Abstract
Abundance of pseudo splice sites in introns can potentially give rise to innumerable pseudoexons, outnumbering the real ones. Nonetheless, these are efficiently ignored by the splicing machinery, a process yet to be understood completely. Although numerous 5′ splice site-like sequences functioning as splicing silencers have been found to be enriched in predicted human pseudoexons, the lack of active pseudoexons pose a fundamental challenge to how these U1snRNP-binding sites function in splicing inhibition. Here, we address this issue by focusing on a previously described pathological ATM pseudoexon whose inhibition is mediated by U1snRNP binding at intronic splicing processing element (ISPE), composed of a consensus donor splice site. Spliceosomal complex assembly demonstrates inefficient A complex formation when ISPE is intact, implying U1snRNP-mediated unproductive U2snRNP recruitment. Furthermore, interaction of SF2/ASF with its motif seems to be dependent on RNA structure and U1snRNP interaction. Our results suggest a complex combinatorial interplay of RNA structure and trans-acting factors in determining the splicing outcome and contribute to understanding the intronic splicing code for the ATM pseudoexon.
Keywords: RNA structure, SF2/ASF, spliceosome, U1snRNP, U2snRNP
Introduction
Deciphering the ‘splicing code' (Fu, 2004; Wang and Cooper, 2007) is taking considerably longer than the elucidation of the genetic code, which can be well attributed to the very degenerate and extended nature of the splicing code. Although each amino acid can be accounted for by a few sequences with moderate degeneration, the splicing code relies on a couple of common dinucleotide sequences (GU/AG) and a vast array of highly degenerated signals that act in a complex combinatorial way (Smith and Valcarcel, 2000; Han et al, 2005; Hertel, 2008). The initial clues for the ‘splicing code' started with the sequencing of the early genes (globins, ovalbumin) (Breathnach et al, 1978; Efstratiadis et al, 1980) but the complexity became very obvious with the discovery of alternative splicing of cellular genes such as calcitonin (Amara et al, 1982) or fibronectin (Kornblihtt et al, 1984; Vibe-Pedersen et al, 1984). Surprisingly, the early observation that exonic sequences were involved in the definition of alternative splicing exons (Mardon et al, 1987) went unnoticed for >5 years. These exonic sequences when further mapped revealed an extended array of short signals (Caputi et al, 1994). The fact that these sequences overlap with the genetic code poses an intriguing evolutionary puzzle (Xing and Lee, 2006). Since the early 1990s, a strong focus has been placed on exonic and nearby intronic sequences as the core of most attempts to elucidate the splicing code (Wang and Burge, 2008; Wang et al, 2008). The reason for this focus being that accurate pre-mRNA splicing is essential for proper gene expression, and derangements of this process accounts for about one fifth of inherited diseases (Solis et al, 2008; Cooper et al, 2009; Tazi et al, 2009).
In the splicing process, from the transcribed pre-mRNA molecule, the introns must be spliced out and the exons correctly ligated with each other to obtain the mature mRNA. Intron removal is carried out by the spliceosome, a multi-component enzymatic complex formed stepwise by the ordered interaction of UsnRNPs and non-snRNP proteins on consensus sequences, known as 5′ splice site (5′ss) and 3′ splice site (3′ss), that define the intron/exon boundaries (Nilsen, 2003; Chen et al, 2007; Matlin and Moore, 2007; Wahl et al, 2009). The basic steps in spliceosome assembly have been known since many years. In general, spliceosome assembly is initiated by the interaction of the U1snRNP with the 5′ss, forming the E complex (Mount et al, 1983; Seraphin and Rosbash, 1989). The latter also contains the 17S U2snRNP, which at this stage associates via a non-base pairing interaction (Das et al, 2000). In a subsequent ATP-dependent step, the U2snRNA base pairs with the branch site of the pre-mRNA, leading to stable association of U2snRNP and formation of the A complex or prespliceosome (Konarska and Sharp, 1987). Finally, the U4/U6.U5 tri-snRNP complex binds generating the B complex, and after a major conformational change the C complex is formed (Will et al, 2002).
The fidelity of this process is severely challenged especially in vertebrate genes, where small exons are interspersed within multiple introns ranging from several hundred to more than one hundred thousand nucleotides. As a result, gene architecture has been recognized as a major influencing factor of the splicing process (Fox-Walsh et al, 2005; Baralle et al, 2006). Besides the gene architecture, an additional complexity is represented by the observation that introns contain many sequences that match the consensus 5′ss and 3′ss motifs as well as authentic sites and contain splicing regulatory sequences that would enhance their inclusion in the mRNA, yet are virtually never used in splicing (Cote et al, 2001). In keeping with this, in silico searches show that these sequences (also known as ‘pseudoexons') are usually very abundant in the introns of most genes (with this term we refer to any nucleotide (nt) sequence between 50 and 2–300 nt in length with apparently viable 5′ss, 3′ss, and branch sites at either end) (Sun and Chasin, 2000). The ability of the splicing machinery to reliably distinguish real exons, that, in some cases are numerically outnumbered by an order of magnitude by pseudoexons is of paramount importance, especially considering that pseudoexon inclusion has been increasingly associated with occurrence of human disease (Buratti et al, 2006). Normally, exclusion of many of these aberrant pseudoexon sequences is achieved by the presence of intrinsic defects in their composition (Sun and Chasin, 2000), by the enrichment of silencer elements (Fairbrother and Chasin, 2000; Sironi et al, 2004; Zhang and Chasin, 2004), or by the formation of inhibiting RNA secondary structures (Zhang et al, 2005).
In humans, among the exonic splicing silencer elements found to be enriched in predicted pseudoexons, intriguingly there are sequence elements bearing strong resemblance to positions +1 through +6 of the human 5′ss consensus sequence (/GTRAGT), where the splice junction is indicated by / and R indicates A or G (Wang et al, 2004). Functional splicing assays using minigenes showed these 5′ss-like sequences as having silencer activity as well as being recognized as 5′ss. However, there is no clear mechanism and very little is known regarding how these intronic 5′ss-like sequences sometimes inhibit pseudoexon splicing, whereas in other cases are used as viable donor sites.
In this work, we have addressed this issue by experimentally focusing on an earlier identified pathological pseudoexon splicing event between exon 20 and 21 of the ATM gene whose exclusion is mediated by U1snRNP-binding site within its sequence (Pagani et al, 2002). Using this model, we have tried to understand the mechanism of U1snRNP-mediated splicing inhibition by analysing in-depth the spliceosomal complex assembly on the inactive (ATM WT) and active (ATMΔ) pseudoexon sequences. We have also observed that a binding site for positive splicing factor SF2/ASF is present both in ATM WT and ATMΔ pseudoexons, but is available for productive interaction only in the case of the active ATMΔ pseudoexon that does not bind U1snRNP. We additionally show that this availability for interaction is dependent on RNA secondary structure for the display of the protein-binding sequence. These observations have allowed us to develop a model of ATM pseudoexon inhibition that highlights the complexity of even seemingly simple splicing decisions. As similar mechanisms are probably operating in normal exons and introns, they should be taken into consideration when constructing splicing code models. In fact, until now all such models have focused on exonic sequences and mainly considered linear sequence parameters.
Results
Earlier, we have described the inclusion of a 65-nt long pseudoexon between ATM exons 20 and 21 in a patient affected by ataxia-telengiectasia (Pagani et al, 2002). This pseudoexon activation event consisted of a 4-nt deletion (GTAA) that occurred within a high-affinity U1snRNP-binding site acting as an internal exonic splicing repressor (Figure 1A). This region was termed intronic splicing processing element (ISPE) (Pagani et al, 2002; Lewandowska et al, 2005). More recently, we have demonstrated that the donor splice site usage in this pseudoexon is also highly influenced by local RNA secondary structure constraints (Buratti et al, 2007).
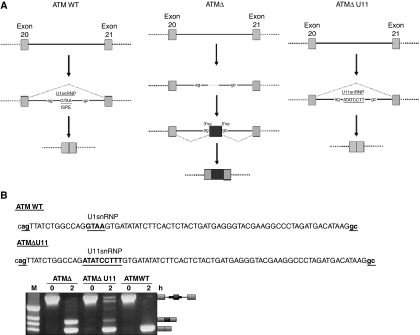
Figure 1.
ATM pre-mRNA splicing of exon 20 and exon 21 and effect on U11snRNP binding as opposed to U1snRNP in the ATM WT sequence. (A) Exons 20 and 21 are shown with light grey boxes, whereas the ATMΔ pseudoexon with black. Introns are depicted with a straight line. ‘ag' and ‘gc' represent the intronic splice sites flanking the pseudoexon. In normal splicing, the ISPE sequence is present and can bind an U1snRNP molecule at this position. In the A-T patient, deletion of a GTAA sequence in the ISPE abrogates U1snRNP binding and activates inclusion of the ATMΔ pseudoexon. Introduction of an U11snRNP-binding site restores inhibition. (B) Schematic representations of the binding positions of U1snRNP and of U11snRNP on the ATM WT and ATMΔ U11 pseudoexons, respectively (the sequences are shown with bold underlined letters). The sequence of the pseudoexon is shown with capital letters whereas splice sites are shown with underlined small bold letters. In vitro splicing and RT–PCR analysis of the ATMΔ, ATMΔ U11, and ATM WT substrates in PY7 minigenes are shown. The scheme of the spliced and unspliced substrates is shown on the right.
In this work, we have attempted to clarify the unusual function of U1snRNP binding to the ATM WT sequence that determines pseudoexon inhibition. First of all, to test whether the inhibitory effect mediated through the ISPE could be attributed to specific functional properties of U1snRNP or to any other massive macromolecular complex of similar size, we placed an U11snRNP-binding site in this position (Figure 1B). As shown in Figure 1B, almost no pseudoexon inclusion could be detected during the processing of the ATMΔ U11RNA when compared with the ATMΔ pseudoexon. In the U11 variant, in fact, the low level of pseudoexon inclusion still observed can probably be attributed to the lower concentration of U11snRNP (1/100th) with respect to U1snRNP in Hela nuclear extracts (Pessa et al, 2008).
To better understand at which stage of the splicing process U1snRNP is acting, it was decided to analyse spliceosomal complex assembly on the ATM WT and ATMΔ pseudoexon RNAs.
Spliceosomal complex assembly on ATM WT and ATMΔ RNAs
To achieve this, we assembled spliceosomal complexes on a biexonic substrate derived from the PY7 minigene. In these constructs, the Exon 2 of α-tropomysin was present upstream to the ATM WT and ATMΔ pseudoexon sequences that also contained the downstream 5′ss (Figure 2A). Spliceosome complexes on these RNAs were then assembled in Hela nuclear extract under splicing conditions and after heparin treatment, splicing complexes were separated on native poly-acrylamide gels. Interestingly, the intron-defined complexes assembled on the ATM WT substrate showed an inefficient/strongly reduced spliceosomal A complex formation (compare lanes 2 and 5, Figure 2B) but not so in the ATMΔ substrate. As the extent of ATP-dependent complex A formation reflects stable binding of U2snRNP on to the 3′ss region, this result suggested an inefficient recruitment of U2snRNP to the ATM WT 3′ss. As expected from these results, in vitro splicing of these biexonic constructs demonstrated that the spliceosomal complexes assembled across the EX2-ATM WT construct resulted in no intron processing (Figure 2C, lanes 1–2), whereas this occurred in the case of the EX2-ATMΔ RNA (Figure 2C, lanes 3–4).
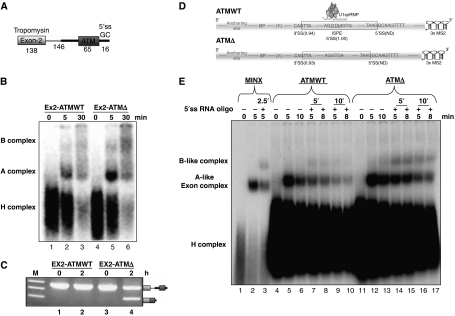
Figure 2.
Assembly of the spliceosomal complexes on ATM WT and ATMΔ pseudo exon substrates. (A) Scheme of the biexonic construct used for assembly of cross-intron spliceosomal complexes upstream to the ATM pseudoexon. (B) Spliceosomal complex assembly at different time-points on the ATM WT and Δ biexonic substrates. The positions of the splicing complexes are shown on the left. (C) In vitro splicing and RT–PCR analyses of the ATM WT and ATMΔ biexonic constructs. (D) Scheme of the ATM WT and ATMΔ single exon substrates used for the assembly of spliceosomal complexes. The 3′ ends are tagged with 3 MS2 repeats. (E) Spliceosomal complex assembly at different time points across both ATM WT and ATMΔ single exon substrates either in the absence or presence of the 5′ssRNA oligo ‘in-trans'.
To confirm these differences further, spliceosomal complexes assembled on single exon substrates derived from the ATM WT and ATMΔ sequences were analysed in vitro. Single exon constructs of ATM WT and ATMΔ RNAs were used carrying the upstream 3′ss, the polypyrimidine/predicted branch point (BP) sequences, 20 nt (named AS, as the anchoring site) upstream to the BP, and the downstream 5′ss (Figure 2D). Under similar splicing condition used in Figure 2B, this analysis showed that complexes were formed both on the WT and Δ RNAs and that these complexes assembled with near equal efficiencies at 5 min (compare lanes 5 and 12, Figure 2E). We termed these ATP-dependent complexes as A-like exon complexes. Interestingly, at longer incubation periods of 10 min, the A-like exon complex on the WT substrate seemed to dissociate or fall apart pointing towards the unproductive nature of this complex. On the other hand, the one assembled on the ATMΔ RNA was still stable at this time point (compare lanes 6 and 13, Figure 2E). Formation of these complexes was dependent on ATP and U2snRNP (data not shown). Furthermore, the long-term stability of the A-like complex in the ATMΔ RNA with respect to the ATM WT RNA (in the absence of additional oligos) was confirmed up to a 30-min incubation time (Supplementary Figure S1B). The approximately three-fold difference in the relative intensity of A-like complex between WT and Δ appears at longer incubation of 30 min, suggested that U2snRNP might be engaged in a unstable unproductive binding at the 3′ss of the WT substrate.
To address the question of whether these A-like exon complexes could be functional or not, a 5′ss RNA oligo ‘in-trans' was added at two separate time points of 5 and 10 min to each substrate, as described earlier (Konforti and Konarska, 1995). Each of the splicing reactions was then incubated further for an additional 5 and 8 min (Figure 2E). Gel analysis showed that in the presence of the 5′ss RNA oligo, the A-like exon complex formed on the ATMΔ RNA could progress well to a B-like complex (lanes 14–17, Figure 2E), whereas it could not do so as efficiently in the case of the ATM WT RNA (lanes 7–10, Figure 2E). A control oligo did not have any effect on B-like complex formation (Supplementary Figure S1A).
Taken together, these observations on both biexonic as well as single exon substrates highlight intrinsic differences between ATM WT and ATMΔ A/A-like complex formation. In this respect, therefore, the reduced ability of A-like to B-like progression observed in ATM WT seems to be the consequence of an inefficient A-like complex in ATM WT (Figure 2E).
U2snRNP association with ATM WT and ATMΔ RNAs
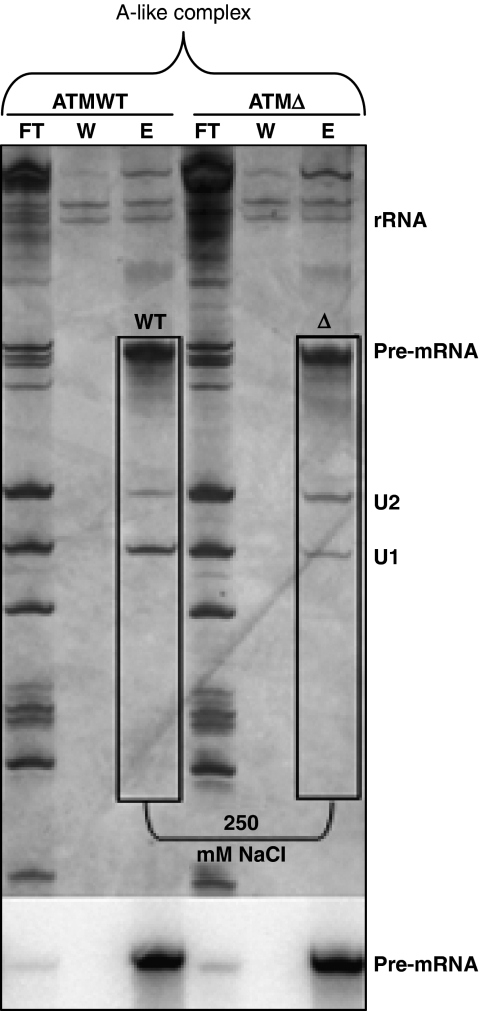
As the A-like exon complex formed on ATM WT and ATMΔ RNAs assembled with almost equal efficiencies at 5 min (Figure 2E, lanes 5 and 12), it was decided to compare their UsnRNP composition. To do this, these complexes were purified taking advantage of an MS2 RNA hairpin-tagged version of the ATM WT and ATMΔ RNAs as described earlier (Deckert et al, 2006) (Figure 2D). We sought to compare the relative binding stabilities of these UsnRNP to high-salt washings. To achieve this, the WT and Δ A-like exon complexes were bound to amylose beads and then washed with 250 mM NaCl. Silver stain of the resulting gels showed stronger signal for U1snRNA in the ATM WT lane as compared with ATMΔ lane, an observation consistent with the fact that the ATM WT pseudoexon has two U1snRNP-binding sites, one at ISPE and the other at the gc 5′ss. Importantly, and also consistent with the defective A-like exon complex, ATM WT RNA showed a substantially reduced signal for the U2snRNA as compared with the ATMΔ RNA (Figure 3). Unstable U2snRNP recruitment in ATM WT can also be better appreciated by observing the ratio of the signal for U1snRNA and U2snRNA (Figure 3). These results allowed us to conclude that U2snRNP is unstably associated with the 3′ss region of the ATM WT RNA as compared with ATMΔ in the A-like exon complexes. It is interesting to point out that although the A-like complexes are formed with almost equal efficiency at 5 min time point (at which point UsnRNPs are analysed), the observed differences for the U2snRNP recruitment under high-salt washings clearly highlight the intrinsic differences in the A-like complexes of ATM WT and Δ, that become apparent at longer incubations (Figure 2E; Supplementary Figure S1B).
Figure 3.
U2snRNP stability at high-salt washings of the ATM WT and ATMΔ A-like exon complexes. High-salt washings of the A-like exon complex assembled on the single exon substrate. Exon complexes that assembled on the MS2-tagged ATM WT and Δ substrate were affinity selected on amylose beads after glycerol gradient ultracentrifugation. The bound complexes were washed with 250 mM NaCl and eluted with maltose. FT lane represents flow through, W lane is for wash, and E lane for elute. RNAs were separated and visualized as described above. The autoradiograph of the same gel is shown below as loading control of the ATM WT and Δ pre-mRNAs. Efficiency of recovery has been estimated at 46% according to CPM counts before and after elution.
Mapping of the SF2/ASF-binding site in the ATMΔ sequence
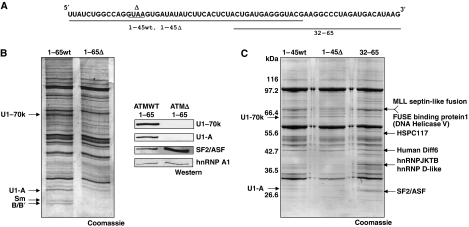
To identify important splicing factors binding to the ATM pseudoexon sequence beside the already identified U1snRNP, we used an affinity pull-down assay protocol (Buratti et al, 2004a). The coomassie gel in Figure 4B (left panel) shows the pull-down profiles of full-length ATM WT and ATMΔ constructs. As shown in this figure, clear differences could only be detected (as determined by mass-spec analysis) at the level of the U1snRNP subunits. These differences were confirmed by western blot analysis (Figure 4B, right panel). At the same time, we also probed the gel for common splicing factors (i.e. hnRNP A1 and SF2/ASF). In these cases, no binding differences could be seen for hnRNP A1 but, rather unexpectedly, they were detected for SF2/ASF. Interestingly, SF2/ASF was also the only SR protein that could bind to the ATMΔ construct, as determined using immunoprecipitation analysis (Supplementary Figure S2). To better map the SF2/ASF-binding site, we then repeated the pull-down analysis using shorter RNA sequences. The pseudoexon sequence was then divided in two halves for both ATM WT and ATMΔ to obtain the three following synthetic RNAs: ATMWT 1–45, ATMΔ 1–45, and ATM 32–65 RNA (Figure 4A). Pull-down and Coomassie stain from these RNAs detected numerous protein-binding profiles. Not many differences were observed for the ATM WT 1–45 and ATMΔ 1–45 except for the additional U1-associated proteins. On the other hand, the ATM 32–65 RNA was found to bind specifically to SF2/ASF. Mass-spec analysis also identified several proteins such as hnRNP D-like, Septins, and FUBP that were preferentially bound to this sequence (Figure 4C). However, as no clear differences in this region could be detected in the pull down using the whole pseudoexon sequences, their significance remains to be tested.
Figure 4.
Identification of trans-acting factors that bind the ATM pseudoexon using adipic acid–agarose beads based pull-down and mass-spec analysis. (A) A scheme of the ATM pseudoexon RNAs used for pull-down analysis is presented. The underlined sequence represents the region of the pseudoexon used for pull-down analysis that spans from nucleotide 1–45 for ATM WT and ATMΔ and from nucleotide 32–65 common to both. (B) Identification of proteins that interact with the ATM WT 1–65 and Δ 1–65 pseudoexon RNAs. Different synthetic RNAs were covalently linked to agarose beads and incubated with Hela nuclear extract under splicing conditions. Proteins that remained bound to the RNAs after washing were separated on 10% SDS–PAGE and detected by Coomassie blue staining. The protein bands identified by mass-spec are mentioned with arrowheads on the sides of the gel. To obtain a better quantitative picture, the right panel contains western blots against the U1-70K, U1-A, SF2/ASF, and hnRNP A1 factors (C) Pull-down analysis using RNAs that span from nucleotide 1–45 for ATM WT and ATMΔ and from nucleotide 32–65 common to both. Protein bands identified by mass-spec are mentioned with arrowheads on the sides of the gel.
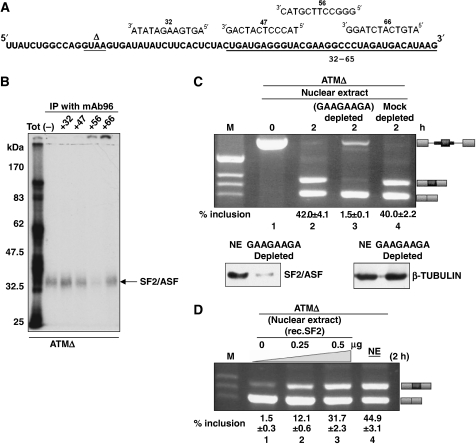
To better map the SF2/ASF-binding site on the 32–65 sequence, we then incubated the entire ATMΔ RNA with a series of 12-mer antisense DNA oligos complementary to the different regions of the 32–65 RNA (Figure 5A). In this experiment, we observed a significantly diminished IP signal only in the presence of the 56 oligo, suggesting that the SF2/ASF-binding site is placed in the 47–66 regions, narrowing down the putative binding sequence to ‘CGAAGGC' (Figure 5B).
Figure 5.
Mapping of the SF2/ASF-binding site on the ATMΔ pseudoexon sequence. (A) Schematic representation of 12-mer antisense DNA oligonucleotides targeting the ATMΔ pseudoexon sequence. (B) SF2/ASF immunoprecipitation with mAb96 antibody in the absence (lane –) or presence of these oligonucleotides. The position of the IP complex for SF2/ASF is shown with an arrowhead. (C) In vitro splicing of the ATMΔ pre-mRNA using SF2/ASF-depleted Hela Nuclear extract (lane 3) and with mock-depleted extract (lane 4). Schematic diagrams of the spliced products are shown on the right. Lower panel of (C) contains western blot showing the level of SF2/ASF depletion using the pull-down affinity procedure together with a tubulin control (D) ATMΔ pre-mRNA was incubated in either dilute nuclear extract (24%) supplemented with increasing quantities of recombinant SF2/ASF (lanes 1–3) or standard nuclear extract (48%) (lane 4) under splicing conditions. The quantities of the SF2/ASF added in μg to the nuclear extract are shown on the top. Schematic diagrams of the spliced products are shown on the right. Quantification of pseudoexon inclusion levels as determined by densitometric analysis are reported under each figure. Standard deviation values from three independent experiments are shown.
To demonstrate the functional involvement of SF2/ASF in ATMΔ pseudoexon splicing, affinity depletion of SF2/ASF was performed from the nuclear extract by incubating it with a short GAAGAAGAC RNA bound to agarose beads. This sequence, from the EDA exon, has been previously shown to be specific binding site for SF2/ASF and to some extent for other SR proteins (Buratti et al, 2004b). The depletion of SF2/ASF from the nuclear extract was confirmed by western blot (Figure 5C, lower panels). In keeping with expectations, in vitro splicing of the ATMΔ RNA in this depleted extract showed complete pseudoexon exclusion (Figure 5C, lane 3) compared with a mock-depleted nuclear extract (Figure 5C, lane 4). In parallel, we also performed an in vitro overexpression experiment by adding recombinant SF2/ASF into the nuclear extract. Under dilute conditions, the pseudoexon spliced poorly (lane 1, Figure 5D). However, addition of increasing amounts of recombinant SF2/ASF led to increased splicing of the ATMΔ pseudoexon in a linear manner (lanes 2–3, Figure 5D). Instead, splicing of ATM WT in the presence of increased amounts SF2/ASF did not result in any pseudoexon inclusion (data not shown).
Relationship between SF2/ASF binding and RNA secondary structure
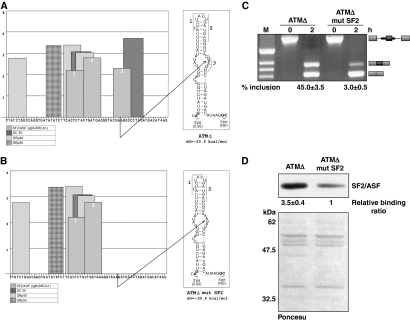
When we compared the predicted SF2/ASF-binding motifs obtained on the entire pseudoexon structure using ESE finder (Cartegni et al, 2003; Smith et al, 2006), the program predicted three high scoring SF2/ASF-binding motifs in the ATMΔ pseudoexon (Figure 6A, numbered 1–3). Notably, the first two SF2/ASF motifs (1 and 2) fell on the opposite sides of the upper stem region of ATMΔ, whereas the third SF2/ASF motif (3) fell on the internal loop conformation. Interestingly, this third motif fell exactly in the region mapped with the antisense DNA oligos in Figure 5B, suggesting a relationship between SF2/ASF binding and RNA secondary structure. A mutation in this sequence was then introduced that abolished the predicted motif for SF2/ASF binding without affecting the RNA secondary structure (Figure 6B). In vitro splicing of the ATMΔ mut SF2 mutant showed almost complete inhibition of pseudoexon inclusion (Figure 6C). The expected reduction in SF2/ASF binding efficiency introduced by this mutation was also confirmed by pull-down analysis followed by a western blot against this protein (Figure 6D).
Figure 6.
Correlation between the real and predicted SF2/ASF-binding site with the RNA secondary structure of ATMΔ. (A) The score matrix of various SR proteins predicted to bind ATMΔ RNA as predicted by ESE finder ver3.0. The SF2/ASF predicted motifs are numbered 1–3. The SF2/ASF-binding site validated experimentally to bind the ATMΔ pseudoexon overlaps with the predicted motif 3 and is shown by an arrow to fall in the internal loop conformation. (B) The predicted abolishment of the no. 3 SF2/ASF-binding site in the ATMΔ mutSF2 mutant. (C) In vitro splicing and RT–PCR of ATMΔ and ATMΔ mutSF2 RNA. Schematic diagrams of the spliced products are shown on the right. Quantification of pseudoexon inclusion levels as determined by densitometric analysis are reported under each figure. Standard deviation values from three independent experiments are shown. (D) Western blot pull-down analysis to confirm the abolishment of the SF2/ASF-binding site in the ATMΔ mutSF2 substrate in comparison to ATMΔ. Quantification of SF2/ASF-binding levels as determined by densitometric analysis are reported under each figure. Standard deviation values from three independent experiments are shown. Ponceau stain of the nitrocellulose membrane is shown for equal protein loading of the sample.
Secondary structure changes modulate the binding of SF2/ASF and additional trans-acting factors on the pseudoexon sequence
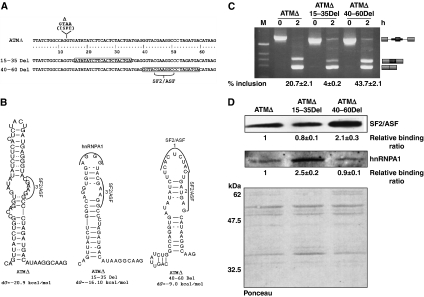
To further explore the relationship between RNA secondary structure and binding of the trans-acting factors, we generated two deletion mutants by randomly deleting 20 nt stretches from the pseudoexon (Figure 7A). In vitro splicing showed that the two deletion mutants gave contrasting splicing outcomes: the ATMΔ 15–35 Del showed strong inhibition of pseudoexon inclusion, whereas the ATMΔ 40–60 Del mutant displayed pseudoexon inclusion with an even greater efficiency than ATMΔ (Figure 7C).
Figure 7.
RNA secondary structure can influence splicing by modulating the display of trans-acting factor binding sites. (A) Scheme of the 20 nt deletions (15–35 and 40–60 Del mutants) made within the ATMΔ pseudoexon. The ATMΔ pseudoexon sequence is numbered starting from the 5′ end. Boxed sequences correspond to 20 nucleotide deleted region of the ATM pseudoexon. The positions of the GTAA deletion and of the SF2/ASF-binding site are also shown. (B) Predicted RNA secondary structure of ATMΔ and of the two deletion mutants. The position of the trans-acting factors (SF2/ASF and the putative hnRNPA1-binding site) is shown with arcs and brackets. (C) In vitro splicing and RT–PCR analysis of the ATMΔ deletion mutants. Schematic diagrams of the spliced products are shown on the right. Quantification of pseudoexon inclusion levels as determined by densitometric analysis are reported under each figure. Standard deviation values from three independent experiments are shown. (D) Pull-down analysis of ATMΔ and the deletion mutants followed by western blotting with mAb96 antibody against SF2/ASF and a polyclonal hnRNPA1 antibody. Quantification of SF2/ASF-binding levels as determined by densitometric analysis are reported under each figure. Standard deviation values from three independent experiments are shown. Ponceau stain of the nitrocellulose membrane is shown for equal protein loading of the sample.
In the case of the 15–35 Del mutant, the puzzling element was represented by the observation that this mutant showed almost complete pseudoexon skipping despite the SF2/ASF-binding site number 3 falling outside the 20 nt deleted region (Figure 7A). In this case, however, it was observed that the RNA secondary structure displayed the SF2/ASF number 3 site in a more constricted internal loop compared with the ATMΔ structure. Moreover, this 15–35 mutant now displayed a ‘AGGG' sequence in a terminal loop conformation that was earlier concealed in the upper stem region, which closely matched with a high-affinity hnRNPA1-binding site (Burd and Dreyfuss, 1994) (Figure 7B).
Second, considering the function of the previously identified SF2/ASF-binding site, it was unclear why the ATMΔ 40–60 Del showed better inclusion despite the fact that the mapped SF2/ASF-binding site was in the 20 nt deleted region. However, in the case of this mutant, the predicted RNA secondary structure now displayed the previously unavailable SF2/ASF-binding site number 1 in a large terminal loop, and the SF2/ASF number 2 site in an internal loop (Figure 7B). Verification of the putative secondary structures for the 15–35 and 40–60 Del mutants was performed using RNAse mapping (Supplementary Figure S3).
Finally, to further validate these considerations, we also performed pull-down analysis to determine SF2/ASF-binding levels. Notably, SF2/ASF binding to the 40–60 Del mutant enhanced by at least two times, whereas for the 15–35 Del mutant it was only slightly reduced (Figure 7D). Moreover, in keeping with the expectations, the 15–35 Del mutant showed more than two times increased binding of hnRNPA1 when compared with both ATMΔ and 40–60 Del RNAs (Figure 7D).
Sterical hindrance of the ISPE region for SF2/ASF binding to ATM WT
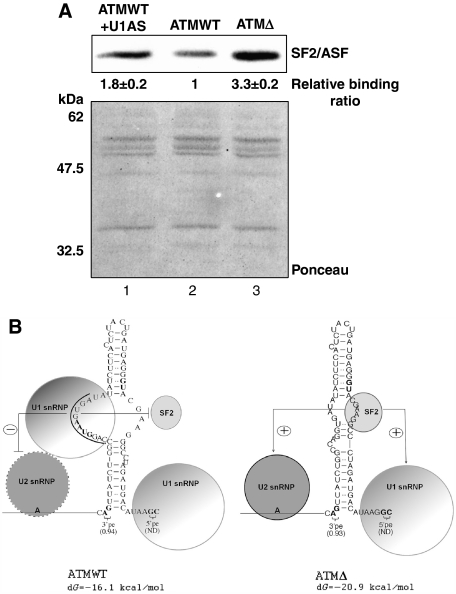
Considering that the SF2/ASF-binding region is also present in an open configuration in the ATM WT RNA, it was interesting to find why the signal of SF2/ASF for the WT 1–65 sequence was reduced with respect to Δ 1–65 (Figure 4B). Pull-down analysis confirmed that SF2/ASF binds more efficiently to the ATMΔ RNA, whereas much weaker binding was observed to the ATM WT RNA (Figure 8A, lanes 2 and 3, respectively). It should be noted, however, that the RNA secondary fold of the ATM WT showed that the U1snRNP binding at ISPE region falls on the bulge just opposite to the one occupied by SF2/ASF (Figure 8B). Therefore, U1snRNP docking at the ISPE would be expected to interfere also with the SF2/ASF deployment on the opposite side of the internal loop. To test this hypothesis, we depleted the nuclear extract with antisense DNA oligo against U1snRNP as described earlier (Raponi et al, 2009) (Supplementary Figure S4). Pull-down analysis with this extract rescued a significant level of SF2/ASF binding to the ATM WT sequences, thereby suggesting that SF2/ASF binding in WT is also modulated by U1snRNP binding at the ISPE (Figure 8A, lane 1).
Figure 8.
Schematic model of the U1snRNP-mediated inhibition in the ATM pseudoexon sequence. (A) Pull-down analysis and subsequent western blotting with mAb96 antibody on ATM WT and ATMΔ pseudoexon RNA in the presence of U1snRNP-depleted nuclear extract. Quantification of SF2/ASF-binding levels as determined by densitometric analysis are reported under each figure. Standard deviation values from three independent experiments are shown. Ponceau stain of the nitrocellulose membrane is shown for equal protein loading of the samples. (B) Model of the U1snRNP-mediated inhibition of ATM WT pseudoexon. In this model, the U1snRNP-mediated inhibition of the ATM WT pseudoexon is constituted by a unproductive less stable recruitment of U2snRNP on the 3′ss region. Irregular boundary of U2snRNP demonstrates defective recruitment. Moreover, it also obstructs SF2/ASF occupancy to its binding site on the opposite side of the stem. In ATMΔ, the U1snRNP is no longer present due to the deletion of the ISPE. This leads to a more efficient binding of SF2/ASF to the enhancer site and stabilization of the U2snRNP interaction.
Discussion
Normally, inclusion of aberrant exon-like sequences (pseudoexons) in mature mRNAs is actively inhibited by the formation of RNA secondary structures that prevent false 5′ and 3′ss recognition (Zhang et al, 2005; Schwartz et al, 2009), by the presence of intrinsic defects in the pseudoexon composition (Sun and Chasin, 2000) and by the enrichment of silencer elements in their vicinity (Fairbrother and Chasin, 2000; Sironi et al, 2004; Zhang and Chasin, 2004).
In particular, very little is known about the trans-acting factors that bind these silencer elements. So far, research on this subject has identified several well-known splicing inhibitory factors, such as PTB (Spellman and Smith, 2006; Sharma et al, 2008). This protein has been found to have an important function in downregulating the inclusion efficiency of a pathological pseudoexon in NF-1 intron 31, independently of the activating mutation that creates a very strong splicing acceptor site (Raponi et al, 2008). This finding supports the hypothesis that silencer sequences may be actively preserved by the evolutionary mechanisms to decrease the probability of randomly activating mutations in generating potentially harmful pseudoexon inclusion. In addition, in the case of non-pathological pseudoexon inclusions, such as one described in the α-tropomyosin gene (Grellscheid and Smith, 2006), specific binding of hnRNP H/F proteins have been recently found to act as repressors of this inclusion event (Coles et al, 2009).
One unusual silencer molecule that has been identified in this search is represented by the U1snRNP, a ribonucleoprotein complex normally associated in 5′ss recognition in the normal splicing process (Mount et al, 1983). Earlier, our laboratory has described a strong U1snRNP-binding site that can inhibit pathological pseudoexon inclusion in intron 20 of the ATM gene (Figure 1A). Inactivation of this element through a 4-nt deletion caused pseudoexon inclusion and occurrence of ataxia telangiectasia in a patient (Pagani et al, 2002). This finding has opened a new research field related to looking at the U1snRNP factor as a pseudoexon inhibitory factor. The fact, this new U1snRNP property may not be limited to a single pseudoexon splicing event comes from the recent finding that binding of hnRNP E1 and U1snRNP to a weak 5′ss can efficiently inhibit pseudoexon inclusion in the GH gene preventing the development of Laron syndrome (Akker et al, 2007). Moreover, examples of 5′ss sequences in splicing suppression have been observed in avian retroviral RNA via negative regulator of splicing (Giles and Beemon, 2005) and the intronic pseudo-5′ss in Drosophila P element transcripts (Siebel et al, 1992). It should also be mentioned, in parallel, that U1snRNP has already been reported to be a strong inhibitor of poly(A) addition (and thus gene expression) when bound to 5′ss-like sequences in the 3′ terminal exon of the papilloma virus (Furth et al, 1994), a property that has been recently exploited to set up a novel gene silencing methodology (Abad et al, 2008).
In this work, we have further characterized the U1snRNP-mediated splicing inhibition that is observed in the ATM gene. Our results have shown that U1snRNP binding to the ATM ISPE sequence inhibits pseudoexon inclusion for two reasons.
First, previous and recent observations concur with the opinion that repression of the 3′ss in ATM WT is a direct consequence of U1snRNP binding in correspondence to the ISPE sequence (Lewandowska et al, 2005; Pastor et al, 2009). From our experiments, it is now evident that this event causes an unproductive U2snRNP association with the 3′ss, which is reflected strongly by the inefficient A complex formation in the ATM WT RNA.
Second, the presence of the U1snRNP molecule bound to the ISPE significantly reduces the binding affinity of SF2/ASF for an enhancer sequence localized far away in the pseudoexon sequence but brought in its vicinity by the effects of RNA secondary structure. In this respect, it is important to note that these results are totally in keeping with earlier results obtained for a well-known enhancer sequence in the EDA exon, where we demonstrated that to have optimal binding of SR proteins, the binding site needs to be displayed in an open configuration (Buratti et al, 2004b). These results are also consistent with recent global studies, which show that enhancer and silencer sequences in a wide variety of experimental systems are significantly more single stranded than expected (Hiller et al, 2007) and that alternative splice site choice is often modified by the presence of evolutionarily conserved structures, which favour the use of one splice site over the competing ones (Shepard and Hertel, 2008). It is also interesting to note that the SF2/ASF-binding site contains a (G)AA(G) motif, which was recently found to be enriched in cryptic exons derived from human transposable elements (Vorechovsky, 2009). Here, it might also be interesting to speculate the critical requirement of such a strong enhancer in the selection of weak 5′ss like the ‘gc' ones, that are represented by only 0.5–1% of all the 5′ss in the human genome (Thanaraj and Clark, 2001). Our observation that alternative SF2/ASF-binding sites identified by in silico analysis using ESE finder are not active, suggests that one way to improve these kinds of programs would be to take local RNA secondary structure into account. In fact, the bioinformatic approaches (although a valuable tool) available provide only a rough approximation to the splicing outcome prediction and anyone looking for exonic enhancers with the available programs need to further validate them experimentally (Hartmann et al, 2008; Houdayer et al, 2008). Furthermore, the use of linear degenerated sequences in the statistical approaches without considering their spatial configuration in the RNA molecule, and the kinetics of RNA synthesis, leave without explanation why multiple intronic sequences with the full credentials to be an exon are not included in the final mRNA molecule.
In conclusion, our results will be useful to better understand how 5′ss-like sequences can inhibit splicing, a field of study that is rapidly emerging due to global analyses of splicing systems, and have further highlighted the function of RNA secondary structure in the display of protein/RNA-binding motifs. Finally, from a therapeutic point of view, our results show that a suitably modified U1snRNP molecule targeted to bind a particular pseudoexon sequence may act as a powerful silencer element. An alternative therapeutic approach, based on our results, would be to affect the concentration of SF2/ASF in patient's cells to decrease pseudoexon inclusion. In this respect, it should be noted that several compounds have been recently developed to achieve such an aim (Soret et al, 2005). The advantage represented by such a molecule over more conventional antisense oligonucleotide approaches (Wood et al, 2007) would be to avoid toxicity, increase the efficiency of nuclear delivery, and to follow a biocompatible degradation process.
Materials and methods
Constructs carrying the ATM wild-type and mutant sequences
The PY7 plasmid carrying ATM WT and ATMΔ pseudoexon sequences have been described in detail elsewhere (Buratti et al, 2007). To obtain ATMΔ U11, a two-step PCR method was used with the following primers: 5′-tctggccagatatcctttgtgatatatcttc-3′ (s) and 5′-gaagatatatcacaaaggatatctggccaga-3′ (as). Similarly, to obtain the ATMΔ mutSF2, the primers used were the following 5′-actgatgagggtaaagatgccctagatgac-3′ (s) and 5′-gtcatctagggcatctttaccctcatcagt-3′ (as). Finally, to obtain the deletion mutants, ATMΔ 15–35 Del and ATMΔ 40–60 Del the following primers 5′-ttatctggccaggtgtgagggtacgaaggc-3′ (s) and 5′-gccttcgtaccctcacacctggccagataa-3′ (as), 5′-cactctactgatgagcataaggcaagtttt-3′ (s) and 5′-aaaacttgccttatgctcatcagtagagtg-3′ (as) were used, respectively. To generate the single exon substrates, the following primer pairs were used: 5′-ctaatacgactcactatagggcttaactgcaacagtggt-3′ (s) and 5′-tatcggatccgtaccacagccttaac-3′ (as). In parallel, the pMINX–MS2 plasmid was used to generate DNA fragments containing three MS2 coat protein RNA-binding sites using the following primers 5′-gctgtggtacggatccgatatccgtac-3′ (s) and 5′-ctatagaactcgactctagag-3′ (as), which was fused with the single exon substrates through overlapping PCR. In addition, ATM WT and Δ biexonic constructs were derived from the original plasmids with the following primers: 5′-ctaatacgactcactatagggaatacaagcttgtcgag-3′ (s) and 5′-ctggtaaaaaacttgccttatgtcat-3′ (as).
Spliceosomal complexes and MS2 affinity purification
Splicing complexes were assembled as described earlier (Behzadnia et al, 2006). Briefly, 10 nM of 32P-labelled capped RNA was incubated with 40% (v/v) of Hela nuclear extract under splicing conditions except for final concentration of 46.5 mM KCl. For the ATP-dependant complexes, at the indicated time, standard splicing reactions were stopped by addition of heparin to a final concentration of 0.1 mg/ml, and the incubation was continued for another 5 min. The splicing complexes were separated and visualized using 4% native PAG according to established protocols (Konarska and Sharp, 1987). Where indicated, a RNA oligonucleotide representing a 5′ss (cuguucagguaaguau, exonic sequence underlined) was added and incubation was continued further as mentioned. The gels were dried under vacuum and visualized by Phosphorimager (Molecular Dynamics). Purification of the pre-mRNP complexes were performed as described before (Deckert et al, 2006), except for the following modifications: the final RNA concentration was 10 nM and splicing reactions were incubated for 5 min at 30°C. The purification of the complexes was performed at 46.5 mM NaCl (Buffer G46.5). Ultracentrifugation in a 10–30% glycerol gradient was performed for 2 h 10 min at 60 K in a sorvall TH660 rotor at 4°C. High-salt washings of amylose bound spliceosomal complexes were performed with 6 column volumes of 250 mM NaCl. RNA was separated on an 8.3 M urea/10% (w/v) denaturing polyacrylamide gel and visualized by silver staining and autoradiography.
Immobilization of RNA on agarose beads and binding assays
The RNAs for bead immobilization were either chemically synthesized in case of ATM 1–45 WT, ATM 1–45Δ, and ATM 32–65 RNA oligonucleotides (IDT Technologies) or obtained by in vitro transcription using T7 RNA polymerase (Stratagene) of PCR-generated DNA templates. To generate the full-length ATM WT and Δ pseudoexon and its various mutants as substrate for pull down, the following primers were used: 5′-taatacgactcactatagggacagttatctggccaggt-3′ (s) and 5′-cttgccttatgtcatctaggg-3′ (as) except for the ATMΔ 40–60 Del where 5′-cttgccttatgctcatcagta-3′ (as) was used. RNAs were covalently linked to agarose beads as described earlier (Buratti et al, 2001) except for few modifications. Buffer D used at all the washing steps had a final concentration of 50 mM KCl. A measure of 100 μl of RNA-agarose beads was incubated with 400 μl of in vitro splicing reaction (Behzadnia et al, 2006) with final KCl concentration of 46.5 mM. The proteins bound to the immobilized RNA were eluted by addition of 60 μl of sodium dodecyl sulfate (SDS)-sample buffer and heated for 5 min at 95°C. Proteins were separated on a 10% SDS–polyacrylamide gel electrophoresis (PAGE) and analysed by coomassie blue staining, Ponceau stain of the transferred nitrocellulose membrane, western blotting, and electronspray mass spectrometry as have been described in detail elsewhere (Buratti et al, 2004a). To inactivate U1snRNP, a small oligonucleotide (5′-ccaggtaagtat-3′, U1AS oligonucleotide) was added to the above reaction mixture at final concentration of 5 ng/μl and reactions were incubated in the presence of RNase H (USB). In silico secondary structure predictions were performed using the mFold program (Zuker, 2003).
IP of SF2/ASF following UV cross-linking and mapping with 12-mer DNA oligos
The ATMΔ RNA probe was obtained by linearizing the ATMΔ PY7 plasmid with BamH1 and transcribed with SP6 RNA polymerase (Promega) in accordance with the standard procedures. UV cross-linking of (α-32P) UTP-labelled RNAs with commercial HeLa nuclear extract (C4; Cil Biotech, Belgium) was performed as described before (Buratti et al, 2004b). To each sample, we added 150 μl of IP buffer (20 mM Tris pH=8.0, 300 mM NaCl, 1 mM EDTA, 0.25% NP-40) together with 1 μg of monoclonal antibody (MAb) and incubated the mixture for 2 h at 4°C on a rotator wheel. Anti-SF2/ASF (mAb96) was purchased from Zymed Laboratories Inc. After 2 h incubation, we added to each sample 30 μl of Protein A/G-Plus Agarose (Santa Cruz Biotechnologies) and incubated the mixture at 4°C overnight. On the following morning, the beads were subjected to four washing cycles with 1.5 ml of IP buffer and then loaded onto an SDS–11% PAGE gel. Gels were run, dried, and then exposed for 4 to 6 days with a BioMax Screen (Kodak).
In vitro splicing assays
In vitro splicing of capped RNAs was performed as described elsewhere (Buratti et al, 2007). When necessary, human recombinant SF2/ASF (Jena Bioscience, Germany) was preincubated with the Hela nuclear extract before the overexpression experiment as described before (Fu et al, 1992). Conversely, SF2/ASF was depleted from the Hela nuclear extract using in vitro transcribed single stranded GAAGAAGAC RNA immobilized on adipic acid dihydrazide agarose beads (Buratti et al, 2001).
Supplementary Material
Supplementary Data
Review Process File
Acknowledgments
We are grateful to Reinhard Lührmann for providing us with mouse monoclonal antibodies against U1-70k and U1-A proteins and also pMINX-MS2 plasmid as a gift. We are also thankful to CWJ Smith for providing the PY7 plasmid. This work was supported by Telethon Onlus Foundation (Italy) (grant no. GGP06147) and by a European community grant (EURASNET-LSHG-CT-2005-518238).
Footnotes
The authors declare that they have no conflict of interest.
References
- Abad X, Vera M, Jung SP, Oswald E, Romero I, Amin V, Fortes P, Gunderson SI (2008) Requirements for gene silencing mediated by U1 snRNA binding to a target sequence. Nucleic Acids Res 36: 2338–2352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akker SA, Misra S, Aslam S, Morgan EL, Smith PJ, Khoo B, Chew SL (2007) Pre-spliceosomal binding of U1 small nuclear ribonucleoprotein (RNP) and heterogenous nuclear RNP E1 is associated with suppression of a growth hormone receptor pseudoexon. Mol Endocrinol 21: 2529–2540 [DOI] [PubMed] [Google Scholar]
- Amara SG, Jonas V, Rosenfeld MG, Ong ES, Evans RM (1982) Alternative RNA processing in calcitonin gene expression generates mRNAs encoding different polypeptide products. Nature 298: 240–244 [DOI] [PubMed] [Google Scholar]
- Baralle M, Skoko N, Knezevich A, De Conti L, Motti D, Bhuvanagiri M, Baralle D, Buratti E, Baralle FE (2006) NF1 mRNA biogenesis: effect of the genomic milieu in splicing regulation of the NF1 exon 37 region. FEBS Lett 580: 4449–4456 [DOI] [PubMed] [Google Scholar]
- Behzadnia N, Hartmuth K, Will CL, Luhrmann R (2006) Functional spliceosomal A complexes can be assembled in vitro in the absence of a penta-snRNP. RNA 12: 1738–1746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breathnach R, Benoist C, O'Hare K, Gannon F, Chambon P (1978) Ovalbumin gene: evidence for a leader sequence in mRNA and DNA sequences at the exon-intron boundaries. Proc Natl Acad Sci USA 75: 4853–4857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratti E, Baralle M, Baralle FE (2006) Defective splicing, disease and therapy: searching for master checkpoints in exon definition. Nucleic Acids Res 34: 3494–3510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratti E, Baralle M, De Conti L, Baralle D, Romano M, Ayala YM, Baralle FE (2004a) hnRNP H binding at the 5′ splice site correlates with the pathological effect of two intronic mutations in the NF-1 and TSHbeta genes. Nucleic Acids Res 32: 4224–4236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratti E, Dhir A, Lewandowska MA, Baralle FE (2007) RNA structure is a key regulatory element in pathological ATM and CFTR pseudoexon inclusion events. Nucleic Acids Res 35: 4369–4383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratti E, Dork T, Zuccato E, Pagani F, Romano M, Baralle FE (2001) Nuclear factor TDP-43 and SR proteins promote in vitro and in vivo CFTR exon 9 skipping. EMBO J 20: 1774–1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratti E, Muro AF, Giombi M, Gherbassi D, Iaconcig A, Baralle FE (2004b) RNA folding affects the recruitment of SR proteins by mouse and human polypurinic enhancer elements in the fibronectin EDA exon. Mol Cell Biol 24: 1387–1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd CG, Dreyfuss G (1994) RNA binding specificity of hnRNP A1: significance of hnRNP A1 high-affinity binding sites in pre-mRNA splicing. EMBO J 13: 1197–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caputi M, Casari G, Guenzi S, Tagliabue R, Sidoli A, Melo CA, Baralle FE (1994) A novel bipartite splicing enhancer modulates the differential processing of the human fibronectin EDA exon. Nucleic Acids Res 22: 1018–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartegni L, Wang J, Zhu Z, Zhang MQ, Krainer AR (2003) ESEfinder: a web resource to identify exonic splicing enhancers. Nucleic Acids Res 31: 3568–3571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YI, Moore RE, Ge HY, Young MK, Lee TD, Stevens SW (2007) Proteomic analysis of in vivo-assembled pre-mRNA splicing complexes expands the catalog of participating factors. Nucleic Acids Res 35: 3928–3944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coles JL, Hallegger M, Smith CW (2009) A nonsense exon in the Tpm1 gene is silenced by hnRNP H and F. RNA 15: 33–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper TA, Wan L, Dreyfuss G (2009) RNA and disease. Cell 136: 777–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cote J, Dupuis S, Jiang Z, Wu JY (2001) Caspase-2 pre-mRNA alternative splicing: identification of an intronic element containing a decoy 3′ acceptor site. Proc Natl Acad Sci USA 98: 938–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R, Zhou Z, Reed R (2000) Functional association of U2 snRNP with the ATP-independent spliceosomal complex E. Mol Cell 5: 779–787 [DOI] [PubMed] [Google Scholar]
- Deckert J, Hartmuth K, Boehringer D, Behzadnia N, Will CL, Kastner B, Stark H, Urlaub H, Luhrmann R (2006) Protein composition and electron microscopy structure of affinity-purified human spliceosomal B complexes isolated under physiological conditions. Mol Cell Biol 26: 5528–5543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efstratiadis A, Posakony JW, Maniatis T, Lawn RM, O'Connell C, Spritz RA, DeRiel JK, Forget BG, Weissman SM, Slightom JL, Blechl AE, Smithies O, Baralle FE, Shoulders CC, Proudfoot NJ (1980) The structure and evolution of the human beta-globin gene family. Cell 21: 653–668 [DOI] [PubMed] [Google Scholar]
- Fairbrother WG, Chasin LA (2000) Human genomic sequences that inhibit splicing. Mol Cell Biol 20: 6816–6825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox-Walsh KL, Dou Y, Lam BJ, Hung SP, Baldi PF, Hertel KJ (2005) The architecture of pre-mRNAs affects mechanisms of splice-site pairing. Proc Natl Acad Sci USA 102: 16176–16181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu XD (2004) Towards a splicing code. Cell 119: 736–738 [DOI] [PubMed] [Google Scholar]
- Fu XD, Mayeda A, Maniatis T, Krainer AR (1992) General splicing factors SF2 and SC35 have equivalent activities in vitro, and both affect alternative 5′ and 3′ splice site selection. Proc Natl Acad Sci USA 89: 11224–11228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furth PA, Choe WT, Rex JH, Byrne JC, Baker CC (1994) Sequences homologous to 5′ splice sites are required for the inhibitory activity of papillomavirus late 3′ untranslated regions. Mol Cell Biol 14: 5278–5289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giles KE, Beemon KL (2005) Retroviral splicing suppressor sequesters a 3′ splice site in a 50S aberrant splicing complex. Mol Cell Biol 25: 4397–4405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grellscheid SN, Smith CW (2006) An apparent pseudo-exon acts both as an alternative exon that leads to nonsense-mediated decay and as a zero-length exon. Mol Cell Biol 26: 2237–2246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han K, Yeo G, An P, Burge CB, Grabowski PJ (2005) A combinatorial code for splicing silencing: UAGG and GGGG motifs. PLoS Biol 3: e158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann L, Theiss S, Niederacher D, Schaal H (2008) Diagnostics of pathogenic splicing mutations: does bioinformatics cover all bases? Front Biosci 13: 3252–3272 [DOI] [PubMed] [Google Scholar]
- Hertel KJ (2008) Combinatorial control of exon recognition. J Biol Chem 283: 1211–1215 [DOI] [PubMed] [Google Scholar]
- Hiller M, Zhang Z, Backofen R, Stamm S (2007) Pre-mRNA secondary structures influence exon recognition. PLoS Genet 3: e204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houdayer C, Dehainault C, Mattler C, Michaux D, Caux-Moncoutier V, Pages-Berhouet S, d'Enghien CD, Lauge A, Castera L, Gauthier-Villars M, Stoppa-Lyonnet D (2008) Evaluation of in silico splice tools for decision-making in molecular diagnosis. Hum Mutat 29: 975–982 [DOI] [PubMed] [Google Scholar]
- Konarska MM, Sharp PA (1987) Interactions between small nuclear ribonucleoprotein particles in formation of spliceosomes. Cell 49: 763–774 [DOI] [PubMed] [Google Scholar]
- Konforti BB, Konarska MM (1995) A short 5′ splice site RNA oligo can participate in both steps of splicing in mammalian extracts. RNA 1: 815–827 [PMC free article] [PubMed] [Google Scholar]
- Kornblihtt AR, Vibe-Pedersen K, Baralle FE (1984) Human fibronectin: molecular cloning evidence for two mRNA species differing by an internal segment coding for a structural domain. EMBO J 3: 221–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewandowska MA, Stuani C, Parvizpur A, Baralle FE, Pagani F (2005) Functional studies on the ATM intronic splicing processing element. Nucleic Acids Res 33: 4007–4015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardon HJ, Sebastio G, Baralle FE (1987) A role for exon sequences in alternative splicing of the human fibronectin gene. Nucleic Acids Res 15: 7725–7733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matlin AJ, Moore MJ (2007) Spliceosome assembly and composition. Adv Exp Med Biol 623: 14–35 [DOI] [PubMed] [Google Scholar]
- Mount SM, Pettersson I, Hinterberger M, Karmas A, Steitz JA (1983) The U1 small nuclear RNA-protein complex selectively binds a 5′ splice site in vitro. Cell 33: 509–518 [DOI] [PubMed] [Google Scholar]
- Nilsen TW (2003) The spliceosome: the most complex macromolecular machine in the cell? Bioessays 25: 1147–1149 [DOI] [PubMed] [Google Scholar]
- Pagani F, Buratti E, Stuani C, Bendix R, Dork T, Baralle FE (2002) A new type of mutation causes a splicing defect in ATM. Nat Genet 30: 426–429 [DOI] [PubMed] [Google Scholar]
- Pastor T, Talotti G, Lewandowska MA, Pagani F (2009) An Alu-derived intronic splicing enhancer facilitates intronic processing and modulates aberrant splicing in ATM. Nucleic Acids Res 37: 7258–7267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pessa HK, Will CL, Meng X, Schneider C, Watkins NJ, Perala N, Nymark M, Turunen JJ, Luhrmann R, Frilander MJ (2008) Minor spliceosome components are predominantly localized in the nucleus. Proc Natl Acad Sci USA 105: 8655–8660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raponi M, Buratti E, Dassie E, Upadhyaya M, Baralle D (2009) Low U1 snRNP dependence at the NF1 exon 29 donor splice site. FEBS J 276: 2060–2073 [DOI] [PubMed] [Google Scholar]
- Raponi M, Buratti E, Llorian M, Stuani C, Smith CW, Baralle D (2008) Polypyrimidine tract binding protein regulates alternative splicing of an aberrant pseudoexon in NF1. FEBS J 275: 6101–6108 [DOI] [PubMed] [Google Scholar]
- Schwartz S, Gal-Mark N, Kfir N, Oren R, Kim E, Ast G (2009) Alu exonization events reveal features required for precise recognition of exons by the splicing machinery. PLoS Comput Biol 5: e1000300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seraphin B, Rosbash M (1989) Identification of functional U1 snRNA-pre-mRNA complexes committed to spliceosome assembly and splicing. Cell 59: 349–358 [DOI] [PubMed] [Google Scholar]
- Sharma S, Kohlstaedt LA, Damianov A, Rio DC, Black DL (2008) Polypyrimidine tract binding protein controls the transition from exon definition to an intron defined spliceosome. Nat Struct Mol Biol 15: 183–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard PJ, Hertel KJ (2008) Conserved RNA secondary structures promote alternative splicing. RNA 14: 1463–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebel CW, Fresco LD, Rio DC (1992) The mechanism of somatic inhibition of Drosophila P-element pre-mRNA splicing: multiprotein complexes at an exon pseudo-5′ splice site control U1 snRNP binding. Genes Dev 6: 1386–1401 [DOI] [PubMed] [Google Scholar]
- Sironi M, Menozzi G, Riva L, Cagliani R, Comi GP, Bresolin N, Giorda R, Pozzoli U (2004) Silencer elements as possible inhibitors of pseudoexon splicing. Nucleic Acids Res 32: 1783–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CW, Valcarcel J (2000) Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem Sci 25: 381–388 [DOI] [PubMed] [Google Scholar]
- Smith PJ, Zhang C, Wang J, Chew SL, Zhang MQ, Krainer AR (2006) An increased specificity score matrix for the prediction of SF2/ASF-specific exonic splicing enhancers. Hum Mol Genet 15: 2490–2508 [DOI] [PubMed] [Google Scholar]
- Solis AS, Shariat N, Patton JG (2008) Splicing fidelity, enhancers, and disease. Front Biosci 13: 1926–1942 [DOI] [PubMed] [Google Scholar]
- Soret J, Bakkour N, Maire S, Durand S, Zekri L, Gabut M, Fic W, Divita G, Rivalle C, Dauzonne D, Nguyen CH, Jeanteur P, Tazi J (2005) Selective modification of alternative splicing by indole derivatives that target serine-arginine-rich protein splicing factors. Proc Natl Acad Sci USA 102: 8764–8769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellman R, Smith CW (2006) Novel modes of splicing repression by PTB. Trends Biochem Sci 31: 73–76 [DOI] [PubMed] [Google Scholar]
- Sun H, Chasin LA (2000) Multiple splicing defects in an intronic false exon. Mol Cell Biol 20: 6414–6425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tazi J, Bakkour N, Stamm S (2009) Alternative splicing and disease. Biochim Biophys Acta 1792: 14–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanaraj TA, Clark F (2001) Human GC-AG alternative intron isoforms with weak donor sites show enhanced consensus at acceptor exon positions. Nucleic Acids Res 29: 2581–2593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vibe-Pedersen K, Kornblihtt AR, Baralle FE (1984) Expression of a human alpha-globin/fibronectin gene hybrid generates two mRNAs by alternative splicing. EMBO J 3: 2511–2516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorechovsky I (2009) Transposable elements in disease-associated cryptic exons. Hum Genet (e-pub ahead of print) [DOI] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Luhrmann R (2009) The spliceosome: design principles of a dynamic RNP machine. Cell 136: 701–718 [DOI] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB (2008) Alternative isoform regulation in human tissue transcriptomes. Nature 456: 470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GS, Cooper TA (2007) Splicing in disease: disruption of the splicing code and the decoding machinery. Nat Rev Genet 8: 749–761 [DOI] [PubMed] [Google Scholar]
- Wang Z, Burge CB (2008) Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA 14: 802–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Rolish ME, Yeo G, Tung V, Mawson M, Burge CB (2004) Systematic identification and analysis of exonic splicing silencers. Cell 119: 831–845 [DOI] [PubMed] [Google Scholar]
- Will CL, Urlaub H, Achsel T, Gentzel M, Wilm M, Luhrmann R (2002) Characterization of novel SF3b and 17S U2 snRNP proteins, including a human Prp5p homologue and an SF3b DEAD-box protein. EMBO J 21: 4978–4988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood M, Yin H, McClorey G (2007) Modulating the expression of disease genes with RNA-based therapy. PLoS Genet 3: e109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y, Lee C (2006) Alternative splicing and RNA selection pressure—evolutionary consequences for eukaryotic genomes. Nat Rev Genet 7: 499–509 [DOI] [PubMed] [Google Scholar]
- Zhang XH, Chasin LA (2004) Computational definition of sequence motifs governing constitutive exon splicing. Genes Dev 18: 1241–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XH, Leslie CS, Chasin LA (2005) Dichotomous splicing signals in exon flanks. Genome Res 15: 768–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31: 3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data
Review Process File