R, Lindsay HD, Michael WM (2002) DNA replication is required for the checkpoint response to damaged DNA in Xenopus egg extracts. J Cell Biol 158: 863–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Nandi S, Osman F, Ahn JS, Jakovleska J, Lorenz A, Whitby MC (2008) The FANCM ortholog Fml1 promotes recombination at stalled replication forks and limits crossing over during DNA double-strand break repair. Mol Cell 32: 118–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai H, Wang RC, Takai KK, Yang H, de Lange T (2007) Tel2 regulates the stability of PI3K-related protein kinases. Cell 131: 1248–1259 [DOI] [PubMed] [Google Scholar]
- Trenz K, Errico A, Costanzo V (2008) Plx1 is required for chromosomal DNA replication under stressful conditions. EMBO J 27: 876–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LC, Stone S, Hoatlin ME, Gautier J (2008) Fanconi anemia proteins stabilize replication forks. DNA Repair (Amst) 7: 1973–1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W (2007) Emergence of a DNA-damage response network consisting of Fanconi anaemia and BRCA proteins. Nat Rev Genet 8: 735–748 [DOI] [PubMed] [Google Scholar]
- Yan S, Michael WM (2009) TopBP1 and DNA polymerase-alpha directly recruit the 9-1-1 complex to stalled DNA replication forks. J Cell Biol 184: 793–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo HY, Kumagai A, Shevchenko A, Dunphy WG (2007) Ataxia-telangiectasia mutated (ATM)-dependent activation of ATR occurs through phosphorylation of TopBP1 by ATM. J Biol Chem 282: 17501–17506 [DOI] [PubMed] [Google Scholar]
- Zachos G, Rainey MD, Gillespie DA (2003) Chk1-deficient tumour cells are viable but exhibit multiple checkpoint and survival defects. EMBO J 22: 713–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachos G, Rainey MD, Gillespie DA (2005) Chk1-dependent S-M checkpoint delay in vertebrate cells is linked to maintenance of viable replication structures. Mol Cell Biol 25: 563–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Piwnica-Worms H (2001) ATR-mediated checkpoint pathways regulate phosphorylation and activation of human Chk1. Mol Cell Biol 21: 4129–4139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L, Cortez D, Elledge SJ (2002) Regulation of ATR substrate selection by Rad17-dependent loading of Rad9 complexes onto chromatin. Genes Dev 16: 198–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L, Elledge SJ (2003) Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300: 1542–1548 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Figure 5
Supplementary Figure 6
Supplementary Information
Review Process File