Abstract
Prion diseases differ from other amyloid-associated protein misfolding diseases (e.g. Alzheimer's) because they are naturally transmitted between individuals and involve spread of protein aggregation between tissues. Factors underlying these features of prion diseases are poorly understood. Of all protein misfolding disorders, only prion diseases involve the misfolding of a glycosylphosphatidylinositol (GPI)-anchored protein. To test whether GPI anchoring can modulate the propagation and spread of protein aggregates, a GPI-anchored version of the amyloidogenic yeast protein Sup35NM (Sup35GPI) was expressed in neuronal cells. Treatment of cells with Sup35NM fibrils induced the GPI anchor-dependent formation of self-propagating, detergent-insoluble, protease-resistant, prion-like aggregates of Sup35GPI. Live-cell imaging showed intercellular spread of Sup35GPI aggregation to involve contact between aggregate-positive and aggregate-negative cells and transfer of Sup35GPI from aggregate-positive cells. These data demonstrate GPI anchoring facilitates the propagation and spread of protein aggregation and thus may enhance the transmissibility and pathogenesis of prion diseases relative to other protein misfolding diseases.
Keywords: amyloid, glycosylphosphatidylinositol anchor, nanotube, protein transfer, Sup35
Introduction
Transmissible spongiform encephalopathies (TSEs), or prion diseases, include Creutzfeldt–Jakob disease and Gerstmann–Sträussler–Scheinker syndrome in humans, chronic wasting disease in cervids, scrapie in sheep, and bovine spongiform encephalopathy in cattle. These diseases are fatal infectious neurodegenerative diseases characterized by the conformational conversion of the normal monomeric cell-associated prion protein (PrPC) into an abnormally folded form called PrPSc. PrPC is a protease-sensitive glycoprotein attached to cell surfaces and intracellular membranes by a glycosylphosphatidylinositol (GPI) anchor. PrPSc, like other proteins associated with protein misfolding diseases, exhibits greater protease resistance and exists in β-sheet-rich multimeric aggregates of varying size, including amyloid fibrils. One key difference between prion diseases and other protein misfolding diseases is that prion diseases are infectious, with PrPSc, perhaps in association with other cofactors, suspected as being the infectious agent/prion. It remains unclear why only prion diseases, among all protein misfolding diseases, are infectious under natural circumstances (Caughey et al, 2009).
Processes that are critical for a misfolded protein aggregate to behave as a naturally transmissible prion in mammals include: (i) aggregate-induced conversion of the non-aggregated precursor protein to the prion-associated conformation; (ii) a net positive growth rate versus clearance for the aggregates; (iii) fragmentation of aggregates to generate new seeding entities; and (iv) horizontal spread of aggregates between cells and tissues. The inherent capacity of different protein aggregates to undergo each of these processes likely varies between proteins. The sum of these factors could determine whether a given protein aggregate can be a prion and influence the phenotype associated with the prion state. It follows that modulation of these factors could make otherwise non-infectious protein aggregates behave as prions. Experimental evidence from yeast prion and polyglutamine protein systems supports these concepts (Borchsenius et al, 2001; Osherovich et al, 2004; Tanaka et al, 2006; Immel et al, 2007).
Yeast prion replication has many features in common with mammalian prion replication (Chien et al, 2004; Ross et al, 2005). One yeast prion, [PSI+], was identified as a non-Mendelian element of inheritance caused by self-propagating aggregates of the translation termination factor Sup35p (Cox, 1965; Wickner, 1994; Patino et al, 1996; Paushkin et al, 1996). The prion-forming activity of Sup35p is controlled by the N-terminal NM domains of the protein. Using recombinant amyloid fibrils of the NM domains of Sup35p (Sup35NM) investigators have shown that, like mammalian prions, Sup35NM prions exhibit strain (Tanaka et al, 2004) and species barrier (Tanaka et al, 2005) properties that are dependent on Sup35NM aggregate conformation.
A unique feature of PrPC relative to yeast prion proteins and amyloidogenic proteins associated with human protein misfolding disorders is that PrPC is tethered to membranes via a GPI anchor. This membrane anchor could affect the biochemical, biophysical, and neurotoxic properties of PrP molecules, and the efficiency of PrPSc formation and propagation (Caughey et al, 2009). This is consistent with data obtained from prion infections of transgenic mice expressing anchorless PrPC (Chesebro et al, 2005) and other studies (Gabizon et al, 1987, 1988; McKinley et al, 1991; Baron et al, 2002, 2006; Fevrier et al, 2004; Silveira et al, 2005). Furthermore, there are cellular mechanisms (e.g. exosomes, membrane microparticles, membrane nanotubes) that mediate the exchange of GPI-anchored and other membrane-bound proteins between cells (Mack et al, 2000; Fevrier et al, 2004; Onfelt et al, 2004; Sherer et al, 2007; Sowinski et al, 2008). This includes a very recent report of intercellular spread of PrPSc via tunnelling nanotubes (Gousset et al, 2009). These exchange processes could provide a mechanism for the efficient spread of membrane-bound protein aggregates to new host cells and tissues, that is, transmitting the ‘infection', at least as defined by the presence of self-propagating protein aggregates.
Cell–cell spread of GPI-anchored protein aggregation is particularly relevant in cases of peripheral mammalian prion infection, the most common natural route. Intercellular transmission of mammalian prions occurs during the neuroinvasion and spread of prions (and PrPSc) along defined neural pathways after peripheral prion infection (Mabbott and MacPherson, 2006). In infected animals at terminal stages of disease, PrPSc is distributed throughout the central nervous system (CNS), with lymphoid tissues involved to varying extents (Mabbott and MacPherson, 2006). This neuroinvasive capacity of PrPSc is a distinctive feature that is an important element of TSE disease transmission between hosts. Such horizontal spreading activity is crucial for a protein aggregate to act as a naturally transmissible prion in mammals, particularly for aggregates whose pathogenic effects require spread to, and within, the CNS. In vitro studies suggest that transmission of PrPSc and infection from dendritic cells to neurons through TNTs may contribute to neuroinvasion (Gousset and Zurzolo, 2009; Gousset et al, 2009). Altogether, this raises important questions about the mechanisms of neuroinvasion and spread of prions within mammals, and the factors controlling these features that appear to be specific to prions among mammalian protein misfolding diseases.
Consideration of these issues prompted the current study to investigate whether membrane anchoring could modulate the induction and intercellular transmission of protein aggregation when applied to another protein capable of forming inducible, misfolded aggregates. Although other modes of membrane anchoring (e.g. transmembrane) may confer this activity, in this initial study we focused on the effect of GPI anchoring because this is the form of membrane anchoring associated with the lone naturally transmissible protein misfolding disorder (i.e. prion diseases). To explore this question, we expressed GPI-anchored forms of the NM domains of Sup35p (Sup35GPI) in N2a mouse neuroblastoma cells. Sup35NM offers several advantages for our investigation. The aggregation properties of Sup35NM are well-characterized and it can aggregate when fused to other proteins (Patino et al, 1996; Li and Lindquist, 2000). It has no mammalian orthologues and would not be expected to participate in physiological interactions between mammalian amyloidogenic proteins and other molecules. This allows us to specifically assess the effects of GPI anchoring and explore questions of broad interest such as whether GPI-anchored, membrane-bound aggregates are directly neurotoxic. In addition, its prion behaviour in yeast is dependent on the appropriately tuned expression of the cytoplasmic chaperone protein Hsp104 (Chernoff et al, 1995), which facilitates [PSI+] prion replication by fragmentation of large aggregates into smaller aggregation-inducing seeds (Chernoff et al, 1995; Cox et al, 2003; Kryndushkin et al, 2003; Shorter and Lindquist, 2006; Satpute-Krishnan et al, 2007). Hsp104 has no known mammalian orthologue with ATP-dependent protein disaggregase activity or equivalent biological activity localized to the cell surface/extracellular environment (Doyle and Wickner, 2009). A very recent report described the yeast prion-like vertical propagation of Sup35NM aggregates in the cytoplasm of mammalian cells, suggesting that Hsp104-like activity may be present in this specific subcellular compartment (Krammer et al, 2009). Thus in the context of our experiments, Sup35NM would not be expected a priori to behave as a prion unless GPI anchoring could somehow compensate for the factors necessary to propagate Sup35p prion aggregates in the yeast cytoplasm. Using this new model we show that self-propagating, detergent-insoluble, protease-resistant aggregates of Sup35GPI can be induced in N2a cells expressing Sup35GPI by treatment with exogenous Sup35NM aggregates. Moreover, spread of aggregation to separate cells can occur through cell-to-cell contact and aggregate shearing. Collectively, our data demonstrate that GPI anchoring can modulate the properties of Sup35NM protein aggregates to allow their replication as prions when expressed, like PrPC, on the surface of mammalian cells. These observations may help explain why among all protein misfolding diseases only prion diseases are recognized as naturally transmissible.
Results
Recombinant Sup35NM fibrils induce acute aggregation of Sup35-GFPGPI
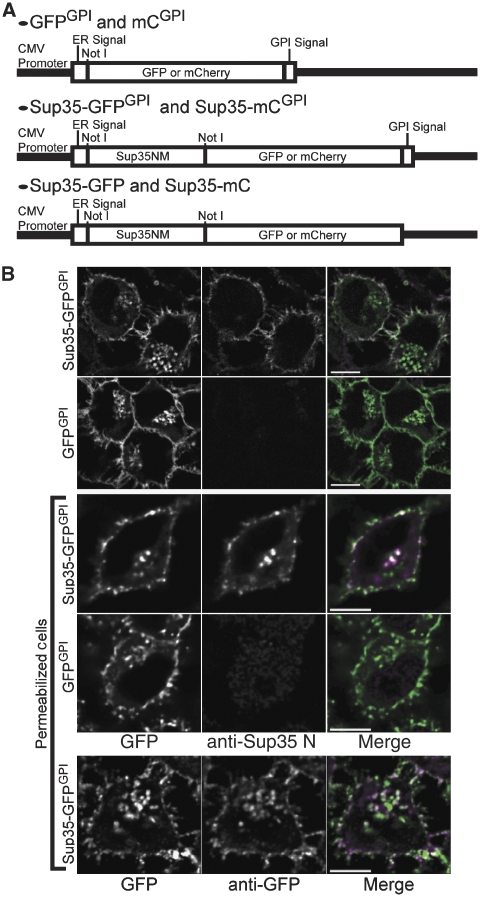
To test the role of GPI anchoring as one mode of membrane association that could modulate protein aggregation, N2a cells stably transfected with either GFPGPI or Sup35-GFPGPI (Figure 1A) were generated. Cells expressing either construct exhibited GFP fluorescence on the cell surface and in a perinuclear region. Immunolabelling of non-permeabilized Sup35-GFPGPI cells using an anti-Sup35 N-domain antibody (anti-Sup35 N) suggested that full-length Sup35-GFPGPI was expressed on the cell surface (Figure 1B, top row). Immunolabelling of permeabilized Sup35-GFPGPI cells with anti-Sup35 N showed labelling of the intracellular population with a comparable efficiency to that observed using anti-GFP antibody (Figure 1B, third row). Together, these data verified that the cells expressed the GFP fusion proteins with a distribution typical of GPI-anchored proteins.
Figure 1.
Plasmids and the expression of Sup35-GFPGPI. (A) Plasmids used to generate stably transfected cell lines. (B) Immunofluorescence using anti-Sup35 N or anti-GFP antibody. Top two rows: Cell-surface labelling of non-permeabilized cells. Bottom three rows: Labelling of permeabilized cells. Merge image: GFP (green) and anti-Sup35 N or anti-GFP (magenta). Colocalized areas appear white. Bars, 10 μm.
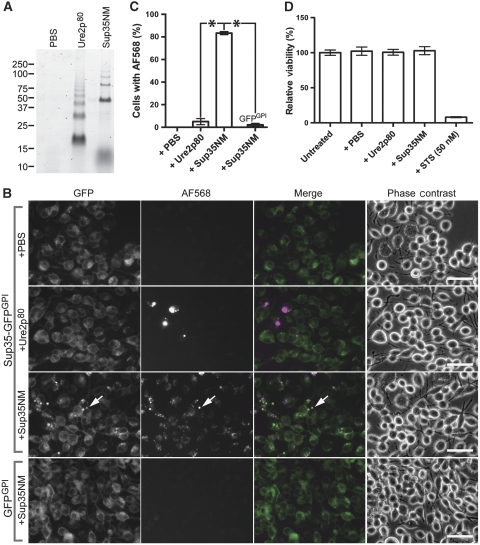
To induce aggregation of Sup35-GFPGPI, we prepared fibrils of recombinant Sup35NM labelled with Alexa Fluor-568 (AF568) (Supplementary Figure S1A and B). We also prepared AF568-labelled recombinant fibrils of the prion domain (residues 1–80) of the yeast prion protein Ure2p (Ure2p80) as a specificity control (Supplementary Figure S1C and D). Sup35-GFPGPI-expressing cells were treated with either PBS or equimolar amounts of AF568-labelled Sup35NM or Ure2p80 fibrils (Figure 2A). Two days after treatment, more AF568-positive cells were present in cultures treated with Sup35NM-AF568 than in the Ure2p80 control culture (Figure 2B and C). Sup35NM-AF568 fibril binding to GFPGPI control cells was negligible (Figure 2B and C). These data suggested that Sup35-GFPGPI specifically bound the Sup35NM-AF568 fibrils, although we cannot ascertain the subcellular localization based on these images. Punctate accumulations of GFP appeared only in Sup35NM fibril-treated cells (arrow; Figure 2B), suggesting that Sup35NM fibril treatment induced acute aggregation of Sup35-GFPGPI. These differences did not result from any toxic effects from the fibril treatments (Figure 2D).
Figure 2.
Sup35NM fibrils bind specifically to Sup35-GFPGPI cells. (A) Fluorescence SDS–PAGE of AF568-labelled fibrils. Laddering in lanes 2 and 3 corresponds to SDS-insoluble oligomers. Lower apparent molecular mass material (∼13 kDa) in lane 3 and to a lesser extent in lane 2 is free dye, which does not detectably associate with cells (Supplementary Figure S2). (B) Wide-field fluorescence microscopy of cells 2 days after adding fibrils. Arrow (third row) indicates newly induced Sup35-GFPGPI aggregate colocalized with Sup35NM-AF568 fibrils. Merge images: GFP (green), AF568 (magenta) and colocalization (white). Bars, 50 μm. (C) Percentage of Sup35-GFPGPI cells associated with AF568-labelled fibrils 2 days after treatment with PBS, Ure2p80-AF568 fibrils, or Sup35NM-AF568 fibrils. For an additional specificity control, the rightmost bar indicates the percentage of cells expressing GFPGPI associated with Sup35NM-AF568 fibrils. Data are mean±standard deviation (s.d.) of n=5316 (PBS), n=5107 (Ure2p80-AF568), n=4746 (Sup35NM-AF568), and n=6388 (GFPGPI cells+Sup35NM-AF568) cells counted from six different fields of view for each treatment (*P<0.0001; unpaired t-test). (D) Cell viability was determined 2 days after fibril treatments and is expressed as a percentage versus untreated controls (mean±s.d., n=12). Staurosporine (STS), positive control. All data are representative of at least two independent experiments performed in triplicate.
Biochemical characterization of self-propagating Sup35-GFPGPI aggregates induced by Sup35NM fibrils
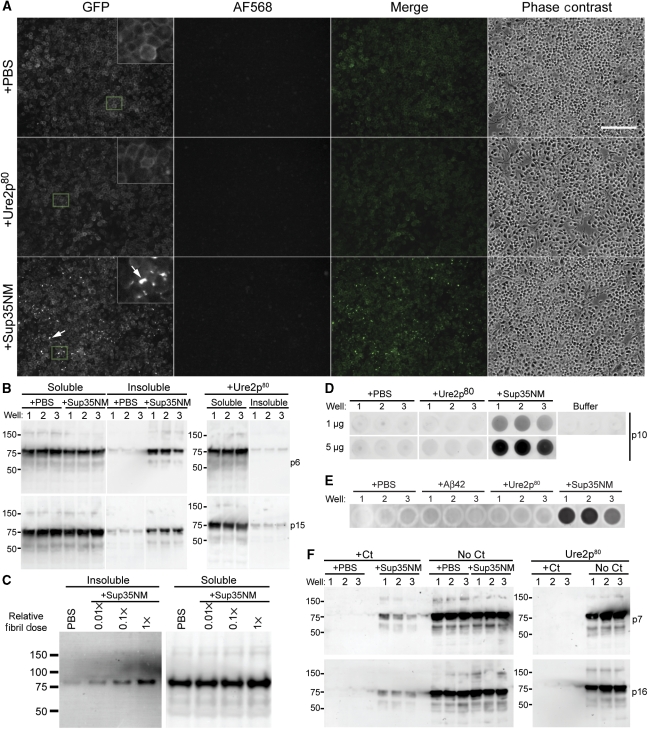
To confirm that Sup35NM fibrils induced Sup35-GFPGPI aggregation, cells were analysed by detergent-insolubility and protease resistance assays after extended passage post-treatment to allow removal of the input Sup35NM fibrils by dilution. After six 10-fold serial passages, only cells treated with Sup35NM fibrils exhibited GFP aggregates (Figure 3A) and detergent (sarkosyl)-insoluble Sup35-GFPGPI (Figure 3B). The level of induced Sup35-GFPGPI aggregation was dependent on the fibril dose (Figure 3C).
Figure 3.
Sup35NM fibrils induce aggregation. (A) Wide-field fluorescence images of fibril-treated Sup35-GFPGPI cells at passage-6 following fibril addition. Arrows: GFP punctae indicative of Sup35-GFPGPI aggregates. Merge: GFP (green) and AF568 (magenta). Insets: Magnified view of the boxed areas. Bar, 200 μm. (B) Western blots of detergent insolubility assays from fibril-treated cell lysates at passage-6 and 15. Ure2p80 fibril-treated samples shown are from the same blot with irrelevant lanes removed. Well number indicates cells were treated in triplicate. (C) Dose-dependent induction of Sup35-GFPGPI aggregation by Sup35NM fibrils. Western blot (anti-Sup35 N antibody) of insoluble and soluble fractions separated by sedimentation of 1% sarkosyl lysates of Sup35-GFPGPI cells treated with serial dilutions of Sup35NM fibrils nine passages after adding fibrils. The relative amounts of fibrils are indicated with ‘1x' corresponding to the amount of fibrils used for fibril addition experiments as described under Materials and methods. Lanes shown are from the same blot with intensity range settings for the soluble lanes 10-fold higher (i.e. less sensitive) than the settings for the insoluble lanes. (D) Filter-trap assay immunoblot at passage-10 showing SDS-insoluble Sup35-GFPGPI present only in Sup35NM-AF568 fibril-treated samples. The total protein amounts filtered per well for each row are indicated. ‘Buffer' wells were loaded with 4% SDS buffer then treated identically to other wells. Wells shown are from the same blot with irrelevant wells removed. (E) Heterologous amyloid fibrils do not induce Sup35-GFPGPI aggregation. Filter-trap assay of Sup35-GFPGPI cells at passage-12 after treatment (in triplicate wells) with AF568-labelled amyloid fibrils of Aβ (1–42) (Aβ42), Ure2p80, or Sup35NM. Total protein filtered per well was 5 μg. (F) Western blots of chymotrypsin-treated cell lysates at passage-7 and 16. All data are representative of two independent experiments performed in triplicate.
To rigorously assess the stability of the Sup35-GFPGPI aggregates, detergent-insolubility assays were performed in a filter-trap format using a strong denaturing detergent (SDS). Others have shown that Sup35NM aggregates and infectious polymers resist solubilization in SDS (Serio et al, 1999; Bagriantsev et al, 2008). SDS-insoluble Sup35-GFPGPI was only detected in cells treated with Sup35NM fibrils (Figure 3D). Cells treated with Alzheimer's amyloid β-peptide (1–42) fibrils (Aβ42) also did not produce Sup35-GFPGPI aggregates (Figure 3E). Together, these data indicated that treatment of Sup35-GFPGPI-expressing cells with Sup35NM fibrils specifically induced the production of aggregated Sup35-GFPGPI.
Protease resistance was assessed by chymotrypsin digestion. The N domain forms the core of Sup35p aggregates and acquires resistance to chymotrypsin digestion after incorporation into Sup35NM fibrils (Serio et al, 2000). Because the M domain lacks chymotrypsin-cleavage sites, the development of chymotrypsin-resistant full-length Sup35NM species is characteristic of aggregated forms of Sup35NM. Only lysates from Sup35NM fibril-treated cells contained chymotrypsin-resistant, full-length Sup35-GFPGPI (Figure 3F). The protease-resistant and detergent-insoluble material observed in the Sup35NM fibril-treated cells was not from residual Sup35NM fibrils because no AF568-labelled material was detectable by fluorescence microscopy (Figure 3A) and on western blots Sup35NM could be distinguished from Sup35-GFPGPI based on apparent molecular mass.
In cell culture models of prion infection, acute induction of PrPSc formation does not necessarily lead to persistent PrPSc formation (i.e. infection) (Vorberg et al, 2004). To be considered stably infected, cultures must maintain PrPSc propagation for multiple passages (usually at least 5–6, which also removes the inoculum by dilution). With Sup35NM fibril-treated Sup35-GFPGPI cells, maintenance of Sup35-GFPGPI aggregates was observed for many passages (Figure 3B and F, and Supplementary Figure S3) and as high as 23 (unpublished data). Similar results were obtained using multiple Sup35-GFPGPI cloned cell lines with different Sup35-GFPGPI expression levels (unpublished data). Collectively, these data indicate that induced Sup35-GFPGPI aggregates were self-propagating.
Sup35-GFPGPI aggregates induced by intercellular contact
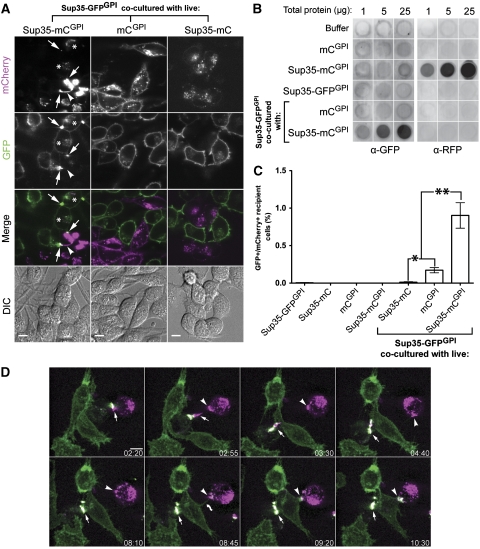
To assess whether GPI-anchored Sup35 aggregates could induce Sup35GPI aggregation in separate cells, Sup35-GFPGPI cells were co-cultured with cells stably expressing a GPI-anchored Sup35NM domain fusion with mCherry red-fluorescent protein (Sup35-mCGPI) and analysed by live-cell confocal microscopy. The Sup35-mCGPI cells persistently produce aggregates, which are often visualized as cell-surface sheet-like accumulations (arrowhead in Figure 4A and Supplementary Figure S4), are detected by filter-trap assay (Figure 4B), and are biochemically indistinguishable from induced Sup35-GFPGPI aggregates. Although we cannot exclude the possibility that a small fraction of the aggregate population may be intracellular, the punctate cell-surface accumulations that we call aggregates invariably correlate with the detection of aggregated Sup35-mCGPI in our biochemical assays. Because mCherry oligomerization can be detected under certain conditions with sensitive assays such as FRET (Piston and Rizzo, 2008; Snapp, 2009), it is possible that GPI-anchored mCherry has a natural tendency to interact with itself to form small oligomeric species undetectable in our assays that can facilitate spontaneous nucleation of associated Sup35NM moieties by bringing them into close proximity. These aggregates were not seen in cells stably expressing either GPI-anchored mCherry protein alone (mCGPI) or secreted GPI-anchorless Sup35NM-mCherry (Sup35-mC), which were used as negative controls (Figure 4A, middle and right panels, respectively, and Figure 4B). After 3 days, GFP aggregates appeared specifically in co-cultures with aggregate-positive Sup35-mCGPI cells (Figure 4A, left panels). Overall cell-surface labelling was decreased for cells making Sup35-GFPGPI aggregates, suggesting that a substantial portion of the cell-surface Sup35-GFPGPI was sequestered in the aggregates (compare GFP distribution in Figure 4A). Interestingly, most of the GFP aggregates (we estimate >90%) colocalized with Sup35-mCGPI material (Figure 4A, arrows), suggesting that Sup35-GFPGPI aggregation was associated with intercellular transfer of Sup35-mCGPI aggregates.
Figure 4.
Acute induction of aggregates on Sup35-GFPGPI cells by co-culture with live Sup35-mCGPI aggregate-positive cells. (A) Live-cell confocal microscopy images of Sup35-GFPGPI cells co-cultured with Sup35-mCGPI aggregate-positive cells for 3 days showed induction of Sup35-GFPGPI aggregates (left column, arrows). Sup35-GFPGPI aggregates were not induced by co-culture with mCGPI or Sup35-mC control cells (middle and right panels). Arrowhead: Cell-surface sheet aggregate of Sup35-mCGPI. Asterisks: Sup35-GFPGPI cells with intracellular Sup35-mCGPI. Images represent single Z planes selected from the middle of cells. DIC: Differential interference contrast microscopy. For panels A and D: Merge: GFP (green), mCherry (magenta), and GFP/mCherry colocalization (white). Bars, 10 μm. (B) Filter-trap assay of FACS-sorted co-cultures of cells at passage-6 post FACS. Immunoblotting was performed using anti-GFP (left three columns) to detect Sup35-GFPGPI aggregates and anti-RFP (right three columns) to demonstrate that Sup35-mCGPI cells were removed by FACS and there were no detectable Sup35-mCGPI aggregates in the co-culture. Rows 2–4 represent control samples not subjected to FACS. Wells shown are from the same respective blot with irrelevant wells removed. (C) Percentage of Sup35-GFPGPI-recipient cells positive for mCherry as determined by flow cytometry. Cells were co-cultured as in panel A prior to analysis. The first four samples represent cells cultured alone to demonstrate the sorting efficiency. The values represent the percentage of GFP-positive cells that are also positive for mCherry. Data are representative of at least three independent experiments in which at least 100 000 events were recorded (*P<0.05, **P<0.001, one-way ANOVA with Bonferroni's post test). (D) Time-lapse imaging of Sup35-GFPGPI (green) and Sup35-mCGPI (magenta) co-culture. Panels were selected from the time-lapse experiment shown in Supplementary Video 1. The elapsed times (hours:minutes) relative to Supplementary Video 1 are indicated. Panels are maximum intensity Z-projection images. Both GFP and mC channels were smoothed by median filtering and gamma-adjusted.
It appeared that more Sup35-mCGPI was transferred to Sup35-GFPGPI cells than either of the control co-cultures (unpublished data). Such transfer was apparent where cell-surface GFP aggregates colocalized with Sup35-mCGPI (Figure 4A, arrows) and in areas containing intracellular Sup35-mCGPI, which usually colocalized with Sup35-GFPGPI (Figure 4A, asterisks). Flow cytometry confirmed this observation showing a higher percentage of mCherry-positive recipient Sup35-GFPGPI cells when co-cultured with Sup35-mCGPI cells as opposed to control cells (Figure 4C). Relative to Sup35-mCGPI cells, both Sup35-mC cells and mCGPI cells released higher amounts (∼20-fold and >2-fold, respectively) of mCherry to the culture media (Supplementary Figure S5A). Since detection of transferred mCherry was limited to co-cultures with cells expressing GPI-anchored mCherry proteins, these data suggest that both GPI anchoring and the Sup35NM domains contribute to efficient intercellular transfer of mCherry. The Sup35-GFPGPI/Sup35-mC co-cultures also provide evidence that exposure to high levels of soluble Sup35NM is not sufficient to induce aggregation of Sup35-GFPGPI.
To assess whether co-culture-induced Sup35-GFPGPI aggregation was self-propagating, Sup35-GFPGPI-expressing cells were isolated from co-cultures by fluorescence-activated cell sorting (FACS). Self-propagating Sup35-GFPGPI aggregates were present only in Sup35-GFPGPI cells that had been co-cultured with aggregate-positive Sup35-mCGPI cells (Figure 4B). Together these data show that Sup35-mCGPI aggregates induced self-propagating Sup35-GFPGPI aggregates in Sup35-GFPGPI cells in association with intercellular transfer of Sup35-mCGPI protein.
Live-cell imaging of Sup35GPI aggregation and spread
Time-lapse imaging of cultures over 1–3 days was used to determine the mechanisms of Sup35-GFPGPI aggregate formation in Sup35-GFPGPI/Sup35-mCGPI co-cultures. Examples of two classes of aggregation-related events observed over the course of six different experiments are shown in Supplementary Videos 1 and 2, with selected frames from Supplementary Video 1 shown in Figure 4D. In the initial frames of Supplementary Video 1, a Sup35-GFPGPI and a Sup35-mCGPI cell shared an aggregate containing GFP and mCherry (Figure 4D, t=02:20, white arrow). Aggregate shearing was mediated by ‘tug of war'-like events due to the divergent movement of cells sharing aggregates (Figure 4D, t=02:55), leaving a portion of the original co-aggregate on each cell (Figure 4D, arrow and arrowhead, t>02:20). It is difficult to determine whether this represented shearing of a single large aggregate versus subdivision of a cluster of smaller aggregates, but in either case these events generated at least two new surfaces for interaction with other cells to initiate new seeding events. In a typical experiment we may observe at least four independent aggregate-shearing events (between different cells). This observation provided a potential mechanism for aggregate fragmentation, a process essential for efficient propagation of prions (Borchsenius et al, 2001; Ness et al, 2002; Kryndushkin et al, 2003; Castilla et al, 2005; Shorter and Lindquist, 2006; Tanaka et al, 2006; Deleault et al, 2007; Satpute-Krishnan et al, 2007). In other cases, as shown at later time points in Figure 4D and Supplementary Video 1, similar interactions resulted in the transfer of entire aggregates between cells. For example, in Supplementary Video 1 an aggregate (Figure 4D and Supplementary Video 1, t>02:55, white arrow) initially associated with cell G1 is transferred to cell G2 and is shared by both G2 and G3 by the end of the time lapse. Altogether, this suggests that Sup35GPI aggregates can exhibit strong attachment to cell surfaces as might be expected due to the multivalent membrane association of a GPI-anchored polymer.
Sheared aggregates exhibited seeding activity, as shown during subsequent induction of Sup35-GFPGPI aggregation by a freshly sheared Sup35-mCGPI aggregate (Figure 4D, t=08:10–10:30, arrowhead, and Supplementary Video 1). Remarkably, the kinetics of induced Sup35-GFPGPI aggregation were very rapid with initial aggregation usually apparent within 35–70 min after initial cell–cell contact. In some instances, the first detectable intercellular contact appeared to be mediated by fine filopodial or membrane nanotube-like projections through space (Figure 4D, t=08:45, bent arrow), structurally akin to those involved in retroviral and PrPSc spread (Sherer et al, 2007; Sowinski et al, 2008; Gousset et al, 2009). Analysis of 3D volume projection images of Sup35-mCGPI/Sup35-GFPGPI co-aggregates indicated that induction of Sup35-GFPGPI aggregation occurred at points of intimate intercellular contact (Supplementary Video 2). In a typical experiment we may observe at least 17 aggregate formation events. These data indicate that intercellular spread of GPI-anchored Sup35 aggregation can occur through direct cell–cell interactions involving contact and either complete or partial transfer of GPI-anchored aggregates. Collectively, these experiments visualize the fundamental processes important for in vivo propagation and spread of PrPSc: formation of new seeding entities via fragmentation of large aggregates; transfer of aggregates between cells; and induction of aggregation in separate cells, thereby resulting in intercellular spread of self-propagating protein aggregation.
Inefficient induction of persistent Sup35-GFPGPI aggregation by fixed Sup35-mCGPI cells
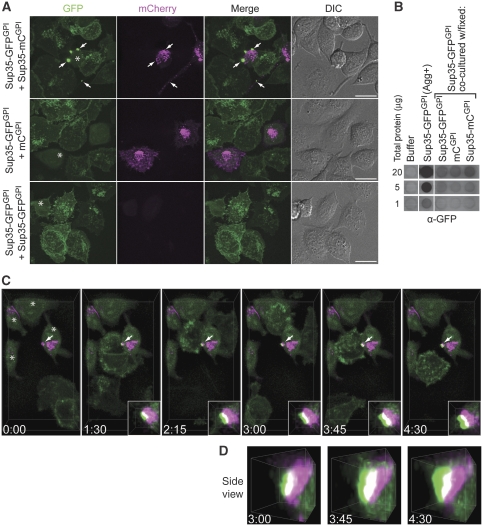
The association between intercellular transfer of Sup35-mCGPI protein to Sup35-GFPGPI cells and induction of Sup35-GFPGPI aggregation raised the question of whether physical transfer of Sup35-mCGPI to recipient cells was necessary. To investigate this issue, we employed a co-culture model analogous to one previously used to study mouse prion infections (Kanu et al, 2002). This involved co-culture of fixed Sup35-mCGPI ‘donor' cells with live Sup35-GFPGPI recipient cells to inhibit transfer of Sup35-mCGPI aggregates. Over 3–4 days of co-culture, new Sup35-GFPGPI aggregates were readily detected in live Sup35-GFPGPI co-cultures with fixed Sup35-mCGPI cells but not in control cultures (Figure 5A). In most cases, GFP aggregates were localized either on Sup35-mCGPI cells or at interfaces between Sup35-GFPGPI and Sup35-mCGPI cells (Figure 5A). After prolonged co-culture (3–4 days), only rare examples of Sup35-GFPGPI cells in distinct possession of GFP aggregates lacking detectable mCherry fluorescence were found (unpublished data). Sup35-GFPGPI cells that contained intracellular Sup35-mCGPI protein were also rare, showing that fixation greatly reduced but did not completely prevent protein transfer from Sup35-mCGPI cells (Supplementary Video 3, t=5:15–6:00). Subsequent passage of the co-cultures was used to eliminate the fixed cells and to assess whether persistent propagation of Sup35-GFPGPI aggregation was induced. Over six passages, only rare examples of aggregate-positive Sup35-GFPGPI cells were found (i.e. at most one Sup35-GFPGPI aggregate might be visualized in a scan of every field of view in a 25-cm2 culture flask) and aggregates from such cultures were near the limit of biochemical detection (Figure 5B). These observations established that fixed Sup35-mCGPI aggregates efficiently induced acute but not persistent Sup35-GFPGPI aggregation.
Figure 5.
Induction of Sup35-GFPGPI aggregation by fixed Sup35-mCGPI cells. (A) Confocal microscopy of co-cultures of live Sup35-GFPGPI cells with aldehyde-fixed Sup35-mCGPI, mCGPI, or Sup35-GFPGPI cells. Images were acquired after 3 days of co-culture and correspond to maximum intensity Z-projections. Arrows: Induced Sup35-GFPGPI aggregates and apparent Sup35-mCGPI seeds. Asterisks: Glutaraldehyde fixation-induced green autofluorescence. GFP (green), mCherry (magenta). Bars, 20 μm. (B) Filter-trap assay of Sup35-GFPGPI cell lysates six passages after co-culture with fixed cells. Sup35-GFPGPI (Agg+): Sup35-GFPGPI cells stably propagating Sup35-GFPGPI aggregates. Wells shown are from the same blot with irrelevant wells removed. (C) Time-lapse imaging of Sup35-GFPGPI aggregate induction by fixed Sup35-mCGPI cell. Sup35-mCGPI cells exhibit mCherry fluorescence (magenta) and diffuse fixation-induced green autofluorescence (asterisks). Arrow: Induced Sup35-GFPGPI aggregate (see insets for magnified view). Panels show 3D volume renderings and the numbers indicate the elapsed time (hours:minutes) relative to Supplementary Video 3. (D) Magnified 3D volume side view (see Supplementary Video 4) of induced Sup35-GFPGPI aggregate (arrow) from panel C. All data are representative of two independent experiments.
Time-lapse imaging of Sup35-GFPGPI aggregate formation revealed that new Sup35-GFPGPI aggregates rapidly assembled onto Sup35-mCGPI aggregates after initial cell contact (Figure 5C and Supplementary Video 3). The growth of the GFP-derived portion of newly induced Sup35-GFPGPI aggregates initiated from small regions of interaction with Sup35-mCGPI seeds (Figure 5A and C, compare 1:30 versus 3:00) strongly suggests that Sup35-GFPGPI aggregation is also seeded by the newly induced Sup35-GFPGPI portion of the co-aggregate seen at t=1:30. In Figure 5C, two successive rounds of Sup35-GFPGPI aggregate assembly on a single pre-existing aggregate were observed. Closer examination of the two aggregation events showed that in the second round (t=3:45–4:30) at least some of the Sup35-GFPGPI added onto the new Sup35-GFPGPI aggregate created in the previous round (t=1:30–3:00) (Figure 5C, inset; Figure 5D, and Supplementary Video 4). In a typical experiment we may observe at least 26 Sup35-GFPGPI aggregate induction events, with 38% of these corresponding to sequential rounds of aggregate assembly seeded by a Sup35-GFPGPI aggregate. This provides direct evidence of the self-propagating nature of Sup35-GFPGPI aggregation. Surprisingly, new Sup35-GFPGPI aggregates were usually retained by fixed Sup35-mCGPI cells when Sup35-GFPGPI cells ultimately disengaged (Figure 5C and Supplementary Video 3). This provides an explanation for the inefficient induction of persistent protein aggregation in Sup35-GFPGPI cells by fixed Sup35-mCGPI cells. The data suggest that intercellular transfer of Sup35-GPI aggregates is important for efficient induction of self-propagating Sup35-GPI aggregation in separate cells.
Sup35 aggregate induction and propagation is dependent on GPI anchoring
To establish that GPI anchoring modulates the inducible aggregation of Sup35NM in N2a cells, a GPI-anchorless construct (Sup35-GFP) was generated (Figure 1A). Subcellular localization of the resulting Sup35-GFP protein was consistent with that of secreted GPI-anchorless proteins (unpublished data) such as anchorless PrPC (Chesebro et al, 2005), including release of high levels of protein to the culture media (Supplementary Figure S5B). Cells stably expressing Sup35-GFP were treated with recombinant Sup35NM-AF568 fibrils. Analysis of the cultures 2 days after fibril treatment showed no evidence of Sup35-GFP aggregates contrary to the Sup35-GFPGPI control (Figure 6A). It appeared by fluorescence microscopy that the Sup35-GFP cells bound less fibrils than Sup35-GFPGPI controls and this was confirmed by quantitative analysis (Supplementary Figure S6). These data suggested that expression of GPI-anchored Sup35NM enhanced the binding of exogenously added Sup35NM fibrils.
Figure 6.
Induction of aggregation is dependent on GPI anchor. (A) Wide-field fluorescence microscopy of Sup35-GFP (top two rows) and Sup35-GFPGPI positive control (bottom row) cells 2 days after treatment with Sup35NM-AF568 fibrils. Arrows: newly induced Sup35-GFPGPI aggregates. Merge: GFP (green), AF568 (magenta), and colocalization (white). Bars, 50 μm. (B) Detergent insolubility assays of fibril-treated cells at passage-6 and 17. Lanes shown are from the same blot for each passage, with irrelevant lanes removed. Cells were treated in triplicate. (C) Filter-trap assay of lysates (5 μg total protein) of fibril-treated Sup35-GFP cells at passage-6 and 10. ‘+' control, Sup35NM-AF568 fibril-treated Sup35-GFPGPI cells at equivalent pass post fibril addition; ‘−' control, 4% SDS buffer. Wells shown are from the same blot with irrelevant wells removed. (D) Filter-trap assay of culture supernatants from cells at passage-6 shown in panel C. Culture supernatant volumes were normalized based on total protein from cells and adjusted to 4% SDS prior to filtering. Positive (‘+') control as described in panel C. Negative (‘−') control is culture medium in 4% SDS. The culture supernatant volume analysed corresponds to approximately six times the cell equivalents loaded in the positive control well. Wells shown are from the same blot with irrelevant wells removed. All data are representative of two independent experiments performed in triplicate.
Extended passage showed that Sup35NM fibril treatment had no effect on Sup35-GFP distribution, suggesting that persistent aggregation was not induced (unpublished data). This observation was confirmed by biochemical analyses, which indicated that there were no detectable Sup35-GFP aggregates in the cell lysates or culture supernatants of Sup35NM fibril-treated cells (Figure 6B–D). Acute co-culture of Sup35-GFP cells with aggregate-positive Sup35-mCGPI cells also failed to induce aggregate formation in Sup35-GFP cells (Supplementary Figure S7). Filter-trap assays verified that Sup35-GFP was capable of specific incorporation into recombinant Sup35NM fibrils (Supplementary Figure S8). These results indicated that, for Sup35NM expressed in the secretory pathway, GPI anchoring was required for induction of self-propagating Sup35NM aggregation.
Discussion
We have shown that Sup35-GFPGPI cells were induced to generate self-propagating aggregates by exposure to Sup35NM amyloid fibrils or cell-associated, GPI-anchored Sup35NM aggregates. In contrast, cells expressing GPI-anchorless Sup35NM produced no detectable aggregates when exposed to the same treatments (Figure 6 and Supplementary Figure S7). This illustrated the role of GPI anchoring in mediating persistent aggregate propagation and ruled out the possibility that aggregate propagation in Sup35-GFPGPI cells was attributed to GPI anchor-independent mechanisms of cell association. These findings were consistent with the inability of GPI-anchorless PrPC to support persistent scrapie infection in cell culture (McNally et al, 2009). Live-cell imaging revealed that new aggregation occurred through intercellular contact between Sup35-GFPGPI cells and aggregates on the surface of Sup35-mCGPI cells (Figures 4 and 5, and Supplementary Videos 1, 2, 3). In addition, inhibition of intercellular aggregate transfer by chemically fixing the aggregate donor Sup35-mCGPI cells significantly decreased the level of persistent Sup35-GFPGPI aggregate induction (Figure 5B), suggesting that efficient intercellular aggregate propagation requires intercellular aggregate transfer. Together, these data demonstrate that GPI anchoring facilitates the propagation and intercellular spread of Sup35NM aggregation and provide direct evidence of fundamental cell–cell aggregate-spreading mechanisms thought to be required for propagation of PrPSc in prion diseases.
Efficient propagation of [PSI+] and other prions in the yeast cytoplasm requires a complex interplay between Hsp104 and other cytoplasmic chaperone proteins (Shorter and Lindquist, 2008; Tipton et al, 2008). The recent report of vertical propagation of cytoplasmic Sup35NM aggregates in N2a cells shows that the mammalian cell cytoplasm contains factors sufficient for propagation of Sup35NM aggregates (Krammer et al, 2009), which could conceivably include orthologues of the corresponding yeast chaperone proteins (Doyle and Wickner, 2009). This is consistent with another very recent report of propagation of polyglutamine aggregates inside mammalian cells (Ren et al, 2009). In our study, the prion-like propagation of GPI-anchored Sup35NM in mammalian cells apparently occurred in a cell-surface/extracellular compartment lacking these cytoplasmic chaperones. This suggests that GPI anchoring of Sup35NM compensated for the absence of these chaperones, perhaps by altering the properties and/or behaviour of Sup35NM aggregates, such as structure and/or propensity, for intercellular spread as discussed below. It is possible that extracellular chaperone-like molecules such as clusterin (Wilson et al, 2008) can assist the propagation and spread of extracellular protein aggregates. However, there are no reports that extracellular chaperones possess the specific aggregate fragmentation activity of Hsp104 required for amplification of seeding-competent particles. Regardless, our findings provide an example of how GPI anchoring can facilitate the replication of a misfolded protein aggregate as a mammalian prion.
Intercellular transfer of PrPC was demonstrated to be dependent on the GPI anchor and cell-to-cell contact, which raised the question of whether GPI anchoring could mediate the transfer of PrPSc (Liu et al, 2002) and other misfolded protein aggregates. Data presented in Figures 4 and 5, and Supplementary Videos 1, 2, 3 show that in our model Sup35 aggregation was induced by cell contact and that stable propagation of new aggregation in a cell depends on the transfer of all or part of the original co-aggregate. Similar to our model, others observed reduced mouse prion infection efficiency initiated by fixed infected cells versus live cells (Kanu et al, 2002; Paquet et al, 2007). Our data provide an explanation for this result, where we observed that fixed aggregates are highly stabilized and less prone to shearing or transfer, thereby favouring the retention of newly induced aggregates by fixed cells. Shearing within the GFP portion of GFP-tagged aggregates induced by Sup35-mCGPI aggregates is possible, but in our many imaging experiments we rarely observed such an event. This may be because GFP-tagged Sup35 aggregates rarely have an opportunity to grow large enough to create a sufficiently frangible surface on which fracture can occur, the size apparently being dependent upon the length of time for which the two cells are in contact. Altogether, our results demonstrate horizontal intercellular spread of Sup35GPI aggregation, a process that presumably allows neuroinvasion and spread of PrPSc that is ultimately essential to transmission of TSE disease between hosts after peripheral infection. By contrast, horizontal spread of cytoplasmic protein aggregates between live cells was either not examined (Krammer et al, 2009) or not clearly detected (Ren et al, 2009). Consequently, propagation of these cytoplasmic protein aggregates either fails to meet an important practical criterion for a mammalian prion in vivo or is much less efficient than mammalian prions. This highlights a key difference between propagation of cytoplasmic and GPI-anchored protein aggregates, and emphasizes the significance of the role of GPI anchoring in aggregate-spreading mechanisms.
Although different mechanisms may be involved, our observation of the importance of cell contact/proximity in intercellular spread of protein aggregates is consistent with previous reports of transmission of mouse prion infection between cells (Kanu et al, 2002; Paquet et al, 2007; Gousset et al, 2009). Furthermore, immunogold electron microscopy studies of scrapie-infected tissues suggest that PrPSc can spread between adjacent cells in vivo (Jeffrey et al, 2009). Other mechanisms of intercellular spread of GPI-anchored proteins that may contribute to PrPSc propagation include membrane painting (Medof et al, 1996) and/or transport on intercellular processes such as exosomes, microparticles, filopodia, and tunnelling nanotubes (Fevrier et al, 2004; Caughey and Baron, 2006; Sherer et al, 2007; Sowinski et al, 2008; Caughey et al, 2009; Gousset et al, 2009). The role of these trafficking pathways in the spread of Sup35GPI aggregates remains to be investigated although we occasionally observed filopodial/membrane nanotube-like projections during initial interactions between aggregate-positive and aggregate-negative cells (Figure 4D).
In addition to effects on intercellular spread, GPI anchoring restricts aggregation to the two-dimensional plane of a membrane thereby impacting aggregate structure. GPI anchoring may limit the formation of large, higher order, highly stable aggregates such as amyloid plaques and promote the formation of smaller oligomeric assemblies that can exhibit enhanced seeding activity, neurotoxic activity, and infectivity (Caughey, 2001; Silveira et al, 2005; Chiti and Dobson, 2006; Tanaka et al, 2006), and might spread readily through the extracellular milieu. Data from in vivo and in vitro studies provide evidence of modulation of PrPSc fibril formation, neurotoxicity, and infectivity via PrP membrane association (Gabizon et al, 1987; McKinley et al, 1991; Chesebro et al, 2005; Silveira et al, 2005; Baron et al, 2006). Similarly, the specific infectivity ([PSI+] induction per mole of Sup35p) and propagation efficiency of Sup35p prion particles in yeast is also higher for smaller particles (Borchsenius et al, 2001; Kryndushkin et al, 2003; Shorter and Lindquist, 2006; Tanaka et al, 2006; Bagriantsev et al, 2008). The upper limit of Sup35-GFPGPI and Sup35-mCGPI aggregate size by fluorescence microscopy varied, but none approached the size of typical amyloid plaques, and other analyses (to be described elsewhere) suggest that these aggregates are not amyloid. Live-cell imaging revealed examples of very small Sup35-mCGPI aggregates inducing the formation of much larger Sup35-GFPGPI aggregates, suggesting that small GPI-anchored Sup35 aggregates can efficiently seed new aggregation (unpublished data). Thus, the potential effects of GPI-anchoring on protein aggregation and transmission of self-propagating aggregates are multifaceted and interrelated.
GPI-anchored PrPC is critical to TSE disease pathogenesis. When mice expressing GPI-anchorless PrPC were inoculated intracerebrally (IC) they did not develop typical scrapie disease (Chesebro et al, 2005). Although PrPSc was produced in the anchorless PrPC mice, the nature and distribution of PrPSc deposits in the brain was very different from that in wild-type mice. Instead of forming diffuse, widespread membrane-bound accumulations as in wild-type mice, GPI-anchorless PrPSc formed large, localized amyloid deposits associated with limited pathology, which resembles that associated with other amyloid diseases (e.g. Alzheimer's disease). It is unclear in this model whether aggregate spread due to PrPSc transport between distant locations is occurring within the CNS, but if it occurs it would seem less pervasive than GPI-anchored PrPSc. In addition, significantly longer incubation times were required to produce TSE infectivity titres and PrPSc levels comparable to those in infected wild-type mice, but these and other phenotypes mentioned above were complemented by introducing a single copy of wild-type prnp to GPI-anchorless PrP mice. This shows that neurodegenerative pathology, the rate of PrPSc formation, and dissemination of PrPSc within the CNS is greatly facilitated by the presence of GPI-anchored PrPC (Chesebro et al, 2005).
There are outstanding questions in regards to scrapie infections of anchorless PrPC mice. Because all the above mentioned experiments were performed using IC inoculations, the distribution of and neuroinvasion by PrPSc in these transgenic mice after peripheral infection remains unknown. The data from anchorless PrPC mice would indicate that with respect to propagation of prions in the context of IC infections, GPI-anchoring may not be essential. However, as discussed in the Introduction, this does not exclude a role for GPI anchoring in modulating the transmissibility of other protein misfolding diseases due to the high likelihood of the multifactorial nature of such processes. It is possible that factors specific to PrP or inherent properties of anchorless PrPSc fibrils (e.g. interactions with membrane proteins or extracellular matrix components, GPI anchor-independent binding to membranes, high frangibility of fibrils) may partially compensate for the absence of the GPI anchor.
The possibility that Alzheimer's amyloid-β (Meyer-Luehmann et al, 2006), polyglutamine (Ren et al, 2009), and tau (Clavaguera et al, 2009; Frost et al, 2009) aggregation are transmissible might raise questions about the argument that GPI anchoring can modulate the transmissibility of protein-misfolding diseases. Despite a lack of epidemiological evidence, this has been noted before in the case of Alzheimer's disease (Goudsmit et al, 1980) and is based in part on an investigation which demonstrated that Alzheimer's disease mouse models developed agent-dependent Aβ-amyloid deposits at accelerated time points after IC inoculation with brain homogenates from Alzheimer's patients or aged Alzheimer's transgenic mice (Meyer-Luehmann et al, 2006). In more relevant models, however, no reproducible clinical signs of disease were observed in 240 primates inoculated IC with brain homogenates from patients with either familial or sporadic Alzheimer's disease (Goudsmit et al, 1980; Brown et al, 1982, 1994). The inability of Aβ-containing brain material to induce cerebral β-amyloidosis when administered by peripheral routes clearly distinguishes this phenomenon from prion transmission (Eisele et al, 2009). The cumulative data support the proposition that TSEs are significantly more transmissible, if not uniquely so, relative to other protein deposition diseases even if there is an element of cell-to-cell propagation of pathology in the brain in other diseases. One author has even proposed a separate terminology (prionoid) for these prion-like disorders (Aguzzi, 2009). The reason for this difference has not been established, but we believe GPI anchoring (or at least membrane anchoring) of the associated misfolded polypeptide could play a critical role in enhancing the spread of protein aggregation from the periphery into and within the CNS. Whether any other protein-misfolding disorder is ultimately found transmissible under special circumstances, it nevertheless remains an important question as to whether GPI anchoring might modulate the associated transmissibility and pathology.
Our data are the first, to our knowledge, to directly demonstrate that GPI-anchored protein aggregation can spread between unrelated cells by cell-to-cell contact and transfer of aggregate seeds. While not excluding additional mechanisms of transfer, we propose that GPI anchor-mediated intercellular spread of PrP aggregates is a central component to prion disease pathogenesis. While such transfer would have implications for prion biogenesis and spread in all circumstances of prion disease, it would be particularly critical for infections resulting from peripheral prion exposure where prion neuroinvasion and propagation in different tissue types occurs over the course of infection. Although no natural examples exist, it will also be important to investigate whether other mechanisms of membrane association (e.g. transmembrane) could support the propagation of misfolded protein aggregates as this may not be limited to GPI anchoring.
Materials and methods
Aggregate induction by Sup35NM-AF568 fibrils
Cells were plated at 10% confluence in a 24-well tissue culture plate. After one day of growth, cells were treated with 2.6 pmole of either recombinant Sup35NM or Ure2p80 fibrils labelled with AF568. Mock controls were treated with equal volumes of PBS that had been processed through the AF568 labelling protocol to account for the possibility of AF568 carry-over. Sup35-GFP cells were treated with PBS or Sup35NM-AF568 fibrils only. Cells were maintained for 2 more days at which point they were split and re-plated at 10% confluence. Cells were subsequently re-plated at 5 or 10% confluence every 4 or 3 days, respectively.
GFP-tagged Sup35 cell co-cultures with aggregate-producing Sup35-mCGPI cells
For flow cytometric and FACS experiments cells were plated at 5% confluence each, grown for 3 days, and then analysed for GFP and mCherry or sorted by FACS to collect GFP-expressing cells only. FACS-sorted GFP cells were grown to confluence and analysed by wide-field fluorescence microscopy for mCherry-expressing cells. Cells were re-plated at 10% confluence and FACS was repeated until no mCherry cells were visible by fluorescence microscopy. Cells were subsequently re-plated at 5 or 10% confluence every 4 or 3 days, respectively. Flow-cytometric data were analysed using the FlowJo software.
Live-cell confocal microscopy of co-cultured cells was performed on cells each plated at either 5 or 25% confluence for live-cell co-cultures. Fixed cell co-cultures were created as described elsewhere (Kanu et al, 2002) with the exception that fixed ‘donor' cells were plated at ∼30% confluence and fixed 2.5 h after plating while live Sup35-GFPGPI ‘recipient' cells were plated at 5% confluence.
Detergent insolubility
Cells were plated at 10% confluence in six-well tissue culture plates and grown to confluence. Cells were washed with phosphate-buffered balanced salts solution (PBBS) (pH 7.4) and lysed with DS buffer (1% sarkosyl, 1 mM EDTA in PBS) supplemented with Complete EDTA-free protease inhibitor cocktail (Roche). Lysates were treated with 25 U of benzonase per millilitre of lysate supplemented with 8 mM MgCl2 for 30 min. After removing aliquots for BCA total-protein analysis, samples were supplemented with EDTA to 50 mM to prevent precipitation of sarkosyl-magnesium salts. Lysates were centrifuged for 30 min at 350 000 × g to separate detergent-insoluble material. The resulting supernatants were methanol-precipitated. Samples were analysed by SDS–PAGE and Western blotting using anti-Sup35 N and anti-rabbit IgG alkaline phosphatase, loading 100 μg total protein equivalents per lane.
Filter-trap assays were performed based on methods described previously (Scherzinger et al, 1997; Xu et al, 2002). Cells plated at 10% confluence and grown to confluence in 24-well plates were lysed in FT buffer (1% SDS, 1 mM EDTA in PBS) supplemented with Complete EDTA-free protease inhibitors and treated with benzonase as described above, except using 75 U of benzonase per millilitre of lysates supplemented with 16 mM MgCl2. EDTA was added to a final concentration of 50 mM after benzonase digestion. Cell lysates were adjusted to 10 and 50 μg/ml protein in 4% SDS with PBS and allowed to incubate at room temperature for 30 min. Culture supernatants were prepared by centrifugation at 3000 × g for 5 min to clear cellular debris. The top 80% of the cleared supernatant was removed and adjusted to 4% SDS in PBS by adding 1 volume of 8% SDS in 2 × PBS. Volumes were normalized for total protein from their respective cell lysates using 4% SDS in PBS, with 50% unconditioned cleared media. All samples were applied to an Immobilon-P 0.45 μm membrane pre-wetted with FT buffer using The Convertible dot blot apparatus (Gibco). Wells were washed five times with 200 μl of FT buffer. Proteins were detected by Western blotting as described above using mouse anti-GFP (Roche) or rabbit anti-RFP (for mCherry; Rockland Immunochemicals) as primary antibodies, and anti-mouse or anti-rabbit IgG alkaline phosphatase as secondary antibody.
Chymotrypsin digests
Cells were plated at 10% confluence and grown to confluence before being lysed in DS buffer and treated with benzonase as described above. The lysates were treated with chymotrypsin at a 1:250 chymotrypsin (Ct)-to-total protein mass ratio for 15 min at 37°C. The reactions were placed on ice and Pefabloc was added to stop the reactions. Samples were methanol-precipitated at −20°C after adding thyroglobulin. The samples were analysed by SDS–PAGE and Western blotting using anti-Sup35 N with anti-rabbit IgG alkaline phosphatase secondary. All lanes were loaded with 25 μg total protein equivalents.
Supplementary Material
Supplementary Video 1
Supplementary Video 2
Supplementary Video 3
Supplementary Video 4
Supplementary Data
Review Process File
Acknowledgments
We thank D Dorward for TEM analysis, R Messer for FACS analysis, K Hasenkrug for flow cytometry interpretation, A Athman for video editing assistance, S Lindquist for anti-Sup35M antibody and pJC45Sup35Stop plasmid, J Weissman for pAEDNMCh plasmid, R Wickner for p530 plasmid and anti-Ure2p antibody, B Nichols and J Lippincott-Schwartz for GFPGPI plasmid, V Boyko for pHIV-gag-mCherry, and R Tsien for permission to use mCherry. We thank M Davis and K Kisiel for helpful input on imaging and image processing. We also thank B Caughey, B Chesebro, and K McNally for critical review of the paper. This research was supported by the Intramural Research Program of the NIH (National Institute of Allergy and Infectious Diseases).
Footnotes
The authors declare that they have no conflict of interest.
References
- Aguzzi A (2009) Cell biology: beyond the prion principle. Nature 459: 924–925 [DOI] [PubMed] [Google Scholar]
- Bagriantsev SN, Gracheva EO, Richmond JE, Liebman SW (2008) Variant-specific [PSI+] infection is transmitted by Sup35 polymers within [PSI+] aggregates with heterogeneous protein composition. Mol Biol Cell 19: 2433–2443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron GS, Magalhaes AC, Prado MA, Caughey B (2006) Mouse-adapted scrapie infection of SN56 cells: greater efficiency with microsome-associated versus purified PrP-res. J Virol 80: 2106–2117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron GS, Wehrly K, Dorward DW, Chesebro B, Caughey B (2002) Conversion of raft associated prion protein to the protease-resistant state requires insertion of PrP-res (PrP(Sc)) into contiguous membranes. EMBO J 21: 1031–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borchsenius AS, Wegrzyn RD, Newnam GP, Inge-Vechtomov SG, Chernoff YO (2001) Yeast prion protein derivative defective in aggregate shearing and production of new ‘seeds'. EMBO J 20: 6683–6691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown P, Gibbs CJ Jr, Rodgers-Johnson P, Asher DM, Sulima MP, Bacote A, Goldfarb LG, Gajdusek DC (1994) Human spongiform encephalopathy: the National Institutes of Health series of 300 cases of experimentally transmitted disease. Ann Neurol 35: 513–529 [DOI] [PubMed] [Google Scholar]
- Brown P, Salazar AM, Gibbs CJ Jr, Gajdusek DC (1982) Alzheimer's disease and transmissible virus dementia (Creutzfeldt–Jakob disease). Ann N Y Acad Sci 396: 131–143 [DOI] [PubMed] [Google Scholar]
- Castilla J, Saa P, Hetz C, Soto C (2005) In vitro generation of infectious scrapie prions. Cell 121: 195–206 [DOI] [PubMed] [Google Scholar]
- Caughey B (2001) Interactions between prion protein isoforms: the kiss of death? Trends Biochem Sci 26: 235–242 [DOI] [PubMed] [Google Scholar]
- Caughey B, Baron GS (2006) Prions and their partners in crime. Nature 443: 803–810 [DOI] [PubMed] [Google Scholar]
- Caughey B, Baron GS, Chesebro B, Jeffrey M (2009) Getting a grip on prions: oligomers, amyloids, and pathological membrane interactions. Annu Rev Biochem 78: 177–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernoff YO, Lindquist SL, Ono B, Inge-Vechtomov SG, Liebman SW (1995) Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+]. Science 268: 880–884 [DOI] [PubMed] [Google Scholar]
- Chesebro B, Trifilo M, Race R, Meade-White K, Teng C, LaCasse R, Raymond L, Favara C, Baron G, Priola S, Caughey B, Masliah E, Oldstone M (2005) Anchorless prion protein results in infectious amyloid disease without clinical scrapie. Science 308: 1435–1439 [DOI] [PubMed] [Google Scholar]
- Chien P, Weissman JS, DePace AH (2004) Emerging principles of conformation-based prion inheritance. Annu Rev Biochem 73: 617–656 [DOI] [PubMed] [Google Scholar]
- Chiti F, Dobson CM (2006) Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem 75: 333–366 [DOI] [PubMed] [Google Scholar]
- Clavaguera F, Bolmont T, Crowther RA, Abramowski D, Frank S, Probst A, Fraser G, Stalder AK, Beibel M, Staufenbiel M, Jucker M, Goedert M, Tolnay M (2009) Transmission and spreading of tauopathy in transgenic mouse brain. Nat Cell Biol 11: 909–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox B, Ness F, Tuite M (2003) Analysis of the generation and segregation of propagons: entities that propagate the [PSI+] prion in yeast. Genetics 165: 23–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox BS (1965) PSI, a cytoplasmic suppressor of super-suppressor in yeast. Heredity 20: 505–521 [Google Scholar]
- Deleault NR, Harris BT, Rees JR, Supattapone S (2007) Formation of native prions from minimal components in vitro. Proc Natl Acad Sci USA 104: 9741–9746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle SM, Wickner S (2009) Hsp104 and ClpB: protein disaggregating machines. Trends Biochem Sci 34: 40–48 [DOI] [PubMed] [Google Scholar]
- Eisele YS, Bolmont T, Heikenwalder M, Langer F, Jacobson LH, Yan ZX, Roth K, Aguzzi A, Staufenbiel M, Walker LC, Jucker M (2009) Induction of cerebral {beta}-amyloidosis: intracerebral versus systemic Aβ inoculation. Proc Natl Acad Sci USA 106: 12926–12931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fevrier B, Vilette D, Archer F, Loew D, Faigle W, Vidal M, Laude H, Raposo G (2004) Cells release prions in association with exosomes. Proc Natl Acad Sci USA 101: 9683–9688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost B, Jacks RL, Diamond MI (2009) Propagation of tau misfolding from the outside to the inside of a cell. J Biol Chem 284: 12845–12852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabizon R, McKinley MP, Groth DF, Kenaga L, Prusiner SB (1988) Properties of scrapie prion protein liposomes. J Biol Chem 263: 4950–4955 [PubMed] [Google Scholar]
- Gabizon R, McKinley MP, Prusiner SB (1987) Purified prion proteins and scrapie infectivity copartition into liposomes. Proc Natl Acad Sci USA 84: 4017–4021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudsmit J, Morrow CH, Asher DM, Yanagihara RT, Masters CL, Gibbs CJ Jr, Gajdusek DC (1980) Evidence for and against the transmissibility of Alzheimer disease. Neurology 30: 945–950 [DOI] [PubMed] [Google Scholar]
- Gousset K, Schiff E, Langevin C, Marijanovic Z, Caputo A, Browman DT, Chenouard N, de CF, Martino A, Enninga J, Olivo-Marin JC, Mannel D, Zurzolo C (2009) Prions hijack tunnelling nanotubes for intercellular spread. Nat Cell Biol 11: 328–336 [DOI] [PubMed] [Google Scholar]
- Gousset K, Zurzolo C (2009) Tunnelling nanotubes: a highway for prion spreading? Prion 3: 94–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Immel F, Jiang Y, Wang YQ, Marchal C, Maillet L, Perrett S, Cullin C (2007) In vitro analysis of SpUre2p, a prion-related protein, exemplifies the relationship between amyloid and prion. J Biol Chem 282: 7912–7920 [DOI] [PubMed] [Google Scholar]
- Jeffrey M, McGovern G, Goodsir CM, Gonzalez L (2009) Strain-associated variations in abnormal PrP trafficking of sheep scrapie. Brain Pathol 19: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanu N, Imokawa Y, Drechsel DN, Williamson RA, Birkett CR, Bostock CJ, Brockes JP (2002) Transfer of scrapie prion infectivity by cell contact in culture. Curr Biol 12: 523–530 [DOI] [PubMed] [Google Scholar]
- Krammer C, Kryndushkin D, Suhre MH, Kremmer E, Hofmann A, Pfeifer A, Scheibel T, Wickner RB, Schatzl HM, Vorberg I (2009) The yeast Sup35NM domain propagates as a prion in mammalian cells. Proc Natl Acad Sci USA 106: 462–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryndushkin DS, Alexandrov IM, Ter-Avanesyan MD, Kushnirov VV (2003) Yeast [PSI+] prion aggregates are formed by small Sup35 polymers fragmented by Hsp104. J Biol Chem 278: 49636–49643 [DOI] [PubMed] [Google Scholar]
- Li L, Lindquist S (2000) Creating a protein-based element of inheritance. Science 287: 661–664 [DOI] [PubMed] [Google Scholar]
- Liu T, Li R, Pan T, Liu D, Petersen RB, Wong BS, Gambetti P, Sy MS (2002) Intercellular transfer of the cellular prion protein. J Biol Chem 277: 47671–47678 [DOI] [PubMed] [Google Scholar]
- Mabbott NA, MacPherson GG (2006) Prions and their lethal journey to the brain. Nat Rev Microbiol 4: 201–211 [DOI] [PubMed] [Google Scholar]
- Mack M, Kleinschmidt A, Bruhl H, Klier C, Nelson PJ, Cihak J, Plachy J, Stangassinger M, Erfle V, Schlondorff D (2000) Transfer of the chemokine receptor CCR5 between cells by membrane-derived microparticles: a mechanism for cellular human immunodeficiency virus 1 infection. Nat Med 6: 769–775 [DOI] [PubMed] [Google Scholar]
- McKinley MP, Meyer RK, Kenaga L, Rahbar F, Cotter R, Servan A, Prusiner SB (1991) Scrapie prion rod formation in vitro requires both detergent extraction and limited proteolysis. J Virol 65: 1340–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally KL, Ward AE, Priola SA (2009) Cells expressing anchorless prion protein are resistant to scrapie infection. J Virol 83: 4469–4475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medof ME, Nagarajan S, Tykocinski ML (1996) Cell-surface engineering with GPI-anchored proteins. FASEB J 10: 574–586 [DOI] [PubMed] [Google Scholar]
- Meyer-Luehmann M, Coomaraswamy J, Bolmont T, Kaeser S, Schaefer C, Kilger E, Neuenschwander A, Abramowski D, Frey P, Jaton AL, Vigouret JM, Paganetti P, Walsh DM, Mathews PM, Ghiso J, Staufenbiel M, Walker LC, Jucker M (2006) Exogenous induction of cerebral beta-amyloidogenesis is governed by agent and host. Science 313: 1781–1784 [DOI] [PubMed] [Google Scholar]
- Ness F, Ferreira P, Cox BS, Tuite MF (2002) Guanidine hydrochloride inhibits the generation of prion ‘seeds' but not prion protein aggregation in yeast. Mol Cell Biol 22: 5593–5605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onfelt B, Nedvetzki S, Yanagi K, Davis DM (2004) Cutting edge: membrane nanotubes connect immune cells. J Immunol 173: 1511–1513 [DOI] [PubMed] [Google Scholar]
- Osherovich LZ, Cox BS, Tuite MF, Weissman JS (2004) Dissection and design of yeast prions. PLoS Biol 2: E86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquet S, Langevin C, Chapuis J, Jackson GS, Laude H, Vilette D (2007) Efficient dissemination of prions through preferential transmission to nearby cells. J Gen Virol 88: 706–713 [DOI] [PubMed] [Google Scholar]
- Patino MM, Liu JJ, Glover JR, Lindquist S (1996) Support for the prion hypothesis for inheritance of a phenotypic trait in yeast. Science 273: 622–626 [DOI] [PubMed] [Google Scholar]
- Paushkin SV, Kushnirov VV, Smirnov VN, Ter-Avanesyan MD (1996) Propagation of the yeast prion-like [psi+] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. EMBO J 15: 3127–3134 [PMC free article] [PubMed] [Google Scholar]
- Piston DW, Rizzo MA (2008) FRET by fluorescence polarization microscopy. Methods Cell Biol 85: 415–430 [DOI] [PubMed] [Google Scholar]
- Ren PH, Lauckner JE, Kachirskaia I, Heuser JE, Melki R, Kopito RR (2009) Cytoplasmic penetration and persistent infection of mammalian cells by polyglutamine aggregates. Nat Cell Biol 11: 219–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross ED, Minton A, Wickner RB (2005) Prion domains: sequences, structures and interactions. Nat Cell Biol 7: 1039–1044 [DOI] [PubMed] [Google Scholar]
- Satpute-Krishnan P, Langseth SX, Serio TR (2007) Hsp104-dependent remodeling of prion complexes mediates protein-only inheritance. PLoS Biol 5: e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherzinger E, Lurz R, Turmaine M, Mangiarini L, Hollenbach B, Hasenbank R, Bates GP, Davies SW, Lehrach H, Wanker EE (1997) Huntingtin-encoded polyglutamine expansions form amyloid-like protein aggregates in vitro and in vivo. Cell 90: 549–558 [DOI] [PubMed] [Google Scholar]
- Serio TR, Cashikar AG, Kowal AS, Sawicki GJ, Moslehi JJ, Serpell L, Arnsdorf MF, Lindquist SL (2000) Nucleated conformational conversion and the replication of conformational information by a prion determinant. Science 289: 1317–1321 [DOI] [PubMed] [Google Scholar]
- Serio TR, Cashikar AG, Moslehi JJ, Kowal AS, Lindquist SL (1999) Yeast prion [psi +] and its determinant, Sup35p. Methods Enzymol 309: 649–673 [DOI] [PubMed] [Google Scholar]
- Sherer NM, Lehmann MJ, Jimenez-Soto LF, Horensavitz C, Pypaert M, Mothes W (2007) Retroviruses can establish filopodial bridges for efficient cell-to-cell transmission. Nat Cell Biol 9: 310–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter J, Lindquist S (2006) Destruction or potentiation of different prions catalyzed by similar Hsp104 remodeling activities. Mol Cell 23: 425–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter J, Lindquist S (2008) Hsp104, Hsp70 and Hsp40 interplay regulates formation, growth and elimination of Sup35 prions. EMBO J 27: 2712–2724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silveira JR, Raymond GJ, Hughson AG, Race RE, Sim VL, Hayes SF, Caughey B (2005) The most infectious prion protein particles. Nature 437: 257–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snapp EL (2009) Fluorescent proteins: a cell biologist's user guide. Trends Cell Biol 19: 649–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowinski S, Jolly C, Berninghausen O, Purbhoo MA, Chauveau A, Kohler K, Oddos S, Eissmann P, Brodsky FM, Hopkins C, Onfelt B, Sattentau Q, Davis DM (2008) Membrane nanotubes physically connect T cells over long distances presenting a novel route for HIV-1 transmission. Nat Cell Biol 10: 211–219 [DOI] [PubMed] [Google Scholar]
- Tanaka M, Chien P, Naber N, Cooke R, Weissman JS (2004) Conformational variations in an infectious protein determine prion strain differences. Nature 428: 323–328 [DOI] [PubMed] [Google Scholar]
- Tanaka M, Chien P, Yonekura K, Weissman JS (2005) Mechanism of cross-species prion transmission: an infectious conformation compatible with two highly divergent yeast prion proteins. Cell 121: 49–62 [DOI] [PubMed] [Google Scholar]
- Tanaka M, Collins SR, Toyama BH, Weissman JS (2006) The physical basis of how prion conformations determine strain phenotypes. Nature 442: 585–589 [DOI] [PubMed] [Google Scholar]
- Tipton KA, Verges KJ, Weissman JS (2008) In vivo monitoring of the prion replication cycle reveals a critical role for Sis1 in delivering substrates to Hsp104. Mol Cell 32: 584–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorberg I, Raines A, Priola SA (2004) Acute formation of protease-resistant prion protein does not always lead to persistent scrapie infection in vitro. J Biol Chem 279: 29218–29225 [DOI] [PubMed] [Google Scholar]
- Wickner RB (1994) [URE3] as an altered URE2 protein: evidence for a prion analog in Saccharomyces cerevisiae. Science 264: 566–569 [DOI] [PubMed] [Google Scholar]
- Wilson MR, Yerbury JJ, Poon S (2008) Potential roles of abundant extracellular chaperones in the control of amyloid formation and toxicity. Mol Biosyst 4: 42–52 [DOI] [PubMed] [Google Scholar]
- Xu G, Gonzales V, Borchelt DR (2002) Rapid detection of protein aggregates in the brains of Alzheimer patients and transgenic mouse models of amyloidosis. Alzheimer Dis Assoc Disord 16: 191–195 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Video 1
Supplementary Video 2
Supplementary Video 3
Supplementary Video 4
Supplementary Data
Review Process File