Abstract
Phosphorylation of the RelA (p65) NF-κB (nuclear factor κB) subunit has been previously shown to modulate its ability to induce or repress transcription. In the present study we have investigated the consequences of Thr435 phosphorylation within the C-terminal transactivation domain of RelA. We confirm that Thr435 is phosphorylated in cells and is induced by TNFα (tumour necrosis factor α) treatment. Mutational analysis of this site revealed gene-specific effects on transcription, with a T435D phosphomimetic mutant significantly enhancing Cxcl2 (CXC chemokine ligand 2) mRNA levels in reconstituted Rela−/− mouse embryonic fibroblasts. Chromatin immunoprecipitation analysis revealed that this mutation results in enhanced levels of histone acetylation associated with decreased recruitment of HDAC1 (histone deacetylase 1). Moreover, mutation of this site disrupted RelA interaction with HDAC1 in vitro. Thr435 phosphorylation of promoter-bound RelA was also detected at NF-κB target genes following TNFα treatment in wild-type mouse embryonic fibroblasts. Phosphorylation at this site therefore provides an additional mechanism through which the specificity of NF-κB transcriptional activity can be modulated in cells.
Keywords: CXC ligand 2 (CXCL2)/macrophage inflammatory protein 2 (MIP2), histone acetylation, histone deacetylase 1 (HDAC1), nuclear factor κB (NF-κB), RelA (p65), tumour necrosis factor α (TNFα)
Abbreviations: CBP, cAMP-response-element-binding protein-binding protein; ChIP, chromatin immunoprecipitation; CK2, casein kinase 2; CXCL2, CXC ligand 2; DBD, DNA-binding domain; EMSA, electrophoretic mobility-shift assay; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; HAT, histone acetyltransferase; HDAC1, histone deacetylase 1; HEK-293 cells, human embryonic kidney cells; IκB, inhibitor of NF-κB; IL, interleukin; LPS, lipopolysaccharide; MEF, mouse embryonic fibroblast; MIP2, macrophage inflammatory protein 2; NF-κB, nuclear factor κB; PARP, poly(ADP-ribose) polymerase; Pol II, RNA polymerase II; PP4, protein phosphatase 4; qPCR, quantitative PCR; TAD, transactivation domain; TAFII31, TATA-box-binding-protein-associated factor 31; TNFα, tumour necrosis factor α
INTRODUCTION
Members of the NF-κB (nuclear factor κB) transcription factor family are known to regulate a variety of cellular processes including inflammatory and immune responses, cell survival, cell differentiation and cell proliferation [1]. Furthermore, dysregulation of NF-κB signalling has been implicated in the development and progression of a multitude of diseases, particularly conditions involving chronic inflammation or compromised immunity and cancer [2,3]. Mammalian NF-κB is a multigene family composed of five members capable of forming a variety of homo- and hetero-dimeric complexes through their highly conserved N-terminal Rel homology domain [4]. In unstimulated cells, NF-κB dimers are primarily found as inactive, cytoplasmic complexes. Classical activation of NF-κB occurs in a well-defined IκB (inhibitor of NF-κB)-kinase-dependent manner, typically culminating in the release of RelA/p50 heterodimers from inhibitory IκB proteins, enabling dimer translocation to the nucleus and transcriptional modulation of NF-κB target genes [5].
The RelA (p65) subunit contains a potent TAD (transactivation domain), which allows recruitment of co-transcriptional regulators and components of the basal transcriptional machinery to gene targets [6]. Numerous post-translational modifications to RelA have been reported and varying effects on transcriptional activity, protein interactions, stability and degradation have been demonstrated [7]. Phosphorylation of sites within the TAD of RelA lead to both increased and decreased levels of transcriptional activity, with the precise effect dependent on the context and gene target [8–20]. The effects these modifications exert on protein–protein interactions has not been studied extensively. However, site-specific phospho-dependent increased binding to the co-transcriptional regulators TAFII31 (TATA-box-binding-protein-associated factor 31) and HDAC1 (histone deacetylase 1) has been shown [9,11]. Additionally, components of the ubiquitin ligase complex COMMD1 and cullin 2 were found to bind to RelA via GCN5 in a phospho-site-specific manner, thereby directing RelA ubiquitination and degradation at certain promoters following TNFα (tumour necrosis factor α) stimulation [13,16].
The mechanisms behind the ability of RelA to specifically regulate endogenous target-gene expression are continually being uncovered. Activation of certain endogenous target genes was recently shown to occur by two distinct modes, one involving the direct interaction of RelA with the Trap-80 mediator complex subunit and subsequent recruitment of Pol II (RNA polymerase II), and the other via RelA's ability to regulate promoter occupancy of secondary transcription factors [21]. The murine chemokine Cxcl2 [CXC ligand 2/MIP2 (macrophage inflammatory protein 2)] was found to be regulated in a Trap-80-independent manner and RelA was shown to regulate the recruitment of the secondary transcription factor SP1 to this promoter [21]. In the present paper, we present evidence for the ability of RelA to induce changes in the acetylation state of histones at the Cxcl2 promoter. Furthermore, we demonstrate that this effect is influenced by phosphorylation at Thr435 within RelA's TAD, which modulates the interaction of RelA with HDAC1.
EXPERIMENTAL
Cell culture
Human U-2 OS osteosarcoma cells and HEK-293 cells (human embryonic kidney cells) were obtained from the A.T.C.C. (American Type Culture Collection). Immortalized MEF (mouse embryonic fibroblast) cells and immortalized Rela−/− MEFs were provided by Professor R. Hay, College of Life Sciences, University of Dundee, Dundee, U.K. RelA-null MEFs were stably reconstituted with vector alone (control), wild-type RelA (RelA), or RelA with Thr435 mutated to an alanine residue (T435A) or an aspartic acid residue (T435D). Stably reconstituted Rela−/− MEFs were selected with 2 μg/ml puromycin (Sigma) and subsequently maintained in 0.5 μg/ml puromycin.
Preparation of viral stocks
Viral stocks for lentiviral infections were generated by transfecting 5 μg of viral envelope (pCMV-VSV-G), packaging (pCMVΔR8.91) and vector (PIRESpuro-deNotI and derivatives) plasmids (provided by Professor R. Hay, College of Life Sciences, University of Dundee, Dundee, U.K., and Professor M. Collins, Department of Immunology, University College London, London, U.K.) into HEK-293 cells. Medium containing the viral stocks was removed after 48 h. The medium was centrifuged at 350 g for 3 min and filtered through a 0.45 μm filter to remove cells. Viral stocks were then divided into aliquots (1ml), snap-frozen and stored at −80 °C.
Generation of stable cells
Infections were performed by mixing 4 μg/ml polybrene (Sigma) with 30 μl of virus stock in a total volume of 3 ml of medium. This was used to infect a 50% confluent 10-cm-diameter plate of Rela−/− MEFs by incubating overnight at 37 °C. On the second day, the medium containing the virus was removed and the plate was washed twice with PBS before the addition of fresh medium. On the third day, the cells were split 1:5. On the fourth day, cells were selected by replacing the medium with puromycin-containing medium (2 μg/ml), and when all cells in the mock infection plate had died the puromycin concentration was lowered to 0.5 μg/ml. This concentration was used to maintain stably expressing cell lines. In all experiments, pools of reconstituted MEF cells were used.
qPCR (quantitative PCR)
qPCRs were performed on Rotor-Gene 3000 and 6000 machines (Corbett Research). Ct values (threshold cycle values) were calculated using Rotor-Gene 6 software and all values were calculated relative to GAPDH (glyceraldehyde-3-phosphate dehydrogenase) or input levels using the Pfaffl method [22].
Primers
RT–PCR (reverse transcription–PCR)
Primers used were: Bcl2l1 (Bcl-xl) (FWD 5′-GATGCAGGTATTGGTGAGTCG-3′, REV 5′-GCTCTCGGCTGCTGCATT-3′); Cxcl1 (KC) (FWD 5′-CTGGGATTCACCTCAAGAAC-3′, REV 5′-GAAGCCAGCGTTCACCAGAC-3′); Cxcl2 (MIP2) (FWD 5′-TCAAGGGCGGTCAAAAAGTT-3′, REV 5′-TCCTCCTTTCCAGGTCAGTTA-3′); Cxcl3 (FWD 5′-CCTACCAAGGGTTGATTT-3′, REV 5′-CGCTCTTCAGTATCTTCTT-3′); Cxcl4 (Pf4) (FWD 5′-GAAAGCGATGGAGATCTTA-3′, REV 5′-TCTTATATAGGGGTGCTTGC-3′); Cxcl5 (GCP-2) (FWD 5′-CATTTCTGTTGCTGTTCA-3′, REV 5′-GGGATCACCTCCAAATTA-3′); Cxcl7 (Ppbp) (FWD 5′-CTTCATAACCTCCAGATCTT-3′, REV 5′-ACACATTCACAAGGGAGATA-3′); Cxcl10 (IP-10) (FWD 5′-CCAAGTGCTGCCGTCATTTTC-3′, REV 5′-GGCTCGCAGGGATGATTTCAA-3′); Cxcl11 (FWD 5′-AGGAAGGTCACAGCCATAG-3′, REV 5′-CTCGATCTCTGCCATTTT-3′); Cxcl12 (FWD 5′-CATCTGAAAATCCTCAACACT-3′, REV 5′-AAGCTTTCTCCAGGTACTCT-3′); Gapdh (FWD 5′-GCTACACTGAGGACCAGGTTG-3′, REV 5′-GCCCCTCCTGTTATTATGGGG-3′); and Tnfaip3 (A20) (FWD 5′-GAACAGCGATCAGGCCAGG-3′, REV 5′-GGACAGTTGGGTGTCTCACATT-3′).
ChIP (chromatin immunoprecipitation)
Primers used were: Cxcl1 (FWD 5′-CTAATCCTTGGGAGTGGAG-3′, REV 5′-CCCTTTTATGCTCGAAAC-3′); Cxcl2 (FWD 5′-CGTGCATAAAAGGAGCTCTC-3′, REV 5′-GTGCCCGAGGAAGCTTGT-3′); and Tnfaip3 (FWD 5′-CGCTGAGAGAGAGACAAAC-3′, REV 5′-TGGCCCTGAAGATTAACT-3′).
Antibodies
Antibodies used were anti-RelA antibody (sc-372; Santa Cruz Biotechnology), anti-Gal4 (DBD; DNA-binding domain) antibody (sc-577; Santa Cruz Biotechnology), anti-β-actin antibody (A5441; Sigma), anti-PARP [poly(ADP-ribose) polymerase] antibody (9542; Cell Signaling Technology), anti-Pol II antibody (sc-56767; Santa Cruz Biotechnology), anti-SP1 antibody (sc-59; Santa Cruz Biotechnology), anti-acetyl histone H3 antibody (06-599; Upstate), anti-acetyl histone H4 antibody (ab-1761; Abcam), anti-HDAC1 antibody (06-720; Upstate), anti-PKTAG/V5-TAG antibody (MCA1360GA; AbD Serotec), and anti-HA-TAG antibody (2367; Cell Signaling Technology). The RelA Thr435 phospho-specific antibody was raised in rabbit by BioGenes. The peptide used had the sequence TQAGEGT*LSEALC (phospho-Thr435 is indicated by *). The antibody was purified using two-step peptide affinity chromatography with the phospho- and non-phospho-peptides. ELISA analysis confirmed that the purified antibody was specific for the phosphorylated epitope.
Plasmids
The Gal4 E1B and 3× κB ConA luciferase reporter plasmids, along with the Gal4–RelA-TAD, RSV RelA and HA–HDAC1 expression plasmids, have been reported previously [18,23]. The viral envelope (pCMV-VSV-G), packaging (pCMVΔR8.91) and vector (PIRESpuro-deNotI) plasmids were obtained from Professor R. Hay (College of Life Sciences, University of Dundee, Dundee, U.K.) and Professor M. Collins (Department of Immunology, University College London London, U.K.). The Cxcl2 (MIP2) luciferase reporter plasmid was obtained from Professor M. Hottiger (Institute of Veterinary Biochemistry and Molecular Biology, University of Zürich, Zürich, Switzerland) [24]. PK-tagged RelA was generated by inserting an N-terminal PK-TAG into pcDNA3.1 vector. All point mutations were generated by PCR overlap extension, except for the Gal4-fusion mutants, which were generated in a single PCR step, and were sequenced prior to use.
Other assays
ChIP, co-immunoprecipitation, reporter-gene assays, Western blot analysis and EMSA (electrophoretic mobility-shift assay) analysis were performed essentially as described previously [18,25,26].
RESULTS
RelA Thr435 is phosphorylated following calyculin A and TNFα treatment
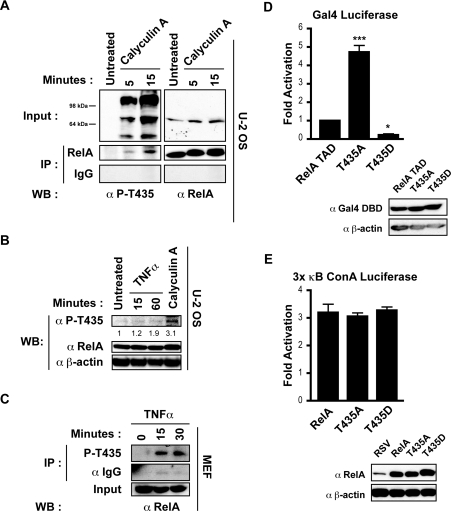
PP4 (protein phosphatase 4)-mediated Thr435 dephosphorylation was previously proposed to enhance RelA-mediated activation following cisplatin treatment, implying that phosphorylation at this site has a negative effect on the activity of RelA [27]. In order to determine the phosphorylation status of Thr435 in cells, a phospho-specific antibody against this site was raised, and ELISA analysis confirmed that the purified antibody was specific for this phosphorylated epitope (results not shown). Treating U-2 OS osteosarcoma cells with the serine/threonine phosphatase inhibitor calyculin A resulted in increasing levels of phosphorylation at this site that were masked by background bands in direct Western blots, but were clearly visible after immunoprecipitation with an anti-RelA antibody (Figure 1A). It was subsequently found that there was weak induction of phosphorylation at this site in U-2 OS cells following stimulation with the well-characterized NF-κB activator TNFα (Figure 1B). However, rapid Thr435 phosphorylation was detected in MEF cells following TNFα stimulation (Figure 1C).
Figure 1. RelA Thr435 is a phospho-site that affects TAD activity.
(A) U-2 OS cells were stimulated for the indicated times with calyculin A (50 nM). A portion of whole cell extract (40 μg) was analysed by Western blotting (WB) using the anti-phospho-Thr435 (α P-T435) or anti-RelA (α RelA) antibodies (upper panel). Whole cell extract (350 μg) was immunoprecipitated (IP) with anti-RelA antibody and samples were analysed by Western blotting with the anti-phospho-Thr435 antibody (lower panel). Membranes were then stripped and reprobed with total RelA antibody. (B) U-2 OS cells were stimulated with calyculin A (50 nM) for 15 min or TNFα (20 ng/ml) as indicated. A portion of whole cell extract (40 μg) was analysed by Western blotting with the anti-phospho-Thr435 antibody. The membrane was then stripped and reprobed with total anti-RelA antibody. Densitometry analysis indicates an increase in intensity of the phospho-band in relation to the untreated sample, normalized to RelA input levels. Anti-β-actin antibody (α β-actin) was used as a loading control. (C) MEF cells were stimulated with TNFα (20 ng/ml) as indicated. Whole cell extract (100 μg) was immunoprecipitated with the anti-phospho-Thr435 antibody and samples were analysed by Western blotting with total anti-RelA antibody. Anti-IgG antibody (α IgG) was used to control for non-specific binding. (D) U-2 OS cells were transfected with 1.5 μg of Gal4 E1B luciferase reporter plasmid and 750 pg of expression plasmid encoding the Gal4 DBD, the Gal4 DBD fused to amino acids 428–551 of RelA (RelA TAD), amino acids 428–551 of RelA with Thr435 mutated to an alanine residue (T435A) or amino acids 428–551 of RelA with Thr435 mutated to an aspartic acid residue (T435D). A Western blot demonstrating expression levels of Gal4 fusion proteins is shown. Luciferase assay results are expressed as fold-activation over Gal4 DBD control, relative to wild-type RelA TAD. Results are means±S.E.M., n=8. ANOVA was performed followed by a Tukey–Kramer multiple comparisons test using Prism 4 software (GraphPad). *P<0.05 and ***P<0.001 compared with wild-type TAD. (E) U-2 OS cells were transfected with 1 μg of 3× κB ConA luciferase reporter plasmid and 0.6 μg of expression plasmid encoding empty RSV vector, full-length wild-type RelA (RelA), RelA with Thr435 mutated to alanine (T435A) or RelA with Thr435 mutated to aspartic acid (T435D). A Western blot demonstrating expression levels of RelA proteins is shown. Luciferase-assay results are expressed as fold-activation over empty RSV plasmid. Results are the means± S.E.M., n=3. ANOVA was performed followed by a Tukey–Kramer multiple comparisons test using Prism 4 software (GraphPad).
Role of Thr435 on RelA transcriptional activity
To investigate potential effects on the transcriptional activity of RelA mediated through the Thr435 residue, both phospho-null (T435A) and phospho-mimicking (T435D) mutations of this site were created. These mutants were initially generated as TAD-fusion proteins with the DBD of Gal4, thereby allowing only TAD-dependent effects to be investigated. Significantly, when transfected into U-2 OS cells, the T435A mutation resulted in a dramatic increase in transcriptional activity, whereas the phospho-mimicking T435D mutation decreased activity, indicating that phosphorylation at this site may repress the transcriptional potential of the RelA TAD (Figure 1D). To further investigate these transcriptional effects, full-length versions of these mutants were created. However, the dramatic differences in activity observed with the Gal4 reporter were not reciprocated in RelA-dependent reporter assays, and no significant differences in transcriptional activity were found following the mutation of Thr435, either in the context of a 3× κB ConA luciferase reporter plasmid (Figure 1E) or DR5, IκB or IL (interleukin)-8 reporter constructs (results not shown). Taken together, these results indicate that strong transcriptional modulation of the TAD occurs through Thr435, but effects mediated through this residue may be specific for certain gene targets.
Creating stably reconstituted wild-type and Thr435-mutant RelA cell lines
To further investigate effects on RelA activity, full-length wild-type and Thr435-mutant forms of RelA were stably introduced into Rela−/− MEFs, using a lentiviral infection approach. This approach also prevented effects of mutant RelA proteins being obscured by endogenous wild-type RelA. Pools of cells stably expressing wild-type and mutant forms of RelA at equivalent levels were generated (Figure 2A).
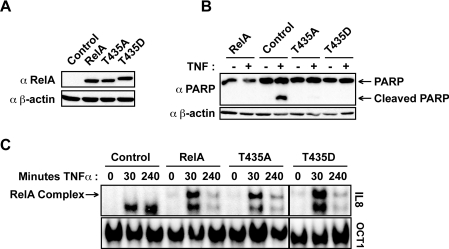
Figure 2. Creation of stable cell lines constitutively expressing wild-type and mutant forms of RelA.
(A) Western blot analysis of RelA protein levels in stably reconstituted Rela−/− MEFs. α RelA, anti-RelA antibody; α β-actin, anti-β-actin antibody. (B) Introduction of RelA protects cells from TNFα-induced cell death. Whole cell extracts were prepared from reconstituted MEF cells stably expressing wild-type and mutant forms of RelA, either unstimulated or stimulated with TNFα (40 ng/ml) for 4 h. Western blot analysis was performed for PARP cleavage. α PARP, anti-PARP antibody. (C) TNFα-induced DNA binding was not affected by mutating Thr435. Nuclear protein extracts were prepared from reconstituted MEF cells stimulated with TNFα (40 ng/ml) for the indicated times, and 5 μg of nuclear extract was subjected to EMSA analysis using 32P-labelled oligonucleotides containing the IL-8 κB element or the Oct1 transcription factor-binding site as a loading control.
It is known that deletion of Rela results in embryonic lethality from extensive foetal hepatocyte apoptosis, due to increased sensitivity to TNFα [28]. Loss of RelA also sensitizes MEFs to TNFα-induced cell death and this can be rescued by stably expressing RelA [29]. Both the wild-type and Thr435-mutant RelA reconstituted cells were rescued from TNFα-induced apoptotic cell death, as revealed by the absence of PARP cleavage, an indicator of caspase-mediated cell death (Figure 2B), indicating that phosphorylation at this site is unlikely to affect this important function of RelA [30].
Stimulation with TNFα induces the activation of the canonical NF-κB pathway, resulting in the rapid translocation of RelA complexes to the nucleus of the cell and enhanced DNA binding to κB sites in responsive genes, such as IL-8 [1,31]. DNA binding in response to canonical pathway activation in the reconstituted cells were investigated by EMSA analyses. Although a low level of basal NF-κB DNA binding to an IL-8 κB oligonucleotide could be seen in unstimulated RelA reconstituted cells, this was significantly induced following TNFα stimulation (Figure 2C). Furthermore, mutating Thr435 did not affect TNFα-induced DNA binding. Therefore these reconstituted cell lines contain functional TNFα-inducible RelA, capable of regulating the expression of the anti-apoptotic genes necessary for rescue from TNFα-induced cell death.
Regulation of NF-κB gene targets
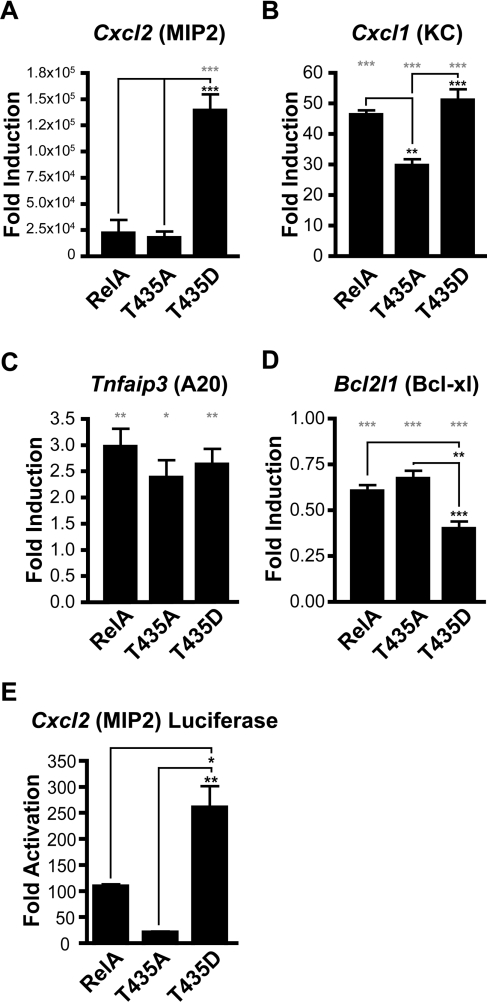
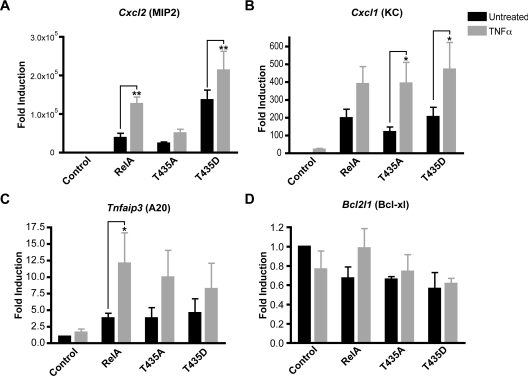
To analyse the potential effects mediated through Thr435 on known NF-κB gene targets, qPCR analysis was performed on cDNA generated from these reconstituted cell lines. Initially, known NF-κB target genes, Bcl2l1 (Bcl-xl), Cxcl1 (KC), Cxcl2 (MIP2) and Tnfaip3 (A20) [32–35], were investigated in unstimulated cells to allow analysis of Thr435 mutation on basal level expression. In agreement with previous studies, expression of the chemokines Cxcl1 and Cxcl2 was severely diminished in the absence of RelA and was found to be enhanced following reconstitution with all forms of RelA (Figures 3A and 3B) [21,33,35]. However, some differences between the Thr435-mutant forms of RelA were observed. Cxcl2 expression was dramatically increased in the phospho-mimicking T435D cell line, indicating that phosphorylation at this site may play a role in the induction of this gene (Figure 3A). In agreement with this, a Cxcl2 promoter luciferase-reporter plasmid was also found to have higher expression levels with the T435D mutant (Figure 3E). Similar enhancement of mRNA expression by the T435D mutation was not observed for other members of the CXC chemokine superfamily whose expression was detectable in these cell lines (see Supplementary Figure S1 at http://www.BiochemJ.org/bj/426/bj4260345add.htm). Expression levels of the CXC chemokine Cxcl1 were significantly reduced in the unphosphorylatable T435A cell line, suggesting that phosphorylation at this site is important for basal expression of this gene (Figure 3B). Levels of the ubiquitin-editing enzyme Tnfaip3 also became elevated following the introduction of RelA, but no significant differences were detected between the wild-type and Thr435-mutant cell lines (Figure 3C). Finally, expression of the anti-apoptotic gene Bcl2l1 was reduced following reconstitution with RelA (Figure 3D).
Figure 3. Regulation of endogenous NF-κB target genes.
(A) Introduction of RelA into Rela−/− MEFs increases expression levels of Cxcl2 (MIP2). Inserting a T435D mutation further enhances this effect. (B) Introduction of RelA into Rela−/− MEFs increases expression levels of Cxcl1 (KC). (C) Introduction of RelA increases expression levels of Tnfaip3 (A20). (D) Introduction of RelA reduces expression levels of Bcl2l1 (Bcl-xl). RNA was extracted from reconstituted MEF cells, total cDNA was prepared and qPCR analysis was performed using primers to mouse Cxcl2, Cxcl1, Tnfaip3, Bcl2l1 and Gapdh control. All results are normalized to Gapdh levels and expressed as fold-induction relative to the level of gene expression in the control cell line. (E) U-2 OS cells were transfected with 0.5 μg of Cxcl2 luciferase reporter plasmid and 0.5 μg of expression plasmid encoding empty RSV vector, full-length wild-type RelA (RelA), RelA with Thr435 mutated to an alanine residue (T435A) and RelA with Thr435 mutated to an aspartic acid residue (T435D). Results are expressed as fold-activation over empty RSV plasmid. Results are the means±S.E.M.; (A–D) n=4, (E) n=3. ANOVA was performed followed by a Tukey–Kramer multiple comparisons test using Prism 4 software (GraphPad). *P<0.05, **P<0.01 and ***P <0.001. Grey asterisks indicate P values relative to the control cell line.
Taken together, these results indicate that the Thr435 residue regulates the transcriptional activity of full-length RelA in a gene-specific manner. To begin a more mechanistic evaluation of these effects, the role played by the T435D mutant in the up-regulation of Cxcl2 expression levels was investigated using ChIP analysis.
Protein occupancy at the Cxcl2 promoter in reconstituted cell lines
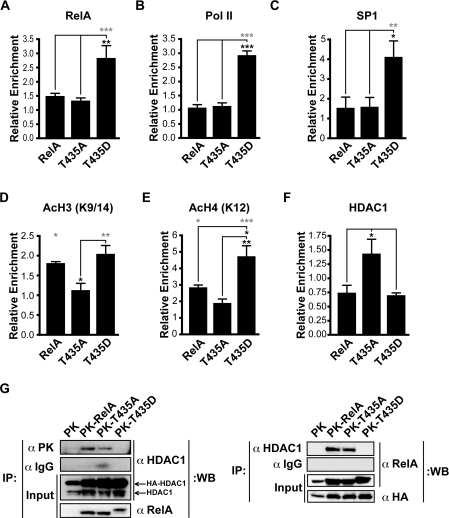
Increased expression of the murine Cxcl2 gene is known to occur in a RelA-dependent manner, following exposure to a variety of NF-κB-inducing stimuli, including TNFα, LPS (lipopolysaccharide) and hypoxia [21,33,36,37]. The Cxcl2 promoter contains not only an NF-κB-binding motif, but also SP1- and AP1-binding motifs, all of which are required for optimal promoter activation [36,38]. ChIP analysis revealed that in unstimulated cells, consistent with enhanced levels of gene expression, elevated levels of RelA, Pol II and SP1 were found at the Cxcl2 promoter in the T435D cell line (Figures 4A–4C). In contrast, enhanced binding of the T435D mutant form of RelA was not observed at either the Cxcl1 or Tnfaip3 promoters (see Supplementary Figures 2A and 2B at http://www.BiochemJ.org/bj/426/bj4260345add.htm). This indicated that Cxcl2-promoter-specific recruitment is regulated by the T435D mutation and that this is not an intrinsic effect on DNA binding, consistent with the earlier EMSA analysis (Figure 2C).
Figure 4. ChIP analysis of the murine Cxcl2 promoter in reconstituted cell lines.
(A–C) Elevated levels of RelA (A), Pol II (B) and SP1 (C) were detected at the Cxcl2 initiation site in the T435D cell line by qPCR. (D, E) Introducing wild-type RelA and T435D increases acetylation (Ac) of both histone H3 (K9/14) and histone H4 (K12). Creating a T435A mutation does not significantly increase the acetylation at this promoter. (F) Enhanced binding of HDAC1 to the Cxcl2 promoter in the T435A cell line. qPCR analysis was performed using primers to the mouse Cxcl2 initiation site. All results were normalized to input levels and control antibodies. Results are the mean enrichment levels relative to the control cell line±S.E.M.; (A) n=5, (B) n=3, (C) n=6, (D) n=3, (E) n=4, (F) n=7. ANOVA was performed followed by a Tukey–Kramer multiple comparisons test using Prism 4 software (GraphPad). *P<0.05, **P<0.01 and ***P<0.001. Grey asterisks indicate P values relative to the control cell line. (G) HDAC1 interaction with RelA is reduced by mimicking phosphorylation at Thr435. Co-immunoprecipitation (IP) analysis of the HDAC1 interaction with wild-type and Thr435-mutant forms of RelA was performed using the antibodies indicated and whole cell extracts (300 μg) from HEK-293 cells transfected with expression plasmid encoding HA–HDAC1 and PK, PK–RelA, PK–T435A or PK–T435D. A portion of whole cell extracts (20 μg) was included as input. WB, Western blot.
Next, we examined effects on histone modification at the Cxcl2 promoter, and enhanced acetylation of histones H3 (Lys9 and Lys14) and H4 (Lys12), both being markers for active regions of transcription, were observed following the introduction of wild-type RelA or the T435D mutant (Figures 4D and 4E). In the T435D cells, acetylated histone H3 levels were higher than those found in the phospho-null T435A cells, but not significantly higher than in the wild-type RelA cells (Figure 4D). However, acetylated histone H4 levels were significantly higher in the T435D cells, compared with both wild-type RelA and T435A-mutant cells, consistent with increased expression of this gene in the T435D cell line (Figure 4E). To determine whether the reduced acetylation detected in the T435A cell line results from enhanced binding of a deacetylase enzyme, levels of HDAC1 binding to the Cxcl2 promoter were determined, and enhanced HDAC1 binding was detected at this promoter (Figure 4F). These results illustrate that mutating Thr435, which is located in the TAD of RelA, influences the ability of RelA and other transcription factors, such as SP1 and Pol II, to bind specifically to the Cxcl2 promoter. Additionally, differences in the acetylation status of histone tails at this promoter were also detected and can be accounted for, at least in part, by elevated levels of HDAC1 binding to the promoter in the T435A cell line.
HDAC1 interaction with RelA
To investigate potential differences in HDAC1 interaction with RelA, co-immunoprecipitation analyses were performed using PK-tagged wild-type and Thr435-mutant forms of RelA, along with HA-tagged HDAC1. Significantly, the phospho-mimicking T435D mutation decreased HDAC1 interaction with RelA (Figure 4G), corroborating results obtained through ChIP analysis of the Cxcl2 promoter (Figure 4F). As TNFα stimulation is known to result in decreased HDAC1 interaction with RelA [39], this result is consistent with Thr435 phosphorylation contributing to this process.
Regulation of NF-κB gene targets following TNFα stimulation
Taken together, these results suggested that the effect of the T435D mutation was to mimic the effect of TNFα stimulation on the RelA-dependent regulation of Cxcl2 expression. Therefore we next investigated the effects of TNFα stimulation on the RelA-reconstituted MEFs. A RelA-dependent TNFα-induced up-regulation of both Cxcl1 and Cxcl2 was observed (Figures 5A and 5B). In the case of Cxcl1, this induction resulted in similar levels of mRNA in the reconstituted cells, bypassing the reduced basal expression seen in the T435A cell line. However, in the case of Cxcl2, there were differences in the induction of this gene between the Thr435 mutants. Cells containing wild-type RelA showed a robust induction in Cxcl2 mRNA levels, reaching levels similar to that found basally in the T435D cell line, suggesting that this mutation did indeed mimic the effects of TNFα stimulation for this NF-κB gene target. This increase was much reduced in cells containing the T435A mutant. The already enhanced levels of expression seen with the T435D mutant were stimulated somewhat further by TNFα treatment, as might be expected from the enhanced translocation of RelA to the nucleus under these conditions. These results are consistent with RelA Thr435 phosphorylation playing an important role in the TNF-dependent induction of Cxcl2 expression.
Figure 5. Expression levels of NF-κB target genes following TNFα treatment.
(A–C) TNFα stimulation differentially increases mRNA levels of Cxcl2, Cxcl1 and Tnfaip3 in the reconstituted cells. (D) TNFα stimulation does not significantly alter the expression of Bcl2l1 in the reconstituted cells. RNA was extracted from reconstituted MEF cells, either unstimulated or stimulated with TNFα (40 ng/ml) for 30 min. Total cDNA was prepared and qPCR analysis was performed using primers to mouse (A) Cxcl2, (B) Cxcl1, (C) Tnfaip3, (D) Bcl2l1 and Gapdh control. All results were normalized to Gapdh levels and are expressed as fold-induction relative to the level of gene expression in the control cell line. Results are the means±S.E.M., n=3. Two-way ANOVA was performed followed by a Bonferroni post-hoc test to compare replicate untreated and TNFα means, using Prism 4 software (GraphPad). *P<0.05 and **P<0.01.
Effects on other NF-κB target genes were also examined. RelA also enhanced Tnfaip3 expression following TNFα treatment, but significant differences were not detected between wild-type and Thr435 mutants (Figure 5C). However, TNFα had little effect on Bcl2l1 mRNA levels (Figure 5D). This result is consistent with ChIP analysis, where we could not observe RelA binding to the Bcl2l1 promoter in the reconstituted MEF cell lines (results not shown), suggesting that in this context this gene is not a RelA target and the effects we see on its expression are indirect. We also examined the expression levels of the other members of the CXC chemokine superfamily expressed in these cells, before and after TNFα stimulation (Supplementary Figure S1). Interestingly, although some effects with Thr435 mutation are seen, these are associated with reductions in the levels of gene expression (e.g. see Cxcl3 and Cxcl7). However, in this analysis, no other genes were found with the enhanced levels of expression seen with Cxcl2 that are associated with Thr435 phosphorylation. This confirmed that, in this context, the effect of Thr435 phosphorylation on RelA-dependent gene expression is highly specific.
Protein occupancy at the Cxcl2 promoter in wild-type MEFs
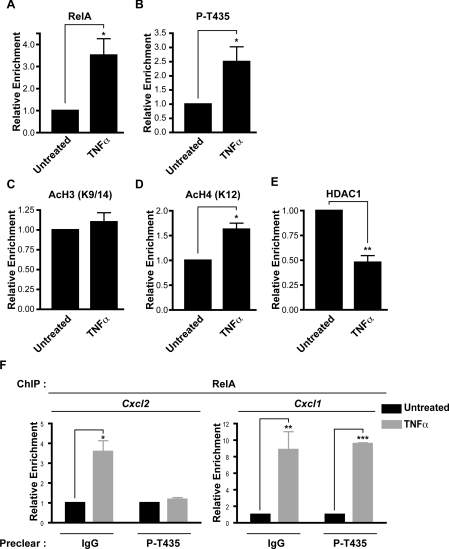
Next, we investigated endogenous RelA binding to the Cxcl2 promoter in wild-type MEFs. As expected, ChIP analysis revealed enhanced RelA binding following TNFα stimulation (Figure 6A). Importantly, and consistent with its predicted role as a regulator of Cxcl2 expression, we also detected enhanced phospho-Thr435 RelA bound to the Cxcl2 promoter following TNFα stimulation (Figure 6B). Levels of RelA binding to the Cxcl2 promoter, both before and after TNFα stimulation, were comparable between the reconstituted MEFs and wild-type MEFs (see Supplementary Figure S3 at http://www.BiochemJ.org/bj/426/bj4260345add.htm). Furthermore, consistent with the earlier results from the reconstituted MEFs, specific Thr435-dependent effects at the Cxcl2 promoter were also observed in the wild-type MEFs after TNFα stimulation, with acetylated histone H4 levels being significantly enhanced, whereas levels of HDAC1 present were reduced. However, some differences were observed between these experimental systems, with acetylated histone H3 levels not being seen to increase in the wild-type MEFs (Figures 6C–6E).
Figure 6. ChIP analysis of the murine Cxcl2 promoter in wild-type MEFs.
(A, B) TNFα stimulation increases binding of RelA and RelA phosphorylated at Thr435 (P-T435) to the Cxcl2 promoter region. (C) TNFα stimulation does not significantly increase levels of acetylated (Ac) histone H3 (K9/14) at the Cxcl2 promoter. (D) Elevated levels of acetylated histone H4 (K12) were found at the Cxcl2 promoter following TNFα treatment. (E) HDAC1 binding to the Cxcl2 promoter was reduced after TNFα treatment. qPCR analysis was performed using primers to the mouse Cxcl2 initiation site. (F) Depleting samples of RelA phosphorylated at Thr435 with phospho-specific antibody prior to performing ChIP analysis negates RelA binding to the Cxcl2 promoter, but not to the Cxcl1 promoter, following TNFα treatment. All results were normalized to input levels and control antibodies. Results are the mean enrichment levels relative to untreated cells±S.E.M.; (A–C) n=7, (D, E) n=4, (F) n=3. Student's t test was performed using Prism 4 software (GraphPad). *P<0.05, **P<0.01 and ***P<0.001.
Since this ChIP analysis indicated that Thr435-modified RelA is bound to the Cxcl2 promoter, we investigated what proportion of total promoter-associated RelA this represented. To determine this, the ChIP samples were immunodepleted with the anti-phospho-Thr435 antibody prior to RelA immunoprecipitation and PCR analysis. Significantly, preclearing Thr435-modified RelA negated the TNFα-induced increase in RelA binding to the Cxcl2 promoter, but had little effect on RelA binding to the Cxcl1 promoter, indicating that a very specific pool of activated RelA binds to and regulates the expression of Cxcl2 (Figure 6F).
Taken together, these results support the data obtained from the reconstituted MEF cell lines and demonstrate that phosphoryation of Thr435 plays an important role in the regulation of Cxcl2 mRNA expression in wild-type MEF cells following TNFα stimulation.
DISCUSSION
Although Thr435 had been identified as a putative phosphorylation site in the TAD of RelA, subject to dephosphorylation by PP4, this had relied on in vitro phosphorylation assays, and no analysis of modification in cells was performed [27]. Therefore we generated a phospho-specific antibody against Thr435 that allowed the detection of endogenous RelA phosphorylation at this site following calyculin A treatment of U-2 OS cells (Figure 1A). Phosphorylation was also detected following TNFα treatment in MEF cells and at the Cxcl1, Cxcl2 and Tnfaip3 promoters following ChIP analysis (Figure 1C, Figure 6B and Supplementary Figure S2). Although this analysis establishes Thr435 phosphorylation as a regulator of RelA transactivation, the identity of the kinase responsible is currently unknown. Phospho-site prediction algorithms identify Thr435 as a potential CK2 (casein kinase 2) or polo-like kinase 1 target. Interestingly, CK2 was previously shown to phosphorylate RelA at Ser529 in response to TNFα treatment [20]. However, phosphorylation of Thr435 by CK2 could not be verified empirically during the course of the present study (results not shown). Identification of the Thr435 kinase is an important question that should be investigated in future studies. Furthermore, Thr435 phosphorylation in response to other NF-κB inducers is also currently unknown and will require further analysis. That this site is only weakly phosphorylated after TNFα stimulation in U-2 OS cells implies a possible cell-type-specific role in vivo.
In contrast with the dramatic transcriptional effects seen with Gal4-fusion proteins (Figure 1D), full-length RelA versions of Thr435 phospho-mutants did not reproduce significant effects on transcription, when used with a selection of standard NF-κB reporter plasmids (see Figure 1E for the 3× κB ConA luciferase reporter plasmid; results not shown for other plasmids). Consistent with this, analysis of endogenous gene expression revealed that, at least in this context, the effects of Thr435 phosphorylation are highly promoter-specific, suggesting that this modification can help to provide the specificity and selectivity required to produce a context-dependent NF-κB transcriptional response to a stimulus. Interestingly, the effects seen with Thr435 mutation on the endogenous Cxcl2 gene were also mirrored with a Cxcl2 promoter luciferase reporter (Figure 3E). In contrast, other members of the CXC superfamily of genes did not display Cxcl2-like sensitivity to RelA Thr435 phosphorylation (Supplementary Figure S1), and gene-expression-array analysis will be required to identify other target genes similarly regulated by modification at this site.
These observations suggest that it is the specific organization of the Cxcl2 promoter that confers Thr435 regulation. Importantly, immunodepletion prior to ChIP analysis revealed that the majority of RelA bound to the Cxcl2 promoter was Thr435-phosphorylated, in contrast with the Cxcl1 promoter, at which most RelA was not modified at Thr435 (Figure 6F). Moreover, the T435D mutant shows elevated levels of Cxcl2 promoter binding, not seen with Cxcl1 or Tnfaip3 (Figure 3A, and Supplementary Figures 2A and 2B), suggesting that this modification facilitates RelA recruitment to the Cxcl2 promoter. Given the location of this motif in the TAD, we consider it unlikely that this effect is due to an alteration in site-specific RelA DNA binding. Rather, it is more likely to arise through effects on transcriptional co-regulators required to appropriately modify Cxcl2 promoter chromatin structure. Such effects may be mediated by the co-repressor HDAC1, whose binding to RelA is inhibited by the T435D mutation (Figure 4G). However, we cannot rule out effects on other interactions and it is entirely possible that Thr435 phosphorylation not only decreases HDAC1 binding, but also enhances binding to an, as yet unidentified, HAT (histone acetyltransferase). We also investigated binding of the CBP (cAMP-response-element-binding protein-binding protein) and TIP60 (Tat-interacting protein, 60kDa) HATs to the Thr435 mutants as a potential explanation for the variations in histone acetylation observed, but no differences in their occupation of the Cxcl2 promoter were detected in the RelA T435D-reconstituted cells and no differences in binding to Thr435-mutant forms of RelA were observed (results not shown). However, other TNFα-induced interactions with additional co-transcriptional proteins, such as SIRT6, Pin1, which is required for optimal Cxcl2 expression, and RPS3 [40–42], were not investigated during the course of the present study and may also emerge as being regulated by Thr435 phosphorylation.
Our investigation of the consequences of RelA Thr435 phosphorylation shows similarities and differences to previous analyses of point mutations to known RelA post-translational modification sites. Phosphorylation at Ser276 is known to play a role in activating RelA and regulating its interaction with either the co-activators CBP/p300 or HDAC co-repressors [43]. Reconstituting Rela−/− MEFs, with a non-phosphorylatable S276A mutant form of human RelA, similarly resulted in distinctive expression profiles of NF-κB target genes, potentially owing to decreased p300 recruitment [44–47]. Furthermore, the S276A mutation was found to severely impair the ability of RelA to rescue Rela−/− MEFs from TNFα-mediated cell death [48], an effect not found with Thr435 mutation. Phosphorylation at Ser536 is known to affect the activity of RelA, and reconstituting Rela−/− MEFs with a non-phosphorylatable S536A mutant form of human RelA was shown to decrease LPS-induced NF-κB activation and also to reduce p300 recruitment, subsequently decreasing TNFα activation [45,49]. Reconstituting Rela−/− MEFs with a phospho-mimicking S536D mutant produced enhanced binding to TAFII31 and enhanced transcriptional activity, reciprocating effects of IL-1-induced NF-κB activation [9]. Finally, phosphorylation at Ser468 has been reported to have both inhibitory and activatory effects on transactivation, and reconstituting Rela−/− MEFs with a non-phosphorylatable S468A mutant form of human RelA has been shown to have a negative effect on both IL-1- and TNFα-induced NF-κB transactivation [19]. TNFα-induced Ser468 phosphorylation was also found to regulate ubiquitin-mediated proteasome-dependent removal of chromatin-bound RelA on certain NF-κB gene targets [13]. However, this effect was not observed for Cxcl2, suggesting that other modifications, such as the Thr435 phosphorylation identified in the present study, will have a primary regulatory role.
Taking these results together with the conclusions of previous studies, it is becoming clearer that phosphorylation of RelA helps to programme its transcriptional functions by determining its activity at select target genes through modulating interactions with co-activators and co-repressors. In the present study we have identified a role for Thr435 phosphorylation, which, following TNFα stimulation, modulates the interactions with HDAC1 and selectively regulates NF-κB-dependent gene expression. This adds to the number of such modulatory phosphorylation events controlling NF-κB activity. Given the possible side effects associated with total NF-κB inhibition as a clinical therapy, drugs that selectively regulate RelA activity by affecting its post-translational modifications have the potential to act as effective therapies for the wide range of diseases associated with aberrant NF-κB activity.
Online data
AUTHOR CONTRIBUTION
John O'Shea performed all experiments and contributed to experimental design, ideas and manuscript writing. Neil Perkins contributed to experimental design, ideas and manuscript writing.
ACKNOWLEDGEMENTS
We thank Mary Collins (Department of Immunology, University College London, London, U.K.), Ron Hay (College of Life Sciences, University of Dundee, Dundee, U.K.), Michael Hottiger (Institute of Veterinary Biochemistry and Molecular Biology, University of Zürich, Zürich, Switzerland) and Tony Kouzarides (The Gurdon Institute, University of Cambridge, Cambridge, U.K.) for supplying reagents, and all members of N.P.'s laboratory for their advice and support. We thank Sonia Rocha and Stefan Roberts for their critical reading of this manuscript.
FUNDING
This work was funded by a Wellcome Trust four-year studentship at the University of Dundee (to J. O'S.) and by Cancer Research UK [grant number C1443/A4201 to J. O'S.] at the University of Bristol.
References
- 1.Hayden M. S., Ghosh S. Shared principles in NF-κB signaling. Cell. 2008;132:344–362. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 2.Courtois G., Gilmore T. D. Mutations in the NF-κB signaling pathway: implications for human disease. Oncogene. 2006;25:6831–6843. doi: 10.1038/sj.onc.1209939. [DOI] [PubMed] [Google Scholar]
- 3.Kumar A., Takada Y., Boriek A. M., Aggarwal B. B. Nuclear factor-κB: its role in health and disease. J. Mol. Med. 2004;82:434–448. doi: 10.1007/s00109-004-0555-y. [DOI] [PubMed] [Google Scholar]
- 4.Gilmore T. D. Introduction to NF-κB: players, pathways, perspectives. Oncogene. 2006;25:6680–6684. doi: 10.1038/sj.onc.1209954. [DOI] [PubMed] [Google Scholar]
- 5.Perkins N. D. Integrating cell-signalling pathways with NF-κB and IKK function. Nat. Rev. Mol. Cell. Biol. 2007;8:49–62. doi: 10.1038/nrm2083. [DOI] [PubMed] [Google Scholar]
- 6.O'Shea J. M., Perkins N. D. Regulation of the RelA (p65) transactivation domain. Biochem. Soc. Trans. 2008;36:603–608. doi: 10.1042/BST0360603. [DOI] [PubMed] [Google Scholar]
- 7.Perkins N. D. Post-translational modifications regulating the activity and function of the nuclear factor κB pathway. Oncogene. 2006;25:6717–6730. doi: 10.1038/sj.onc.1209937. [DOI] [PubMed] [Google Scholar]
- 8.Adli M., Baldwin A. S. IKK-i/IKKε controls constitutive, cancer cell associated NF-κB activity via regulation of Ser-536 p65/RelA phosphorylation. J. Biol. Chem. 2006;281:26976–26984. doi: 10.1074/jbc.M603133200. [DOI] [PubMed] [Google Scholar]
- 9.Buss H., Dorrie A., Schmitz M. L., Hoffmann E., Resch K., Kracht M. Constitutive and interleukin-1-inducible phosphorylation of p65 NF-κB at serine 536 is mediated by multiple protein kinases including IκB kinase (IKK)-α, IKKβ, IKKε, TRAF family member-associated (TANK)-binding kinase 1 (TBK1), and an unknown kinase and couples p65 to TATA-binding protein-associated factor II31-mediated interleukin-8 transcription. J. Biol. Chem. 2004;279:55633–55643. doi: 10.1074/jbc.M409825200. [DOI] [PubMed] [Google Scholar]
- 10.Buss H., Dorrie A., Schmitz M. L., Frank R., Livingstone M., Resch K., Kracht M. Phosphorylation of serine 468 by GSK-3β negatively regulates basal p65 NF-κB activity. J. Biol. Chem. 2004;279:49571–49574. doi: 10.1074/jbc.C400442200. [DOI] [PubMed] [Google Scholar]
- 11.Campbell K. J., Rocha S., Perkins N. D. Active repression of antiapoptotic gene expression by RelA(p65) NF-κB. Mol. Cell. 2004;13:853–865. doi: 10.1016/s1097-2765(04)00131-5. [DOI] [PubMed] [Google Scholar]
- 12.Chovolou Y., Watjen W., Kampkotter A., Kahl R. Downregulation of NF-κB activation in a H4IIE transfectant insensitive to doxorubicin-induced apoptosis. Toxicology. 2007;232:89–98. doi: 10.1016/j.tox.2006.12.012. [DOI] [PubMed] [Google Scholar]
- 13.Geng H., Wittwer T., Dittrich-Breiholz O., Kracht M., Schmitz M. L. Phosphorylation of NF-κB p65 at Ser468 controls its COMMD1-dependent ubiquitination and target gene-specific proteasomal elimination. EMBO Rep. 2009;10:381–386. doi: 10.1038/embor.2009.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang X., Takahashi N., Matsui N., Tetsuka T., Okamoto T. The NF-κB activation in lymphotoxin β receptor signaling depends on the phosphorylation of p65 at serine 536. J. Biol. Chem. 2003;278:919–926. doi: 10.1074/jbc.M208696200. [DOI] [PubMed] [Google Scholar]
- 15.Mattioli I., Geng H., Sebald A., Hodel M., Bucher C., Kracht M., Schmitz M. L. Inducible phosphorylation of NF-κB p65 at serine 468 by T cell costimulation is mediated by IKKε. J. Biol. Chem. 2006;281:6175–6183. doi: 10.1074/jbc.M508045200. [DOI] [PubMed] [Google Scholar]
- 16.Mao X., Gluck N., Li D., Maine G. N., Li H., Zaidi I. W., Repaka A., Mayo M. W., Burstein E. GCN5 is a required cofactor for a ubiquitin ligase that targets NF-κB/RelA. Genes Dev. 2009;23:849–861. doi: 10.1101/gad.1748409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O'Mahony A. M., Montano M., Van Beneden K., Chen L.-F., Greene W. C. Human T-cell lymphotropic virus type 1 tax induction of biologically active NF-κB requires IκB kinase-1-mediated phosphorylation of RelA/p65. J. Biol. Chem. 2004;279:18137–18145. doi: 10.1074/jbc.M401397200. [DOI] [PubMed] [Google Scholar]
- 18.Rocha S., Campbell K. J., Perkins N. D. p53- and Mdm2-independent repression of NF-κB transactivation by the ARF tumor suppressor. Mol. Cell. 2003;12:15–25. doi: 10.1016/s1097-2765(03)00223-5. [DOI] [PubMed] [Google Scholar]
- 19.Schwabe R. F., Sakurai H. IKKβ phosphorylates p65 at S468 in transactivaton domain 2. FASEB J. 2005;19:1758–1760. doi: 10.1096/fj.05-3736fje. [DOI] [PubMed] [Google Scholar]
- 20.Wang D., Westerheide S. D., Hanson J. L., Baldwin A. S., Jr. Tumor necrosis factor α-induced phosphorylation of RelA/p65 on Ser529 is controlled by casein kinase II. J. Biol. Chem. 2000;275:32592–32597. doi: 10.1074/jbc.M001358200. [DOI] [PubMed] [Google Scholar]
- 21.van Essen D., Engist B., Natoli G., Saccani S. Two modes of transcriptional activation at native promoters by NF-κB p65. PLoS Biol. 2009;7:e73. doi: 10.1371/journal.pbio.1000073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pfaffl M. W. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anderson L. A., Perkins N. D. The large subunit of replication factor C interacts with the histone deacetylase, HDAC1. J. Biol. Chem. 2002;277:29550–29554. doi: 10.1074/jbc.M200513200. [DOI] [PubMed] [Google Scholar]
- 24.Hassa P. O., Haenni S. S., Buerki C., Meier N. I., Lane W. S., Owen H., Gersbach M., Imhof R., Hottiger M. O. Acetylation of poly(ADP-ribose) polymerase-1 by p300/CREB-binding protein regulates coactivation of NF-κB-dependent transcription. J. Biol. Chem. 2005;280:40450–40464. doi: 10.1074/jbc.M507553200. [DOI] [PubMed] [Google Scholar]
- 25.Barré B., Perkins N. D. A cell cycle regulatory network controlling NF-κB subunit activity and function. EMBO J. 2007;26:4841–4855. doi: 10.1038/sj.emboj.7601899. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Chapman N. R., Webster G. A., Gillespie P. J., Wilson B. J., Crouch D. H., Perkins N. D. A novel form of the RelA nuclear factor κB subunit is induced by and forms a complex with the proto-oncogene c-Myc. Biochem. J. 2002;366:459–469. doi: 10.1042/BJ20020444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yeh P. Y., Yeh K. H., Chuang S. E., Song Y. C., Cheng A. L. Suppression of MEK/ERK signaling pathway enhances cisplatin-induced NF-κB activation by protein phosphatase 4-mediated NF-κB p65 Thr dephosphorylation. J. Biol. Chem. 2004;279:26143–26148. doi: 10.1074/jbc.M402362200. [DOI] [PubMed] [Google Scholar]
- 28.Beg A. A., Sha W. C., Bronson R. T., Ghosh S., Baltimore D. Embryonic lethality and liver degeneration in mice lacking the RelA component of NF-κB. Nature. 1995;376:167–170. doi: 10.1038/376167a0. [DOI] [PubMed] [Google Scholar]
- 29.Beg A. A., Baltimore D. An essential role for NF-κB in preventing TNF-α-induced cell death. Science. 1996;274:782–784. doi: 10.1126/science.274.5288.782. [DOI] [PubMed] [Google Scholar]
- 30.Kaufmann S. H., Desnoyers S., Ottaviano Y., Davidson N. E., Poirier G. G. Specific proteolytic cleavage of poly(ADP-ribose) polymerase: an early marker of chemotherapy-induced apoptosis. Cancer Res. 1993;53:3976–3985. [PubMed] [Google Scholar]
- 31.Kunsch C., Rosen C. A. NF-κB subunit-specific regulation of the interleukin-8 promoter. Mol. Cell. Biol. 1993;13:6137–6146. doi: 10.1128/mcb.13.10.6137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen C., Edelstein L. C., Gelinas C. The Rel/NF-κB family directly activates expression of the apoptosis inhibitor Bcl-xL. Mol. Cell. Biol. 2000;20:2687–2695. doi: 10.1128/mcb.20.8.2687-2695.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoffmann A., Leung T. H., Baltimore D. Genetic analysis of NF-κB/Rel transcription factors defines functional specificities. EMBO J. 2003;22:5530–5539. doi: 10.1093/emboj/cdg534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krikos A., Laherty C. D., Dixit V. M. Transcriptional activation of the tumor necrosis factor α-inducible zinc finger protein, A20, is mediated by κB elements. J. Biol. Chem. 1992;267:17971–17976. [PubMed] [Google Scholar]
- 35.Ouaaz F., Li M., Beg A. A. A critical role for the RelA subunit of nuclear factor κB in regulation of multiple immune-response genes and in Fas-induced cell death. J. Exp. Med. 1999;189:999–1004. doi: 10.1084/jem.189.6.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee K. W., Lee Y., Kwon H. J., Kim D. S. Sp1-associated activation of macrophage inflammatory protein-2 promoter by CpG-oligodeoxynucleotide and lipopolysaccharide. Cell. Mol. Life Sci. 2005;62:188–198. doi: 10.1007/s00018-004-4399-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zampetaki A., Mitsialis S. A., Pfeilschifter J., Kourembanas S. Hypoxia induces macrophage inflammatory protein-2 (MIP-2) gene expression in murine macrophages via NF-κB: the prominent role of p42/p44 and PI3 kinase pathways. FASEB J. 2004;18:1090–1092. doi: 10.1096/fj.03-0991fje. [DOI] [PubMed] [Google Scholar]
- 38.Widmer U., Manogue K. R., Cerami A., Sherry B. Genomic cloning and promoter analysis of macrophage inflammatory protein (MIP)-2, MIP-1 α, and MIP-1 β, members of the chemokine superfamily of proinflammatory cytokines. J. Immunol. 1993;150:4996–5012. [PubMed] [Google Scholar]
- 39.Ashburner B. P., Westerheide S. D., Baldwin A. S., Jr The p65 (RelA) subunit of NF-κB interacts with the histone deacetylase (HDAC) corepressors HDAC1 and HDAC2 to negatively regulate gene expression. Mol. Cell. Biol. 2001;21:7065–7077. doi: 10.1128/MCB.21.20.7065-7077.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kawahara T. L., Michishita E., Adler A. S., Damian M., Berber E., Lin M., McCord R. A., Ongaigui K. C., Boxer L. D., Chang H. Y., Chua K. F. SIRT6 links histone H3 lysine 9 deacetylation to NF-κB-dependent gene expression and organismal life span. Cell. 2009;136:62–74. doi: 10.1016/j.cell.2008.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kuboki S., Sakai N., Clarke C., Schuster R., Blanchard J., Edwards M. J., Lentsch A. B. The peptidyl-prolyl isomerase, Pin1, facilitates NF-κB binding in hepatocytes and protects against hepatic ischemia/reperfusion injury. J. Hepatol. 2009;51:296–306. doi: 10.1016/j.jhep.2009.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wan F., Anderson D. E., Barnitz R. A., Snow A., Bidere N., Zheng L., Hegde V., Lam L. T., Staudt L. M., Levens D., et al. Ribosomal protein S3: a KH domain subunit in NF-κB complexes that mediates selective gene regulation. Cell. 2007;131:927–939. doi: 10.1016/j.cell.2007.10.009. [DOI] [PubMed] [Google Scholar]
- 43.Zhong H., May M. J., Jimi E., Ghosh S. The phosphorylation status of nuclear NF-κB determines its association with CBP/p300 or HDAC-1. Mol. Cell. 2002;9:625–636. doi: 10.1016/s1097-2765(02)00477-x. [DOI] [PubMed] [Google Scholar]
- 44.Anrather J., Racchumi G., Iadecola C. cis-Acting element-specific transcriptional activity of differentially phosphorylated nuclear factor-κB. J. Biol. Chem. 2005;280:244–252. doi: 10.1074/jbc.M409344200. [DOI] [PubMed] [Google Scholar]
- 45.Chen L.-F., Williams S. A., Mu Y., Nakano H., Duerr J. M., Buckbinder L., Greene W. C. NF-κB RelA phosphorylation regulates RelA acetylation. Mol. Cell. Biol. 2005;25:7966–7975. doi: 10.1128/MCB.25.18.7966-7975.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dong J., Jimi E., Zhong H., Hayden M. S., Ghosh S. Repression of gene expression by unphosphorylated NF-κB p65 through epigenetic mechanisms. Genes Dev. 2008;22:1159–1173. doi: 10.1101/gad.1657408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nowak D. E., Tian B., Jamaluddin M., Boldogh I., Vergara L. A., Choudhary S., Brasier A. R. RelA Ser276 phosphorylation is required for activation of a subset of NF-κB-dependent genes by recruiting cyclin-dependent kinase 9/cyclin T1 complexes. Mol. Cell. Biol. 2008;28:3623–3638. doi: 10.1128/MCB.01152-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Okazaki T., Sakon S., Sasazuki T., Sakurai H., Doi T., Yagita H., Okumura K., Nakano H. Phosphorylation of serine 276 is essential for p65 NF-κB subunit-dependent cellular responses. Biochem. Biophys. Res. Commun. 2003;300:807–812. doi: 10.1016/s0006-291x(02)02932-7. [DOI] [PubMed] [Google Scholar]
- 49.Yang F., Tang E., Guan K., Wang C. Y. IKK β plays an essential role in the phosphorylation of RelA/p65 on serine 536 induced by lipopolysaccharide. J. Immunol. 2003;170:5630–5635. doi: 10.4049/jimmunol.170.11.5630. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.