Abstract
Objective
To investigate the most susceptible genetic loci in systemic sclerosis (SSc) with genome-wide association study (GWAS).
Methods
A genome-wide association study was performed in 137 patients with systemic sclerosis and 564 controls from Korea using the Affymetrix Human SNP Array 5.0. After fine mapping study, the results were replicated in 1,107 SSc patients and 2,747 controls from a US Caucasian population.
Results
The SNPs (rs3128930, rs7763822, rs7764491, rs3117230 and rs3128965) of HLA-DPB1 and –DPB2 on chromosome 6 formed a distinctive peak with log p-values (p =8.16 × 10−13) for association with SSc susceptibility. Subtyping analysis of HLA-DPB1 showed that DPB1*1301 (p = 7.61×10−8) and DPB1*0901 (p = 2.56×10−5)were the most susceptible subtypes for SSc in Koreans. In US Caucasians, two pairs of SNPs, rs7763822/rs7764491 and rs3117230/rs3128965, showed strong association with SSc patients who had either circulating anti-DNA topoisomerase I (p = 7.58 × 10−17/4.84 × 10−16) or anti-centromere autoantibodies (p = 1.12 × 10−3/3.2 × 10−5), respectively.
Conclusion
Our GWAS in Koreans revealed that the region of HLA-DPB1 and –DPB2 contains the most susceptible loci to Korean SSc. The confirmatory studies in US Caucasians indicated that specific SNPs of the HLA-DPB1 and/or –DPB2 were strongly associated with US Caucasian SSc patients who were positive to anti-topoisomerase I or anti-centromere autoantibodies.
Keywords: Systemic sclerosis, Genome wide association study, HLA-DPB1, Anti-topoisomerase I antibody
Introduction
Systemic sclerosis (SSc) is a rare and complex connective tissue disease of unknown etiology characterized by fibrosis and vasculopathy of skin and internal organs, as well as several, mutually exclusive, disease- specific circulating autoantibodies (1,2). SSc can be clinically sub-classified based on patterns of skin fibrosis into limited and diffuse forms (3). In addition, the majority of SSc patients (90%) have circulating anti-nuclear autoantibodies (ANA) (2). The three most common autoantibodies (auto-Abs) are anti-DNA topoisomerase I (topo I), anti-RNA polymerase III, and anti-centromere antibodies, in which the first two auto-Abs tend to be associated with diffuse SSc (2,4), the last one being strongly correlated with limited SSc, although these associations are not complete (2,5).
Genetic predisposition is widely believed to contribute to SSc. However, the low prevalence of SSc (approximately 0.0007--0.049%) (6,7) and clinical/serological heterogeneity make genetic studies of SSc difficult with some differing results reported for the same genes in different ethnic groups. Examples of such discrepancies are the reports of the genes of connective tissue growth factor (CTGF) (8,9), protein tyrosine phosphatase non-receptor 22 (PTPN22) (10–13) and transforming growth factor (TGF-β) (14–16) in association with SSc. Although some of these reported genes might have susceptibility markers for SSc in specific ethnic populations, the candidate gene approach used in the studies might miss other genes that could be more important to SSc susceptibility. Herein, we used GWAS approach to conduct a two-step genetic association study in four independent populations to identify the susceptibility markers for SSc.
Material and Methods
Study Subjects
We examined 4 different ethnic populations (Koreans, Caucasians, African Americans and Hispanics). Korean study population was composed of 151 SSc patients diagnosed according to the ACR preliminary criteria for SSc (17). All Korean patients were enrolled from Seoul National University Hospital between January 1998 and 2007. Genomic DNA was extracted from whole blood using standard methods. A total of 137 cases which passed the DNA quality check were entered into the GWAS using Affymetrix Genome-Wide Human SNP Array 5.0. A total of 133 cases which showed >95% of call rates, were finally entered into the case-control analysis. The mean age at diagnosis was 42 years ranging from 4 to 74 years. Mean duration of the disease was 10 years and the mean time from diagnosis to blood sampling was 5 years. Anti-Topo-I antibodies were measured with ELISA and anti-centromere antibodies were determined by passive immnunodiffusion using HEp-2 cell line. There were 79 positive vs 48 negative for anti-TopoI (It was not determined in 6 cases) and 16 positive vs 117 negative for anti-centromere antibodies. There were 66 diffuse and 67 limited form of SSc patients, according to SSc classification (3).
The 600 healthy controls were randomly selected from 10,000 healthy Koreans belonging to Korean Association Resource (KARE) Project, based on the frequency-matching on sex with the cases. The mean age of the controls was 52.5 years. The same platform (Affymetrix Genome-Wide Human SNP Array 5.0) was used for the whole-genome scan of the controls. After excluding cases with low call rate less than 95%, mismatched sex and potential relatives, a total of 557 controls were finally entered into the case-control analysis. The institutional review board of Seoul National University Hospital approved the study and all patients and controls provided written consents.
There were 1,107 Caucasians, 70 African Americans (AA) and 61 Hispanics who met the ACR criteria for SSc and corresponding controls (447 Caucasians, 90 of each for AA and Hispanics) were enrolled in the Division of Rheumatology, University of Texas Health Science Center at Houston (UTHSC-Houston). In addition, 2,300 Caucasian controls were selected from NIH data base of Genotype and Phenotype (dbGaP) found at http://www.ncbi.nlm.nih.gov/gap. Enrolled SSc patients were routinely examined for circulating anti-topoisomerase I (topo I) and anti-centromere protein autoatibodies using passive immunodiffusion against calf thymus extracts or indirect immunofluorescence patterns on HEp-2 cells, respectively. There were 183 positive vs. 917 negative for anti-topo I, and 316 positive vs. 784 negative for anti-centromere antibodies (7 cases are undetermined). There were 419 diffuse and 654 limited form of SSc (3). The study was approved by the institutional review boards of the University of Texas Health Science Center at Houston. All the individual patients and controls provided written consent.
GWAS analysis
Among the 500,568 SNPs present in Affymetrix Whole Genome Human SNP Array 5.0, 440,734 SNPs were accessible after excluding hidden SNPs. Q-Q plot was obtained in the condition of p-value > 0.0001 for Hardy-Weinberg equilibrium and call-rate > 0.95 (Figure 1). The most significant SNPs were determined to be rs3128930, re7763822, rs7764491, rs3117230 and rs3128965 and they went into the fine mapping process.
Figure 1.
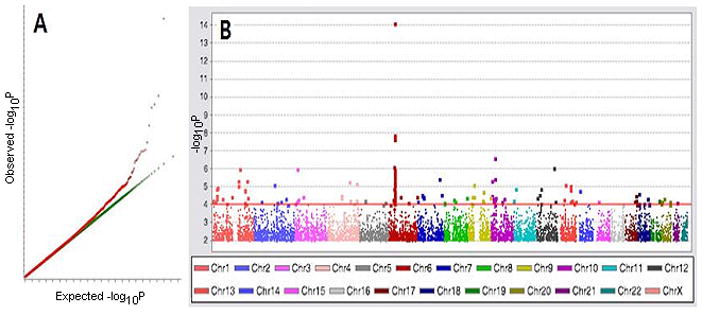
Identification of the major locus associated with SSc in genome wide scan of Koreans. A total of 440,734 SNPs were evaluated in 133 patients with SSc and 557 healthy controls. A) Quantile-quantile plot compares the distributions of observed P-values with expected null P values. B) Distribution of –Log10 P values are plotted against chromosomes.
Fine mapping
For Koreans, we performed a fine mapping focusing on HLA-DPB1 and -DPB2 regions with 137 SSc cases and 548 healthy controls in whom DNA was available. For HLA-DPB1, highly variable exon 2 of the gene was DNA-sequenced to determine subtype of HLA-DPB1. For the other region including HLA-DPB2, total 22 tag SNPs were selected with r2 threshold of 0.8 and minor allele frequency over 5% in HapMap Japanese panel data (release 22) using the Haploview version 4.1 (18,19). In tag SNPs, 17 SNPs which are included in Affymetrix SNP chip were forced to be included.
For replication studies, TaqMan Assays were used for SNPs rs3128930, re7763822, rs7764491, rs3117230 and rs3128965 genotyping with an ABI 7900HT Fast Real-Time PCR System in Caucasian, African American and Hispanic populations. Genotyping results of all five SNPs passed quality tests for Hardy-Weinberg equilibrium (p > 0.001) and calling rate (>95%). Two groups of Caucasian controls from the NIH data base and UTHSC-Houston local controls showed concordant association with Caucasian SSc patients.
Statistical analysis
The association of specific SNPs with the disease or a subset of the disease was analyzed by the comparison of minor allele frequencies of the cases and controls, with significance determined by p-values of chi-square tests. The odds ratio of cases’ having a selected SNP compared with the controls’ and its relevant 95% confidence intervals were also determined. For GWAS, The threshold for declaring significance after Bonferroni correction for adjusting multiple tests was p<1.43×10−7. It was p<2.2×10−3 for the fine mapping study (n=22) and p<0.01 for the replication study (n=5). All the association tests were based on the comparison of alleles. PLINK (20) and SAS 9.1.3 (SAS Institute Inc., Cary, NC, USA) were used for the statistical analysis. Linkage disequilibrium analysis for HLA-DPB1 and –DPB2 regions was performed with Haploview, version 4.1 (19).
Results
We first examined a Korean population who are relatively homogenous in which 62.2% of patients are positive for anti-topo I auto-Abs (based on Korean patients enrolled in this project). We performed a GWAS in 137 Korean SSc patients and 564 sex-matched Korean controls using the Affymetrix Human SNP Array 5.0 containing 440,734 accessible human SNPs. The GWAS showed a distinctive peak of SNP log p-value (p = 8.16 × 10−13 for association with SSc) (Figure 1). The peak was formed with the SNPs rs3128930, rs7763822, rs7764491, rs3117230 and rs3128965, which were located in the region of HLA-DPB1 and –DPB2 (a psuedogene) on chromosome 6p (Figure 1, 2). Fine mapping analysis of this region confirmed that rs3128930, rs7763822 and rs7764491 were the SNPs accounting for associations with Korean SSc (Figure 2, Table 1). Interestingly, the association was even stronger in patients who were positive for anti-topo I auto-Abs (rs3128930, p = 1.70 × 10−22, OR 5.15, 95% CI 3.62–7.34) (Table 1). These SNPs also were associated with the diffuse form of SSc, but not the limited form of SSc (Supplementary Table 1). Subtyping of HLA-DPB1 showed that HLA-DPB1*1301 (21.0% in SSc vs. 5.5% in controls), DPB1*0901 (12.0% vs. 2.6%) and DPB1*030101 (10.0% vs. 4.3%) were significantly more represented in anti-topo I positive patients than controls (Table 2). SNPs corresponding to previous SSc associated reported genes, such as PTPN22 (10), AIF1(21), TNF(22), CTLA-4 (23), and CTGF (8), fell within the significance thresholds of 10−5–10−6 advocated for gene-based scans, as well as the Bonferroni correction for multiple comparisons (24).
Figure 2.
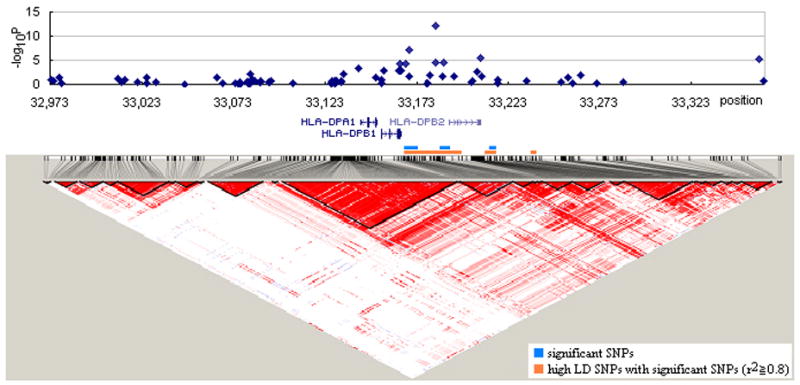
HLA-DPB as a culprit region for susceptibility to systemic sclerosis. –Log10P values are depicted around the HLA-DPB region. All genes in the region are also displayed above the linkage disequilibrium (LD) map. LD between pairs of SNPs is depicted with linkage blocks in which bright red block represents disequilibrium coefficient (D’) of 1.0 and white block D’ of 0.0. Orange bar above the LD map represents high LD SNPs which show significant association (r2>0.8) with culprit SNPs.
Table 1.
Association of SNPs rs3128930, rs7764491, rs7763822, rs3128965 and rs3117230 with Korean SSc patients with and without autoantibodies to DNA topoisomerase I and centromere protein.
| Alleles (Minor allele) | Systemic sclerosis |
Controls | ||||
|---|---|---|---|---|---|---|
| Anti-topoisomerase I Ab | Anti-centromere Ab | Total | ||||
| ve(+) (n=79) | ve(−) (n=48) | ve(+) (n=16) | ve(−) (n=117) | (n=133) | (n=557) | |
| rs3128930 (A) | ||||||
| Frequency | 0.47 | 0.12 | 0.13 | 0.37 | 0.34 | 0.15 |
| P-value* | 8.26×10−22 | 0.47 | 0.73 | 4.38×10−15 | 8.16×10−13 | - |
| OR (95% CI)* | 5.21(3.63,7.49) | 0.79 (0.41–1.51) | 0.83(0.29–2.39) | 3.45(2.50–4.75) | 3.00(2.20–4.10) | - |
| rs7764491 (C) | ||||||
| Frequency | 0.21 | 0.065 | 0.063 | 0.17 | 0.15 | 0.056 |
| P-value* | 7.29×10−12 | 0.71 | 0.87 | 8.24×10−9 | 7.64×10−8 | - |
| OR (95% CI)* | 4.58(2.87–7.33) | 1.18(0.50–2.81) | 1.13(0.26–4.83) | 3.42(2.20–5.30) | 3.09(2.01–4.75) | - |
| rs7763822 (T) | ||||||
| Frequency | 0.21 | 0.065 | 0.063 | 0.17 | 0.15 | 0.056 |
| P-value* | 7.29×10−12 | 0.71 | 0.87 | 8.24×10−9 | 7.64×10−8 | |
| OR (95% CI)* | 4.58(2.87–7.33) | 1.18(0.50–2.81) | 1.13(0.26–4.83) | 3.42(2.20–5.30) | 3.09(2.01–4.75) | |
| rs3128965 (A) | ||||||
| Frequency | 0.25 | 0.054 | 0.063 | 0.20 | 0.18 | 0.093 |
| P-value* | 6.28×10−9 | 0.21 | 0.56 | 4.37×10−6 | 4.47×10−5 | - |
| OR(95% CI)* | 3.31(2.17–5.04) | 0.56 (0.22–1.41) | 0.65(0.15–2.76) | 2.44(1.65–3.59) | 2.18(1.49–3.18) | - |
| rs3117230 (G) | ||||||
| Frequency | 0.25 | 0.054 | 0.063 | 0.20 | 0.18 | 0.092 |
| P-value* | 4.51×10−9 | 0.22 | 0.57 | 3.37×10−6 | 3.52×10−5 | |
| OR (95% CI)* | 3.34(2.19–5.10) | 0.57(0.22–1.43) | 0.66(0.15–2.79) | 2.46(1.67–3.64) | 2.20(1.50–3.22) | |
Each subset versus controls
Missing information of anti-topoisomerase I antibody status in 6 patients; Ab, antibody; CI, confidence interval
Table 2.
Association of HLA-DPB1 allelic subtypes with systemic sclerosis in Koreans.
| HLA alleles | Systemic sclerosis |
Controls | ||||
|---|---|---|---|---|---|---|
| Anti-topoisomerase-I Ab† | Anti-centromere Ab | Total | ||||
| (+) (n=82) | (−) (n=49) | (+) (n=16) | (−) (n=121) | (n=137) | (n=548) | |
| *1301 | ||||||
| Frequency | 0.21 | 0.05 | 0.031 | 0.17 | 0.15 | 0.055 |
| P-value* | 4.05×10−12 | 0.88 | 0.56 | 3.25×10−9 | 7.61×10−8 | - |
| OR (95% CI)* | 4.52(2.86–7.14) | 0.93(0.36–2.37) | 0.56(0.074–4.15) | 3.42(2.23–5.24) | 3.04(1.99–4.63) | - |
| *0901 | ||||||
| Frequency | 0.12 | 0.01 | 0.031 | 0.087 | 0.08 | 0.026 |
| P-value* | 2.44×10−8 | 0.33 | 0.87 | 7.55×10−6 | 2.55×10−5 | - |
| OR (95% CI)* | 4.82(2.64–8.82) | 0.38(0.05–2.82) | 1.19(0.16–8.99) | 3.50(1.96–6.24) | 3.21(1.82–5.69) | - |
| *030101 | ||||||
| Frequency | 0.10 | 0.02 | 0.031 | 0.083 | 0.077 | 0.043 |
| P-value* | 9.47×10−4 | 0.28 | 0.75 | 0.01 | 0.021 | - |
| OR (95% CI)* | 2.58(1.44–4.61) | 0.47(0.11–1.94) | 0.72(0.096–5.39) | 2.01(1.17–3.46) | 1.85(1.09–3.16) | - |
Each subset versus controls
Missing information of anti-topoisomerase I antibody status in 6 patients; Ab, antibody; CI, confidence interval
To confirm these results, we used TaqMan Assays to reexamine the 5 SNPs with the strongest association from the Korean GWAS screen in 1,107 US Caucasian SSc patients (collected from US), of whom 16% were positive for anti-topo I, and 2,747 normal controls, of which 447 were from our local collections (Houston, Texas, US.) and 2,300 were from the NIH data base of Genotype and Phenotype (dbGaP) (http://www.ncbi.nlm.nih.gov/gap). The SNPs rs7763822 and rs7764491 showed highly significant associations with SSc patients who were positive for anti-topo I auto-Abs (p = 7.58 × 10−17 and 4.84 × 10−16, respectively) (Table 3). The HLA-DPB1*1301 allele which occurs in only 3% of US Caucasians was found in 25% of anti-topo I positive patients and conferred the strongest risk by exact logistic regression (p=0.0001, OR=14) of any HLA class II allele (unpublished results). The SNPs rs3128965 and rs3117230 showed strong associations with SSc patients who were positive for anti-centromere auto-Abs (p = 3.20 × 10−5 and 1.12 × 10−3, respectively) (Table 3). In addition, the pair of anti-topo I associated SNPs also showed a weaker association with the diffuse form of SSc (p = 0.0070 and 0.014 for rs7763822 and rs7764491 respectively) (Supplementary Table 2). The genetic concordance of the patients with anti-topo I positivity and the diffuse form of SSc supports clinical observations that these two traits within SSc commonly overlap (2, 4). However, the anti-centromere associated SNP pairs did not show strong associations with the limited form of SSc that usually occurs in patients with anti-centromere auto-Abs (2, 5). The SNP rs3128930 showed only a marginal p value of 0.041 in the limited form of SSc patients who were positive for anti-centromere auto-Abs (Supplementary table 2). Further analysis of Caucasian patients who are negative for anti-topo I or anti-centromere autoantibodies indicated that they have marginal or no association with the genotypes of all the 5 SNPs (Table 3). In contrast, highly significant differences were observed in the comparisons of patients with and without anti-topo I or anti-centromere autoantibodies using corresponding anti-topo I or anti-centromere associated SNP pairs (rs7763822 and rs7764491 pair, p ≤ 2.86×10−18 or rs3128965 and rs3117230 pair, 4.77×10−4 respectively) (Supplementary Table 3). Interestingly, marginally significant differences (0.05 > p ≥ 0.01) were also observed in the comparison of patients with the limited and the diffuse forms of SSc using both pairs of SNPs but the autoantibody associations were the strongest, perhaps because HLA alleles are specific immune response genes (Table 3, and supplementary Table 2).
Table 3.
Association of SNPs of HLA-DPB1 and –DPB2 with systemic sclerosis in Caucasians.
| Alleles (Minor allele) | Systemic sclerosis† |
Controls | ||||
|---|---|---|---|---|---|---|
| Anti-topoisomerase I Ab | Anti-centromere Ab | Total | ||||
| (+) (n=183) | (−) (n=917) | (+) (n=316) | (−) (n=784) | (n=1,107) | (n=2,731) | |
| rs3128930 (A) | ||||||
| Frequency | 0.31 | 0.28 | 0.30 | 0.28 | 0.29 | 0.27 |
| P-value* | 0.117 | 0.30 | 0.056 | 0.47 | 0.14 | - |
| OR (95% CI)* | 1.20(0.95–1.51) | 1.01(0.95–1.20) | 1.19(1.00–1.43) | 1.05(0.92–1.19) | 1.09(0.97–1.21) | - |
| rs7764491 (C) | ||||||
| Frequency | 0.14 | 0.03 | 0.025 | 0.058 | 0.049 | 0.043 |
| P-value* | 4.84×10−16 | 0.018 | 0.036 | 0.016 | 0.29 | |
| OR (95% CI)* | 3.56(2.57–4.94) | 0.70(0.52–0.94) | 0.58(0.35–0.97) | 1.36(1.06–1.75) | 1.14(0.90–1.44) | - |
| rs7763822 (T) | ||||||
| Frequency | 0.14 | 0.031 | 0.024 | 0.059 | 0.049 | 0.043 |
| P-value* | 7.58×10−17 | 0.021 | 0.023 | 0.0076 | 0.24 | - |
| OR (95% CI)* | 3.65(2.64–5.04) | 0.71(0.53–0.95) | 0.55(0.32–0.93) | 1.40(1.09–1.80) | 1.15(0.91–1.46) | - |
| rs3128965 (A) | ||||||
| Frequency | 0.14 | 0.19 | 0.25 | 0.16 | 0.18 | 0.18 |
| P-value* | 0.024 | 0.39 | 3.20×10−5 | 0.010 | 0.99 | - |
| OR (95% CI)* | 0.70(0.52–0.96) | 1.06(0.93–1.21) | 1.50(1.24–1.82) | 0.82(0.70–0.95) | 1.00 (0.88–1.14) | - |
| rs3117230 (G) | ||||||
| Frequency | 0.17 | 0.26 | 0.29 | 0.22 | 0.24 | 0.24 |
| P-value* | 0.0077 | 0.055 | 1.12×10−3 | 0.32 | 0.42 | - |
| OR (95% CI)* | 0.69(0.52–0.91) | 1.13(1.00–1.28) | 1.36(1.13–1.63) | 0.93(0.81–1.07) | 1.05(0.93–1.18) | - |
Each subset versus controls
Missing information of autoantibody status in 6 patients; Ab, antibody; CI confidence interval
Finally, we studied these same SNPs in African Americans and Hispanics with limited numbers of cases and controls (70 cases vs. 90 controls and 61 cases vs. 90 controls, respectively). Although, the numbers of subjects examined in these two populations were small, the SNPs rs7764491 and rs7763822 showed a consistently strong association with anti-topo I positive SSc in both African Americans (p=9.05×10−3, OR=4.23, 95% CI=1.33–13.48 for both SNPs) and Hispanics (7.98×10−4, OR=5.51, 95% CI=1.85–16.43 and p=7.21×10−4, OR=5.57, 95% CI=1.87–16.62, respectively) (Supplementary Table 4, Supplementary Table 5).
Discussion
In studies of SSc, a human complex disease, there have been inconsistent reports of genetic associations from different study populations (8–16). The rareness and the heterogeneity of SSc may contribute at least in part to such discrepancies. In the studies herein, we applied a two-step approach, in which a GWAS was first conducted in a relatively homogenous Korean SSc population, and then was followed by a confirmatory study in other populations. Our findings of SSc-associated HLA-DPB1 and -DPB2 on the basis of autoantibodies, represent the first replicable report in Asians, Caucasians, African-Americans and Hispanics. These complementary studies in four independent populations provide strong support for these identified genetic makers to confer susceptibility to subgroups of SSc patients.
HLA-DPB1, located centromeric to other HLA class II molecules, shows relatively low linkage disequilibrium with other extended MHC haplotypes (25). Although it was not studied as extensively as HLA-A, -B, -C or -DR, it has similar antigen-presentation function to activate CD4+ T cells (26). Its genetic polymorphisms have been found to be associated with chronic berylliosis (27), graft-vs-host disease (28), juvenile rheumatoid arthrtitis (29), insulin dependent diabetes mellitus (30) and sarcoidosis (31). Some studies have suggested the possible roles of HLA-DPB1 in SSc, in the context of HLA-A, B, C and DR molecules (32–34). Typically, HLA-DPB1 *1301 was previously reported to associate with anti-topo I in SSc patients in several independent studies, although each of these studies appeared to have less statistical power with smaller sample size (34–36). The results of Korean GWAS and US confirmatory studies herein identified that specific SNPs of HLA-DPB1 and –DPB2 are strongly associated with SSc, especially to those patients with auto-antibodies to topo I or centromere protein, and that the HLA-PDB1 may be the most important susceptibility gene to Korean SSc.
Our studies indicate that sub-classification and population origin may be two critical factors in identifying genetic susceptibility markers for SSc. Such is evidenced in our lack of association in Caucasian SSc with any of the 5 SNPs found in the Korean GWAS until we took into account those subgroups defined by clinically diffuse and limited forms, but especially by specific autoantibody status (anti-topo I and anti-centromere positives). In addition, this notion may also explain previous inconsistent reports of SSc associated genes in different populations, as well as the fact that these genes were not identified in our GWAS of Koreans.
Importantly, the genetic differentiations of SSc with distinctive serological and clinical features in our studies suggest that SSc should not be considered as a single disease. While SSc, like other human complex diseases, is currently incurable, sub-classification of SSc on the basis of genetic polymorphisms for disease susceptibility may provide a new dimension for exploring pathogenesis and treatment of this disease. Our results strongly indicate that sub-classification of SSc on the basis of autoantibodies against topo I and centromeric proteins is important in defining disease susceptibility genes in this heterogeneous disease.
Despite the successful identification of SSc susceptibility genes of HLA-DPB1 and –DPB2 in our studies, it has several limitations. First, the number of SSc patients for GWAS may not be large enough to distinguish some other true disease risk SNPs from the statistical noise of false positive SNPs by using stringent statistical threshold in the studies. The power of GWAS calculated with the assumption of 10% difference of minor allele frequency was 75.0% (37). Second, majority of Korean SSc patients in the studies were positive for anti-topo-I autoantibodies. Accordingly, genetic predisposition factors for SSc with other SSc autoantibodies may be better unveiled by a GWAS in other populations. Third, the numbers of Hispanics and African American may not be enough for adequate power of the study.
Nonetheless, the SNPs of HLA-DPB1 and –DPB2 showed a distinctively strong association with anti-topo-I positive SSc in our two-step approach of genetic studies. The results confirmed previous reports of this genetic susceptibility region to SSc, and raised the importance of sub-classification of SSc in understanding of disease pathogenesis.
Supplementary Material
Acknowledgments
This study was supported by Korea Health 21 R&D Project, Ministry of Health, Welfare & Family Affairs, R.O.K. (AO30001); NIH/NIAMS P50 AR054144, NO1-AR02251 and UL1RR024148, Department of the Army, Medical Research Acquisition Activity: PR064803 and PR064851. This work cannot be accomplished without the generous provision of GWAS data and DNAs for healthy Koreans from Korean Center for Disease Control. We also thank Hua Dong for her valuable help in statistical analysis
References
- 1.Korn JH. Pathogenesis of systemic sclerosis. In: Koopman WJ, editor. Arthritis and Allied Conditions. 15. Philadelphia: Lippincott Williams & Wilkins; 2005. p. 1621. [Google Scholar]
- 2.Bunn CC, Black CM. Systemic sclerosis: an autoantibody mosaic. Clin Exp Immunol. 1999;117(2):207–8. doi: 10.1046/j.1365-2249.1999.00990.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.LeRoy EC, Black C, Fleischmajer R, Jablonska S, Krieg T, Medsger TA, Jr, Rowell N, Wollheim F. Scleroderma (systemic sclerosis): classification, subsets and pathogenesis. J Rheumatol. 1988;15(2):202–5. [PubMed] [Google Scholar]
- 4.Jarzabek-Chorzelska M, Blaszczyk M, Jablonska S, Chorzelski T, Kumar V, Beutner EH. Scl 70 antibody--a specific marker of systemic sclerosis. Br J Dermatol. 1986;115(4):393–401. doi: 10.1111/j.1365-2133.1986.tb06233.x. [DOI] [PubMed] [Google Scholar]
- 5.Moroi Y, Peebles C, Fritzler MJ, Steigerwald J, Tan EM. Autoantibody to centromere (kinetochore) in scleroderma sera. Proc Natl Acad Sci U S A. 1980;77(3):1627–31. doi: 10.1073/pnas.77.3.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chifflot H, Fautrel B, Sordet C, Chatelus E, Sibilia J. Incidence and prevalence of systemic sclerosis: a systematic literature review. Semin Arthritis Rheum. 2008;37(4):223–35. doi: 10.1016/j.semarthrit.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 7.Mayes MD, Lacey JV, Jr, Beebe-Dimmer J, Gillespie BW, Cooper B, Laing TJ, et al. Prevalence, incidence, survival, and disease characteristics of systemic sclerosis in a large US population. Arthritis Rheum. 2003;48(8):2246–55. doi: 10.1002/art.11073. [DOI] [PubMed] [Google Scholar]
- 8.Fonseca C, Lindahl GE, Ponticos M, Sestini P, Renzoni EA, Holmes AM, et al. A polymorphism in the CTGF promoter region associated with systemic sclerosis. N Engl J Med. 2007;357(12):1210–20. doi: 10.1056/NEJMoa067655. [DOI] [PubMed] [Google Scholar]
- 9.Rueda B, Simeon C, Hesselstrand R, Herrick A, Worthington J, Ortego-Centeno N, et al. A large multicenter analysis of CTGF −945 promoter polymorphism does not confirm association with Systemic Sclerosis susceptibility or phenotype. Ann Rheum Dis. 2008 doi: 10.1136/ard.2008.100180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gourh P, Tan FK, Assassi S, Ahn CW, McNearney TA, Fischbach M, et al. Association of the PTPN22 R620W polymorphism with anti-topoisomerase I- and anticentromere antibody-positive systemic sclerosis. Arthritis Rheum. 2006;54(12):3945–53. doi: 10.1002/art.22196. [DOI] [PubMed] [Google Scholar]
- 11.Kyogoku C, Langefeld CD, Ortmann WA, Lee A, Selby S, Carlton VE, et al. Genetic association of the R620W polymorphism of protein tyrosine phosphatase PTPN22 with human SLE. Am J Hum Genet. 2004;75(3):504–7. doi: 10.1086/423790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Viken MK, Amundsen SS, Kvien TK, Boberg KM, Gilboe IM, Lilleby V, et al. Association analysis of the 1858C>T polymorphism in the PTPN22 gene in juvenile idiopathic arthritis and other autoimmune diseases. Genes Immun. 2005;6(3):271–3. doi: 10.1038/sj.gene.6364178. [DOI] [PubMed] [Google Scholar]
- 13.Wipff J, Allanore Y, Kahan A, Meyer O, Mouthon L, Guillevin L, et al. Lack of association between the protein tyrosine phosphatase non-receptor 22 (PTPN22)*620W allele and systemic sclerosis in the French Caucasian population. Ann Rheum Dis. 2006;65(9):1230–2.5. doi: 10.1136/ard.2005.048181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crilly A, Hamilton J, Clark CJ, Jardine A, Madhok R. Analysis of transforming growth factor beta1 gene polymorphisms in patients with systemic sclerosis. Ann Rheum Dis. 2002;61(8):678–81. doi: 10.1136/ard.61.8.678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee EB, Kim JY, Lee YJ, Abdallah A, Lympany P, Song YW. Transforming growth factor-beta1 polymorphisms in Korean patients with systemic sclerosis. Tissue Antigens. 2004;63(5):491–5. doi: 10.1111/j.1399-0039.2004.00185.x. [DOI] [PubMed] [Google Scholar]
- 16.Sugiura Y, Banno S, Matsumoto Y, Niimi T, Yoshinouchi T, Hayami Y, et al. Transforming growth factor beta1 gene polymorphism in patients with systemic sclerosis. J Rheumatol. 2003;30(7):1520–37. [PubMed] [Google Scholar]
- 17.Preliminary criteria for the classification of systemic sclerosis (scleroderma) Subcommittee for scleroderma criteria of the American Rheumatism Association Diagnostic and Therapeutic Criteria Committee. Arthritis Rheum. 1980;23(5):581–90. doi: 10.1002/art.1780230510. [DOI] [PubMed] [Google Scholar]
- 18.Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449(7164):851–61. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 20.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alkassab F, Gourh P, Tan FK, McNearney T, Fischbach M, Ahn C, Arnett FC, Mayes MD. An allograft inflammatory factor 1 (AIF1) single nucleotide polymorphism (SNP) is associated with anticentromere antibody positive systemic sclerosis. Rheumatology (Oxford) 2007;46(8):1248–51. doi: 10.1093/rheumatology/kem057. [DOI] [PubMed] [Google Scholar]
- 22.Pandey JP, Takeuchi F. TNF-alpha and TNF-beta gene polymorphisms in systemic sclerosis. Hum Immunol. 1999 Nov;60(11):1128–30. doi: 10.1016/s0198-8859(99)00105-6. [DOI] [PubMed] [Google Scholar]
- 23.Takeuchi F, Kawasugi K, Nabeta H, Mori M, Tanimoto K. Association of CTLA-4 with systemic sclerosis in Japanese patients. Clin Exp Rheumatol. 2002 Nov-Dec;20(6):823–8. [PubMed] [Google Scholar]
- 24.Thomas DC, Clayton DG. Betting odds and genetic associations. J Natl Cancer Inst. 2004;96(6):421–3. doi: 10.1093/jnci/djh094. [DOI] [PubMed] [Google Scholar]
- 25.Begovich AB, McClure GR, Suraj VC, Helmuth RC, Fildes N, Bugawan TL, et al. Polymorphism, recombination, and linkage disequilibrium within the HLA class II region. J Immunol. 1992;148(1):249–58. [PubMed] [Google Scholar]
- 26.McCanlies EC, Kreiss K, Andrew M, Weston A. HLA-DPB1 and chronic beryllium disease: a HuGE review. Am J Epidemiol. 2003;157(5):388–98. doi: 10.1093/aje/kwg001. [DOI] [PubMed] [Google Scholar]
- 27.Richeldi L, Sorrentino R, Saltini C. HLA-DPB1 glutamate 69: a genetic marker of beryllium disease. Science. 1993;262(5131):242–4. doi: 10.1126/science.8105536. [DOI] [PubMed] [Google Scholar]
- 28.Shaw BE, Potter MN, Mayor NP, Pay AL, Smith C, Goldman JM, et al. The degree of matching at HLA-DPB1 predicts for acute graft-versus-host disease and disease relapse following haematopoietic stem cell transplantation. Bone Marrow Transplant. 2003;31(11):1001–8. doi: 10.1038/sj.bmt.1704029. [DOI] [PubMed] [Google Scholar]
- 29.Begovich AB, Bugawan TL, Nepom BS, Klitz W, Nepom GT, Erlich HA. A specific HLA-DP beta allele is associated with pauciarticular juvenile rheumatoid arthritis but not adult rheumatoid arthritis. Proc Natl Acad Sci U S A. 1989;86(23):9489–93. doi: 10.1073/pnas.86.23.9489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cruz TD, Valdes AM, Santiago A, Frazer de Llado T, Raffel LJ, Zeidler A, et al. DPB1 alleles are associated with type 1 diabetes susceptibility in multiple ethnic groups. Diabetes. 2004;53(8):2158–63. doi: 10.2337/diabetes.53.8.2158. [DOI] [PubMed] [Google Scholar]
- 31.Lympany PA, Petrek M, Southcott AM, Newman Taylor AJ, Welsh KI, du Bois RM. HLA-DPB polymorphisms: Glu 69 association with sarcoidosis. Eur J Immunogenet. 1996;23(5):353–9. doi: 10.1111/j.1744-313x.1996.tb00008.x. [DOI] [PubMed] [Google Scholar]
- 32.Kuwana M, Inoko H, Kameda H, Nojima T, Sato S, Nakamura K, et al. Association of human leukocyte antigen class II genes with autoantibody profiles, but not with disease susceptibility in Japanese patients with systemic sclerosis. Intern Med. 1999;38(4):336–44. doi: 10.2169/internalmedicine.38.336. [DOI] [PubMed] [Google Scholar]
- 33.Tikly M, Rands A, McHugh N, Wordsworth P, Welsh K. Human leukocyte antigen class II associations with systemic sclerosis in South Africans. Tissue Antigens. 2004;63(5):487–90. doi: 10.1111/j.0001-2815.2004.00199.x. [DOI] [PubMed] [Google Scholar]
- 34.Reveille JD, Brady J, MacLeod-St Clair M, Durban E. HLA-DPB1 alleles and autoantibody subsets in systemic lupus erythematosus, Sjogren’s syndrome and progressive systemic sclerosis: a question of disease relevance. Tissue Antigens. 1992;40(1):45–8. doi: 10.1111/j.1399-0039.1992.tb01957.x. [DOI] [PubMed] [Google Scholar]
- 35.Rihs HP, Conrad K, Mehlhorn J, May-Taube K, Welticke B, Frank KH, Baur X. Molecular analysis of HLA-DPB1 alleles in idiopathic systemic sclerosis patients and uranium miners with systemic sclerosis. Int Arch Allergy Immunol. 1996;109(3):216–22. doi: 10.1159/000237240. [DOI] [PubMed] [Google Scholar]
- 36.Gilchrist FC, Bunn C, Foley PJ, Lympany PA, Black CM, Welsh KI, du Bois RM. Class II HLA associations with autoantibodies in scleroderma: a highly significant role for HLA-DP. Genes Immun. 2001;2(2):76–81. doi: 10.1038/sj.gene.6363734. [DOI] [PubMed] [Google Scholar]
- 37.Lachin JM. Introduction to sample size determination and power analysis for clinical trials. Control Clin Trials. 1981;2:93–113. doi: 10.1016/0197-2456(81)90001-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


