Summary
Combinatorial libraries built with severely restricted chemical diversity have yielded highly functional synthetic binding proteins. Structural analyses of these minimalist binding sites have revealed the dominant role of large tyrosine residues for mediating molecular contacts and of small serine/glycine residues for providing space and flexibility. The concept of using limited residue types to construct optimized binding proteins mirrors findings in the field of small molecule drug development, where it has been proposed that most drugs are built from a limited set of side chains presented by diverse frameworks. The physicochemical properties of tyrosine make it the amino acid that is most effective for mediating molecular recognition, and protein engineers have taken advantage of these characteristics to build tyrosine-rich protein binding sites that outperform natural proteins in terms of affinity and specificity. Knowledge from preceding studies can be used to improve current designs, and thus, synthetic protein libraries will continue to evolve and improve. In the near future, it seems likely that synthetic binding proteins will supersede natural antibodies for most purposes, and moreover, synthetic proteins will enable many new applications beyond the scope of natural proteins.
Introduction
Protein-protein interactions are fundamental to biology 1-3. Consequently, there is great interest in understanding factors governing protein-protein interactions, as this knowledge would enable accurate prediction of natural binding partners and would also aid the efficient design of engineered proteins and chemical compounds for capturing proteins and perturbing their cellular functions.
Numerous studies of natural protein systems have served to advance our understanding of the fundamental principles governing protein-protein interactions. Traditional investigations have proceeded along similar lines and have relied on the following major steps: (i) biophysical characterization of the interaction in terms of affinity and specificity, ii) determination of the three-dimensional structures of the protein-protein complex, and ideally, of each component alone, (iii) comparison of homologous sequences, and (iv) systematic mutagenesis to dissect the energetics at the interface. Studies of this type are exemplified by the extensive studies of the interactions between human growth hormone and its receptor 4-6 and β–lactamase and its inhibitor 7, 8. These studies have revealed details that are specific for each system, but more importantly, they have also contributed to a general understanding of the importance of shape and electrostatic complementarity in natural protein-protein interactions.
Most natural proteins have been shaped under biological selection pressure over the course of evolution. However, there are vast combinations of amino acids even for a relatively small interaction interface, and even over the long times spans of natural evolution, nature has sampled only a small fraction of the theoretical sequence space. Furthermore, natural interfaces have been optimized for specialized biological functions, and nature does not necessarily maximize either affinity or specificity. Thus, it is unlikely that natural protein complexes represent optimal solutions, when considering protein function from a purely biophysical perspective. This view is supported by numerous studies in which the affinities and specificities of natural protein-protein interactions have been improved through protein engineering 9-14. Considering the constraints and complexities inherent to natural systems, we have come to believe that a full understanding of the chemistry and physics of protein-protein interactions necessitates the establishment of simplified systems that are not constrained by biology.
Advances in the understanding of protein function and in protein engineering technologies have enabled the generation of “synthetic” binding proteins with binding sites built by combinatorial design rather than from natural diversity 15-18. Synthetic binding proteins are generated by selection from combinatorial libraries using molecular display technologies. In synthetic libraries, the composition of amino acid diversity and the locations at which diversity is introduced are precisely defined. To a first approximation, functional molecules derived from such libraries using in vitro methods are selected according to their functional properties devoid of ill-defined biological biases. Thus, the analysis of synthetic proteins is likely to provide a less biased view of the mechanisms governing molecular recognition and may also lead to a better understanding of natural proteins.
In recent years, minimalist synthetic proteins have emerged as a particularly intriguing product of protein engineering investigations aimed at defining the minimal requirements for specific protein-protein interactions 18. By severely restricting the amino acid diversity used in libraries, these investigations aim to uncover principles that are obscured in natural systems due to high levels of “evolutionary noise”. Outcomes reviewed in this article prove that this approach is indeed highly effective. Of particular importance has been the finding that tyrosine residues are exceptionally versatile for mediating contacts at interfaces, which helps to rationalize trends found in natural interfaces and provides guidelines for engineering highly functional synthetic binding proteins.
Enabling technologies for synthetic protein engineering
The engineering of synthetic binding proteins has been enabled by technologies that allow for the generation and testing of large libraries of combinatorially mutated binding sites built on defined frameworks. Phage display has been the most commonly used technology, but other methods such as yeast, ribosome and mRNA display have been developed as viable alternatives 17, 19, 20. As exemplified by phage display (Figure 1), molecular display technologies establish unambiguous physical linkage between the phenotype of the protein and the genotype of the encoding DNA. Mutations introduced into the DNA are translated into diverse libraries of proteins, which can be used in binding selections to isolate members with affinity and specificity of interest. At the end of the selection process, the sequences of binding proteins can be deduced by sequencing the encoding DNA. These technologies provide an in vitro version of classic Darwinian evolution with precise control over the library design and the selection process.
Figure 1. Binding protein selection from phage display libraries.
Libraries of binding proteins (e.g. FN3 domains, colored) are displayed on the surfaces of phage particles as fusions to a coat protein (black). Each phage particle displays a unique protein and encapsulates the encoding DNA. Highly diverse libraries (>1010) are represented as phage pools, which are used in selections for binding to immobilized antigen. Bound phage are retained and the nonbinding phage are washed away. Bound phage can be amplified by infection of an Escherichia coli host, and the amplified pool can be used for further rounds of selection to enrich the population for binding clones. Individual phage clones can be subjected to DNA sequence analysis to determine the sequences of the displayed proteins.
The most common scaffolds for synthetic binding proteins have been antibody fragments, including heterodimeric antigen-binding fragments (Fabs) 21-25 monomeric single-chain variable fragments (scFvs) 26-28 and heavy-chain variable (VH) domains 29. However, other proteins have been adapted as scaffolds to produce binding proteins with affinities comparable to those of antibodies (Figure 2) 15, 30, 31. Notably, structural analysis has shown that different scaffolds impose different constraints on the interface shape. The binding sites of Fabs and scFvs are typically flat, but cavities, clefts or protrusions can be created by different combinations of frameworks and hypervariable loops 32, 33. Single-domain β-sandwich proteins, such as VH and fibronectin type III (FN3) domains, form a convex surface 34, 35, while ankyrin repeat proteins provide a slightly concave shape 36-38 and the lipocalin fold presents a deep cavity that can accommodate small molecules 39. Protein engineers can take advantage of this diverse array of scaffolds to design synthetic binding proteins with functional properties tailored for specific demands.
Figure 2. Structures of synthetic binding proteins built with distinct molecular scaffolds.
The respective binding partners are not shown, but the Cα atoms in the binding site are shown as spheres (residues within 5 Å of the binding partner). (a) Fab binding to human VEGF (PDB ID: 1TZI). (b) VHH domain binding to lysozyme (PDB ID: 1JTT). (c) FN3 domain binding to MBP (PDB ID: 2OBG). (d) Ankyrin repeat protein binding to MBP (PDB ID: 1SVX). (e) Lipocalin binding to the small molecule digoxigenin (PDB ID: 1LNM).
Lessons from natural interfaces
Synthetic proteins produced using molecular display technologies are still made of natural amino acids, and thus, they obey the same rules that govern natural proteins. Consequently, the knowledge gained from analyses of natural protein-ligand interactions has provided valuable guidelines for designing strategies to generate synthetic binding proteins.
Structural studies of many protein-ligand complexes have shown that proteins use all types of structural motifs (i.e. loops, turns, helices and sheets) to interact with other molecules. The underlying principle is that proteins accomplish specific recognition of cognate ligands by achieving shape and electrostatic complementarity. In the case of tight protein-protein interactions, the interface is contiguous and quite large with an average buried surface of ~1,600 Å2 on both sides 40. In general, interfaces are also closely packed 41, and systematic mutagenesis studies of high-affinity interfaces have revealed a characteristic spatial organization for the contacting residues. Binding interfaces tend to have “hot spots” that are intolerant to substitutions, but outside the hot spots the interfaces are remarkably plastic to amino acid substitutions 8, 42. A significant bias in amino acid composition was found amongst interface residues 40, and hot spots are particularly enriched in tyrosine, tryptophan and arginine 43.
The features of natural proteins have been formed by evolution, and thus it is difficult to define the minimum requirements for effective interactions. Recent systematic mutagenesis analyses have shown that natural interfaces can be further simplified. An “alanine shaving” analysis of human growth hormone showed that over half of the binding site residues could be simultaneously changed to alanine without significantly affecting affinity for its receptor, so long as most of the hot spot residues were maintained 44. Complementary to this study, affinity maturation of a single-domain antibody with a small binding site resulted in hot spot residues constituting 76% of the binding surface and complete elimination of neutral contacts 12. These “semi-synthetic” studies strongly suggest that protein binding sites can be minimized to an essential cluster of hot spot residues and that natural protein-protein interfaces are larger and more complex than necessitated by biophysical principles.
Minimalist synthetic antibodies
Examination of natural antigen-binding sites has shown that tyrosine is particularly abundant, as it accounts for ~10% of the total composition of the complementarity-determining region (CDR) loops and ~25% of the antigen contacts in functional antibodies 45. Moreover, analysis of naïve diversity in the third heavy-chain CDR (CDR-H3), which dominates most antigen-binding sites, predicts an even more extreme bias prior to antigen recognition and affinity maturation, as ~40% of the sequence is predicted to be tyrosine and ~30% is predicted to be small amino acids (serine, glycine, alanine and threonine) 46. While these biases may be the coincidental result of genetic biases in antibody genes, we and others believe that tyrosine, together with small residues that provide conformational freedom and space, is uniquely suited for mediating favorable contacts for antigen recognition 22, 45-47. Studies with minimalist synthetic antibodies have proven this hypothesis and have provided a path towards designed antibodies with functions beyond those of natural antibodies.
In the first direct demonstration of the special role of tyrosine in naïve antigen recognition, a synthetic library was constructed using a single Fab framework with CDR diversity restricted to only for amino acid types (tyrosine, serine, alanine and aspartate) 22. Despite the simple design, the library yielded high affinity antibodies against human vascular endothelial growth factor (VEGF). Although the four amino acid types were used equally at the primary sequence level, structural analysis of two distinct antibodies revealed that tyrosine plays a dominant role in the binding interfaces, as the bulky side chains contributed ~50% of the intermolecular contacts and ~70% of the buried surface area on the antibody side of the interfaces (Figure 3A).
Figure 3. Tyrosine plays a dominant role in minimalist synthetic antibodies.
(a) Composition of anti-VEGF binding sites derived from a four-amino-acid code 22. The graph shows the percentage that each of the amino acid types allowed in the CDR loops represent in terms of the primary sequence (black bars), the residues that contact antigen (grey bars) and the surface area buried upon binding to antigen (white bars). (b) The interface between DR5 and an Fab derived from a binary tyrosine/serine code (PDB IB: 1ZA3) 48. (Top) Contacts between DR5 and the CDR-H3 loop. DR5 is shown as a molecular surface with residues within 5 Å of an Fab atom colored pink. The CDR-H3 main chain is colored yellow, and tyrosine and serine side chains are colored blue or green, respectively. (Bottom) The amino acid composition of the interface. Each circle represents the surface area on the Fab or DR5 that becomes buried upon complex formation, and the colors indicate the proportion of the buried surface area contributed by each amino acid type. (c) The CDR side chains of a minimalist Fab that contact the antigen VEGF (PDB ID: 2QR0). The antigen is shown as a molecular surface with residues within 5 Å of an Fab atom colored pink. The randomized CDR loops and contact side chains are shown and colored as follows: CDR-L3 (purple), CDR-H1 (yellow), CDR-H2 (orange), CDR-H3 (red). A glycine residue in CDR-H3, which adapts a conformation that is crucial for antigen recognition, is shown as a red sphere.
In a subsequent study, diversity was simplified even further to a binary combination of tyrosine and serine, and remarkably, the resulting libraries remained highly functional and yielded specific antibodies against a number of protein antigens 48. The structure of a binary Fab in complex with the human death receptor DR5 revealed a chemically homogenous antigen-binding site dominated by tyrosine. In particular, the long CDR-H3 loop, which contributed ~40% of the antibody surface buried upon antigen binding, formed a “biphasic” helix with tyrosine and serine residues clustered on opposite faces and the tyrosine face in contact with the antigen (Figure 3B). Consequently, all of the buried surface area on CDR-H3 was contributed by tyrosine residues, which constitute more than 50% of the buried surface area overall. On the antigen side of the interface, there was no corresponding bias in chemical composition, showing that tyrosine is able to mediate productive binding interactions with a diverse of array of amino acid types. Furthermore, the tyrosine-rich antigen-binding sites were highly specific for their cognate antigens and showed no evidence of “sticky” behavior, as evidenced by excellent performance in cell-based assays 48, 49.
Binary tyrosine/serine libraries provide an ideal background against which the impact of additional diversity can be tested in a systematic manner. In particular, the role of additional chemical diversity can be precisely gauged, as exemplified by a study that explored the intrinsic contributions of glycine and arginine to molecular recognition 50. The two amino acids were added to the tyrosine/serine background and glycine was enriched in functional antibodies, suggesting that the flexibility and expanded range of backbone conformations afforded by this small residue impacts favorably on molecular recognition. In contrast, arginine was depleted in functional antibodies, and somewhat surprisingly, high content of this positively charged residue was correlated with increased levels of non-specific binding. Consequently, it was concluded that glycine is a useful addition to naïve libraries while arginine is generally detrimental to naïve interactions. In the future, analogous studies can be designed to explore the contributions of other amino acids to the function of antibodies and other binding proteins.
In a practical application of this approach, additional diversity was added to the tyrosine/serine background to generate a simple yet highly functional antibody library that has proven to be a rich source of high-affinity Fabs. The so-called “library D” was constructed to be heavily biased in favor of tyrosine and serine, but additional chemical diversity was added to the CDR-H3 loops, which were also varied in length 51. In addition, limited diversity was introduced at buried residues that may influence the conformations of the CDR loops, and length diversity was introduced into the third light-chain CDR (CDR-L3). Library D was incorporated into a high-throughput pipeline for antibody generation and yielded highly functional antibodies with affinities in the single-digit nanomolar range against a panel of 14 antigens. The structure of a high affinity Fab in complex with VEGF again showed the dominance of tyrosine for mediating antigen recognition, but it also revealed that a glycine residue that does not contact the antigen is nonetheless required for the CDR-H3 loop to attain a conformation that enables high affinity binding (Figure 3C).
In subsequent studies, library D has been applied to difficult molecular recognition tasks that have pushed synthetic antibody technology into applications that are beyond the scope of typical natural antibodies. In one case, Fabs were generated to specifically recognize different cross-linked forms of ubiquitin with exquisite specificity (Figure 4A) 52, and in another, Fabs were designed to discriminate between the non-active and active conformations of caspase-1 53. Library D has also been effective against nucleic acids, which have been recalcitrant to natural antibody repertoires, and it has yielded a highly specific Fab that has been used to solve the first structure of an antibody in complex with structured RNA (Figure 4B) 54. In an exciting recent development, Fabs from library D have been used to obtain the structure of the full-length KcsA potassium channel, providing the first view of this integral membrane protein in its native form (Figure 4C) 55. Taken together, these results show that minimalist synthetic antibodies can be applied to many challenging tasks that promise to expand significantly our capacity to address important questions about mechanisms of molecular recognition in biological systems.
Figure 4. Synthetic Fabs in complex with diverse antigens.
The light and heavy chains of the Fabs are colored cyan or blue, respectively. The following structures are shown: (a) dimeric ubiquitin cross-linked through lysine-63 of one monomer (orange) and the C terminus of the other (red) (PDB ID: 3DVN), (b) a structured RNA (orange), ΔC209 P4-P6 domain derived from the Tetrahymena group I intron (PDB ID: 2R8S), (c) the full-length KcsA potassium channel from Streptomyces lividans (PDB ID: 3EFF). The four subunits of the KcsA homotetramer are shown in different shades of orange and red.
Minimalist binding proteins built on an FN3 scaffold
The general effectiveness of tyrosine/serine binary interfaces has been demonstrated in non-antibody scaffolds using the small FN3 domain, which is similar in size to a single immunoglobulin domain (Figure 2B) 35. Synthetic FN3 binding proteins that recognize targets with affinities in the low-mid nanomolar range were derived using only tyrosine/serine diversity at as few as 16 positions in just two loops. These results set an exceedingly low threshold of chemical complexity for functional interfaces.
As with synthetic antibodies, an expansion of the tyrosine/serine code was also found to be effective in the FN3 scaffold. Binders were identified from a combinatorial library that was highly enriched for tyrosine and serine but also contained seven additional amino acid types 56. Crystal structures of a model antigen, maltose binding protein (MBP), were solved in complex with an FN3 molecule from the binary tyrosine/serine library or with one from the library with expanded diversity, providing a unique opportunity to directly investigate the role of chemical diversity in a protein-protein interface (Figure 5). In both complexes, tyrosine dominated the contacts from the FN3 domain side of the interface and other amino acid types appeared to be important for conformation. The FN3 domain from the expanded library bound with higher affinity, a slower off rate and a more favorable enthalpic contribution than the one from the binary library, suggesting that additional chemical diversity endowed the interface with better shape complementarity. Notably, the FN3 domain from the expanded library was more tolerant to mutations, suggesting an additional role for amino acid diversity in maintaining evolutionary robustness.
Figure 5. Comparison of the structures of two anti-MBP FN3 domains generated from different minimalist libraries.
YS1 (PDB ID: 3CSG) and YSX1 (PDB ID: 3CSB) refer to an FN3 domain from a tyrosine/serine binary library or from a library that was biased toward tyrosine/serine but included additional amino acids, respectively. (a) Overall views of YS1 (green) and YSX1 (cyan) bound to MBP (surface representation). The two structures were superimposed using the MBP molecules. (b) Structural details of the YS1 (left) and YSX1 (right) interfaces. The FN3 backbones are shown as yellow curves, and the side chain atoms of contacting residues of FN3 are shows as sticks with carbon in green, nitrogen in blue and oxygen in red. The MBP surfaces are shown with residues within 5 Å of an FN3 atom colored pink. (c) The surface area of individual residues in the BC and FG loops of YS1 (left) or YSX1 (right) buried at the interface with MBP. In both complexes, residues in these two loops made predominant contributions to antigen contacts.
The importance of being tyrosine
The above described studies lead to an inevitable question: What physicochemical properties of tyrosine make this amino acid exceptionally effective for mediating recognition in protein-protein interfaces? Long before these studies of synthetic binding proteins unequivocally established the importance of tyrosine for molecular recognition, the favorable properties of this versatile residue in the binding sites of natural antibodies were succinctly summarized 45:
“Amphipathic amino acids could readily tolerate the change of environment from hydrophilic to hydrophobic that occurs upon antibody-antigen complex formation. Residues that are large and can participate in a wide variety of van der Waals' and electrostatic interactions would permit binding to a range of antigens. Amino acids with flexible side-chains could generate a structurally plastic region, i.e. a binding site possessing the ability to mould itself around the antigen to improve complementarity of the interacting surfaces. Hence, antibodies could bind to an array of novel antigens using a limited set of residues interspersed with more unique residues to which greater binding specificity can be attributed.”
The tyrosine side chain is indeed amphipathic, large and capable of forming nonpolar, hydrogen-bonding and cation-π interactions. While the side chain is not highly flexible, we contend that its rigidity is an advantage for achieving affinity, rather than a disadvantage, because a small loss of conformational entropy is encountered when a side chain is immobilized in the binding interface. Furthermore, the lack of flexibility likely contributes to specificity, because interfaces that are rich in rigid tyrosine residues are unlikely to accommodate non-specific interactions. Consequently, it appears that tyrosine endows binding sites with characteristics that are important for both high affinity and specificity.
The versatility of tyrosine in forming intermolecular contacts has also been exploited in surface engineering to promote protein crystallization 57. Surface-exposed, “high entropy” residues, such as glutamate and lysine, were replaced with tyrosine, alanine, histidine, serine or threonine. Tyrosine replacements produced crystals under the largest number of crystallization conditions. These results are consistent with our view that the low side chain entropy of tyrosine is advantageous for forming interaction interfaces, although it is difficult to dissect the separate contributions of reduced conformational entropy and charge removal.
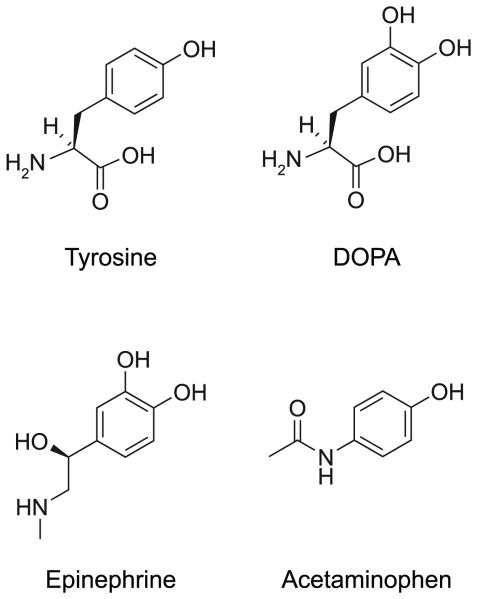
Consistent with the notion that tyrosine is particularly versatile amongst the genetically encoded amino acids for mediating molecular interactions, tyrosine-like molecules are used extensively for molecular recognition in natural systems (Figure 6). For example, proteins with high contents of dihydroxyphenylalanine (DOPA) are used by marine mussels to adhere securely to a wide variety of substrates 58, 59. Copolymers containing a high level of DOPA were found to be highly effective as reversible dry/wet adhesives, directly demonstrating the ability of DOPA to form molecular interactions with a variety of substrates 60. Furthermore, DOPA derivatives such as dopamine, epinephrine and norepinephrine act as neutotransmitters and hormones and the commonly used drug acetaminophen (Tylenol©) resembles tyrosine, suggesting that tyrosine-like molecules are predisposed to form specific interactions with protein receptors.
Figure 6.
The chemical structures of tyrosine and tyrosine-like molecules used for molecular recognition in natural systems.
In summary, we contend that the physicochemical character of tyrosine makes it the genetically encoded amino acid that is most effective for mediating molecular recognition. A particularly illustrative example in support of our contention is provided by a study in which a di-tyrosine motif in thrombin-binding synthetic proteins was converted into an active tetrapeptide 61. These results suggest that it may be possible to develop small-molecule lead compounds by transferring minimalist tyrosine-rich motifs to non-protein scaffolds. Notably, the concept of using limited residue types to mediate molecular recognition in synthetic protein interfaces parallels wisdom that has been gained in small molecule drug development. A systematic analysis of common features among known drug molecules revealed that 11,000 of 15,000 drugs are constructed using “top 20” side chains 62. These results indicate that the chemical diversity of small molecule drugs is limited, and similar to how the minimalist synthetic protein interfaces present tyrosines in distinct conformations, these drugs present similar side chains in diverse conformations using a variety of molecular frameworks 63. In both proteins and small molecules, it appears that conformational diversity trumps chemical diversity for efficient molecular recognition.
Future Perspectives
It is now evident that tyrosine-based minimalist synthetic libraries can produce highly functional recognition interfaces against a remarkable range of proteins. These findings raise the question of just how far we can push this minimalist approach. Because most of the studies to date have focused on the recognition of globular proteins, little is known about the capacity of tyrosine-based interfaces to recognize flexible peptides and nonprotein targets such as small molecules and sugars. Also, as tryptophan shares many attributes with tryosine, the question remains whether tryptophan may be able to replace tyrosine as the main contributor to interface energetics. The significant enhancement of library performance by supplementing the tyrosine/serine binary code with additional diversity suggests that we might be able to find the “ultimate” amino acid composition that balances a high content of tyrosine and tailored addition of other amino acids to produce optimal shape and electrostatic complementarity.
Clearly, further investigation is needed to answer these outstanding questions. However, synthetic protein engineering employs highly controlled library designs and selection procedures, ensuring that we will be able to gain such knowledge in a comprehensive and objective manner. Furthermore, in vitro selection systems are set up to benefit from knowledge gained from preceding experiments, and thus, synthetic protein libraries will continue to evolve. Therefore, we are optimistic that in the near future synthetic binding proteins will supersede natural antibodies for most purposes and will enable many new applications beyond the scope of natural proteins.
References
- 1.Carpenter LR, Yancopoulos GD, Stahl N. General mechanisms of cytokine receptor signaling. In: Wells JA, editor. Advances in Protein Chemistry. Vol. 52. Academic Press; London: 1999. pp. 109–131. [DOI] [PubMed] [Google Scholar]
- 2.Gascoigne NR, Zal T. Molecular interactions at the T cell-antigen-presenting cell interface. Curr. Opin. Immunol. 2004;16:114–119. doi: 10.1016/j.coi.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 3.Pawson T. Specificity in signal transduction from phosphotyrosine-SH2 domain interactions to complex cellular systems. Cell. 2004;116:191–203. doi: 10.1016/s0092-8674(03)01077-8. [DOI] [PubMed] [Google Scholar]
- 4.Cunningham BC, Wells JA. Comparison of a structural and a functional epitope. J. Mol. Biol. 1993;234:554–563. doi: 10.1006/jmbi.1993.1611. [DOI] [PubMed] [Google Scholar]
- 5.Clackson T, Wells JA. A hot spot of binding energy in a hormone-receptor interface. Science. 1995;267:383–386. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- 6.Pal G, Kouadio JL, Artis DR, Kossiakoff AA, Sidhu SS. Comprehensive and quantitative mapping of energy landscapes for protein-protein interactions by rapid combinatorial scanning. J Biol Chem. 2006;281(31):22378–85. doi: 10.1074/jbc.M603826200. [DOI] [PubMed] [Google Scholar]
- 7.Reichmann D, Rahat O, Albeck S, Meged R, Dym O, Schreiber G. The modular architecture of protein-protein binding interfaces. Proc Natl Acad Sci U S A. 2005;102(1):57–62. doi: 10.1073/pnas.0407280102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reichmann D, Rahat O, Cohen M, Neuvirth H, Schreiber G. The molecular architecture of protein-protein binding sites. Curr Opin Struct Biol. 2007;17(1):67–76. doi: 10.1016/j.sbi.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 9.Boder ET, Midelfort KS, Wittrup KD. Directed evolution of antibody fragments with monovalent femtomolar antigen-binding affinity. Proc Natl Acad Sci U S A. 2000;97(20):10701–5. doi: 10.1073/pnas.170297297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holler PD, Holman PO, Shusta EV, O'Herrin S, Wittrup KD, Kranz DM. In vitro evolution of a T cell receptor with high affinity for peptide/MHC. Proc Natl Acad Sci U S A. 2000;97(10):5387–92. doi: 10.1073/pnas.080078297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kelley RF, Totpal K, Lindstrom SH, Mathieu M, Billeci K, DeForge L, Pai R, Hymowitz SG, Ashkenazi A. Receptor-selective mutants of apoptosis-inducing ligand 2/tumor necrosis factor-related apoptosis-inducing ligand reveal a greater contribution of death receptor (DR) 5 than DR4 to apoptosis signaling. J. Biol. Chem. 2005;280:2205–2212. doi: 10.1074/jbc.M410660200. [DOI] [PubMed] [Google Scholar]
- 12.Koide A, Tereshko V, Uysal S, Margalef K, Kossiakoff AA, Koide S. Exploring the capacity of minimalist protein interfaces: interface energetics and affinity maturation to picomolar KD of a single-domain antibody with a flat paratope. J Mol Biol. 2007;373(4):941–53. doi: 10.1016/j.jmb.2007.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li B, Fuh G, Meng G, Xin X, Gerritsen ME, Cunningham B, de Vos AM. Receptor-selective variants of human vascular endothelial growth factor. Generation and characterization. J. Biol. Chem. 2000;275(38):29823–8. doi: 10.1074/jbc.M002015200. [DOI] [PubMed] [Google Scholar]
- 14.Lowman HB, Wells JA. Affinity maturation of human growth hormone by monovalent phage display. J. Mol. Biol. 1993;234:564–578. doi: 10.1006/jmbi.1993.1612. [DOI] [PubMed] [Google Scholar]
- 15.Binz HK, Pluckthun A. Engineered proteins as specific binding reagents. Curr. Opin. Biotechnol. 2005;16:459–469. doi: 10.1016/j.copbio.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 16.Sidhu SS, Fellouse FA. Synthetic therapeutic antibodies. Nature. Chem. Biol. 2006;2(12):682–688. doi: 10.1038/nchembio843. [DOI] [PubMed] [Google Scholar]
- 17.Sidhu SS, Koide S. Phage display for engineering and analyzing protein interaction interfaces. Curr Opin Struct Biol. 2007;17(4):481–7. doi: 10.1016/j.sbi.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 18.Sidhu SS, Kossiakoff AA. Exploring and designing protein function with restricted diversity. Curr. Opin. Chem. Biol. 2007;11(3):347–354. doi: 10.1016/j.cbpa.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 19.Boder ET, Wittrup KD. Yeast surface display for directed evolution of protein expression, affinity, and stability. Methods Enzymol. 2000;328:430–44. doi: 10.1016/s0076-6879(00)28410-3. [DOI] [PubMed] [Google Scholar]
- 20.Lipovsek D, Pluckthun A. In-vitro protein evolution by ribosome display and mRNA display. J Immunol Methods. 2004;290(12):51–67. doi: 10.1016/j.jim.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 21.Barbas CF, 3rd, Amberg W, Simoncsits A, Jones TM, Lerner RA. Selection of human anti-hapten antibodies from semisynthetic libraries. Gene. 1993;137(1):57–62. doi: 10.1016/0378-1119(93)90251-w. [DOI] [PubMed] [Google Scholar]
- 22.Fellouse FA, Wiesmann C, Sidhu SS. Synthetic antibodies from a four amino acid code: A dominant role for tyrosine in antigen recognition. Proc. Natl. Acad. Sci. USA. 2004 doi: 10.1073/pnas.0401786101. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Griffiths AD, Williams SC, Hartley O, Tomlinson IM, Waterhouse P, Crosby WL, Kontermann RE, Jones PT, Low NM, Allison TJ, Prospero TD, Hoogenboom HR, Nissim A, Cox JPL, Harrison JL, Zaccolo M, Gherardi E, Winter G. Isolation of high affinity human antibodies directly from large synthetic repertoires. EMBO J. 1994;13:3245–3260. doi: 10.1002/j.1460-2075.1994.tb06626.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoet RM, Cohen EH, Kent RB, Rookey K, Schoonbroodt S, Hogan S, Rem L, Frans N, Daukandt M, Pieters H, van Hegelsom R, Neer NC, Nastri HG, Rondon IJ, Leeds JA, Hufton SE, Huang L, Kashin I, Devlin M, Kuang G, Steukers M, Viswanathan M, Nixon AE, Sexton DJ, Hoogenboom HR, Ladner RC. Generation of high-affinity human antibodies by combining donor-derived and synthetic complementarity-determining-region diversity. Nature Biotechnol. 2005;23(3):344–348. doi: 10.1038/nbt1067. [DOI] [PubMed] [Google Scholar]
- 25.Lee CV, Liang WC, Dennis MS, Eigenbrot C, Sidhu SS, Fuh G. High-affinity human antibodies from phage-displayed synthetic Fab libraries with a single framework scaffold. J. Mol. Biol. 2004;340(5):1073–1093. doi: 10.1016/j.jmb.2004.05.051. [DOI] [PubMed] [Google Scholar]
- 26.Knappik A, Ge L, Honegger A, Pack P, Fischer M, Wellnhofer G, Hoess A, Wolle J, Pluckthun A, Virnekas B. Fully synthetic human combinatorial antibody libraries (HuCAL) based on modular consensus frameworks and CDRs randomized with trinucleotides. J. Mol. Biol. 2000;296:57–86. doi: 10.1006/jmbi.1999.3444. [DOI] [PubMed] [Google Scholar]
- 27.Sidhu SS, Li B, Chen Y, Fellouse FA, Eigenbrot C, Fuh G. Phage-displayed antibody libraries of synthetic heavy chain complementarity determining regions. J. Mol. Biol. 2004;338(2):299–310. doi: 10.1016/j.jmb.2004.02.050. [DOI] [PubMed] [Google Scholar]
- 28.Silacci M, Brack S, Schirru G, Marlind J, Ettorre A, Merlo A, Viti F, Neri D. Design, construction, and characterization of a large synthetic human antibody phage display library. Proteomics. 2005;5(9):2340–2350. doi: 10.1002/pmic.200401273. [DOI] [PubMed] [Google Scholar]
- 29.Jespers L, Schon O, James LC, Veprintsev D, Winter G. Crystal structure of HEL4, a soluble, refoldable human VH single domain with a germ-line scaffold. J. Mol. Biol. 2004;337:893–903. doi: 10.1016/j.jmb.2004.02.013. [DOI] [PubMed] [Google Scholar]
- 30.Hosse RJ, Rothe A, Power BE. A new generation of protein display scaffolds for molecular recognition. Protein Sci. 2006;15:14–27. doi: 10.1110/ps.051817606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mathonet P, Fastrez J. Engineering of non-natural receptors. Curr. Opin. Struct. Biol. 2004;14:505–511. doi: 10.1016/j.sbi.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 32.Almagro JC. Identification of differences in the specificity-determining residues of antibodies that recognize antigens of different size: implications for the rational design of antibody repertoires. J. Mol. Recogn. 2004;17:132–143. doi: 10.1002/jmr.659. [DOI] [PubMed] [Google Scholar]
- 33.MacCallum RM, Martin AC, Thornton JM. Antibody-antigen interactions: contact analysis and binding site topography. J Mol Biol. 1996;262(5):732–45. doi: 10.1006/jmbi.1996.0548. [DOI] [PubMed] [Google Scholar]
- 34.De Genst E, Silence K, Decanniere K, Conrath K, Loris R, Kinne J, Muyldermans S, Wyns L. Molecular basis for the preferential cleft recognition by dromedary heavy-chain antibodies. Proc. Natl. Acad. Sci. USA. 2006;103:4586–4591. doi: 10.1073/pnas.0505379103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koide A, Gilbreth RN, Esaki K, Tereshko V, Koide S. High-affinity single-domain binding proteins with a binary-code interface. Proc Natl Acad Sci U S A. 2007;104(16):6632–7. doi: 10.1073/pnas.0700149104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Binz HK, Amstutz P, Kohl A, Stumpp MT, Briand C, Forrer P, Grutter MG, Pluckthun A. High-affinity binders selected from designed ankyrin repeat protein libraries. Nature. Biotechnol. 2004;22:575–582. doi: 10.1038/nbt962. [DOI] [PubMed] [Google Scholar]
- 37.Kawe M, Forrer P, Amstutz P, Pluckthun A. Isolation of intracellular proteinase inhibitors derived from designed ankyrin repeat proteins by genetic screening. J. Biol. Chem. 2006;281:40252–40263. doi: 10.1074/jbc.M602506200. [DOI] [PubMed] [Google Scholar]
- 38.Schweizer A, Roschitzki-Voser H, Amstutz P, Briand C, Gulotti-Georgieva M, Prenosil E, Binz HK, Capitani G, Baici A, Pluckthun A. Inhibition of caspase-2 by a designed ankyrin repeat protein: specificity, structure, and inhibition mechansim. Structure. 2007;15:625–636. doi: 10.1016/j.str.2007.03.014. [DOI] [PubMed] [Google Scholar]
- 39.Korndorfer IP, Schlehuber S, Skerra A. Structural mechanism of specific ligand recognition by a lipocalin tailored for the complexation of digoxigenin. J. Mol. Biol. 2003;330:385–396. doi: 10.1016/s0022-2836(03)00573-4. [DOI] [PubMed] [Google Scholar]
- 40.Lo Conte L, Chothia C, Janin J. The atomic structure of protein-protein recognition sites. J Mol Biol. 1999;285(5):2177–98. doi: 10.1006/jmbi.1998.2439. [DOI] [PubMed] [Google Scholar]
- 41.Lawrence MC, Colman PM. Shape complementarity at protein/protein interfaces. J Mol Biol. 1993;234(4):946–50. doi: 10.1006/jmbi.1993.1648. [DOI] [PubMed] [Google Scholar]
- 42.DeLano WL. Unraveling hot spots in binding interfaces: progress and challenges. Curr Opin Struct Biol. 2002;12(1):14–20. doi: 10.1016/s0959-440x(02)00283-x. [DOI] [PubMed] [Google Scholar]
- 43.Bogan AA, Thorn KS. Anatomy of hot spots in protein interfaces. J Mol Biol. 1998;280(1):1–9. doi: 10.1006/jmbi.1998.1843. [DOI] [PubMed] [Google Scholar]
- 44.Kouadio JL, Horn JR, Pal G, Kossiakoff AA. Shotgun alanine scanning shows that growth hormone can bind productively to its receptor through a drastically minimized interface. J Biol Chem. 2005;280(27):25524–32. doi: 10.1074/jbc.M502167200. [DOI] [PubMed] [Google Scholar]
- 45.Mian IS, Bradwell AR, Olson AJ. Structure, function and properties of antibody binding sites. J. Mol. Biol. 1991;217:133–151. doi: 10.1016/0022-2836(91)90617-f. [DOI] [PubMed] [Google Scholar]
- 46.Zemlin M, Klinger M, Link J, Zemlin C, Bauer K, Engler JA, Schroeder HWJ, Kirkham PM. Expressed murine and human CDR-H3 intervals of equal length exhibit distinct repertoires that differ in their amino acid composition and predicted range of structures. J. Mol. Biol. 2003;334:733–749. doi: 10.1016/j.jmb.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 47.Villar HO, Kauvar LM. Amino acid preferences at protein binding sites. FEBS Lett. 1994;349(1):125–30. doi: 10.1016/0014-5793(94)00648-2. [DOI] [PubMed] [Google Scholar]
- 48.Fellouse FA, Bing L, Compaan DM, Peden AA, Hymowitz SG, Sidhu SS. Molecular recognition by a binary code. J. Mol. Biol. 2005;348:1153–1162. doi: 10.1016/j.jmb.2005.03.041. [DOI] [PubMed] [Google Scholar]
- 49.Fellouse FA, Barthelemy PA, Kelley RF, Sidhu SS. Tyrosine plays a dominant functional role in the paratope of a synthetic antibody derived from a four amino acid code. Journal of Molecular Biology. 2006;357(1):100–114. doi: 10.1016/j.jmb.2005.11.092. [DOI] [PubMed] [Google Scholar]
- 50.Birtalan S, Zhang Y, Fellouse FA, Shao L, Schaefer G, Sidhu SS. The intrinsic contributions of tyrosine, serine, glycine and arginine to the affinity and specificity of antibodies. J. Mol. Biol. 2008;377:1518–1528. doi: 10.1016/j.jmb.2008.01.093. [DOI] [PubMed] [Google Scholar]
- 51.Fellouse FA, Esaki K, Birtalan S, Raptis D, Cancasci VJ, Koide A, Jhurani P, Vasser M, Wiesmann C, Kosiakoff AA, Koide S, SIdhu SS. High-throughput generation of synthetic antibodies from highly functional minimalist phage-displayed libraries. J. Mol. Biol. 2007;373:924–940. doi: 10.1016/j.jmb.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 52.Newton K, Matsumoto ML, Wertz IE, Kirkpatrick DS, Lill JR, Tan J, Dugger D, Gordon N, Sidhu SS, Fellouse FA, Komuves L, French DM, Ferrando RE, Lam C, Compaan D, Yu C, Bosanac I, Hymowitz SG, Kelley RF, Dixit VM. Ubiquitin chain editing revealed by polyubiquitin linkage-specific antibodie. Cell. 2008;134:668–678. doi: 10.1016/j.cell.2008.07.039. [DOI] [PubMed] [Google Scholar]
- 53.Gao J, Sidhu SS, Wells JA. Two-state selection of conformation-specific antibodies. Proc. Natl. Acad. Sci. USA. 2009;106:3071–3076. doi: 10.1073/pnas.0812952106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ye J-D, Tereshko V, Frederiksen JK, Koide A, Fellouse FA, Sidhu SS, Koide S, Kossiakoff AA, Piccirilli JA. Synthetic antibodies for specifiction recognition and crystallization of structured RNA. Proc Natl Acad Sci USA. 2008;105:82–87. doi: 10.1073/pnas.0709082105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Uysal S, Vasquez V, Tereshko V, Esaki K, Fellouse FA, Sidhu SS, Koide K, Perozo E, Kossiakoff AA. The crystal structure of closed conformation of the full-length KcsA potassium channel. Proc Natl Acad Sci USA. 2009 In Press. [Google Scholar]
- 56.Gilbreth RN, Esaki K, Koide A, Sidhu SS, Koide S. A dominant conformational role for amino acid diversity in minimalist protein-protein interfaces. J Mol Biol. 2008;381:407–418. doi: 10.1016/j.jmb.2008.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cooper DR, Boczek T, Grelewska K, Pinkowska M, Sikorska M, Zawadzki M, Derewenda Z. Protein crystallization by surface entropy reduction: optimization of the SER strategy. Acta Crystallogr D Biol Crystallogr. 2007;63(Pt 5):636–45. doi: 10.1107/S0907444907010931. [DOI] [PubMed] [Google Scholar]
- 58.Waite JH, Tanzer ML. Polyphenolic Substance of Mytilus edulis: Novel Adhesive Containing L-Dopa and Hydroxyproline. Science. 1981;212(4498):1038–1040. doi: 10.1126/science.212.4498.1038. [DOI] [PubMed] [Google Scholar]
- 59.Papov VV, Diamond TV, Biemann K, Waite JH. Hydroxyarginine-containing polyphenolic proteins in the adhesive plaques of the marine mussel Mytilus edulis. J Biol Chem. 1995;270(34):20183–92. doi: 10.1074/jbc.270.34.20183. [DOI] [PubMed] [Google Scholar]
- 60.Lee H, Lee BP, Messersmith PB. A reversible wet/dry adhesive inspired by mussels and geckos. Nature. 2007;448(7151):338–41. doi: 10.1038/nature05968. [DOI] [PubMed] [Google Scholar]
- 61.Rajagopal S, Meyer SC, Goldman A, Zhou M, Ghosh I. A minimalist approach toward protein recognition by epitope transfer from functionally evolved beta-sheet surfaces. J Am Chem Soc. 2006;128(44):14356–63. doi: 10.1021/ja064885b. [DOI] [PubMed] [Google Scholar]
- 62.Bemis GW, Murcko MA. Properties of known drugs. 2. Side chains. J Med Chem. 1999;42(25):5095–9. doi: 10.1021/jm9903996. [DOI] [PubMed] [Google Scholar]
- 63.Bemis GW, Murcko MA. The properties of known drugs. 1. Molecular frameworks. J Med Chem. 1996;39(15):2887–93. doi: 10.1021/jm9602928. [DOI] [PubMed] [Google Scholar]