Abstract
How tumour suppressor p53 bifurcates cell cycle arrest and apoptosis and executes these distinct pathways is not clearly understood. We show that BAX and PUMA promoters harbour an identical MAR element and are transcriptional targets of SMAR1. On mild DNA damage, SMAR1 selectively represses BAX and PUMA through binding to the MAR independently of inducing p53 deacetylation through HDAC1. This generates an anti-apoptotic response leading to cell cycle arrest. Importantly, knockdown of SMAR1 induces apoptosis, which is abrogated in the absence of p53. Conversely, apoptotic DNA damage results in increased size and number of promyelocytic leukaemia (PML) nuclear bodies with consequent sequestration of SMAR1. This facilitates p53 acetylation and restricts SMAR1 binding to BAX and PUMA MAR leading to apoptosis. Thus, our study establishes MAR as a damage responsive cis element and SMAR1–PML crosstalk as a switch that modulates the decision between cell cycle arrest and apoptosis in response to DNA damage.
Keywords: BAX, HDAC1, PML, PUMA, SMAR1
Introduction
The tumour suppressor p53 is the cellular sentinel of the mammalian cell cycle and an indispensable component of the DNA damage response pathway. Activation of p53 in response to DNA damage results in either cell cycle arrest or apoptosis. Although genes that regulate cellular processes such as arrest and apoptosis are essentially p53 targets, activation of p53 always results in specific and selective transcription of p53-regulated genes (Riley et al, 2008). Thus, it is likely that unique sets of p53-regulated genes operate in tandem to bring about a desired outcome in response to specific stimuli. How p53 executes these two distinct functions in a promoter-specific manner remains largely unclear. Recent reports suggest that activation of specific promoters by p53 is achieved through its interaction with heterologous transcription factors such as Hzf, human cellular apoptosis susceptibility (hCAS)/CSE1L and ASPP family proteins (Samuels-Lev et al, 2001; Das et al, 2007; Tanaka et al, 2007). In addition, under conditions of stress, different phosphorylation and acetylation modules stabilize p53 enhancing its sequence-specific DNA binding and transcriptional activity (Sakaguchi et al, 1998). Although phosphorylation at amino-terminus is required for p53 stability, acetylation at carboxyl-terminus has been shown to be indispensable for p53 transcriptional activation (Tang et al, 2008).
The tumour suppressor promyelocytic leukaemia (PML) protein is involved in the regulation of p53-dependent and -independent apoptosis. The PML protein localizes to multi-protein sub-nuclear structures termed as PML nuclear bodies (PML-NBs), which act as sensors of DNA damage and cellular stress. On genotoxic stress, the PML protein functions as a transcriptional co-activator of p53 by recruiting it to the PML-NBs, wherein PML facilitates p53 phosphorylation and acetylation through recruitment of factors such as HIPK2 and CBP (Bernardi and Pandolfi, 2007). Further, the PML gene itself is a transcriptional target of p53 (de Stanchina et al, 2004).
Scaffold/matrix attachment regions (S/MARs) are regulatory DNA sequences mostly present upstream of various promoters. Matrix attachment region-binding proteins (MARBPs), which bind to such regulatory sequences, interact with numerous chromatin modifying factors and facilitate transcription in response to diverse stimuli (Zaidi et al, 2005). SMAR1 is an MARBP identified in mouse double positive thymocytes, wherein it binds to MARβ sequence at TCRβ locus and affects V(D)J recombination (Chattopadhyay et al, 2000; Kaul-Ghanekar et al, 2005). Subsequently, SMAR1 has been characterized as a tumour suppressor by virtue of its ability to interact with p53 and delay tumour growth in mouse melanoma model (Kaul et al, 2003; Jalota et al, 2005). The tumour suppressor p53 is also an MARBP that associates with the nuclear matrix and this association increases after DNA damage (Jiang et al, 2001). The PML-NBs are also associated with the nuclear matrix indirectly. However, the biological and functional significances of nuclear matrix-bound p53 and nuclear matrix-associated PML-NBs in context to p53 regulation are not known.
In this study, we show that p53 target gene SMAR1 modulates the cellular response to genotoxic stress by a dual mechanism. First, SMAR1 interacts with p53 and facilitates p53 deacetylation through recruitment of HDAC1. Second, SMAR1 represses the transcription of BAX and PUMA by binding to an identical 25 bp MAR element in their promoters. We show that on mild DNA damage, SMAR1 generates an anti-apoptotic response by promoting p53 deacetylation and specifically repressing Bax and Puma expression. Reducing the expression of SMAR1 by shRNA leads to significant increase in p53-dependent apoptosis. However, severe DNA damage results in sequestration of SMAR1 into the PML-NBs. This facilitates p53 acetylation and transactivation of BAX and PUMA leading to apoptosis in cancer cells. Silencing of PML by specific siRNA abrogates DNA damage-induced apoptosis through transrepression of BAX and PUMA by SMAR1. Thus, sequestration of SMAR1 into the PML-NBs acts as a molecular switch to dictate p53-dependent cell arrest and apoptosis on DNA damage and we suggest that SMAR1 may be a new molecular target for cancer therapy.
Results
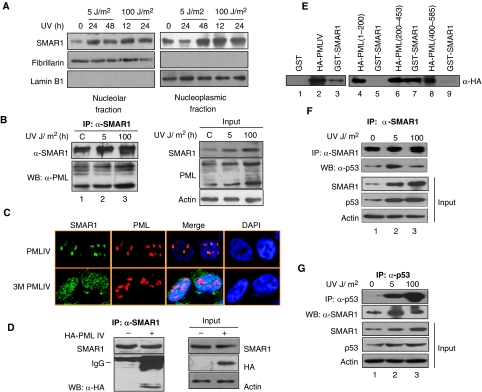
SMAR1 deacetylates p53 through association with HDAC1
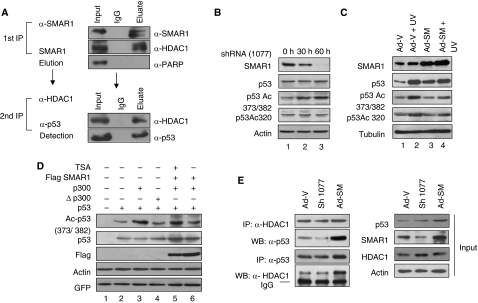
Earlier, we have shown that SMAR1 interacts with p53 and stabilizes it in the nucleus (Jalota et al, 2005). We showed that SMAR1 inhibits Cyclin D1 expression by recruitment of HDAC1–mSin3A repressor complex on the promoter MAR (Rampalli et al, 2005). As SMAR1 and p53 are both matrix-associated transcription factors and both interact with HDAC1 independently, we investigated whether they are associated together in a complex. Double co-immunoprecipitation assay in HCT116 p53+/+ cells revealed the presence of detectable SMAR1–HDAC1–p53 complexes in vivo (Figure 1A). However, in the same eluate, SMAR1 did not show binding to another MAR protein PARP, thus revealing the specificity of the interaction. As p53 exists in a complex with HDAC1 along with SMAR1, it is possible that SMAR1 might deacetylate p53 endogenously by recruiting HDAC1 and thus keep its transactivation potential under check. To validate this, we investigated the levels of p53 acetylation on SMAR1 knockdown. SMAR1-specific shRNAs (sh745 and sh1077) were generated targeting two different regions of SMAR1 mRNA. The knockdown and specificity of the shRNAs were checked by western blotting (Supplementary Figure S1). Notably, knockdown of SMAR1 significantly induced p53 acetylation at Lys 373/382 (Figure 1B). However, total p53 level and p53 acetylation at Lys 320 were not affected. To understand the physiological relevance of this regulation in response to genotoxic stress, HCT116 p53+/+ cells were transduced with GFP expressing control and SMAR1 adenoviruses followed by UV (100 J/m2) treatment. Interestingly, SMAR1 overexpression strongly inhibited p53 acetylation at Lys 373/382, which was otherwise induced significantly on UV treatment (Figure 1C, panel 3, lane 2 versus lanes 3 and 4). Further, we performed an in vivo deacetylation assay. HCT116 p53−/− cells were transfected with p53 alone or in combination with p300 and SMAR1 in the presence or absence of Trichostatin A (TSA). Although p300 induced p53 acetylation at Lys-373/382 (Figure 1D, lane 3), presence of SMAR1 efficiently reduced p53 acetylation (lane 6), which was reversed on treatment with HDAC-inhibitor TSA (lane 5). Notably, although total p53 levels are high in SMAR1-transfected cells as SMAR1 is involved in p53 stabilization (Jalota et al, 2005), p53 acetylation levels are reduced in the presence of SMAR1 and the absence of TSA (lane 6). Finally, to confirm whether SMAR1 imparts deacetylase activity on p53 through recruitment of HDAC1, we carried out reversible co-immunoprecipitation to evaluate the association of p53 with HDAC1 in the presence and absence of SMAR1. Although knockdown of SMAR1 reduced the amount of p53 pulled with HDAC1, overexpression resulted in strong increase in their association (Figure 1E). This suggests that SMAR1-mediated p53 deacetylation is through recruitment of HDAC1.
Figure 1.
SMAR1 interacts with and regulates p53 acetylation endogenously. (A) Double co-immunoprecipitation assay to check the in vivo association of SMAR1–HDAC1 complex with p53. One milligram of cell lysate from HCT116 p53+/+ cells was immunoprecipitated sequentially with SMAR1 and HDAC1 antibodies. The eluted fraction was probed with p53 antibody. (B) HCT116 p53+/+ cells were transfected with SMAR1 shRNA (sh1077). Western blotting shows endogenous acetylation status of p53 at lys 373/382 and total p53 level 30 and 60 h after transfection. (C) HCT116 p53+/+ cells transduced with GFP expressing control and SMAR1 adenoviruses and treated with UV (100 J/m2) for 24 h. The levels of p53 and p53 acetylation status in comparison with SMAR1 expression are shown. (D) In vitro deacetylation assay of p53 by SMAR1. HCT116 p53−/− cells were transfected with p53, p300 expression plasmids in different combinations and treated with TSA (200 nM, 16 h) as given in the figure. GFP expression plasmid was transfected to monitor transfection efficiency. (E) Reversible co-immunoprecipitation assay in HCT116 p53+/+ cells showing differential association of p53 with HDAC1 in SMAR1 knockdown and overexpressed cells (left panel). Input controls are shown in right panel.
BAX and PUMA are transcriptional targets of SMAR1
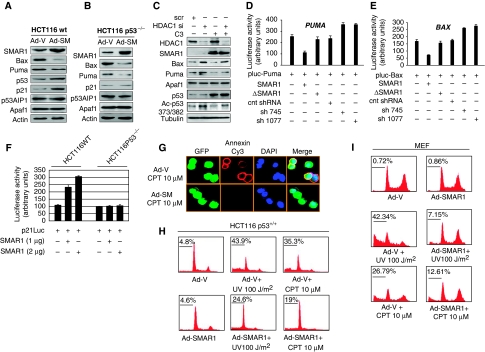
As SMAR1 modulates p53 acetylation through HDAC1, we evaluated the expression of p53 target genes. Overexpression of SMAR1 resulted in significant downregulation of Bax and Puma, whereas the levels of p53AIP1 and Apaf1, which are also p53 targets remained unchanged (Figure 2A). However, the levels of total p53 and p21 increased significantly corroborating our earlier data that overexpression of SMAR1 induces cell cycle arrest through p53 Ser 15 phosphorylation (Jalota et al, 2005). Thus, on one hand, SMAR1 enhances the expression of p21 and, on the other, it inhibits the expression of apoptotic genes BAX and PUMA. Interestingly, SMAR1 overexpression in HCT116 p53−/− cells also repressed Bax and Puma, but failed to induce p21 (Figure 2B). As SMAR1 interacts with HDAC1 and exists in a complex with p53, it is possible that SMAR1 deacetylates p53 through HDAC1. We, therefore, investigated whether knockdown of HDAC1 alters p53 acetylation status and alleviates the repression of Bax and Puma by SMAR1. Silencing of HDAC1 not only abrogated SMAR1-mediated repression of Bax and Puma, but also induced their basal expression. This suggests that SMAR1 negatively regulates the expression of Bax and Puma through HDAC1. In addition, HDAC1 knockdown induced p53 acetylation corroborating with increased Bax and Puma levels (Figure 2C) indicating that deacetylation of p53 by SMAR1 is dependent on HDAC1. Apaf1 expression, however, remained unaltered on HDAC1 knockdown. Reporter assays using full-length BAX and PUMA promoters in HCT116 p53−/− cells exhibited strong inverse correlation on SMAR1 ectopic expression and knockdown (Figure 2D and E), whereas p53AIP1 promoter did not show any significant change (Supplementary Figure S2). Again, p21 reporter was induced by SMAR1 in HCT116 p53+/+ cells, but failed to show read out in p53−/− cells (Figure 2F). Thus, induction of p21 by SMAR1 is p53 dependent, whereas transrepression of BAX and PUMA is p53 independent. Finally, to evaluate the biological significance of SMAR1-mediated repression of Bax and Puma, HCT116 p53+/+ cells were transduced with GFP expressing control adenovirus (Ad-V) and SMAR1 adenovirus (Ad-SM) followed by UV (100 J/m2) and Camptothecin (10 μM) treatment 48 h post-transduction. Surface staining using annexin-V-conjugated Cy3 revealed significant decrease in apoptotic population in SMAR1 overexpressed cells (Figure 2G). The statistical representation of percentage apoptosis is given in Supplementary Figure S2B. Propidium iodide staining in HCT116 p53+/+ cells (Figure 2H) and mouse embryonic fibroblast (Figure 2I) transduced with SMAR1 adenovirus and treated with UV and Camptothecin showed that SMAR1 can significantly protect these cells from genotoxic stress-induced apoptosis. Thus, the protective function of SMAR1 on genotoxic stress is most likely attributed to its function as a negative regulator of Bax and Puma.
Figure 2.
SMAR1 regulates the expression of key apoptotic molecules Bax and Puma and inhibits apoptosis. (A) HCT116 p53+/+ and (B) HCT116 p53−/− cells were transduced with GFP expressing control adenovirus (Ad-V) and SMAR1 Adenovirus (Ad-SM). Forty eight hours post-transduction, the levels of p53, p21, Bax, Puma, Apaf1 and p53AIP1 were determined. (C) HCT116 p53+/+ cells were transfected with HDAC1 siRNA (Santacruz) and levels of Bax, Puma and p53 acetylation were determined in the presence and absence of SMAR1 (C3). (D, E) Luciferase activity of full-length promoters of BAX and PUMA on SMAR1 overexpression and knockdown using two different shRNAs (sh745 and sh1077) in HCT116 p53−/− cells. A deletion mutant of SMAR1 lacking the DNA binding and the protein interacting domain (ΔSMAR1) was used as a control. Bars indicate s.d. from three independent experiments. (F) p21 luciferase assay on SMAR1 overexpression in HCT116 p53+/+ and p53−/− cells. (G) Annexin staining of HCT116 p53+/+ cells treated with Camptothecin (10 μM, 12 h) after transduction of GFP expressing control and SMAR1 adenoviruses. Cell cycle analysis in (H) HCT116 p53+/+ cells and (I) MEFs transduced with GFP expressing control (Ad-V) and SMAR1 (Ad-SM) adenoviruses. Forty eight hours post-transduction, cells were treated with UV (100 J/m2, 24 h) and Campthothecin (10 μM, 12 h) and thereafter stained with propidium iodide.
SMAR1 induces an anti-apoptotic signal in response to mild DNA damage
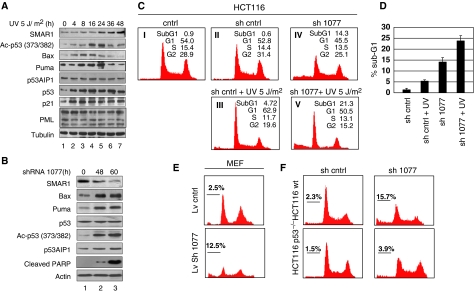
The tumour suppressor p53 induces cell cycle arrest after mild DNA damage and apoptosis after severe irreparable damage. Mild DNA damage results in transient acetylation of p53 and causes cell cycle arrest, whereas severe damage promotes prolonged accumulation and sustained p53 acetylation levels leading to apoptosis (Aylon and Oren, 2007). To further decipher the protective function of SMAR1, we investigated its responsiveness to mild DNA damage. Exposure of HCT116 p53+/+ cells to low-dose UV (5 J/m2) led to a steady increase in SMAR1 expression 24 h onwards (Figure 3A, panel 1). However, p53 acetylation levels increased from 4 h and peaked at 16 h, which corroborated with increased Bax and Puma levels at these time points. Conversely, p53 acetylation, Bax and Puma expression were reduced at 36–48 h, the time during which SMAR1 is induced (Figure 3A, panels 2, 3 and 4). This is in agreement with our earlier data that SMAR1 facilitates p53 deacetylation and represses Bax and Puma expression. Both p53AIP1 and PML, which are p53 targets were not induced at low UV dose suggesting that other post-translational modifications of p53 are required for their transactivation (Oda et al, 2000). Although Bax and Puma were inhibited by 48 h with concomitant deacetylation of p53, p21 levels increased and remain unaltered suggesting the induction of cell cycle arrest. Knockdown of SMAR1 resulted in robust increase in p53 acetylation, Bax and Puma expression with subsequent PARP cleavage (Figure 3B). Cell cycle analysis by propidium iodide staining showed that low-dose UV treatment did not induce significant cell death (∼4%), but caused distinct G1 arrest (Figure 3C, II and III). Notably, SMAR1 knockdown alone induced significant cell death (∼13%), which further increased (∼20%) on treatment with low-dose UV (Figure 3C, IV and V). Statistical representation of percentage sub-G1 population from three independent experiments is shown in Figure 3D. Similar results were observed in HEK 293 and MCF-7 cells (Supplementary Figure S3A and S3B). SMAR1 knockdown in MEFs also induced significant cell death through induction of Bax and Puma (Figure 3E; Supplementary Figure S3C). However, although shRNA-mediated knockdown of SMAR1 in HCT116 p53−/− cells results in a modest increase in Bax and Puma expression (Supplementary Figure S3D), no significant cell death was observed (Figure 3F). Thus, p53 is required for driving the cells towards apoptosis. Recently, it has been shown that acetylation is indispensable for p53-dependent transactivation and, therefore, expression of p53 targets Bax and Puma (Tang et al, 2008). Strikingly, in Figure 3A, we find that the levels of Bax and Puma increase well before increase in acetylated p53. It is possible that under this condition, activation of factors such as hCAS and ASPP family of proteins (Samuels-Lev et al, 2001; Tanaka et al, 2007), which can modulate p53 activity, may also contribute significantly to the expression of BAX and PUMA. Taken together, our results suggest that knockdown of SMAR1 affects the cellular apoptotic response by enhancing p53 acetylation at Lys 373/382 and increasing the expression of Bax and Puma.
Figure 3.
SMAR1 induces an anti-apoptotic signal in response to low-dose DNA damage. (A) HCT116 p53+/+ cells were UV irradiated with 5 J/m2 UV and cells collected at different time points as indicated. Immunoblot analysis was carried out as shown. (B) Expression levels of Bax, Puma, p53, acetylated p53 and PARP cleavage on shRNA (sh1077)-mediated knockdown of SMAR1 in HCT116 p53+/+ cells. (C) Propidium iodide staining in control HCT116 p53+/+ cells (48 h), cells treated with UV (5 J/m2, 48 h) and SMAR1 knockdown UV-treated cells (5 J/m2, 48 h) showing percentage apoptosis represented by sub-G1 population. (D) Statistical representation of percentage sub-G1 population on low-dose UV treatment and SMAR1 knockdown from three independent experiments. Error bars represent standard deviation. (E) Propidium iodide staining in MEFs transduced with control and SMAR1 lentivirus (F) Knockdown of SMAR1 by transient transfection in HCT116 p53+/+ and HCT116 p53−/− cells showing differential sub-G1 population.
SMAR1 drives p53 apoptotic target gene specificity through MARs
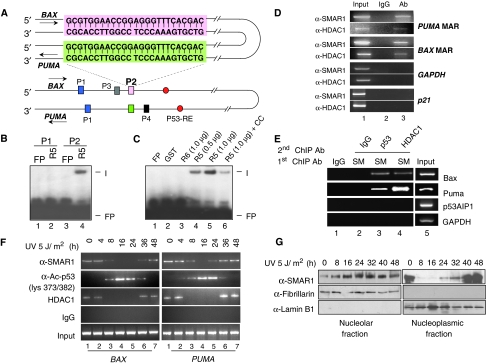
Earlier, we have shown that only BAX and PUMA and not other p53 targets were specifically repressed by SMAR1 overexpression. As SMAR1 is an MARBP and these proteins have propensity to bind AT-rich sequences, which often flank various promoters, we analysed the promoter sequences of p53 inducible genes. Computational analysis using MARWIZ software (Singh et al, 1997) predicted potential MARs in BAX and PUMA promoter within ∼700 bp upstream of p53 response element (p53RE), but not in p53AIP1 (Supplementary Figure S4A–S4C). Sequence alignment of the promoters up to ∼700 bp upstream of the p53RE revealed two stretches of sequences P1 (30 mer) and P2 (25 mer), which are identical in BAX and PUMA promoter (Figure 4A, blue and green boxes). Although the sequence encompassing the region P1 is located outside the MAR, the region P2 lies within the MAR region of both promoters (Supplementary Figure S4A and S4B). We then evaluated whether SMAR1 binds to these sequences. For this, two probes of 40 mer each were designed corresponding to the two segments P1 and P2. For binding reactions, bacterially expressed recombinant protein R5 (GST 350–548 aa) corresponding to the DNA-binding region of SMAR1 and recombinant protein R6 representing the protein interaction domain (GST 160–350 aa) were used. Interestingly, although P1 failed to form any complex, P2 formed nucleoprotein complex with R5 (Figure 4B, lane 4). The binding specificity was further showed by competition experiments showing loss of binding with the addition of 10-fold molar excess cold P2 (Figure 4C, lane 6). Under similar experimental conditions, GST and R6 failed to form any complex with P2 indicating the specific DNA-binding activity of SMAR1 (Figure 4C, lanes 2 and 3). To further check the specificity of SMAR1 binding to P2, we also designed two more similar sized probes P3 and P4 that lie proximal to P2 (Figure 4A, black boxes). Electrophoretic mobility shift assay (EMSA) studies with probes P3 and P4 did not show any complex formation (data not shown). Thus, SMAR1 binds to a specific and identical MAR such as sequence P2 present in both BAX and PUMA promoters. As the repressor activity of SMAR1 is attributed to its association with HDAC1, we further confirmed the binding of SMAR1 and recruitment of HDAC1 by chromatin immunoprecipitation (ChIP) assay. Cross-linked chromatin from HCT116 p53+/+ cells were pulled with SMAR1 and HDAC1 antibodies, respectively, and PCR amplified for BAX and PUMA promoters using primers spanning their respective MAR regions. Both BAX and PUMA promoters gave strong signal in PCR, whereas under similar experimental conditions, p21 and GAPDH promoters were not amplified (Figure 4D). Thus, SMAR1 recruits HDAC1 to the MAR regions of both BAX and PUMA, but not to p21 promoter. Notably, the MAR sites and the p53RE are juxtaposed on BAX and PUMA promoters. As SMAR1–HDAC1 complex interacts with p53, we reasoned that this complex might be associated with the nuclear matrix and more specifically to these MARs. To verify the co-occupancy of SMAR1/HDAC1 and SMAR1/p53 complexes on these MARs, we isolated the nuclear matrix from HCT116 p53+/+cells and performed sequential ChIP using SMAR1/HDAC1 and SMAR1/p53 antibodies. The purity of the isolated nuclear matrix was verified using Lamin B1 and histone H1 antibodies, respectively (Supplementary Figure S4D). Although BAX and PUMA showed detectable amplification, p53AIP1 and GAPDH promoters were not amplified (Figure 4E, lanes 3 and 4). Further, to study the occupancy and recruitment of SMAR1 on these MARs, cross-linked chromatin from UV-treated (5 J/m2) HCT116 p53+/+ cells at different time intervals was pulled with SMAR1, HDAC1 and acetylated p53 lys373/382 antibodies. Although BAX (Figure 4F, left panel) and PUMA (right panel) promoters were PCR detected in anti-SMAR1 pulled fraction, no amplification was observed in p53AIP1 (Supplementary Figure S4E). Furthermore, the kinetics of SMAR1 occupancy showed an oscillatory pattern with initial endogenous-bound SMAR1 slowly getting disappeared 4 h after UV treatment and reappeared by around 24 h. Strikingly, the occupancy of SMAR1 and HDAC1 inversely correlated with p53 acetylation status at these loci (Figure 4F, panels 1, 2 and 3). Interestingly, immunostaining showed strong translocation of SMAR1 into the nucleolus at 12 and 24 h after UV irradiation (Supplementary Figure S4F, white arrows). To verify this observation, we isolated nucleoplasmic and nucleolar fractions from HCT116 p53+/+ cells after irradiation with 5 J/m2 at different time points and probed for SMAR1 in these compartments. Our data shows that SMAR1 is present in the nucleolar fraction of un-irradiated cells albeit at very low levels, but expression increases significantly 8 h onwards after UV stress. Interestingly, although SMAR1 level increases in the nucleolar fraction, it decreases in the nucleoplasmic fraction (Figure 4G). This suggests that SMAR1 translocates into the nucleolus on UV stress. This can possibly explain the disappearance of SMAR1 from BAX and PUMA promoter MAR (Figure 4F) after 8 h of low-dose UV treatment leading to increased expression of Bax and Puma at 8 h (Figure 3A). However, 24 h after UV stress when SMAR1 is induced, it reappears in the nucleoplasmic fraction as well as in the nucleolus. This is again consistent with our ChIP data, wherein SMAR1 binding to the MAR element is restored after 24 h (Figure 4F), the time points after which Bax and Puma expression is maximally repressed as evident by immunoblot (Figure 3A). In the nucleolus, SMAR1 inhibits ribosomal gene transcription to cause cell cycle arrest (unpublished data). The nucleolar translocation of SMAR1 facilitates p53 acetylation, which then unleashes its transactivation potential on BAX and PUMA promoter resulting in pronounced expression of Bax and Puma. What post-translational modifications cause SMAR1 translocation into the nucleolus and binding to the MAR is currently under investigation. Together, these data suggest that nucleolar sequestration of SMAR1 may partly facilitate p53 acetylation and allow acetylated p53 to activate BAX and PUMA. Nonetheless, once SMAR1 is induced, it facilitates p53 deacetylation through HDAC1 and inhibits the transcription of BAX and PUMA. Thus, occupancy of SMAR1 on BAX and PUMA MAR selectively regulates their expression in response to DNA damage.
Figure 4.
SMAR1 regulates BAX and PUMA through MAR. (A) Schematic representation of BAX and PUMA promoter aligned with complementary strands showing the locations of two identical sequences P1 (blue) and P2 (pink in BAX and green in PUMA) in proximity to p53 response element (red). The sequence of P2 is boxed. (B) EMSA showing specific binding of SMAR1 (R5) to probe P2 (lane 4), but not to probe P1. (C) Purified GST (lane 2), R6 (lane 3) and R5 in increasing doses (lanes 4 and 5) were incubated with probe P2. Formation of complex (I) was visualized by autoradiography. Binding specificity of the complex in the presence of 10-fold molar excess of cold competitor (cold probe P2) is shown in lane 6. Free probe is denoted as FP. (D) Chromatin from HCT116 p53+/+ cells was immunoprecipitated with SMAR1 and HDAC1 antibodies (lane 3). PCR amplification was performed on MAR regions of BAX and PUMA. GAPDH and p21 promoters were used as control. Parallel immunoprecipitation with control IgG antibody is shown in lane 2. Lane 1 denotes input control. (E) Sequential ChIP on BAX and PUMA promoter MARs using anti-SMAR1/p53 (lane 3) and anti-SMAR1/HDAC1 (lane 4) antibodies in nuclear matrix fraction of HCT116 cells. Lanes 1 and 5 shows IgG and input control, respectively. (F) ChIP showing occupancy of SMAR1 on BAX (left panel) and PUMA (right panel) promoter on low-dose UV irradiation. Cross-linked chromatins from UV-irradiated HCT116 p53+/+ cells were pulled with SMAR1, p53, acetylated p53 lys373/382 and HDAC1 antibodies and bound chromatin fragments were detected by PCR. (G) Western blotting of SMAR1 in nucleolar and nucleoplasmic fraction of UV-irradiated (5 J/m2) HCT116 p53+/+ cells at different time points.
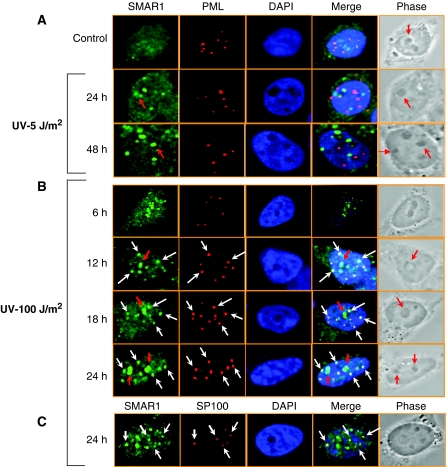
Apoptotic DNA damage translocates SMAR1 into PML-NBs
As low-dose UV DNA damage induced cell cycle arrest in which an apoptotic signal triggered by p53 acetylation was superseded by an anti-apoptotic signal through induction of SMAR1, we now treated the cells with 100 J/m2 UV to evaluate the effect of SMAR1 on p53 acetylation. At this dose of DNA damage, cells invariably goes towards apoptosis. Immunostaining of UV-irradiated cells showed that SMAR1 formed discrete speckled structures co-localizing with PML-NBs. PML-NBs are sub-nuclear domains involved in the regulation of p53-dependent and -independent DNA damage-induced cellular apoptosis (Wang et al, 1998). PML–SMAR1 co-localization was very weak and not distinct in control cells and in cells treated with low-dose UV (Figure 5A). Notably, co-localization of SMAR1 and PML becomes distinctly visible after 12 h of UV (100 J/m2) treatment when the size and number of PML-NBs increase (Figure 5B, white arrows). SMAR1–PML co-localization was further confirmed by targeting SP100, an integral component of PML-NBs (Figure 5C). Interestingly, SMAR1 also translocates to the nucleolus in cells exposed to high-dose UV as in case with low-dose UV irradiation (Figure 5A and B, red arrows). This was further confirmed by sucrose gradient fractionation assays, wherein we find that SMAR1 levels increase in both the nucleolar and nucleoplasmic fraction on UV irradiation (Figure 6A). The increase in the number of PML-NBs at high apoptotic dose raised the possibility that increase in PML protein levels enhances PML–SMAR1 complex formation with subsequent sequestration of SMAR1 into the PML-NBs. To evaluate this, HCT116 p53+/+ cells were treated with low- and high-dose UV and binding complexes were immunoprecipitated with SMAR1 antibody and analysed by western blotting with PML monoclonal antibody. Control and low-dose UV-treated cells showed similar amount of PML protein immunoprecipitated, although the level of SMAR1 was higher at low dose (Figure 6B, lanes 1 and 2). However, only at apoptotic UV dose (100 J/m2) when the level of PML increases, we find a modest increase in SMAR1–PML complex formation (Figure 6B, lane 3). To further validate this finding, we overexpressed PML-IV by transient transfection and found that SMAR1 completely translocates to the PML-NBs on PML-IV overexpression (Figure 6C, upper panel). Thus, stochiometric increase in PML protein level leads to SMAR1 sequestration. This suggests that the level of PML determines the sequestration of SMAR1 into PML-NBs. As sumoylation of PML has been shown to be important for nuclear body formation and its interaction with various transcription factors (Zhong et al, 2000), we used a triple sumoylation mutant PML construct to check its ability to sequester SMAR1. As expected, ectopic expression of this mutant failed to sequester SMAR1 into nuclear bodies and, therefore, showed a diffused distribution pattern (Figure 6C, lower panel). We further confirmed this specific interaction by transiently overexpressing HA-PML-IV and then immunoprecipitating the binding complexes with SMAR1 polyclonal antibody. Immunoblot analysis with HA antibody revealed specific interaction of SMAR1 and PML in HA-PML overexpressed cells (Figure 6D, lane 2). To identify the domain of PML involved in interaction with SMAR1, pull-down assays were performed with recombinant GST-SMAR1 and various HA-tagged deletion mutants of PML-IV. HA-PML-IV (Figure 6E, lanes 2 and 3) and HA-PML-IV (200–453) (Figure 6E, lanes 6 and 7), encompassing the coiled-coil domain of PML, showed strong binding to GST-SMAR1. As at a high apoptotic UV dose (100 J/m2), p53 is strongly acetylated to bring about cell death, we suspected that during this condition, SMAR1 dissociates from p53 to facilitate p53 acetylation. To evaluate the interaction of SMAR1 with p53, we performed a reciprocal quantitative co-immunoprecipitation after treatment of cells with both low (48 h)- and high-dose (24 h) UV. We found that SMAR1–p53 complex increases in cells treated with low-dose 5 J/m2 UV (Figure 6F and G, lanes 1 and 2), but at higher dose of 100 J/m2 UV, the interaction remains same as in control cells (lanes 1 and 3). Interestingly, PML also sequesters p53 into the PML-NBs on γ-irradiation to facilitate its acetylation (Guo et al, 2000). As SMAR1 negatively regulates p53 acetylation, we reasoned that PML sequesters SMAR1 and not p53 into the PML-NBs after apoptotic UV DNA damage. Indeed, immunostaining of PML and p53 after mild and apoptotic UV damage failed to show p53 in the distinct PML punctuate structures (Supplementary Figure S5). This highlights the fact that translocation of p53 to PML-NBs is dispensable for its acetylation at least in response to UV damage. Functionally, these data suggest that PML negatively regulates the activity of SMAR1 by sequestering it into the nuclear bodies to positively regulate the effector functions of p53. Thus, tumour suppressor PML sequesters SMAR1 and in turn potentiates p53 function.
Figure 5.
Apoptotic DNA damage translocates SMAR1 into PML nuclear bodies. Immunofluorescence analysis showing SMAR1–PML co-localization in HCT116 p53+/+ cells at (A) low-dose 5 J/m2, (B) high-dose 100 J/m2 UV and (C) Co-localization of SMAR1 with Sp100 at high dose. Cells were stained with SMAR1 (green), PML (red) and Sp100 (red) antibodies and analysed by confocal microscopy. Nuclei were stained with DAPI (blue). Co-localization of SMAR1 and PML bodies are shown in white arrows. Red arrows indicate SMAR1 in nucleolus. Images shown are representative of >50 images (n>50) taken in different fields from two independent experiments.
Figure 6.
SMAR1 interacts strongly with PML after apoptotic DNA damage. (A) Western blotting of SMAR1 in nucleolar and nucleoplasmic fractions of low and high dose UV-irradiated HCT116 p53+/+ cells at different time points. (B) Two hundred microgram of total cell extract from HCT116 p53+/+ control and UV-irradiated cells were immunoprecipitated (IP) with SMAR1 antibody and complexes detected by immunoblotting (IB) with antibody against PML (left panel). A total of 20% of total cell lysate was used for input (right panel). (C) Immunostaining showing SMAR1–PML co-localization in HCT116 p53+/+ cells after overexpression of PML-IV and PML-IV sumoylation mutant (3 M PML-IV) constructs. PML is stained with red (Cy3) and SMAR1 is stained with green (FITC). Nucleus is stained with blue (DAPI). (D) Co-immunoprecipitation of SMAR1 and PML in HA-PML-IV-transfected HCT116 p53+/+ cells as detected by HA antibody. (E) GST pull-down assays were performed using purified GST and GST-SMAR1 in HCT116 p53+/+ cells overexpressing full-length HA-PML-IV and truncated HA-PML-IV proteins as shown in the figure. Specific binding was detected using anti-HA. (F, G) HCT116 p53+/+ cell extracts (0.5 mg) from control and UV treated with 5 and 100 J/m2 were immunoprecipitated with SMAR1 and p53 antibodies and amount of p53 and SMAR1 in the immunoprecipitated complex were detected by antibodies against p53 and SMAR1, respectively.
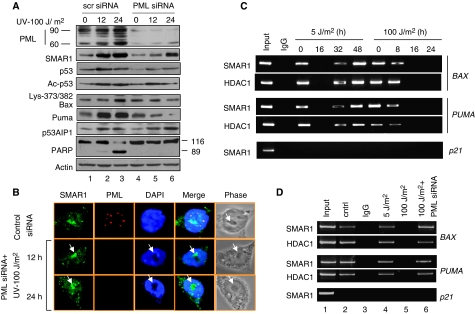
Silencing of PML expression enhances SMAR1-mediated transrepression of BAX and PUMA
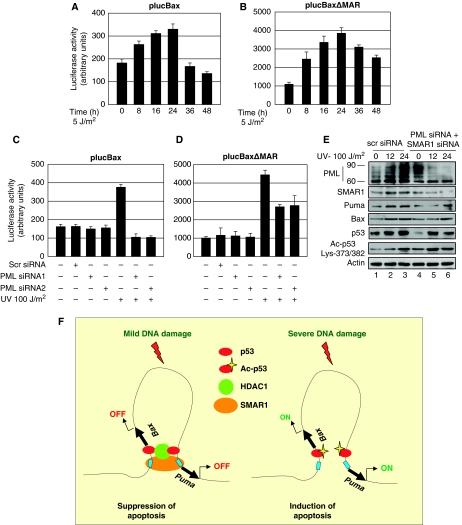
To establish whether compartmentalization of SMAR1 into the PML-NBs is indeed indispensable for induction of SMAR1–p53 target genes BAX and PUMA, endogenous PML was depleted by specific siRNA and then treated with UV (Figure 7A, panel 1, lanes 4, 5 and 6). We found that silencing of PML by specific siRNA does not affect p53 acetylation up to 12 h of UV treatment as reported earlier (Bernardi et al, 2004) (panel 5, lanes 2 and 5). However, SMAR1 levels were lower in PML knockdown cells compared with control (Figure 7A, panel 2). This could be because PML regulates the activities of many transcription factors such as Sp1, which might regulate SMAR1 basal transcription. Our data also shows lower levels of p53 in PML knockdown cells (Figure 7A, panel 4). This can be explained by the fact that PML regulates p53 protein levels and stability (Bernardi et al, 2004). In addition, SMAR1 is a p53 target gene (Singh et al, 2007). Together, these findings could account for the lower levels of SMAR1 and p53. Consequently, on PML knockdown, p53 acetylation levels should be comparatively less. This is, however, not observed and can be explained as being compensated by lower levels of SMAR1. However, SMAR1 induced at around 24 h after UV treatment (panel 2, lane 6) not only reduced p53 acetylation (panel 5, lane 6), but also inhibited Bax and Puma expression (panels 6 and 7, lane 6). The inhibition of Bax and Puma was reflected in reduced apoptotic population as shown by PI staining (Supplementary Figure S6). Interestingly, we found that PML was strongly induced after apoptotic UV DNA damage. Of note, the 60 kDa band corresponding to PML isoform IV was significantly induced in comparison with 90 kDa band representing PML isoform III (Figure 7A, panel 1, lanes 1, 2 and 3). Other isoforms of PML close to the 60 and 90 kDa also increased, but not to significantly high levels. This corroborated with earlier published data suggesting the involvement of PML III and PML IV in p53 regulation. Surprisingly, earlier studies (Seker et al, 2003) did not find changes in PML protein level after UV stress. This could be because these studies were carried out at low-dose UV and also for lesser time points, which is consistent with our own data, wherein no significant induction of PML protein was found at 5 J/m2 UV dose. Even at high apoptotic dose, the increase in PML protein and the number of nuclear bodies were significant only after 12 h. Notably, the observed increase in PML and SMAR1 was p53-dependent (Supplementary Figure S7). Moreover, apoptotic dose of UV treatment in PML knockdown cells did not result in speckled distribution of SMAR1, although SMAR1 was found to be translocated to the nucleolus (Figure 7B, white arrow). In addition, when cross-linked chromatin derived from low-dose UV-treated cells were immunoprecipitated with anti-SMAR1 and anti-HDAC1 antibodies, we found that SMAR1 was selectively bound onto the MAR regulatory regions of BAX and PUMA promoter along with HDAC1 at 32 and 48 h, but not at 16 h when SMAR1 translocates into the nucleolus as explained earlier. However, on apoptotic UV DNA damage, SMAR1 did not show binding to the MAR element after 8 h (Figure 7C). Interestingly, the binding of SMAR1 and recruitment of HDAC1 onto the MAR element was restored on apoptotic DNA damage when PML was knocked down using siRNA (Figure 7D, lane 6). Thus, PML sequesters SMAR1 to regulate the expression of Bax and Puma. This again explains why SMAR1 is unable to transcriptionally repress BAX and PUMA after apoptotic DNA damage, even though the level of SMAR1 increases. Under similar conditions, SMAR1 did not show binding to the p21 promoter, which lacks the MAR element. To further show the repressive effect of the MAR element, a deletion mutant of BAX promoter lacking the MAR region, but having intact p53-binding site and other core elements, was used to test its transactivation potential on low- and high-dose UV irradiation in comparison with the full-length promoter. Although the full-length BAX promoter (plucBax) was effectively repressed even below its basal level, the MAR-deleted mutant of BAX promoter (plucBaxΔMAR) was mildly repressed and was still activated (>two-fold) on low-dose 5 J/m2 UV irradiation (Figure 8A and B). This mild repression of the reporter lacking the MAR could be because of factors such as Hdm2, which can also regulate p53 acetylation through interaction with HDAC1 (Kobet et al, 2000; Ito et al, 2002). Similarly, on apoptotic-dose (100 J/m2) UV DNA damage, the full-length BAX promoter harbouring the MAR (plucBax) exhibited greater degree of repression in comparison with the MAR-deleted reporter (plucBaxΔMAR) in the absence of PML (Figure 8C and D). Nonetheless, on PML knockdown, the plucBaxΔMAR reporter showed mild repression similar to that seen in case of low-dose irradiation. This is because, recruitment of p53 cofactors such as CBP and p300, which are recruited by PML into PML-NBs to facilitate p53 acetylation, are impaired in the absence of PML (Hofmann and Will, 2003). Notably, the basal activity of the MAR-deleted promoter (plucBaxΔMAR) is ∼five-fold stronger than the full-length harbouring the MAR (plucBax) again highlighting the repressive function of the MAR. Collectively, these data suggests that knockdown of PML restores SMAR1 occupancy onto the BAX and PUMA promoter MAR and hence the repression. Finally, to confirm the requirement of SMAR1 for repression through MAR, we knocked down both PML and SMAR1 using specific siRNAs and evaluated the expression pattern of Bax and Puma on apoptotic UV DNA damage. As expected, double knockdown resulted in increased expression of Bax and Puma (Figure 8E). Taken together, these results confirm that PML is required for complete transactivation of BAX and PUMA by releasing the negative effects of SMAR1 through sequestration into the PML-NBs.
Figure 7.
Silencing of PML facilitates transrepression of BAX and PUMA by SMAR1. (A) Comparative expression levels of SMAR1, Ac-p53 and p53 in HCT116 p53+/+ cells transfected with scrambled siRNA (left half) versus PML siRNA (right panel) on UV (100 J/m2) treatment. (B) Confocal staining of SMAR1 and PML in HCT116 p53+/+ cells transfected with PML siRNA and treated with UV. Images are representative of >50 fields (n>50) from two independent experiments. (C) ChIP assay in HCT116 p53+/+ cells exposed to low dose (5 J/m2) and high dose (100 J/m2) UV irradiation. (D) ChIP showing SMAR1 occupancy on BAX and PUMA promoter after low dose (5 J/m2, 48 h) and at high dose (100 J/m2, 24 h) in PML knockdown HCT116 p53+/+ cells.
Figure 8.
Repression of Bax and Puma is specific through the MAR by SMAR1. (A, B) Luciferase activity of the full-length BAX (plucBax) and MAR-deleted (plucBaxΔMAR) reporters in HCT116 p53+/+ cells treated with low dose 5 J/m2 UV. (C, D) Promoter read out of plucBax and plucBaxΔMAR reporters in HCT116 p53+/+ cells transfected with two different PML siRNAs and analysed 24 h after treatment with high dose UV (100 J/m2). (E) Expression analyses of Bax and Puma on double knockdown of PML and SMAR1 in HCT116 p53+/+ cells. (F) Model showing the MAR element (blue box) of BAX and PUMA promoter juxtaposed to each other causing the looping out of the intervening sequence. SMAR1 (orange) binds to this identical MAR element along with HDAC1 (green) and p53 (red) forming a repressor complex switching off transcription after mild DNA damage (suppression of apoptosis). After severe apoptotic DNA damage, SMAR1 is no longer bound to the MAR element facilitating p53 acetylation (p53-Ac) and transcription of BAX and PUMA (induction of apoptosis).
Discussion
In this study using a low- and high-dose UV stress, we have deciphered the molecular functionality of MARBP SMAR1 during p53-dependent cell cycle arrest and apoptosis. We show that p53 target gene SMAR1 is induced during UV DNA damage, but is compartmentalized into the PML-NBs at high-dose DNA damage and, therefore, have different functional outcome. Unlike proteins such as Mdm2, Pirh2 and COP1, which regulate basal levels of p53 through direct ubiquitination (Brooks and Gu, 2006), SMAR1 modulates basal p53 acetylation status by recruiting HDAC1. This kind of basal regulation of p53 acetylation is important to prevent sudden cell death arising out of mild metabolic stresses, which the cells always encounter. Although, transcription factors such as Hzf, hCAS and ASPP family members modulate p53 function by directly binding to selective p53 target genes under conditions of stress, SMAR1 modulates the transactivation potential of p53 by associating with HDAC1 apart from imparting specific regulation of p53 apoptotic targets Bax and Puma through MAR element. We show for the first time that both BAX and PUMA promoter harbour a similar 25 bp MAR element through which SMAR1 anchors these genes to the nuclear matrix making them transcriptionally inactive. Although, mild DNA damage triggers p53 acetylation and transcription of p53 apoptotic targets BAX and PUMA, induction of SMAR1 results in p53 deacetylation through increased association of p53 with SMAR1–HDAC1 complex. This prevents prolonged and sustained accumulation of p53 acetylation levels and inhibits apoptosis. Further, SMAR1 is recruited to BAX and PUMA promoter MAR resulting in abatement of their transcription. Thus, induction of SMAR1 generates an anti-apoptotic signal that prevents cellular apoptosis after mild DNA damage. Although low-dose UV DNA damage induced around 5% cell death, the percentage of apoptosis increased to 21% in SMAR1 knockdown cells treated with low-dose UV. In addition, SMAR1 knockdown itself induced significant apoptosis. Knockdown of SMAR1 resulted in prolonged and robust accumulation of p53 acetylation levels and, therefore, promotes apoptosis. Thus, we establish SMAR1 as an important apoptotic check point regulator. It is intriguing as to why SMAR1 needs to repress BAX and PUMA by binding to an MAR element when it can inhibit p53 acetylation through HDAC1 and thereby block p53 downstream effectors. One possible explanation is that p53 deacetylation occurs at protein level and is kinetically a much slower process compared with direct repression through MAR. The MAR site in BAX and PUMA promoter acts as a negative regulatory element that ensures timely repression and safeguard mechanism from commitment of cells towards apoptosis when damage is repairable. This is the reason why after mild DNA damage, p21 and SMAR1 remain induced till 48 h, whereas BAX and PUMA are suppressed considering that all of them are p53 target genes. This suggests that the drastic repression of BAX and PUMA is a direct effect of the MAR element coupled to p53 deacetylation. Moreover, genes which harbour MARs and are attached to the nuclear matrix are more repressed than genes in matrix without MARs (Rudd et al, 2004). Thus, MARs serve as an additional layer of transcriptional regulation for BAX and PUMA besides p53 in response to DNA damage. Interestingly, Bax and Puma are mitochondrial proteins, genes of which are located in the same locus 19q13.3 and both are transcriptionally regulated by a single MAR element present in both promoters. Thus, it is possible that binding of SMAR1 to the identical MAR element brings the promoters of BAX and PUMA in close proximity of each other to confer regulatory effects by allowing the intervening sequence (1.7 Mb) between the promoters to form intrachromosomal loops (Figures 4A and 8B). Such kind of long-range intrachromosomal interactions have been reported for TH2 cytokine locus (Spilianakis and Flavell, 2004).
It is shown that in PML−/− cells, induction of proapoptotic targets such as BAX is impaired (Guo et al, 2000). However, mechanism how PML regulates the expression of Bax is not known. In this study, we showed that PML is induced at high-dose 100 J/m2 UV in a p53-dependent manner. PML induction is associated with sequestration of SMAR1 with concomitant increase in p53 acetylation. We have shown that SMAR1 forms a repressor complex with p53 and HDAC1, and this repressor complex is associated with BAX and PUMA MAR. Therefore, sequestration of SMAR1 by PML not only facilitates p53 acetylation, but also enhances the expression of Bax and Puma. Further, the occupancy of SMAR1 on BAX and PUMA promoter MAR in UV-treated PML knockdown cells abrogates UV-induced apoptosis. In this context, it is worth mentioning that tumour suppressor PML protein is downregulated in most of the cancers (Scaglioni et al, 2006). Although we have earlier shown that SMAR1 is downregulated in higher grades of breast cancer in which p53 function becomes defective, SMAR1 responds positively to various DNA damaging and chemotherapeutic agents in cells harbouring wild-type p53 (Singh et al, 2007). Thus, it is tempting to speculate that loss of PML expression coupled to the responsiveness of SMAR1 to various drugs in tumours harbouring wild-type p53 might explain their increased refractory nature to various chemotherapeutic treatment modules. Thus, SMAR1 can be a potential target for cancer therapy in which p53 function is not compromised.
Materials and methods
Cell lines, cell culture, plasmids and reagents
HCT116 p53+/+, HCT116 p53−/− (kind gift from Dr Bert Vogelstein, John Hopkins University), HEK 293, MCF-7 and MEFs were cultured in DMEM (Invitrogen) supplemented with 10% foetal bovine serum (Invitrogen). pBK CMV-SMAR1, Flag-SMAR1, GST-SMAR1, GST-R5(350–548), GST-R6(160–350) and custom synthesized siRNA for SMAR1 (Ambion) were used as described (Rampalli et al, 2005). Full-length HA-PML-IV and truncations HA-PML-IV (1–200 aa), HA-PML-IV (200–453 aa) and HA-PML-IV (400–585 aa) were kind gifts from Prof. Carl Maki (University of Chicago, USA). The triple PML SUMO mutant (3MPML) was a kind gift from Prof. PP Pandolffi (Harvard University, USA). p300 expression plasmid was provided by Xuan Liu (University of California, USA). PML siRNAs and control siRNA (Ambion) were used at 300 nM concentration for 24 h. UV treatment was given using UVP cross-linker (Amersham) at two doses: 5 and 100 J/m2. All transfections were carried out using Lipofectamine 2000 (Invitrogen).
Adenovirus, shRNA and lentiviral construction
Replication deficient recombinant SMAR1 adenovirus Ad-SMAR1 was generated as described (He et al, 1998). The 1.9 kb SMAR1 cDNA was cloned in pAdTrack-CMV vector followed by homologous recombination with pAdEasy vector in BJ5183 strain of Escherichia coli. The recombinant clones were screened for insert, linearized with Pac-I (New England Biolabs) and transfected into HEK 293 cells for packaging of virus. Constructs expressing shRNA against SMAR1 (5′-ACGCGTAAAAAAGCCAGAACA-3′ and 5′-GGATCAAGCAGAGCATTGA-3′) and scrambled sequence (5′-ACCGAAGGCAAGCAAAGCTT-3′) were cloned in pSIREN RetroQ-Zs Green plasmid (Clontech). For making SMAR1 lentivirus, the SMAR1 shRNA sequence (5′-GGATCAAGCAGAGCATTGA-3′) was cloned in pCRILV (generated in house); the resultant construct was transfected in HEK293 cells for viral packaging.
Luciferase reporter and apoptosis assays
For luciferase assays, the promoters of BAX (plucBax), PUMA (plucPuma) and p53AIP1 (plucp53AIP1) (∼650 bp) containing the core promoter elements and TATA box were PCR amplified from genomic DNA using promoter-specific primers and cloned in pGL3 enhancer reporter vector. The MAR-deleted (plucBaxΔMAR) reporter (∼600 bp) was cloned using primers with compatible restriction site overhangs in pGL3 enhancer. Cells were co-transfected with indicated reporter plasmids and with GFP plasmid as an internal control. After 24 h of transfection, cells were analysed for Luciferase activity using Luclite substrate (Perkin Elmer) and assay performed using Top-Count luminometer (Packard Life sciences). The results were normalized to GFP expression using Fluoroskan Ascent Luminometer (Lab Systems). All assays were carried out in triplicates. For apoptosis assays, propidium iodide (Roche) staining was carried out after ethanol fixation. The acquisition and analysis was carried out by FACS Calibur (Becton Dickinson) using the Cell Quest software programme. For live cell staining, cells were stained with annexin-Cy3 following manufacturer's protocol (BD Biosciences). Images were captured in confocal laser microscope (LSM 510 version 2.01; Zeiss, Thornwood, NY).
Co-immunoprecipitation, immunoblot and immunoflourescence analysis
Co-immunoprecipitation of proteins was performed after the lysis of cells in TNN buffer (50 mM Tris–Cl pH 7.5, 1% NP-40, 150 mM NaCl and 1 mM DTT) supplemented with complete protease-inhibitor cocktail (Roche). For immunoblotting, 50 μg of total protein was subjected to SDS–PAGE. The following primary antibodies were used: p53 DO-1, PML (PG-M3), HA and GFP, LaminB (Santa Cruz), Histone H1, acetylated p53 373/382 (Upstate), SMAR1 rabbit polyclonal (raised in house), SMAR1/BANP (Bethyl), Bax, Puma, p21, PARP, HDAC1 and p53ser-15 (Cell Signalling), β-actin, PML (clone 97) and Flag (sigma), p53AIP1 and APAF1 (Chemicon). For confocal analysis, paraformaldehyde-fixed and permeabilized cells were blocked in 5% FCS and incubated with desired primary antibodies. Cells were then washed five times in PBS and incubated with goat anti-mouse Cy3 and donkey anti-rabbit FITC (Chemicon). Coverslips were mounted in UltraCruz mounting media with DAPI (Santa Cruz) and examined under confocal laser microscope (LSM 510 version 2.01; Zeiss, Thornwood, NY).
ChIP and Re-ChIP assays
ChIP assays were performed as described earlier (Rampalli et al, 2005) using ChIP assay kit (Upstate Biotechnology) following manufacturer's instructions. In Re-ChIP experiments, complexes were eluted by incubation for 30 min at 37°C in 250 μl of Re-IP buffer (2 mM DTT, 1% Triton-X-100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris–HCl, pH 8.0) and then diluted two-fold in Re-IP dilution buffer (1% Triton-X-100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris–HCl pH 8.0 and protease inhibitors). PCR were performed using primers given in Supplementary Table 1.
Electrophoretic mobility shift assay
Probes were labelled using [α-32P] dATP by kleenowing for 45 min and purified by probequant G-50 columns (Amersham). Binding reactions were set up as described earlier (Rampalli et al, 2005). The reaction mixture was incubated for 20 min at room temperature and loaded in 10% polyacrylamide gel. Probe sequence of P1, P2, P3 and P4 is given in Supplementary Table 2.
Preparation of nuclear matrix. Nuclear matrices were prepared according to a well-established protocol (Cockerill et al, 1986).
Purification of nucleoli. Nucleoli were purified by using a procedure described earlier by Andersen et al, 2002. The crude nuclear pellet isolated using a hypotonic buffer A (10 mM HEPES (pH 7.9), 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT) was resuspended in 3 ml of 0.25 M sucrose containing 10 mM MgCl2 and protease inhibitors, followed by a spin at 1200 g for 10 min through a 0.88 M sucrose cushion (4 ml) containing 0.05 mM MgCl2 and protease inhibitors. The purified nuclear pellet was resuspended in 3 ml of 0.34 M sucrose solution containing 0.05 mM MgCl2 and sonicated on ice for several bursts of 30 s with 5 min intervals. Nucleoli were then purified from the resulting homogenate by centrifugation at 2000 g for 20 min through a 0.88 M sucrose cushion (4 ml) containing 0.05 mM MgCl2 and protease inhibitors. The supernatant containing the nucleoplasmic fraction devoid of nucleoli was harvested and further concentrated using Quick Spin columns (Qiagen). The pellet containing purified nucleoli was lysed in TNN buffer and used for analysis.
Supplementary Material
Supplementary Information
Review Process File
Acknowledgments
We thank Dr GC Mishra, Director, National Centre for Cell Science. This work is funded by NCCS, Department of Biotechnology (DBT), University Grants Commission (UGC), Indian Council of Medical Research (ICMR) and Council for Scientific and Industrial Research (CSIR), Government of India.
Footnotes
The authors declare that they have no conflict of interest.
References
- Andersen JS, Lyon CE, Fox AH, Leung AK, Lam YM, Steen H, Mann M, Lamond AI (2002) Directed proteomic analysis of the human nucleolus. Curr Biol 12: 1–11 [DOI] [PubMed] [Google Scholar]
- Aylon Y, Oren M (2007) Living with p53, dying of p53. Cell 130: 597–600 [DOI] [PubMed] [Google Scholar]
- Bernardi R, Pandolfi PP (2007) Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat Rev Mol Cell Biol 8: 1006–1016 [DOI] [PubMed] [Google Scholar]
- Bernardi R, Scaglioni PP, Bergmann S, Horn HF, Vousden KH, Pandolfi PP (2004) PML regulates p53 stability by sequestering Mdm2 to the nucleolus. Nat Cell Biol 6: 665–672 [DOI] [PubMed] [Google Scholar]
- Brooks CL, Gu W (2006) p53 ubiquitination: Mdm2 and beyond. Mol Cell 21: 307–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay S, Kaul R, Charest A, Housman D, Chen J (2000) SMAR1, a novel, alternatively spliced gene product, binds the scaffold/matrix-associated region at the T cell receptor beta locus. Genomics 68: 93–96 [DOI] [PubMed] [Google Scholar]
- Cockerill PN, Garrard WT (1986) Chromosomal loop anchorage of the kappa immunoglobulin gene occurs next to the enhancer in a region containing topoisomerase II sites. Cell 44: 273–282 [DOI] [PubMed] [Google Scholar]
- Das S, Raj L, Zhao B, Kimura Y, Bernstein A, Aaronson SA, Lee SW (2007) Hzf determines cell survival upon genotoxic stress by modulating p53 transactivation. Cell 130: 624–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Stanchina E, Querido E, Narita M, Davuluri RV, Pandolfi PP, Ferbeyre G, Lowe SW (2004) PML is a direct p53 target that modulates p53 effector functions. Mol Cell 13: 523–535 [DOI] [PubMed] [Google Scholar]
- He TC, Zhou S, da Costa LT, Yu J, Kinzler KW, Vogelstein B (1998) A simplified system for generating recombinant adenoviruses. Proc Natl Acad Sci 95: 2509–2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann TG, Will H (2003) Body language: the function of PML nuclear bodies in apoptosis regulation. Cell Death Differ 10: 1290–1299 [DOI] [PubMed] [Google Scholar]
- Guo A, Salomoni P, Luo J, Shih A, Zhong S, Gu W, Pandolfi PP (2000) The function of PML in p53-dependent apoptosis. Nat Cell Biol 2: 730–736 [DOI] [PubMed] [Google Scholar]
- Ito A, Kawagychi Y, Lai CH, Kovacs JJ, Higashimoto Y, Appella E, Yao TP (2002) MDM2-HDAC1-mediated deacetylation of p53 is required for its degradation. EMBO J 21: 6236–6245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalota A, Singh K, Pavithra L, Kaul-Ghanekar R, Jameel S, Chattopadhyay S (2005) Tumor suppressor SMAR1 activates and stabilizes p53 through its arginine-serine-rich motif. J Biol Chem 280: 16019–16029 [DOI] [PubMed] [Google Scholar]
- Jiang M, Axe T, Holgate R, Rubbi CP, Okorokov AL, Mee T, Milner J (2001) p53 binds the nuclear matrix in normal cells: binding involves the proline-rich domain of p53 and increases following genotoxic stress. Oncogene 20: 5449–5458 [DOI] [PubMed] [Google Scholar]
- Kaul-Ghanekar R, Majumdar S, Jalota A, Gulati N, Dubey N, Saha B, Chattopadhyay S (2005) Abnormal V(D)J recombination of T cell receptor beta locus in SMAR1 transgenic mice. J Biol Chem 280: 9450–9459 [DOI] [PubMed] [Google Scholar]
- Kaul R, Mukherjee S, Ahmed F, Bhat MK, Chhipa R, Galande S, Chattopadhyay S (2003) Direct interaction with and activation of p53 by SMAR1 retards cell-cycle progression at G2/M phase and delays tumor growth in mice. Int J Cancer 103: 606–615 [DOI] [PubMed] [Google Scholar]
- Kobet E, Zeng X, Zhu Y, Keller D, Lu H (2000) MDM2 inhibits p300-mediated p53 acetylation and activation by forming a ternary complex with the two proteins. PNAS 97: 12547–12552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda K, Arakawa H, Tanaka T, Matsuda K, Tanikawa C, Mori T, Nishimori H, Tamai K, Tokino T, Nakamura Y, Taya Y (2000) p53AIP1, a potential mediator of p53-dependent apoptosis, and its regulation by Ser-46-phosphorylated p53. Cell 102: 849–862 [DOI] [PubMed] [Google Scholar]
- Rampalli S, Pavithra L, Bhatt A, Kundu TK, Chattopadhyay S (2005) Tumor suppressor SMAR1 mediates cyclin D1 repression by recruitment of the SIN3/histone deacetylase 1 complex. Mol Cell Biol 25: 8415–8429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley T, Sontag E, Chen P, Levine A (2008) Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol 9: 402–412 [DOI] [PubMed] [Google Scholar]
- Rudd S, Frisch M, Grote K, Meyers BC, Mayer K, Werner T (2004) Genome-wide in silico mapping of scaffold/matrix attachment regions in Arabidopsis suggests correlation of intragenic scaffold/matrix attachment regions with gene expression. Plant Physiol 135: 715–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi K, Herrera JE, Saito S, Miki T, Bustin M, Vassilev A, Anderson CW, Appella E (1998) DNA damage activates p53 through a phosphorylation-acetylation cascade. Genes Dev 12: 2831–2841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels-Lev Y, O'Connor DJ, Bergamaschi D, Trigiante G, Hsieh JK, Zhong S, Campargue I, Naumovski L, Crook T, Lu X (2001) ASPP proteins specifically stimulate the apoptotic function of p53. Mol Cell 8: 781–794 [DOI] [PubMed] [Google Scholar]
- Scaglioni PP, Yung TM, Cai LF, Erdjument-Bromage H, Kaufman AJ, Singh B, Teruya-Feldstein J, Tempst P, Pandolfi PP (2006) A CK2-dependent mechanism for degradation of the PML tumor suppressor. Cell 126: 269–328 [DOI] [PubMed] [Google Scholar]
- Seker H, Rubbi C, Linke SP, Bowman ED, Garfield S, Hansen L, Borden KL, Milner J, Harris CC (2003) UV-C-induced DNA damage leads to p53-dependent nuclear trafficking of PML. Oncogene 22: 1620–1628 [DOI] [PubMed] [Google Scholar]
- Singh K, Mogare D, Giridharagopalan RO, Gogiraju R, Pande G, Chattopadhyay S (2007) p53 target gene SMAR1 is dysregulated in breast cancer: its role in cancer cell migration and invasion. PLoS One 2: e660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh GB, Kramer JA, Krawetz SA (1997) Mathematical model to predict regions of chromatin attachment to the nuclear matrix. Nucleic Acids Res 25: 1419–1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spilianakis GC, Flavell RA (2004) Long-range intrachromosomal interactions in the T helper type 2 cytokine locus. Nat Immunol 5: 1017–1027 [DOI] [PubMed] [Google Scholar]
- Tanaka T, Ohkubo S, Tatsuno I, Prives C (2007) hCAS/CSE1L associates with chromatin and regulates expression of select p53 target genes. Cell 130: 638–650 [DOI] [PubMed] [Google Scholar]
- Tang Y, Zhao W, Chen Y, Zhao Y, Gu W (2008) Acetylation is indispensable for p53 activation. Cell 133: 612–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZG, Ruggero D, Ronchetti S, Zhong S, Gaboli M, Rivi R, Pandolfi PP (1998) PML is essential for multiple apoptotic pathways. Nat Genet 20: 266–272 [DOI] [PubMed] [Google Scholar]
- Zaidi SK, Young DW, Choi JY, Pratap J, Javed A, Montecino M, Stein JL, van Wijnen AJ, Lian JB, Stein GS (2005) The dynamic organization of gene-regulatory machinery in nuclear microenvironments. EMBO Rep 6: 128–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, Muller S, Ronchetti S, Freemont SP, Dejean A, Pandolfi PP (2000) Role of SUMO-1 modified PML in nuclear body formation. Blood 95: 2748–2752 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
Review Process File