Abstract
Aim
To investigate whether intra-personal variation in serum uric acid concentration is influenced by genes that were described to be associated with serum uric acid levels in cross-sectional studies.
Methods
The study included 92 participants from the isolated community of the Croatian island of Vis. For each participant, two uric acid concentration measurements were available, one from 2002 and one from 2003. Changes in uric acid concentration were correlated with a set of 8 genes known to affect it: PDZK1, GCKR, SLC2A9, ABCG2, LRRC16A, SLC17A3, SLC16A9, and SLC22A12.
Results
Thirteen participants (14%) had uric acid concentration change greater than 130 μmol/L. Greater variability of uric acid concentration was recorded in women (coefficient of variation 49% vs 12% in men). Two SNPs belonging to SLC17A3 gene (rs9393672 and rs942379) yielded significant association with serum uric acid concentration changes in women. These two single-nucleotide polymorphisms (SNP) explained 0.2%-1.3% of variance for 2002 or 2003 uric acid measurement and 1.1%-1.8% of variance for the average value of these two measurements.
Conclusions
Repeated measurements offer a possibility to enrich the percent of explained variance and contribute to the understanding of the “missing heritability” concept. Although a number of genes have been shown to affect serum uric acid concentration, SLC17A3 seems to have a major role in determination of serum uric acid repeated measurements variation.
Serum uric acid in humans is a quantitative trait that has lately received a lot of research attention (1). This is primarily due to the current disagreement whether it is a disease risk or a protective factor (2-5). While older studies often refer to it as a cause of gout and associate it with other metabolic diseases, some recent studies suggest that it might be among the most potent anti-oxidants in the human body (6,7). Furthermore, substantial progress in our understanding of its metabolism and biology has recently been made, due to description of a number of genes that affect it (4). However, clinical importance of uric acid and the possibility to use it as a disease marker still remain a matter of dispute (5).
Genetic background of uric acid determination has most commonly been described in genome-wide association studies, which correlate a single uric acid measurement to a large number of genetic markers (8,9). Although this approach has yielded a number of interesting results, it relies on the key assumption that a single measurement of uric acid concentration is a good proxy for this trait. However, studies have reported that serum uric acid concentrations show substantial variation even within a single day (10) or during periods as long as years (11-15).
Theoretically, studies of phenotypic traits that are not very reproducible suffer from a lack of statistical power and consequently have greater chances of producing false positive or false negative results. Furthermore, such underpowered studies may produce underestimated percentages of variance attributable to genetic factors in phenotypic trait determination. This is often seen in genome-wide association studies that manage to explain no more than a few percents of variance for majority of human quantitative traits, despite the fact that they are sometimes based on more than a hundred thousand samples (16,17). The difference between high heritability and low percent of explained variance of genetic factors was entitled the “missing heritability,” and it is currently one of the central issues in human genetics (16,18,19). Therefore, the aim of this study was to investigate the repeatability of serum uric acid concentration measurements in an isolated island population, to examine whether there is a genetic background of serum uric acid concentration changes, and to investigate the possibility to use multiple phenotypic measurements in genetic epidemiology.
Population and methods
Study population
Data for the study were obtained from a larger genetic epidemiology program performed on some of the Croatian Adriatic islands (20-24). The study sample included participants from the island of Vis who took part in the pilot study in 2002 and were later recruited to a larger gene mapping effort (25-28). In both cases, a population-based approach was applied, which included first informing the local community and then sending out the postal invitations. Participation was open for all individuals who would respond to it. Sampling frame comprised 200 participants in 2002 and 1057 in 2003. Those who responded to both invitations were included in this study.
The population of the island is an interesting isolate, due to genetic, geographical, and even partial linguistic isolation (21,29). After almost a decade of detailed work in this population, a number of specific characteristics were described in some population genetics features (29-35) and epidemiological, environmental, and behavioral disease risk factors (36-38), making the island a large biomedical research resource (39).
All participants were given detailed study information prior to the enrolment and gave signed consent to participate in the study. The study was approved by the Ethics Committee of the Medical School, University of Zagreb and the Multi-Centre Research Ethics Committee for Scotland. Further details on this program are available elsewhere (21,29).
Measurements
Blood samples were taken from all participants, immediately frozen, and transported frozen to the laboratory that performed the measurement. Both uric acid measurements, from 2002 and 2003, were performed in a single laboratory, which did not change quantification methodology during that period. Sequential analyses and laboratory quality controls check-ups were within the expected and certified ranges. Serum uric acid measurements were based on the uricase UV photometry, performed by Olympus AU400 (Olympus Corp, Tokyo, Japan), according to the manufacturer’s instructions.
The initial analysis of uric acid concentrations was aimed toward understanding the pattern of annual variation. In order to provide estimates for this, we calculated coefficient of variation, which was calculated for the entire sample and for sex-stratified sub-samples. Furthermore, we performed Bland-Altman agreement analysis (40), which uses graphical output to indicate the agreement between two measurements based on the comparison of the difference and the means of two measurements. This approach offers a substantial methodological advancement over the calculation of correlation coefficients between the two measurements (41).
Furthermore, all participants were classified into two groups according to the extent of serum uric acid concentration change. Participants with absolute value of difference between the two measurements higher than 130 μmol/L (roughly a third of the upper laboratory range limit) were compared to those with difference between the two measurements lower than 130 μmol/L.
Besides uric acid, we used blood samples for DNA extraction. After extraction, all samples were genotyped using Illumina HumanHap 300 panel, version 1, (Illumina Inc., San Diego, CA, USA), with 317 508 single-nucleotide polymorphisms (SNP). Among them, 16 SNPs belonging to 8 genes that were previously associated with uric acid (8,9), were selected and used in the analysis. These included rs1797052 and rs1298954 from PDZK1 gene, rs780094 from GCKR gene, rs13129697, rs4447863, rs13131257, rs6449213, rs1014290, and rs733175 from SLC2A9, rs2231142 from ABCG2, rs9358856 from LRRC16A, rs9393672 and rs942379 from SLC17A3, rs2242206 from SLC16A9, and rs2078267 and rs505802 from SLC22A12 gene. Further information on these SNPs and genes is provided elsewhere (9).
Statistical analysis
Numerical data were presented as means and standard deviations, since the assumption of normality was satisfied, according to the Kolmogorov-Smirnov test. Categorical data were presented as numbers and percentages. The data were analyzed using χ2 test or Fisher exact test; the latter was used when the expected values in more than 20% of cells were less than 5. Numerical variables were analyzed with t-test for dependent samples (comparison of two uric acid concentration measurements). Additionally, general linear modeling was employed to calculate the percent of variance that was explained by genetic markers. Selected single-nucleotide polymorphisms (SNP) were used as predictors, while uric acid concentration was used as an outcome variable. Three different values of uric acid were used: measurement from 2002, measurement from 2003, and the average value of these two measurements. Linkage disequilibrium between genetic markers was calculated using PLINK (42), but only for the selected two SNPs. Analysis was performed in R (43), except for Fisher exact test that was performed using Simple Interactive Statistical Analysis (SISA) (44). Since 16 SNPs were included in the analysis, a Bonferroni correction was applied. Calculation of the P value correction was based on 16 comparisons and the initial value of P < 0.05. The final outcome of the P value correction was that significance limit was set to P < 0.0031.
Results
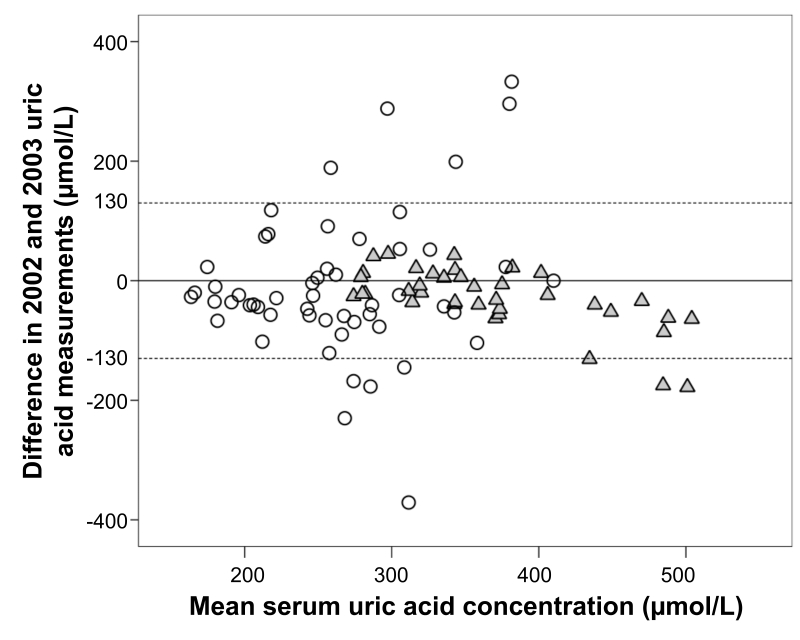
The study included 92 samples from 38 men and 53 women (Table 1). Average serum uric acid concentration was greater in 2003 than in 2002, with a largely different coefficient of variation in men and women (Table 1). When all measurements were ranked according to the greatest change between the two sampling periods, there were 8 women and only 2 men in top 10 samples. The use of Bland-Altman agreement approach indicated that there were directional changes in uric acid concentrations in men, while it showed seemingly equal dispersal in women (Figure 1). This was further confirmed using the t-test for repeated samples, where the entire sample showed significant change between the 2002 and 2003 measurements (P < 0.001). This was due to the changes in men (P < 0.001) but not in women (P = 0.916).
Table 1.
Basic characteristics of the study participants
| Men (n = 38) | Women (n = 53) | Total | |
|---|---|---|---|
| Age (years); mean ± standard deviation |
59.6 ± 12.3 |
59.3 ± 13.4 |
59.4 ± 12.9 |
| Uric acid, 2002 measurement (μmol/L); mean ± standard deviation |
353.3 ± 55.0 |
259.0 ± 94.5 |
298.4 ± 92.7 |
| Uric acid, 2003 measurement (μmol/L); mean ± standard deviation |
381.7 ± 88.7 |
268.9 ± 76.4 |
316.0 ± 98.7 |
| Coefficient of variation between 2002 and 2003 measurement; % | 11.5 | 49.3 | 39.9 |
Figure 1.
Bland-Altman chart for the uric acid concentration changes in men and women; dashed lines represent a cut-off value for uric acid serum concentration of 130 μmol/L. Gray triangle – men; open circle – women
The changes of serum uric acid concentration greater than 130 μmol/L were then correlated with a set of 16 SNPs that were previously associated with the serum uric acid concentrations in cross-sectional studies. The analysis yielded 2 SNPs that were significantly associated with these changes, both belonging to a single gene, SLC17A3 (Table 2). The subsequent analysis suggested that these 2 SNPs were in a strong linkage disequilibrium (r2 = 0.923). Despite overall small sample size, the total sample was additionally stratified by sex in order to investigate possible sex-specific differences. This revealed that the changes of uric acid concentration were not associated with these 2 SNPs (rs9393672 and rs942379) in men (P = 0.073 and P = 0.061, respectively; Fisher exact test), but were in women (P = 0.003 and P = 0.001, respectively).
Table 2.
The association of the uric acid changes higher and lower than 130 μmol/L with the set of 16 loci implicated in the uric acid metabolism
| Uric acid changes |
||||
|---|---|---|---|---|
| Gene and short nucleotide polymorphism; n (%) | Genotype | higher than 130 μmol/L | lower than 130 μmol/L | P |
| PDZK1 |
||||
| rs1797052 |
AG |
1 (14.3) |
6 (85.7) |
0.734* |
| GG |
12 (14.5) |
71 (85.5) |
||
| rs1298954 |
AA |
1 (14.3) |
6 (85.7) |
0.999† |
| AG |
6 (14.3) |
36 (85.7) |
||
| GG |
6 (14.6) |
35 (85.4) |
||
| GCKR |
||||
| rs780094 |
AA |
1 (9.1) |
10 (90.9) |
0.059* |
| AG |
7 (13) |
47 (87) |
||
| GG |
5 (20.8) |
19 (79.2) |
||
| SLC2A9 |
||||
| rs13129697 |
AA |
6 (23.1) |
20 (76.9) |
0.024* |
| AC |
6 (13) |
40 (87) |
||
| CC |
1 (5.6) |
17 (94.4) |
||
| rs4447863 |
AA |
1 (11.1) |
8 (88.9) |
0.191† |
| AG |
8 (22.9) |
27 (77.1) |
||
| GG |
4 (8.7) |
42 (91.3) |
||
| rs13131257 |
AA |
0 (0) |
9 (100) |
0.385† |
| AG |
6 (14.3) |
36 (85.7) |
||
| GG |
7 (17.9) |
32 (82.1) |
||
| rs6449213 |
AA |
8 (20.5) |
31 (79.5) |
0.269† |
| AG |
5 (12.2) |
36 (87.8) |
||
| GG |
0 (0) |
8 (100) |
||
| rs1014290 |
AA |
7 (22.6) |
24 (77.4) |
0.015* |
| AG |
6 (13) |
40 (87) |
||
| GG |
0 (0) |
13 (100) |
||
| rs733175 |
AA |
9 (23.1) |
30 (76.9) |
0.096† |
| AG |
4 (9.5) |
38 (90.5) |
||
| GG |
0 (0) |
9 (100) |
||
| ABCG2 |
||||
| rs2231142 |
AC |
1 (11.1) |
8 (88.9) |
0.616* |
| CC |
12 (14.8) |
69 (85.2) |
||
| LRRC16A |
||||
| rs9358856 |
AA |
1 (50) |
1 (50) |
0.062* |
| AG |
3 (17.6) |
14 (82.4) |
||
| GG |
9 (12.7) |
62 (87.3) |
||
| SLC17A3 |
||||
| rs9393672 |
AA |
5 (50) |
5 (50) |
0.003† |
| AC |
5 (11.6) |
38 (88.4) |
||
| CC |
3 (8.1) |
34 (91.9) |
||
| rs942379 |
AA |
5 (62.5) |
3 (37.5) |
<0.001† |
| AG |
5 (11.9) |
37 (88.1) |
||
| GG |
3 (7.7) |
36 (92.3) |
||
| SLC16A9 |
||||
| rs2242206 |
AA |
1 (20) |
4 (80) |
0.087* |
| AC |
4 (14.8) |
23 (85.2) |
||
| CC |
8 (13.8) |
50 (86.2) |
||
| SLC22A12 |
||||
| rs2078267 |
AA |
4 (21.1) |
15 (78.9) |
0.050* |
| AG |
6 (13.3) |
39 (86.7) |
||
| GG |
3 (11.5) |
23 (88.5) |
||
| rs505802 |
AA |
6 (14.6) |
35 (85.4) |
0.062* |
| AG |
7 (17.1) |
34 (82.9) |
||
| GG |
0 (0.0) |
8 (100.0) |
||
| Total; n (%) | - | 13 (14.3) | 78 (85.7) | - |
*Fisher exact test.
†χ2 test.
Lastly, uric acid concentration was used as the target variable in 3 general linear models aiming to estimate the percent of variance that was attributable to the genetic markers. Each of the 2 selected SNPs were entered in the model as predictors separately, with 3 outcome variables – uric acid concentration measured in 2002, measured in 2003, and average value of these 2 measurements. The results indicated that the percent of variance for SNP rs9393672 was 0.2% for 2002 measurement, 0.7% for 2003 measurement, and 1.1 for average uric acid concentration; rs942379 yielded a total of 0.7% of variance for 2002 measurement, 1.3% for 2003 measurement, and 1.8% for the average uric acid concentration.
Discussion
This study suggests that gene SLC17A3 (solute carrier family 17 [sodium phosphate] member 3, also known as NPT4 - Na(+)/PI cotransporter 4) may have a strong effect on substantial changes of serum uric acid concentration in repeated measurements, while other proposed genes were not significantly associated with these changes. Furthermore, although women showed overall greater variability of serum uric acid concentration, this study suggests that the effects of SCL17A3 were present in women but not in men.
SLC17A3 belongs to the large family of solute carriers, involved in the urinary urate reabsorption (4). However, its exact role in the urate metabolism remains to be investigated (4). Currently, it seems that mutations in this gene that involve the existence of AA genotype have a strong effect on substantial changes of the uric acid concentration, and may even be involved in the variability of serum uric acid. A step forward in this line of research could be the sequential investigation of uric acid concentration in participants with all 3 genotypes (AA, A/C, or CC for rs9393672 and AA, A/G, and GG for rs942379), based on multiple uric acid concentration measures, which could provide additional information on the extent of variation. If such a study is performed on a sex-stratified sample, it could even shed more light on the sex-dependent determination of uric acid concentration, which was suggested in a number of previous studies (3,6,8,9,45).
Serum uric acid concentration seems to be a highly complex trait, affected by a number of described genes and environmental factors (1,4,8,9,46). One of the very interesting research areas investigates the relationship of the uric acid, gout, nephrolithiasis, and metabolic syndrome (4). This is even more interesting knowing that some of the genes associated with uric acid have been independently associated with gout (47), while the other have not shown any association with either of the metabolic or even cardiovascular diseases (48). These results suggest that we lack the proper information on uric acid determination, which is why some characteristics of isolated populations could be considered as an important research resource (49-51).
The use of average value of two or more phenotypic measurement instead a single measurement seems to provide an interesting way of increasing the percent of SNP-attributable explained variance in quantitative trait analysis. The amount of variance in this study increased for over one half when only two uric acid measurements were averaged compared with any single measurement. This offers an exciting possibility that a fair share of genome-wide association studies, which were as a rule based on a single measurement and yielded only a small percent of variance (52), could be substantially improved by the provision of an additional phenotypic measurement. This could also contribute to the solving one of the key contemporary issues in genetic epidemiology, a problem of missing heritability, ie, the finding that genetic markers, such as SNPs, fail to explain a substantial amount of heritability for any given trait (16). The examples for missing heritability are numerous, including heritability of height which is close to 95%, while huge genetic mapping efforts managed to identify over 40 variants which all together explain as much as 5% of total height variance (26,53-56), or type-2 diabetes where 18 loci explain as much as 6% of total variance (16). The possible causes for this discrepancy include a number of reasons, among which is also the possibility that phenotypic measurements are imprecise (19). If this is a true problem in genetic epidemiology, then we could expect to see a gradient of strength of association between genetic markers and traits that are highly repeatable and easy to measure (ie, height) toward less likely significant results in less repeatable and more variable traits (ie, serum measures, especially hormones). Since we do not often see such a gradient, especially not in height genetics (55), it could be hypothesized that this mechanism is not particularly strong or prevalent. Furthermore, such problems are likely to be overcome in meta-analytic studies, which serve as a potent mechanism for random variation removal or control, with substantial enrichment of significant results (9), compared with results from a single study (8). Nevertheless, the results presented here suggest that phenotyping inaccuracy may introduce certain amount of variance dissolution, and that the averaging of two measurements of the same trait may increase the percent of explained variance.
The limitations of this study include a rather small sample size, which may have caused the large coefficient of variation. Due to this, one of next steps is to replicate these results in an independent population or populations. Furthermore, the samples originated from an isolated population with possibly unique genetic make-up and specific environmental determinants, making these results perhaps less generalizable. Furthermore, certain levels of relatedness were expected among participants, which could have affected the results. The extent of variation recorded among women in this study was several orders of magnitude greater than that in a previously published one (11), which could also be regarded as a local population feature, acting in favor of detecting a gene associated with strong serum uric acid changes. Despite all these limitations, this study suggests that uric acid concentration changes in longitudinal measurements are under strong effect of the SLC17A3 gene. If similar results are obtained in other studies, it could be hypothesized that repeated phenotypic measurements may provide a substantial contribution toward understanding of such a fundamental questions of modern genetic epidemiology as the missing heritability.
Acknowledgments
This study was supported through the grants from the Medical Research Council UK to Alan F. Wright, Harry Campbell, and Igor Rudan; and Ministry of Science, Education and Sport of the Republic of Croatia to I.R. (No. 216-1080315-0302). The authors collectively thank a large number of individuals for their individual help in obtaining funding support, organizing, planning, and carrying out the field work related to the project, and for assistance with data management, analysis, and interpretation: the teams from the Medical Research Council Human Genetics Unit in Edinburgh and The University of Edinburgh, UK, led by Professors Nicholas D. Hastie, Alan Wright, and Harry Campbell, including Caroline Hayward and Veronique Vitart (for funding support, intellectual input in study design, and their role in data management, analysis, and interpretation); Professor Pavao Rudan and the staff of the Institute for Anthropological Research in Zagreb, Croatia (organization of the field work, anthropometric and physiological measurements, and DNA extraction); Professor Ariana Vorko-Jović and the staff and medical students of the Andrija Štampar School of Public Health of the Medical School, University of Zagreb, Croatia (questionnaires, genealogical reconstruction, and data entry); Dr Branka Salzer from the biochemistry lab “Salzer,” Croatia (measurements of biochemical traits); local general practitioners and nurses (recruitment and communication with the study population); and the employees of several other Croatian institutions who participated in the field work, including but not limited to the University of Rijeka; Croatian Institute of Public Health; Institutes of Public Health in Split and Dubrovnik. SNP Genotyping of the Vis samples was carried out by the Genetics Core Laboratory at the Wellcome Trust Clinical Research Facility, WGH, Edinburgh.
References
- 1.Johnson RJ, Titte S, Cade JR, Rideout BA, Oliver WJ. Uric acid, evolution and primitive cultures. Semin Nephrol. 2005;25:3–8. doi: 10.1016/j.semnephrol.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 2.Dehghan A, van Hoek M, Sijbrands EJ, Hofman A, Witteman JC. High serum uric acid as a novel risk factor for type 2 diabetes. Diabetes Care. 2008;31:361–2. doi: 10.2337/dc07-1276. [DOI] [PubMed] [Google Scholar]
- 3.Johnson RJ, Gaucher EA, Sautin YY, Henderson GN, Angerhofer AJ, Benner SA. The planetary biology of ascorbate and uric acid and their relationship with the epidemic of obesity and cardiovascular disease. Med Hypotheses. 2008;71:22–31. doi: 10.1016/j.mehy.2008.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Riches PL, Wright AF, Ralston SH. Recent insights into the pathogenesis of hyperuricaemia and gout. Hum Mol Genet. 2009;18:R177–84. doi: 10.1093/hmg/ddp369. [DOI] [PubMed] [Google Scholar]
- 5.Feig DI, Kang DH, Johnson RJ. Uric acid and cardiovascular risk. N Engl J Med. 2008;359:1811–21. doi: 10.1056/NEJMra0800885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alatalo PI, Koivisto HM, Hietala JP, Bloigu RS, Niemela OJ. Gender-dependent impacts of body mass index and moderate alcohol consumption on serum uric acid–an index of oxidant stress status? Free Radic Biol Med. 2009;46:1233–8. doi: 10.1016/j.freeradbiomed.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 7.Baillie JK, Bates MG, Thompson AA, Waring WS, Partridge RW, Schnopp MF, et al. Endogenous urate production augments plasma antioxidant capacity in healthy lowland subjects exposed to high altitude. Chest. 2007;131:1473–8. doi: 10.1378/chest.06-2235. [DOI] [PubMed] [Google Scholar]
- 8.Vitart V, Rudan I, Hayward C, Gray NK, Floyd J, Palmer CN, et al. SLC2A9 is a newly identified urate transporter influencing serum urate concentration, urate excretion and gout. Nat Genet. 2008;40:437–42. doi: 10.1038/ng.106. [DOI] [PubMed] [Google Scholar]
- 9.Kolz M, Johnson T, Sanna S, Teumer A, Vitart V, Perola M, et al. Meta-analysis of 28,141 individuals identifies common variants within five new loci that influence uric acid concentrations. PLoS Genet. 2009;5:e1000504. doi: 10.1371/journal.pgen.1000504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Devgun MS, Dhillon HS. Importance of diurnal variations on clinical value and interpretation of serum urate measurements. J Clin Pathol. 1992;45:110–3. doi: 10.1136/jcp.45.2.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu KH, Luo SF, Tsai WP, Huang YY. Intermittent elevation of serum urate and 24-hour urinary uric acid excretion. Rheumatology. 2004;43:1541–5. doi: 10.1093/rheumatology/keh379. [DOI] [PubMed] [Google Scholar]
- 12.Goldstein RA, Becker KL, Moore CF. Serum urate in healthy men. Intermittent elevations and seasonal effect. N Engl J Med. 1972;287:649–50. doi: 10.1056/NEJM197209282871308. [DOI] [PubMed] [Google Scholar]
- 13.Rahe RH, Rubin RT, Arthur RJ. The three investigators study. Serum uric acid, cholesterol, and cortisol variability during stresses of everyday life. Psychosom Med. 1974;36:258–68. doi: 10.1097/00006842-197405000-00009. [DOI] [PubMed] [Google Scholar]
- 14.Rahe RH, Rubin RT, Arthur RJ, Clark BR. Serum uric acid and cholesterol variability. A comprehensive view of underwater demolition team training. JAMA. 1968;206:2875–80. doi: 10.1001/jama.206.13.2875. [DOI] [PubMed] [Google Scholar]
- 15.Kuzuya M, Ando F, Iguchi A, Shimokata H. Effect of aging on serum uric acid levels: longitudinal changes in a large Japanese population group. J Gerontol A Biol Sci Med Sci. 2002;57:M660–4. doi: 10.1093/gerona/57.10.m660. [DOI] [PubMed] [Google Scholar]
- 16.Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–53. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dupuis J, Langenberg C, Prokopenko I, Saxena R, Soranzo N, Jackson AU, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet. 2010;42:105–16. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wray NR, Goddard ME, Visscher PM. Prediction of individual genetic risk of complex disease. Curr Opin Genet Dev. 2008;18:257–63. doi: 10.1016/j.gde.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 19.Maher B. Personal genomes: The case of the missing heritability. Nature. 2008;456:18–21. doi: 10.1038/456018a. [DOI] [PubMed] [Google Scholar]
- 20.Rudan I. The land of 1000 islands. Croat Med J. 2006;47:523–5. [Google Scholar]
- 21.Rudan I. Health effects of human population isolation and admixture. Croat Med J. 2006;47:526–31. [PMC free article] [PubMed] [Google Scholar]
- 22.Rudan I, Biloglav Z, Carothers AD, Wright AF, Campbell H. Strategy for mapping quantitative trait loci (QTL) by using human metapopulations. Croat Med J. 2006;47:532–42. [PMC free article] [PubMed] [Google Scholar]
- 23.Rudan I, Biloglav Z, Vorko-Jovic A, Kujundzic-Tiljak M, Stevanovic R, Ropac D, et al. Effects of inbreeding, endogamy, genetic admixture, and outbreeding on human health: a (1001 Dalmatians) study. Croat Med J. 2006;47:601–10. [PMC free article] [PubMed] [Google Scholar]
- 24.Rudan I, Campbell H, Rudan P. Genetic epidemiological studies of eastern Adriatic Island isolates, Croatia: objective and strategies. Coll Antropol. 1999;23:531–46. [PubMed] [Google Scholar]
- 25.Repapi E, Sayers I, Wain LV, Burton PR, Johnson T, Obeidat M, et al. Genome-wide association study identifies five loci associated with lung function. Nat Genet. 2010;42:36–44. doi: 10.1038/ng.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johansson A, Marroni F, Hayward C, Franklin CS, Kirichenko AV, Jonasson I, et al. Common variants in the JAZF1 gene associated with height identified by linkage and genome-wide association analysis. Hum Mol Genet. 2009;18:373–80. doi: 10.1093/hmg/ddn350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Knezevic A, Polasek O, Gornik O, Rudan I, Campbell H, Hayward C, et al. Variability, heritability and environmental determinants of human plasma N-glycome. J Proteome Res. 2009;8:694–701. doi: 10.1021/pr800737u. [DOI] [PubMed] [Google Scholar]
- 28.Pattaro C, Aulchenko YS, Isaacs A, Vitart V, Hayward C, Franklin CS, et al. Genome-wide linkage analysis of serum creatinine in three isolated European populations. Kidney Int. 2009;76:297–306. doi: 10.1038/ki.2009.135. [DOI] [PubMed] [Google Scholar]
- 29.Vitart V, Biloglav Z, Hayward C, Janicijevic B, Smolej-Narancic N, Barac L, et al. 3000 years of solitude: extreme differentiation in the island isolates of Dalmatia, Croatia. Eur J Hum Genet. 2006;14:478–87. doi: 10.1038/sj.ejhg.5201589. [DOI] [PubMed] [Google Scholar]
- 30.Campbell H, Carothers AD, Rudan I, Hayward C, Biloglav Z, Barac L, et al. Effects of genome-wide heterozygosity on a range of biomedically relevant human quantitative traits. Hum Mol Genet. 2007;16:233–41. doi: 10.1093/hmg/ddl473. [DOI] [PubMed] [Google Scholar]
- 31.McQuillan R, Leutenegger AL, Abdel-Rahman R, Franklin CS, Pericic M, Barac-Lauc L, et al. Runs of homozygosity in European populations. Am J Hum Genet. 2008;83:359–72. doi: 10.1016/j.ajhg.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rudan I, Carothers AD, Polasek O, Hayward C, Vitart V, Biloglav Z, et al. Quantifying the increase in average human heterozygosity due to urbanisation. Eur J Hum Genet. 2008;16:1097–102. doi: 10.1038/ejhg.2008.48. [DOI] [PubMed] [Google Scholar]
- 33.Carothers AD, Rudan I, Kolcic I, Polasek O, Hayward C, Wright AF, et al. Estimating human inbreeding coefficients: comparison of genealogical and marker heterozygosity approaches. Ann Hum Genet. 2006;70:666–76. doi: 10.1111/j.1469-1809.2006.00263.x. [DOI] [PubMed] [Google Scholar]
- 34.Navarro P, Vitart V, Hayward C, Tenesa A, Zgaga L, Juricic D, et al. Genetic comparison of a Croatian isolate and CEPH European founders. Genet Epidemiol. 2010;34:140–5. doi: 10.1002/gepi.20443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pulanic D, Polasek O, Petrovecki M, Vorko-Jovic A, Pericic M, Lauc LB, et al. Effects of isolation and inbreeding on human quantitative traits: an example of biochemical markers of hemostasis and inflammation. Hum Biol. 2008;80:513–33. doi: 10.3378/1534-6617-80.5.513. [DOI] [PubMed] [Google Scholar]
- 36.Ivkovic V, Vitart V, Rudan I, Janicijevic B, Smolej-Narancic N, Skaric-Juric T, et al. The Eysenck personality factors: Psychometric structure, reliability, heritability and phenotypic and genetic correlations with psychological distress in an isolated Croatian population. Pers Individ Dif. 2007;42:123–33. doi: 10.1016/j.paid.2006.06.025. [DOI] [Google Scholar]
- 37.Smoljanovic A, Vorko-Jovic A, Kolcic I, Bernat R, Stojanovic D, Polasek O. Micro-scale socioeconomic inequalities and health indicators in a small isolated community of Vis Island, Croatia. Croat Med J. 2007;48:734–40. [PMC free article] [PubMed] [Google Scholar]
- 38.Kolcic I, Polasek O, Rudan I. Gender differences in spousal household material status estimation. J Epidemiol Community Health. 2009;63:175–6. doi: 10.1136/jech.2008.077123. [DOI] [PubMed] [Google Scholar]
- 39.Rudan I, Marusic A, Jankovic S, Rotim K, Boban M, Lauc G, et al. “10001 Dalmatians:” Croatia launches its national biobank. Croat Med J. 2009;50:4–6. doi: 10.3325/cmj.2009.50.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bland JM, Altman DG. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet. 1986;1:307–10. [PubMed] [Google Scholar]
- 41.Altman DG, Bland JM. Measurement in medicine: the analysis of method comparison studies. Statistician. 1983;32:307–17. doi: 10.2307/2987937. [DOI] [Google Scholar]
- 42.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.The R. Foundation for Statistical Computing. The R Project for Statistical Computing. Available from: http://www.r-project.org/index.html Accessed: February 2, 2010.
- 44.Simple Interactive Statistical Analysis. Fisher 2 by 5. Available from: http://www.quantitativeskills.com/sisa/statistics/five2hlp.htm. Accessed: February 2, 2010.
- 45.Brandstatter A, Kiechl S, Kollerits B, Hunt SC, Heid IM, Coassin S, et al. Sex-specific association of the putative fructose transporter SLC2A9 variants with uric acid levels is modified by BMI. Diabetes Care. 2008;31:1662–7. doi: 10.2337/dc08-0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van der Harst P, Bakker SJ, de Boer RA, Wolffenbuttel BH, Johnson T, Caulfield MJ, et al. Replication of the five novel loci for uric acid concentrations and potential mediating mechanisms. Hum Mol Genet. 2010;19:387–95. doi: 10.1093/hmg/ddp489. [DOI] [PubMed] [Google Scholar]
- 47.Dehghan A, Kottgen A, Yang Q, Hwang SJ, Kao WL, Rivadeneira F, et al. Association of three genetic loci with uric acid concentration and risk of gout: a genome-wide association study. Lancet. 2008;372:1953–61. doi: 10.1016/S0140-6736(08)61343-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stark K, Reinhard W, Grassl M, Erdmann J, Schunkert H, Illig T, et al. Common polymorphisms influencing serum uric acid levels contribute to susceptibility to gout, but not to coronary artery disease. PLoS One. 2009;4:e7729. doi: 10.1371/journal.pone.0007729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kolcic I, Vorko-Jovic A, Salzer B, Smoljanovic M, Kern J, Vuletic S. Metabolic syndrome in a metapopulation of Croatian island isolates. Croat Med J. 2006;47:585–92. [PMC free article] [PubMed] [Google Scholar]
- 50.Polasek O, Kolcic I, Smoljanovic A, Stojanovic D, Grgic M, Ebling B, et al. Demonstrating reduced environmental and genetic diversity in human isolates by analysis of blood lipid levels. Croat Med J. 2006;47:649–55. [PMC free article] [PubMed] [Google Scholar]
- 51.Hicks AA, Pramstaller PP, Johansson A, Vitart V, Rudan I, Ugocsai P, et al. Genetic determinants of circulating sphingolipid concentrations in European populations. PLoS Genet. 2009;5:e1000672. doi: 10.1371/journal.pgen.1000672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wray NR, Goddard ME, Visscher PM. Prediction of individual genetic risk to disease from genome-wide association studies. Genome Res. 2007;17:1520–8. doi: 10.1101/gr.6665407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lettre G, Jackson AU, Gieger C, Schumacher FR, Berndt SI, Sanna S, et al. Identification of ten loci associated with height highlights new biological pathways in human growth. Nat Genet. 2008;40:584–91. doi: 10.1038/ng.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gudbjartsson DF, Walters GB, Thorleifsson G, Stefansson H, Halldorsson BV, Zusmanovich P, et al. Many sequence variants affecting diversity of adult human height. Nat Genet. 2008;40:609–15. doi: 10.1038/ng.122. [DOI] [PubMed] [Google Scholar]
- 55.Visscher PM. Sizing up human height variation. Nat Genet. 2008;40:489–90. doi: 10.1038/ng0508-489. [DOI] [PubMed] [Google Scholar]
- 56.Sanna S, Jackson AU, Nagaraja R, Willer CJ, Chen WM, Bonnycastle LL, et al. Common variants in the GDF5-UQCC region are associated with variation in human height. Nat Genet. 2008;40:198–203. doi: 10.1038/ng.74. [DOI] [PMC free article] [PubMed] [Google Scholar]