Abstract
Aim
To investigate possible interactions between genetic variants in glucose transporter type 9 (SLC2A9) gene and dietary habits in serum uric acid regulation.
Methods
Participants for this study were recruited from two isolated Croatian island communities of Vis (n = 918) and Korčula (n = 898). Three single nucleotide polymorphisms (SNP) from the SLC2A9 gene (rs1014290, rs6449213, rs737267) were correlated with dietary habits and uric acid.
Results
A significant decrease in uric acid levels was recorded with increasing consumption of milk, sour cream, duck and turkey, and eggs. The only significant interaction was found between potato consumption and rs737267 and a near-significant interaction was found between soft drinks and rs1014290 (interaction P = 0.068). Increased consumption of soft drinks interacting with the TT genotype at rs1014290 increased serum uric acid. No significant interactions were observed between food products consumption and rs6449213.
Conclusion
There is a certain extent of interaction between SLC2A9 and dietary patterns in serum uric acid determination. The metabolic effect of soft drinks seems to be determined by the underlying genotype of rs1014290.
Gout is a disorder of purine metabolism that presents as inflammatory arthritis caused by urate crystallizing in joints following chronic hyperuricemia. It affects 1%-2% of the adult population in the Western world, especially elderly men. Diet and, more recently, genetic polymorphisms have been recognized as the most important factors causatively associated with serum uric acid levels and gout (1). High protein intake and purine-rich products, like meat and seafood, have been known to increase serum uric acid and the risk of gout, while vegetables rich in purine content showed no effect (2). On the other hand, the protective effect of certain foods has been identified. For example, dairy products, vitamin C, and coffee have been reported to decrease serum uric acid and prevalence of gout (1).
Serum urate concentration is a highly variable trait among humans. It is greatly affected by environmental factors (diet), but shows high heritability of about 60% (3). Not surprisingly, genome-wide association studies identified genetic polymorphisms that substantially affect urate concentrations and gout. Polymorphisms in a transporter gene GLUT9 (SLC2A9) have been shown to explain 1.7%-5.3% of the variance in serum uric acid concentration (4,5). Recently, polymorphisms in 3 additional genes, URAT1 (SLC22CA12), ABCG2, and SLC17A3, have also been associated with uric acid concentrations and the risk of gout (6-8).
SLC2A9 was first described as a glucose transporter and only later as a urate transporter. The study investigating whether glucose or fructose influenced urate transport in African clawed frog oocytes (Xenopus laevis) demonstrated that SLC2A9-mediated urate transport was facilitated by glucose and, to a lesser extent, fructose (9). We wanted to investigate if we could observe the same effect in vivo in humans and whether diet in general had a different impact on serum uric acid depending on the genetic background of an individual. To our knowledge, this is the first study on uric acid and gout that investigates genotype-environment interaction, more specifically, the interaction between previously reported genetic polymorphisms in the SLC2A9 gene and diet in humans.
Materials and methods
Samples
In the total sample of 1822 individuals, 924 were recruited on the island of Vis, Croatia, in 2002 and 2003, and 898 participants on the island of Korčula, Croatia, in 2006 and 2007 (10-13). There were 1109 women and 713 men (60% women), with mean age of 56 years (age range: 18-98 years). In both of these isolated communities genetic and environment specificities were found (12,14-21), which gave rise to a number of interesting research findings (4,22-28).
All participants gave written informed consent. The Vis and Korčula studies were approved by the Ethics Committee of the University of Zagreb Medical School and the Multi-Centre Research Ethics Committee for Scotland.
Body and biochemical measures
For all participants, height and weight were recorded and body-mass index (BMI) was calculated as weight (kg) divided by squared height (m2). Uric acid was measured as a part of classical biochemistry analysis and was available for 1732 individuals in our sample (29).
Food intake
All participants filled in a detailed food frequency questionnaire, developed for the specific purposes of this study. The questionnaire enquired about the habit of consumption for 50 different food products (Table 1) and the consumption frequency was first recorded as daily, 2-3 times per week, once per week, rarely, and never. In the subsequent analysis, original categorical variables were replaced by values 30, 10, 4, 1, and 0, respectively, to reflect the monthly frequency of intake for a given product. No information was available on the serving size of consumed food products.
Table 1.
List of all food products and derived variables tested for association with serum uric acid levels and gout
|
Dairy |
Vegetables |
Meat |
| Milk |
Leafy |
Pork |
| Yoghurt |
Root |
Beef |
| Sour cream |
Flowery |
Veil |
| Cottage cheese |
Fruity |
Lamb |
| Hard cheese |
Vegetable Total |
Bacon |
| Dairy total |
Sausages and salami |
|
| Venison |
||
|
Bread, pasta, rice |
Other |
Chicken |
| White bread |
Beans |
Duck and turkey |
| Brown bread |
Potato |
Meat total |
| Muesli |
Mushrooms |
Eggs |
| Pasta and rice |
Fruit |
|
| Carbs total |
Nuts |
|
|
Desert |
Fish |
Drinks |
| Cakes |
White fish |
Fruit drinks |
| Chocolate |
Blue fish |
“Cedevita” |
| Biscuits |
Sea food |
Soft drinks |
| Candy |
Squid and octopus |
Strong drinks |
| Jam |
Salted fish |
Coffee |
| Deserts total |
Fish cans |
Tea |
| Fish total |
The intake frequencies of food products from the same group were combined to create group totals: “Dairy total” (milk, yoghurt, sour cream, cottage cheese, and hard cheese); “Carbs Total” (for food rich in carbohydrates, including white and brown bread, muesli, pasta, and rice); “Deserts Total” (cakes, chocolate, biscuits, candies, or jam); “Vegetable Total” (all vegetables); “Fish Total” for all fish; and “Meat Total” for all meat products.
The maximum frequency of consumption for any individual food product was 30 (the attributed value if the product was consumed daily). However, for derived variables that summed the frequencies of all products from the same group the score could be much higher.
When we analyzed the effect of soft drinks, the intake was recoded as “low” for never, “medium” for rarely and once a week, and “high” for daily consumption or consumption 2-3 times a week.
Genotyping
The genotype results from genome-wide SNP array typing were available. DNA samples were genotyped according to the manufacturer’s instructions on Illumina Infinium HumanHap 300 v1 for Vis and 370CNV for Korčula sample (Illumina Inc., San Diego, CA, USA). Genotypes were determined using Illumina BeadStudio software. Samples with a call rate below 97% were excluded from the analysis. We analyzed 3 SNPs in SLC2A9 gene (chromosome 4p16.1) previously reported for strong association with uric acid levels: rs1014290, rs6449213, and rs737267 (4). Genotypes for 3 SNPs of interest were extracted from the original data set and used in the subsequent analysis.
Statistical analysis
Descriptive statistics was performed using R (http://www.r-project.org) and Excel. The analysis and management of the genetic data was done with R and GenABEL package in R (30). Polygenic function in GenABEL implements a linear mixed effects model, where the relatedness structure of the samples is modeled as random effects, and the residuals are adjusted for relatedness. A simple regression was done with uric acid residuals from polygenic model adjusted for age and sex as the outcome, to test the effect of different food products and groups of products and 3 selected SNPs (rs1014290, rs6449213, and rs737267) on serum uric acid. Next, we investigated if there was any interaction between polymorphisms in the newly identified urate transporter gene (SLC2A9) and diet, each previously described to affect uric acid levels. In each model, the outcome variables were uric acid residuals from the polygenic model and the explanatory variables were BMI, intake frequency of a given food product, genotype at a chosen SNP locus, and their interaction (SNP*food product). A total of 56 models were tested, 50 models for the individual food products and another 6 models for derived food categories (dairy, fish, meat, vegetables, carbohydrates, and sweets). Statistical significance and the effect estimate of a food product, the selected SNP, and their interaction in the linear regression model were assessed.
Results
Genotyping data were available for 1816 individuals. Minor allele frequencies for rs1014290, rs6449213, and rs737267 were 26.07, 20.69, and 25.37%, respectively. Genotype and allele frequencies are presented in Table 2. For all 3 investigated SNPs, the minor allele was associated with the decrease in serum uric acid levels.
Table 2.
Genotype and allele frequencies for 3 selected single nucleotide polymorphism (SNP), rs1014290, rs6449213, and rs737267, in our sample
| SNP | Genotypes | Allele frequencies | |||
|---|---|---|---|---|---|
| rs1014290 | TT | TC | CC | T | C |
| (n = 1816) | 1014 | 657 | 145 | ||
| 55.84% | 36.18% | 7.98% | 73.93% | 26.07% | |
| rs6449213 | TT | TC | CC | T | C |
| (n = 1815) | 1163 | 553 | 99 | ||
| 64.08% | 30.47% | 5.45% | 79.31% | 20.69% | |
| rs737267 | GG | GT | TT | G | T |
| (n = 1815) | 1029 | 651 | 135 | ||
| 56.69% | 35.87% | 7.44% | 74.63% | 25.37% | |
Food frequency data were available for 1755 participants (97.7%). Mean total dairy products consumption, expressed as portions/mo (±standard deviation) was 38.5 ± 25.8, for meat products 35.1 ± 20.0, for fish products 19.9 ± 15.2, for all vegetables 42.7 ± 27.0, for bread, rice, and pasta 44.0 ± 15.3, and for deserts 23.6 ± 24.5 portions/mo. Main characteristics of individuals according to the intake for selected food groups are presented in Table 3.
Table 3.
Characteristics of individuals according to intake for selected food groups: dairy, meat, fish, vegetables, carbohydrate-rich foods, and deserts. P value for the difference in serum uric acid residuals from polygenic model between the high and low consumption groups (values above or below the mean for a given food group) are shown (t-test)
| Part of consumption distribution of food product | n | Mean consumption frequency (servings/mo) | Mean age (years) | Mean uric acid (mg/dL) | P |
|---|---|---|---|---|---|
| Dairy: | |||||
| upper | 929 | 19.46 | 56.23 | 5.14 | 0.071 |
| lower | 822 | 59.92 | 56.23 | 5.00 | |
| Meat: | |||||
| upper | 1027 | 22.63 | 60.10 | 5.08 | 0.108 |
| lower | 721 | 52.83 | 50.77 | 5.09 | |
| Fish: | |||||
| upper | 1024 | 11.46 | 56.06 | 4.99 | 0.152 |
| lower | 724 | 31.92 | 56.51 | 5.21 | |
| Vegetables: | |||||
| upper | 1168 | 27.64 | 55.57 | 5.08 | 0.649 |
| lower | 584 | 72.69 | 57.56 | 5.08 | |
| Bread, pasta, and rice: | |||||
| upper | 1117 | 35.25 | 57.11 | 5.11 | 0.782 |
| lower | 636 | 59.32 | 54.69 | 5.02 | |
| Sweets: | |||||
| upper | 1125 | 9.46 | 57.72 | 5.15 | 0.031 |
| lower | 627 | 48.88 | 53.60 | 4.95 |
The individuals who consumed dairy products more often (values above the mean consumption) had lower mean uric acid levels than those who consumed dairy products less often (below the mean) (5.00 vs 5.14 mg/dL) but the difference was not significant (P = 0.07). Individuals with above-the-mean deserts intake had significantly lower uric acid levels than those with below-the-mean intake (4.95 vs 5.15; P = 0.031).
For the food products tested, we observed a significant decrease in uric acid levels with increasing consumption of milk, sour cream, duck and turkey, pork, salted fish, eggs, and coffee, while significant increase in uric acid levels was observed with increasing consumption of veil, sausages, and tea. As expected, the 3 selected SNPs were confirmed to significantly affect serum uric acid.
The only significant interaction was found between potato consumption and rs737267 (P = 0.046); but suggestive interactions were observed with the other 2 tested SNPs, rs1014290 and rs6449213 (P = 0.083 and P = 0.141, respectively).
The other interaction close to the nominal significance threshold of P = 0.05 was the one between soft drinks (consumption levels: low, medium, and high) and rs1014290, where the interaction P was 0.068 (Table 4). No significant interaction was observed between food products consumption frequency and rs6449213.
Table 4.
Effect estimates for each explanatory variable (food product, single nucleotide polymorphism rs1014290 and their interaction) and P values are shown. Effect estimates are shown for residuals of uric acid from the polygenic model and are presented to enable the comparison of food product, single nucleotide polymorphism and interaction effect, but cannot be translated into uric acid levels (analysis is done on residuals from polygenic model). The unit in which food products are measured is servings/mo. Genotypes are coded as 0, 1, and 2, where 2 corresponds to minor allele homozygote
| Food product | rs1014290 | Interaction | ||||
|---|---|---|---|---|---|---|
|
effect estimate |
P |
effect estimate |
P |
effect estimate |
P |
|
| Milk | -0.009 | 0.002 | -0.439 | 4.7E-21 | 0.002 | 0.504 |
| Sour cream | -0.018 | 0.017 | -0.402 | 1.9E-20 | -0.002 | 0.960 |
| Pork | -0.017 | 0.023 | -0.397 | 2.5E-20 | -0.003 | 0.950 |
| Veil | 0.023 | 0.046 | -0.359 | 4.8E-21 | -0.011 | 0.284 |
| Sausages | 0.018 | 0.019 | -0.385 | 1.2E-20 | -0.009 | 0.791 |
| Duck and turkey | -0.011 | 0.032 | -0.393 | 4.6E-21 | -0.008 | 0.719 |
| Salted fish | -0.014 | 0.018 | -0.397 | 1.2E-20 | -0.004 | 0.857 |
| Eggs | -0.016 | 8.2E-05 | -0.377 | 2.1E-20 | -0.004 | 0.674 |
| Soft drinks | 0.004 | 0.215 | -0.390 | 9.4E-21 | -0.004 | 0.081 |
| Coffee | -0.005 | 0.004 | -0.386 | 8.6E-21 | -0.001 | 0.397 |
| Tea | 0.006 | 4.2E-04 | -0.385 | 1.5E-21 | -0.002 | 0.440 |
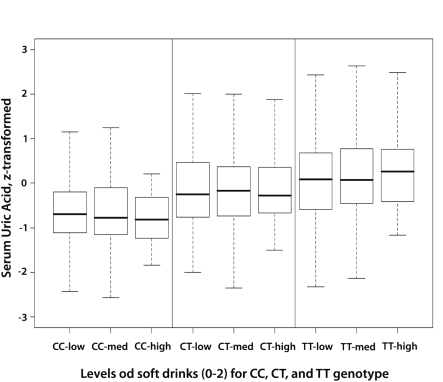
The observed interaction between rs1014290 and soft drinks was investigated further and showed that, while increased consumption of soft drinks slightly lowered serum uric acid levels in individuals with CC and CT genotype, in individuals with TT genotype, the group with highest soft drinks intake had the highest levels of uric acid (Figure 1).
Figure 1.
The effect of soft drinks consumption on serum uric acid levels depending on the genotype at rs1014290 is shown. While inverse association is observed for CC genotype (increasing consumption being associated with the decreasing serum uric acid), the opposite is observed for TT genotype (increasing consumption of soft drinks is associated with the increase in serum uric acid).
Discussion
To our knowledge, this is the first study investigating the genotype-environment interaction between diet and identified genetic polymorphisms in SLC2A9 associated with serum uric acid level. One near-significant interaction shown to affect serum uric acid level was the one between soft drinks and SLC2A9 polymorphism rs1014290, but since the P value did not reach the nominal significance threshold of 0.05, the null hypothesis of no interaction cannot be rejected. The only significant interaction was found between potato consumption and rs737267. We could not find literature data to clarify this interaction. No other interactions between a food product (or a group of products) with the genotype at the 3 selected loci showed a significant effect on uric acid.
The increasing consumption of soft drinks increased serum uric acid in the interaction with TT genotype at rs1014290, however, it also decreased serum uric acid in homozygotes for the minor allele (CC) and almost no effect could be noted in heterozygotes (CT). This shows that the metabolic effect of soft drinks at this locus is determined by the underlying genotype. The interaction between soft drinks and rs6449213 and rs737267 showed the same pattern, but these results were not significant (data not shown).
Soft drinks with added sugar (sucrose, glucose, or fructose) are strongly associated with gout risk (31) and both glucose and fructose have been shown to facilitate SLC2A9-mediated urate transport (9). Moreover, SLC2A9 has been shown to exchange extracellular glucose for intracellular urate, which occurs in renal tubules where urate is to be excreted and glucose reabsorbed from the tubules. However, we found the opposite effect depending on rs1014290 genotype, namely in TT homozygotes.
Since no interaction was observed, it could mean that serum uric acid is simply, in the additive manner, the result of urate influx from the metabolism and the genetically modified transporter capacity. The observed interaction with glucose and fructose might suggest that the identified polymorphisms in SLC2A9 primarily affect protein domains concerning fructose transport, which is only consequentially reflected to uric acid levels because SLC2A9 exchanges glucose for urate. If polymorphisms affected binding or transport of urate, there is no reason to expect anything other than an additive effect of genotype-modified transporter capacity and uric acid levels of an individual.
This newly recognized effect of genotype on metabolic response to soft drinks could prove important in dietary recommendation and for the understanding of gout, diabetes type 2, and metabolic syndrome. It is possible that we failed to recognize other food products with the similar interaction effect on serum uric acid due to the incompleteness of our data; since we relied only on consumption frequency, we lack the important information on the amount of consumed products. More accurate answers could be provided by studies that would calculate the total daily intake for every dietary compound, eg, daily purines, glucose, and fructose consumption.
It is very difficult to obtain accurate diet measurements in free-living participants. Some overestimation of “healthy foods” might be expected, as has been reported for dairy products (32). Our data were based on a questionnaire recording the consumption frequency, rather than portion sizes. If a participant consumes a food item daily, it cannot be determined if they consume it one or more times a day.
We confirmed the associations between diet and serum uric acid found in previous studies. Our finding that consumption of milk and sour cream, and the overall consumption of dairy products, decreased serum uric acid levels was previously described by other researchers (1,2). We also observed significant association between coffee consumption and decreasing levels of uric acid. Such association was described in detail in a study that compared individuals who took 4 or 5 cups of coffee a day with those who did not drink coffee at all (2). The lack of stronger statistical evidence in our study could be due to the fact that we did not have the information about the amount of coffee taken, and therefore could not differentiate further in the category of “daily” consumption.
Interestingly, the same study investigated the effect of tea on uric acid level but found no association (2), while we observed the increasing levels of uric acid with increasing tea consumption. This effect may be relatively specific for the populations we tested and caused by a particular tea commonly drank in the islands of Vis and Korčula. It is likely that different sorts of tea can affect serum uric acid in different ways.
Although a positive association between meat intake and gout has been described before, we did not observe a significant difference in uric acid levels between the groups with the highest and lowest overall meat intake. The individuals in high meat consumption category were younger (mean age 50.7 vs 60.1 years), which could have diminished the difference between the groups because uric acid is known to increase with age. Of the individual meat products, consumption of veal and sausages significantly increased uric acid levels, while consumption of duck, turkey, pork, salted fish, and eggs decreased uric acid levels. Higher frequency of consumption of bread, pasta, and rice was associated with lower uric acid levels although not significantly; however, the inverse association with bread consumption has been reported previously (33).
This study was performed in two isolated island populations. Although certain benefits of such setting are well recognized (12,16,19,34-39), it also has some shortcomings, primarily the possibility that the results might not be representative for other populations (40). Furthermore, in our study there is a lack of information on the number of food products consumed to compliment the available data on frequency of consumption. Although more detailed dietary data might further clarify diet-genotype interactions, we find our dietary data reliable since we replicated some of the previously reported effects of diet on serum uric acid. Some of the reported P values did not exceed the nominal significance level and the null-hypothesis of no interaction cannot be excluded.
Our study is, to the best of our knowledge, the first to investigate the interaction between diet and genetic polymorphisms and our findings provide new insights into uric acid excretion and emphasize the importance of other molecules, like glucose and fructose, for the serum uric acid and gout. New evidence about the intertwining of urate and simple sugars metabolism might prove important in understanding the connection between the elevated serum uric acid concentration and metabolic syndrome.
Acknowledgments
The studies in the Croatian islands were supported through the grants from the Medical Research Council UK to A. F. Wright, H. Campbell, and I. Rudan; and Ministry of Science, Education and Sport of the Republic of Croatia to I. R. (No. 216-1080315-0302). The authors collectively thank a large number of individuals for their individual help in obtaining funding support, organizing, planning, and carrying out the field work related to the project, and for assistance with data management, analysis, and interpretation: the teams from the Medical Research Council Human Genetics Unit in Edinburgh and The University of Edinburgh, UK, led by Professors N. D. Hastie, A. Wright, and H. Campbell, including Caroline Hayward and Veronique Vitart (for funding support, intellectual input in study design, and their role in data management, analysis, and interpretation); Professor Pavao Rudan and the staff of the Institute for Anthropological Research in Zagreb, Croatia (organization of the field work, anthropometric and physiological measurements, and DNA extraction in Vis); Professor Stipan Janković and the staff of the University of Split Medical School (organization of the field work, anthropometric and physiological measurements, and DNA extraction in Korčula); Professor Ariana Vorko-Jović and the staff and medical students of the Andrija Štampar School of Public Health of the Medical School, University of Zagreb, Croatia (questionnaires, genealogical reconstruction, and data entry in Vis and Korčula); Dr Branka Salzer from the biochemistry lab “Salzer,” Croatia (measurements of biochemical traits in Vis and Korčula); local general practitioners and nurses (recruitment and communication with the study population); and the employees of several other Croatian institutions who participated in the field work, including but not limited to the University of Rijeka; Croatian Institute of Public Health; Institutes of Public Health in Split and Dubrovnik. SNP Genotyping of the Vis samples was carried out by the Genetics Core Laboratory at the Wellcome Trust Clinical Research Facility, WGH, Edinburgh; of the Korčula samples by the Genotyping Institute in Munich, Germany.
References
- 1.Richette P, Bardin T. Gout. Lancet. 2010;375:318–28. doi: 10.1016/S0140-6736(09)60883-7. [DOI] [PubMed] [Google Scholar]
- 2.Choi HK, Curhan G. Coffee, tea, and caffeine consumption and serum uric acid level: the third national health and nutrition examination survey. Arthritis Rheum. 2007;57:816–21. doi: 10.1002/art.22762. [DOI] [PubMed] [Google Scholar]
- 3.Yang Q, Guo CY, Cupples LA, Levy D, Wilson PW, Fox CS. Genome-wide search for genes affecting serum uric acid levels: the Framingham Heart Study. Metabolism. 2005;54:1435–41. doi: 10.1016/j.metabol.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 4.Vitart V, Rudan I, Hayward C, Gray NK, Floyd J, Palmer CN, et al. SLC2A9 is a newly identified urate transporter influencing serum urate concentration, urate excretion and gout. Nat Genet. 2008;40:437–42. doi: 10.1038/ng.106. [DOI] [PubMed] [Google Scholar]
- 5.Doring A, Gieger C, Mehta D, Gohlke H, Prokisch H, Coassin S, et al. SLC2A9 influences uric acid concentrations with pronounced sex-specific effects. Nat Genet. 2008;40:430–6. doi: 10.1038/ng.107. [DOI] [PubMed] [Google Scholar]
- 6.Guan M, Zhang J, Chen Y, Liu W, Kong N, Zou H. High-resolution melting analysis for the rapid detection of an intronic single nucleotide polymorphism in SLC22A12 in male patients with primary gout in China. Scand J Rheumatol. 2009;38:276–81. doi: 10.1080/03009740802572483. [DOI] [PubMed] [Google Scholar]
- 7.Dehghan A, Köttgen A, Yang Q, Hwang SJ, Kao WL, Rivadeneira F, et al. Association of three genetic loci with uric acid concentration and risk of gout: a genome-wide association study. Lancet. 2008;372:1953–6. doi: 10.1016/S0140-6736(08)61343-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kolz M, Johnson T, Sanna S, Teumer A, Vitart V, Perola M, et al. Meta-analysis of 28,141 individuals identifies common variants within five new loci that influence uric acid concentrations. PLoS Genet. 2009;5:e1000504. doi: 10.1371/journal.pgen.1000504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caulfield MJ, Munroe PB, O'Neill D, Witkowska K, Charchar FJ, Doblado M, et al. SLC2A9 is a high-capacity urate transporter in humans. PLoS Med. 2008;5:E197. doi: 10.1371/journal.pmed.0050197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rudan I, Campbell H, Rudan P. Genetic epidemiological studies of eastern Adriatic Island isolates, Croatia: objective and strategies. Coll Antropol. 1999;23:531–46. [PubMed] [Google Scholar]
- 11.Polasek O, Marusic A, Rotim K, Hayward C, Vitart V, Huffman J, et al. Genome-wide association study of anthropometric traits in Korcula Island, Croatia. Croat Med J. 2009;50:7–16. doi: 10.3325/cmj.2009.50.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vitart V, Biloglav Z, Hayward C, Janicijevic B, Smolej-Narancic N, Barac L, et al. 3000 years of solitude: extreme differentiation in the island isolates of Dalmatia, Croatia. Eur J Hum Genet. 2006;14:478–87. doi: 10.1038/sj.ejhg.5201589. [DOI] [PubMed] [Google Scholar]
- 13.Rudan I, Marusić A, Janković S, Rotim K, Boban M, Lauc G, et al. “10001 Dalmatians:” Croatia launches its national biobank. Croat Med J. 2009;50:4–6. doi: 10.3325/cmj.2009.50.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rudan I, Carothers AD, Polasek O, Hayward C, Vitart V, Biloglav Z, et al. Quantifying the increase in average human heterozygosity due to urbanisation. Eur J Hum Genet. 2008;16:1097–102. doi: 10.1038/ejhg.2008.48. [DOI] [PubMed] [Google Scholar]
- 15.Campbell H, Carothers AD, Rudan I, Hayward C, Biloglav Z, Barac L, et al. Effects of genome-wide heterozygosity on a range of biomedically relevant human quantitative traits. Hum Mol Genet. 2007;16:233–41. doi: 10.1093/hmg/ddl473. [DOI] [PubMed] [Google Scholar]
- 16.Rudan I, Biloglav Z, Vorko-Jović A, Kujundzić-Tiljak M, Stevanović R, Ropac D, et al. Effects of inbreeding, endogamy, genetic admixture, and outbreeding on human health: a (1001 Dalmatians) study. Croat Med J. 2006;47:601–10. [PMC free article] [PubMed] [Google Scholar]
- 17.Smoljanovic A, Vorko-Jovic A, Kolcic I, Bernat R, Stojanovic D, Polasek O. Micro-scale socioeconomic inequalities and health indicators in a small isolated community of Vis Island, Croatia. Croat Med J. 2007;48:734–40. [PMC free article] [PubMed] [Google Scholar]
- 18.Kolcic I, Vorko-Jovic A, Salzer B, Smoljanovic M, Kern J, Vuletic S. Metabolic syndrome in a metapopulation of Croatian island isolates. Croat Med J. 2006;47:585–92. [PMC free article] [PubMed] [Google Scholar]
- 19.Polasek O, Kolcic I, Smoljanovic A, Stojanovic D, Grgic M, Ebling B, et al. Demonstrating reduced environmental and genetic diversity in human isolates by analysis of blood lipid levels. Croat Med J. 2006;47:649–55. [PMC free article] [PubMed] [Google Scholar]
- 20.Navarro P, Vitart V, Hayward C, Tenesa A, Zgaga L, Juricic D, et al. Genetic comparison of a Croatian isolate and CEPH European founders. Genet Epidemiol. 2010;34:140–5. doi: 10.1002/gepi.20443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McQuillan R, Leutenegger AL, Abdel-Rahman R, Franklin CS, Pericic M, Barac-Lauc L, et al. Runs of homozygosity in European populations. Am J Hum Genet. 2008;83:359–72. doi: 10.1016/j.ajhg.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dupuis J, Langenberg C, Prokopenko I, Saxena R, Soranzo N, Jackson AU, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet. 2010;42:105–16. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Repapi E, Sayers I, Wain LV, Burton PR, Johnson T, Obeidat M, et al. Genome-wide association study identifies five loci associated with lung function. Nat Genet. 2010;42:36–44. doi: 10.1038/ng.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johansson A, Marroni F, Hayward C, Franklin CS, Kirichenko AV, Jonasson I, et al. Common variants in the JAZF1 gene associated with height identified by linkage and genome-wide association analysis. Hum Mol Genet. 2009;18:373–80. doi: 10.1093/hmg/ddn350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pattaro C, Aulchenko YS, Isaacs A, Vitart V, Hayward C, Franklin CS, et al. Genome-wide linkage analysis of serum creatinine in three isolated European populations. Kidney Int. 2009;76:297–306. doi: 10.1038/ki.2009.135. [DOI] [PubMed] [Google Scholar]
- 26.Hicks AA, Pramstaller PP, Johansson A, Vitart V, Rudan I, Ugocsai P, et al. Genetic determinants of circulating sphingolipid concentrations in European populations. PLoS Genet. 2009;5:e1000672. doi: 10.1371/journal.pgen.1000672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heard-Costa NL, Zillikens MC, Monda KL, Johansson A, Harris TB, Fu M, et al. NRXN3 is a novel locus for waist circumference: a genome-wide association study from the CHARGE Consortium. PLoS Genet. 2009;5:e1000539. doi: 10.1371/journal.pgen.1000539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aulchenko YS, Ripatti S, Lindqvist I, Boomsma D, Heid IM, Pramstaller PP, et al. Loci influencing lipid levels and coronary heart disease risk in 16 European population cohorts. Nat Genet. 2009;41:47–55. doi: 10.1038/ng.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zemunik T, Boban M, Lauc G, Janković S, Rotim K, Vatavuk Z, et al. Genome-wide association study of biochemical traits in Korcula Island, Croatia. Croat Med J. 2009;50:23–33. doi: 10.3325/cmj.2009.50.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aulchenko YS, Ripke S, Isaacs A, van Duijn CM. GenABEL: an R library for genome-wide association analysis. Bioinformatics. 2007;23:1294–6. doi: 10.1093/bioinformatics/btm108. [DOI] [PubMed] [Google Scholar]
- 31.Hak AE, Choi HK. Lifestyle and gout. Curr Opin Rheumatol. 2008;20:179–86. doi: 10.1097/BOR.0b013e3282f524a2. [DOI] [PubMed] [Google Scholar]
- 32.Martin GS, Tapsell LC, Batterham MJ, Russell KG. Relative bias in diet history measurements: a quality control technique for dietary intervention trials. Public Health Nutr. 2002;5:537–45. doi: 10.1079/PHN2002329. [DOI] [PubMed] [Google Scholar]
- 33.Loenen HM, Eshuis H, Lowik MR, Schouten EG, Hulshof KF, Odink J, et al. Serum uric acid correlates in elderly men and women with special reference to body composition and dietary intake (Dutch Nutrition Surveillance System). J Clin Epidemiol. 1990;43:1297–303. doi: 10.1016/0895-4356(90)90095-7. [DOI] [PubMed] [Google Scholar]
- 34.Rudan I, Campbell H, Carothers AD, Hastie ND, Wright AF. Contribution of consanguinuity to polygenic and multifactorial diseases. Nat Genet. 2006;38:1224–5. doi: 10.1038/ng1106-1224. [DOI] [PubMed] [Google Scholar]
- 35.Rudan I. Effects of inbreeding on late-onset diseases [PhD Thesis]. Edinburgh (UK): University of Edinburgh; 2007. [Google Scholar]
- 36.Pulanic D, Polasek O, Petrovecki M, Vorko-Jovic A, Pericic M, Lauc LB, et al. Effects of isolation and inbreeding on human quantitative traits: an example of biochemical markers of hemostasis and inflammation. Hum Biol. 2008;80:513–33. doi: 10.3378/1534-6617-80.5.513. [DOI] [PubMed] [Google Scholar]
- 37.Saftic V, Rudan D, Zgaga L. Mendelian diseases and conditions in Croatian island populations: historic records and new insights. Croat Med J. 2006;47:543–52. [PMC free article] [PubMed] [Google Scholar]
- 38.Rudan I. Health effects of human population isolation and admixture. Croat Med J. 2006;47:526–31. [PMC free article] [PubMed] [Google Scholar]
- 39.Rudan I, Biloglav Z, Carothers AD, Wright AF, Campbell H. Strategy for mapping quantitative trait loci (QTL) by using human metapopulations. Croat Med J. 2006;47:532–42. [PMC free article] [PubMed] [Google Scholar]
- 40.Kolcic I, Polasek O, Rudan I. Gender differences in spousal household material status estimation. J Epidemiol Community Health. 2009;63:175-6.40. [DOI] [PubMed]