Abstract
Neurodegeneration in Parkinson's disease (PD) affects mainly dopaminergic neurons in the substantia nigra, where age-related, increasing percentages of cells lose detectable respiratory activity associated with depletion of intact mitochondrial DNA (mtDNA). Replenishment of mtDNA might improve neuronal bioenergetic function and prevent further cell death. We developed a technology (“ProtoFection”) that uses recombinant human mitochondrial transcription factor A (TFAM) engineered with an N-terminal protein transduction domain (PTD) followed by the SOD2 mitochondrial localization signal (MLS) to deliver mtDNA cargo to the mitochondria of living cells. MTD–TFAM (MTD = PTD + MLS = “mitochondrial transduction domain”) binds mtDNA and rapidly transports it across plasma membranes to mitochondria. For therapeutic proof-of-principle we tested ProtoFection technology in Parkinson's disease cybrid cells, using mtDNA generated from commercially available human genomic DNA (gDNA; Roche). Nine to 11 weeks after single exposures to MTD–TFAM + mtDNA complex, PD cybrid cells with impaired respiration and reduced mtDNA genes increased their mtDNA gene copy numbers up to 24-fold, mtDNA-derived RNAs up to 35-fold, TFAM and ETC proteins, cell respiration, and mitochondrial movement velocities. Cybrid cells with no or minimal basal mitochondrial impairments showed reduced or no responses to treatment, suggesting the possibility of therapeutic selectivity. Exposure of PD but not control cybrid cells to MTD–TFAM protein alone or MTD–TFAM + mtDNA complex increased expression of PGC-1α, suggesting activation of mitochondrial biogenesis. ProtoFection technology for mitochondrial gene therapy holds promise for improving bioenergetic function in impaired PD neurons and needs additional development to define its pharmacodynamics and delineate its molecular mechanisms. It also is unclear whether single-donor gDNA for generating mtDNA would be a preferred therapeutic compared with the pooled gDNA used in this study.
Introduction
Mitochondrial diseases comprise a diverse group of illnesses affecting mainly high-energy, postmitotic tissues (brain, retina, heart, and skeletal muscle) of children and adults. Many of these conditions arise from point mutations or deletions of mitochondrial DNA (mtDNA), a circular 16.5-kb genome that is maternally inherited and present in hundreds to thousands of copies per cell. Other mitochondrial diseases can appear from mutations in nuclear genes that contribute to proteins of the electron transport chain (ETC), ETC macrocomplex assembly, or mtDNA replication (Dimauro and Schon, 2008; Wallace, 2002, 2005).
Aging in humans is associated with bioenergetic deficiencies in the same high-energy tissues and may arise from progressive damage to mtDNA. Isolated midbrain pigmented substantia nigra neurons have age-related increases in deleted mtDNA species that can account for >80% of mtDNA content in elderly persons (Bender et al., 2006; Kraytsberg et al., 2006). Most of these mtDNA deletions are unique and have break points with direct repeat regions (Bender et al., 2006; Reeve et al., 2008), suggesting that they may arise by “skipping” of the mtDNA polymerase γ during repair of damaged mtDNA (Reeve et al., 2008). However, the exact mechanisms producing these deleted mtDNAs are not known.
Neurons with a high abundance of deleted mtDNA are more likely to lose detectable cytochrome oxidase (CO) histochemical activity (Bender et al., 2006; Kraytsberg et al., 2006) and thus be unable to engage in significant respiratory ATP synthesis. Persons dying of Parkinson's disease (PD), in addition to marked overall loss of these dopaminergic nigral neurons, show an ≈3-fold increase in abundance of CO-negative nigral neurons among the survivors (Bender et al., 2006). This finding suggests that PD pathogenesis, at least in its later stages, includes clonal expansion of uniquely deleted mtDNAs within aging neurons, leading to loss of bioenergetic capacity. Such neurons would be impaired in normal synaptic function and vulnerable to death.
Restorative therapy of bioenergetic function in neurons with mtDNA-derived loss of respiration could arise if technology existed to introduce exogenous, intact mtDNA into cells. In this paper we describe our initial results with mitochondrial gene therapy using ProtoFection (for “protein-mediated transfection”), a technology that uses recombinant mitochondrial transcription factor A (TFAM) (Garstka et al., 2003; Ekstrand et al., 2004; Kang et al., 2007) engineered to include an N-terminal, 11-arginine protein transduction domain (PTD) followed by a mitochondrial localization signal (MLS) (Khan and Bennett, 2004). We refer to the combination of PTD plus MLS as a “mitochondrial transduction domain” (MTD) and used MTD–TFAM alone or in combination with circular human mtDNA to stimulate mitochondrial physiology in a cybrid model of sporadic PD.
Our cybrid cell model of sporadic PD is based on the expression of donor platelet mtDNA in a clonal host human neural cell line (SH-SY5Y neuroblastoma) devoid of endogenous mtDNA (Swerdlow et al., 1996; Gu et al., 1998; Schapira, 1998; Ghosh et al., 1999; Swerdlow, 2007). PD cybrids show significant reduction of ETC complex I and nonsignificant reduction of complex IV catalytic activities (Swerdlow et al., 1996), can spontaneously form Lewy body inclusions (Trimmer et al., 2004), have increased oxidative stress (Swerdlow et al., 1996; Cassarino et al., 1997; Schapira et al., 1998; Kosel et al., 1999), and show respiratory impairment that correlates with reduction of fully assembled complex I and mtDNA levels (Borland et al., 2009). The underlying logic of the cybrid model is that differences among cybrid cell lines from various donors arise from expression of different donor mtDNAs. The cybrid model does not provide information about what those mtDNA differences are but can provide a platform for therapy development targeted to improvement of mitochondrial physiology.
Materials and Methods
Cybrid creation
Cybrids were created as described (Swerdlow et al., 1996) by fusing platelets from PD or control (CTL) donors to SH-SY5Y ρ0 cells that had been chronically treated with ethidium bromide to remove mtDNA. Metabolic selection by removal of pyruvate/uridine supplements promoted survival of cells with mtDNA repopulation.
Production of recombinant MTD–TFAM
MTD–TFAM was produced initially as a 6× His-tagged SUMO (small ubiquitin-related modifier) derivative to increase solubility (Iyer et al., 2009). The SUMO group was removed by treatment with SUMO protease and isolated on a nickel fast protein liquid chromatography (FPLC) column. The MTD–TFAM was treated with Benzonase to remove contaminating DNA, purified further by FPLC, and stored in Tris-buffered 50% glycerol at −20°C. Its DNA-binding capacity was assayed by electrophoretic mobility shift assay (EMSA) based on retardation of circular DNA. LabelIT plasmid (Mirus Bio, Madison, WI) or human mtDNA (250 ng; see below) was mixed with various volumes of MTD–TFAM solution and a 1 × final concentration of Expand long template buffer 3 (Roche Applied Science, Indianapolis, IN), incubated for 30 min at 37°C, and then electrophoresed to determine the amount of MTD–TFAM solution that completely retarded gel penetration of the circular DNA.
Human mitochondrial DNA
Roche human genomic DNA is manufactured from buffy coats pooled from 80 to 100 donors of both genders. Circular mitochondrial DNA (mtDNA) was prepared by treating human Roche genomic DNA with limiting amounts of Plasmid-Safe ATP-dependent DNase (Epicentre Biotechnologies, Madison, WI), which selectively digests all forms of DNA but does not affect closed circular or nicked circular double-stranded DNA. The protocols were obtained from Epicentre Biotechnologies and were suggested by the previous work of Mukai and colleagues (1973). In brief, genomic DNA was digested with limiting amounts of ATP-dependent DNase and further purified according to protocols for the UltraClean GelSpin DNA purification kit (MO BIO Laboratories, Carlsbad, CA). The DNA solution was quantified with a DNA Quant-iT assay kit (Invitrogen, Carlsbad, CA) and the linear band was visualized on a 0.8% agarose gel. Further verification was performed by real-time quantitative polymerase chain reaction (qPCR) with mitochondrial DNA markers as probes.
Treatment of cybrid cells with MTD–TFAM complexed with human mtDNA
Cells were grown to ≈70% confluency in 25-cm2 flasks and growth medium was removed. A 3.5-μg amount of freshly prepared (<24 hr) and purified human mtDNA (see above) was mixed with a 2- to 3-fold excess of MTD–TFAM (based on EMSA) and 1 × Roche Expand long template buffer 3 in a total volume of 0.4–0.5 ml, incubated for 30 min at 37°C, diluted 10-fold with Dulbecco's modified Eagle's medium (DMEM), and applied to the cybrid cells for 4 hr. The MTD–TFAM + mtDNA solution was then removed and cells were rinsed with DMEM and then placed into growth medium. Other flasks of cells were simultaneously treated with an equivalent amount of MTD–TFAM or vehicle, Roche buffer 3, and DMEM and carried forward in culture.
Real-time quantitative PCR
We used both SYBR green detection of individual double-stranded DNA PCR products and multiplex PCR simultaneous detection of up to four PCR products.
iScript reverse transcriptase, iQ SYBR Green PCR mix, and iQ Powermix were obtained from Bio-Rad (Hercules, CA). All PCR primers were designed with Beacon Designer software (PREMIER Biosoft International, Palo Alto, CA) and custom synthesized by Sigma-Genosys (The Woodlands, TX), Operon Biotechnologies (Huntsville, AL), or Invitrogen. Sequences of primers and probes are available on request (J.P.B.). Equal amounts of total RNA (1 μg) from cybrid cells were reverse transcribed simultaneously into cDNA, using the same batch of iScript and random hexamer primers. Each PCR assay for individual mitochondrial genes or D-loop was carried out in a 25-μl volume in triplicate or quadruplicate and used a full-length human mtDNA generated by PCR using primers at the BamHI site in NADH dehydrogenase subunit 6 (ND6) and the Roche Expand long template PCR mix. The substrate was Roche human genomic DNA. The purified 16.5-kb band was assayed with a Quant-iT DNA kit and converted to copy number. The qPCR conditions for mtDNA genes included activation at 95°C for 5 min followed by 50 cycles of 95°C (melting) for 10 sec and 50°C (annealing/extension) for 1 min. Primers and probes were used at 250 nM. Amplicons (λ2 kb) of the mtDNA coding region used primers A–H as described by Bannwarth and colleagues (2005, 2006). qPCR conditions for these amplicons used SYBR green detection and 25 nM primers; cycle conditions were 95°C for 3 min followed by 50 cycles of 95°C for 30 sec, 57°C for 30 sec, 72°C for 3 min, and 72°C for 1 min; after this pictures were taken. Circular mtDNA standards for qPCR analysis of amplicons from these primers were made from Roche human genomic DNA treated with Plasmid-Safe exonuclease followed by purification on MO BIO columns. Human fetal brain cDNA and Roche genomic DNA served as standards for qPCR assays of PGC (peroxisome proliferator-activated receptor [PPAR]-γ-related cofactor)-1α and 18S rRNA gene, respectively.
Surveyor nuclease assay for mtDNA mismatches
The concentration of DNA in amplicons generated with primers A–H was estimated after separation on a 0.8% electrophoresis gel and band intensities were compared, using Gel Doc (Bio-Rad), with that of the bands in a 1-kb ladder, the concentrations of which were known. About 400 ng of amplicon DNA was mixed with 2 μl of “Enhancer” and 2 μl of Surveyor nuclease and added to 0.2-ml PCR tubes on ice, which were then mixed and incubated at 42°C for 60 min. Tubes were then transferred to ice and 2 μl of stop solution was added. Controls (undigested amplicons) were run alongside their corresponding digests. DNAs were separated by automated electrophoresis (Experion; Bio-Rad) and 12K DNA chips. For analysis of heteroplasmies each lane was normalized in intensity to the DNA ladder standards for a given chip. The detection parameters were adjusted for each lane so that all visible bands were identified and no spurious lanes were called. The Experion software then identified each band as to its size and DNA concentration.
Western blot of mitochondria for ETC proteins
Total cell protein (125 μg) was loaded onto 12% Bis-Tris Criterion precast gels (Bio-Rad) and separated. The proteins were then transferred to nitrocellulose membranes, using the iBlot transfer system (Invitrogen). Complex I subunits were detected by immunoblotting with the following antibodies purchased from MitoSciences (Eugene, OR): MS111 against subunit NDUFA9 at 1.125 μg/ml, MS110 against subunit NDUFS3 at 0.5 μg/ml, MS109 against an 8-kDa subunit at 1 μg/ml, MS107 against subunit NDUFB4 at 0.5 μg/ml, and MS105 against subunit NDUFB8 followed by an IRDye 800 goat anti-mouse secondary antibody at 1:15,000 (LI-COR, Lincoln, NE). Subunits from complexes II–V were detected by immunoblotting, using a MitoProfile total OXPHOS anti-human complexes antibody cocktail detection kit (diluted 1:575; MitoSciences) followed by an IRDye 800-conjugated goat anti-mouse secondary antibody diluted 1:25,000 (LI-COR). Mitofilin was assayed as an estimate of mitochondrial mass in each sample and was detected by immunoblotting with MSM02 antibody at 2 μg/ml (purchased from MitoSciences) followed by an IRDye 800-conjugated goat anti-mouse secondary antibody (LI-COR). β-Actin was used as a loading control and was detected by immunoblotting with a polyclonal β-actin antibody purchased from Abcam (Cambridge, UK) followed by an IRDye 680-conjugated goat anti-rabbit secondary antibody diluted 1:15,000 (LI-COR). The membranes were visualized and bands were quantitated with an Odyssey infrared imaging system (LI-COR).
Measurement of respiration of intact cybrid cells
PD and CTL undifferentiated cybrid cells were grown to 80–90% confluence and harvested with trypsin. Aliquots of 2 to 5 million cells/ml in serum-free, high-glucose DMEM were studied in an Oxygraph II respirometer (Oroboros Instruments, Innsbruck, Austria) in the intact state, using a “high-resolution respirometry” approach (Hutter et al., 2006). The Oxygraph instrument and software provide real-time output of both oxygen levels and oxygen consumption rates in each of the two 2.0-ml analysis chambers. After stabilization, baseline respiration rates were determined, followed by measurement of respiration in the presence of oligomycin (2 μg/ml) to inhibit ATP synthase. Pulsed 0.5-μl aliquots of 100 μM carbonyl cyanide-p-(trifluoromethoxy)phenylhydrazone (FCCP) were then added every 30 sec to promote gradual uncoupling of respiration to the maximal uncoupled state, followed by addition of 0.1 μM rotenone (final concentration) to inhibit complex I, and then by 2.5 μM antimycin A/0.5 μM myxothiazole to inhibit complex III. Aliquots of cells were saved for measurement of mitochondrial mass on the basis of cardiolipin external standards, using MitoTracker green (MTG) dye (Invitrogen). Dilutions of cell suspensions were incubated for 120 min at 37°C in 32 nM MTG/0.1% Triton X-100/PBS and then read on a Tecan plate reader (excitation, 485 nm; emission, 535 nm; Tecan Group, Maennedorf, Switzerland). Cardiolipin external standards gave a linear response over the range of 2 to 10 μg/ml. Respiration rates under each condition were calculated on a per-million live cells and per-milligram cardiolipin basis.
Estimation of affinity of respiration for oxygen (Km)
Intact cell respiration was carried out as described previously under basal conditions (no inhibitors). Respiration was continued while chamber oxygen levels were allowed to fall to about zero. The respiration rates and oxygen levels were extracted and fit to a Michaelis–Menten equation, using GraphPad Prism (GraphPad Software, San Diego, CA), yielding apparent Km values for oxygen.
Determination of mitochondrial movement velocities
PD59 cybrid was differentiated into nondividing neurons and mitochondrial movement velocities determined as described by Trimmer and Borland (2005). Statistical comparison among movement velocities of individual mitochondria was done by nonparametric analysis of variance (ANOVA) (Kruskal–Wallis) followed by post-hoc Dunn's test, because the distribution of data for basal PD59 was not normally distributed.
Results
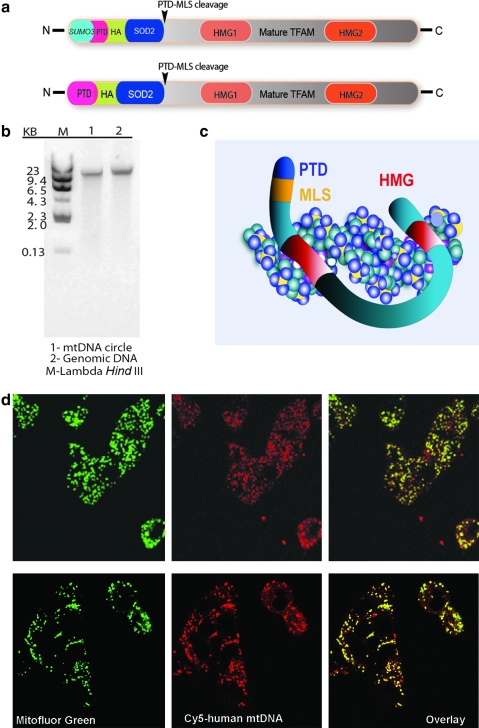
The structure of MTD–TFAM we used is shown in Fig. 1a. It is similar conceptually but different in detail from the original structure we proposed earlier (Khan and Bennett, 2004). In that original structure no SUMO group was used during synthesis, and a malate dehydrogenase mitochondrial localization signal was used instead of the currently used superoxide dismutase-2 (SOD2). Incubation of MTD–TFAM with labeled human mtDNA led to rapid (within ≈40 min) and persistent (up to ≈6 hr) colocalization of the mtDNA with the mitochondrial compartment of SH-SY5Y cybrids (Fig. 1d), presumably by binding to mtDNA with its high-mobility group (HMG) domains (Fig. 1c).
FIG. 1.
(a) Structure of recombinant MTD–TFAM when it is initially produced with the N-terminal (6 × His) SUMO group (top) and after removal of the SUMO group with SUMO protease (bottom). PTD, 11-arginine protein transduction domain; HA, hemagglutinin epitope; SOD2, mitochondrial matrix localization signal (MLS) of superoxide dismutase-2; HMG, high mobility group domains; TFAM, mitochondrial transcription factor A. Arrowheads indicate location of cleavage by mitochondrial endopeptidase of MLS after importation. (b) Agarose gel electrophoresis image of Roche human genomic DNA before (lane 2) and after (lane 1) incubation with Plasmid Safe ATP-dependent exonuclease. A 200-ng amount of DNA was added to each lane. (c) Diagram of proposed binding of MTD–TFAM to mtDNA. (d) Uptake of Cy5-labeled human mtDNA like that shown in (b) after binding to MTD–TFAM and incubation with SH-SY5Y cybrid cells made from platelets of a patient carrying a high abundance of the G11778A mutation causing Leber's hereditary optic neuropathy (LHON). Mitochondria are labeled with MitoFluor green. Top: Image was acquired after about 40 min. Bottom: Image was acquired after about 240 min of incubation with MTD–TFAM–Cy5 mtDNA complex. Color images available online at www.liebertonline.com/hum.
The demographics of subjects donating platelets to create the cybrids used in this study are shown in Supplementary Table 1 (see www.liebertonline.com/hum). All of the PD subjects had sporadically occurring disease, and we selected for this study several PD cybrid lines that spontaneously form Lewy bodies (Trimmer et al., 2004), cytoplasmic inclusions essential for PD pathological diagnosis. One PD subject (PD59) had advanced disease at the time of platelet donation and yielded a cybrid line with the most impaired respiration of all cybrid lines we have created.
We treated six cybrid lines (four PD, two CTL) with MTD–TFAM, MTD–TFAM complexed with circular human mtDNA (generated by exonuclease treatment of commercial human genomic DNA; MTD–TFAM + mtDNA) (Fig. 1b), or buffer vehicle control solutions. Cells from the three treatment arms of each cybrid line were carried forward in parallel in culture and examined at later time points.
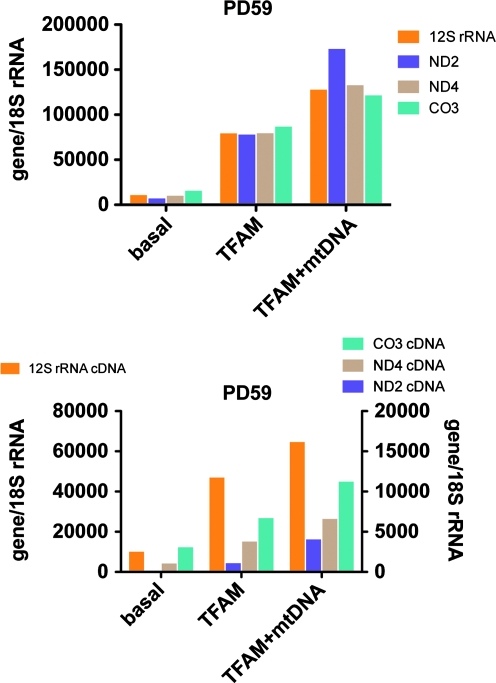
We assayed by multiplex qPCR the levels of four mtDNA genes (12S rRNA, ND2, CO3, and ND4) in genomic DNA samples (see Supplementary Fig. 1 at www.liebertonline.com/hum), and the cDNAs (see Supplementary Fig. 2 at www.liebertonline.com/hum) from these same four mtDNA genes created from their respective total RNA samples isolated from cybrid lines 9–11 weeks after a single 4-hr exposure to MTD–TFAM, MTD–TFAM complexed with mtDNA, or vehicle controls. All PD cybrid lines showed trends toward increased levels of mtDNA gene copy numbers after exposure to MTD–TFAM + mtDNA, with lesser responses to MTD–TFAM alone. For cybrid cell line PD59, which showed the lowest basal level of mtDNA genes, we found a 24-fold increase in ND2 copy number after treatment with MTD–TFAM complexed with mtDNA and a 14-fold increase after exposure to MTD–TFAM alone (Fig. 2). Neither CTL cybrid line showed increases in mtDNA gene copy numbers after MTD–TFAM or MTD–TFAM + mtDNA treatment (Supplementary Fig. 1). The mitochondrial genome cDNAs followed similar trends, except for PD66, where the cDNAs were not increased after treatment. For PD59 ND2 was also increased the most, 35-fold after MTD–TFAM + mtDNA and 10-fold after MTD–TFAM only (Fig. 2).
FIG. 2.
Mitochondrial DNA genes and gene expression in PD59 cybrid, 10 weeks after a single treatment with MTD–TFAM or MTD–TFAM complexed with mtDNA. Top: mtDNA gene copy numbers normalized to 18S rRNA gene levels for basal conditions and after treatment with MTD–TFAM alone or with MTD–TFAM complexed with human mtDNA. Total genomic DNA was analyzed by qPCR. Bottom: Same as above, except showing gene expression data from cDNA generated from total RNA samples. Data are normalized to levels of 18S rRNA. Color images available online at www.liebertonline.com/hum.
These occasionally dramatic increases in mitochondrial gene copy numbers and gene expression levels were reflected in changes in multiple electron transport chain (ETC) proteins and mitofilin, a marker for outer mitochondrial membrane, assayed by Western blots. In five of six cybrid lines mitofilin levels were variably increased from ≈30 to 100% by treatment with either MTD–TFAM or MTD–TFAM + mtDNA (see Supplementary Fig. 3 at www.liebertonline.com/hum). Complex I subunits increased with both treatments, sometimes more than 20-fold, mainly in cybrids PD59, PD61, and PD63 (Supplementary Fig. 3). PD59 and PD63 showed substantial baseline reductions of complex IV subunit-2 (mtDNA encoded), and in both cybrid lines this complex IV subunit was increased by single treatments with MTD–TFAM or MTD–TFAM +mtDNA (see Supplementary Fig. 4 at www.liebertonline.com/hum).
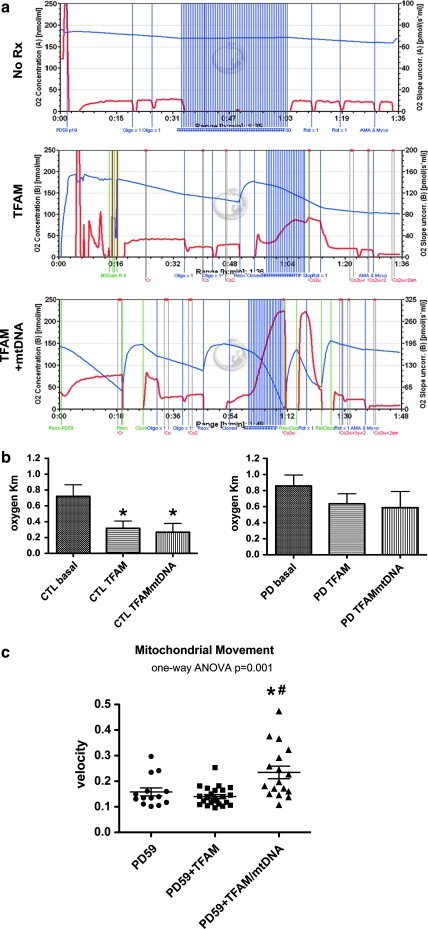
Using intact cells metabolizing glucose, we studied respiration by the “high-resolution respirometry” approach (Hutter et al., 2006), in which metabolite flux control systems are intact in contrast to typical isolated mitochondrial or permeabilized cell respiration experiments, in which ETC-specific substrates are provided. The most dramatic effect of treatment on the intact cell respiration rate was in PD59, which was functionally anaerobic at baseline with low mtDNA gene copy numbers. Ten weeks after treatment with MTD–TFAM + mtDNA the respiration rate was restored to normal. Treatment with MTD–TFAM restored respiration only to an intermediate level (Fig. 3a). Neither of the two CTL cybrid lines and two of the four PD cybrid lines did not show increased respiration rates after treatment (see Supplementary Fig. 5 at www.liebertonline.com/hum).
FIG. 3.
(a) Intact cell respiration of PD59 cybrid under basal conditions (top) and 10 weeks after a single 5-hr treatment with MTD–TFAM alone (middle) or complexed with human mtDNA (bottom). In each tracing the single blue lines show chamber oxygen levels and the red lines show respiration rates. The closely spaced vertical purple lines indicate repetitive injections of the protonophore carbonyl cyanide-p-(trifluoromethoxy)phenylhydrazone (FCCP) to disperse the proton gradient and increase respiration to its maximal uncoupled state. B, basal; o, +oligomycin; r, plus rotenone to inhibit complex I; am, plus antimycin A/myxothiazole to inhibit complex III. (b) Apparent oxygen Km values (μM) determined by curve fitting of respiration versus oxygen level during normoxic–anoxic transitions in control (CTL) and PD cybrids under basal conditions and after treatment with MTD–TFAM or MTD–TFAM + mtDNA. Shown are mean ± SEM composite results from CTL (n = 5 lines, 22 Km assays) and PD (n = 7 lines, 27 Km assays) studied under basal conditions; from CTL (n = 2 lines, 4 Km assays) and PD (n = 4 lines, 9 Km assays) after treatment with MTD–TFAM; and from CTL (2 lines, 5 Km assays) and PD (n = 4 lines, 12 Km assays) after treatment with MTD–TFAM + mtDNA. *p < 0.05 compared with basal CTL values. (c) Mitochondrial movement velocities in processes of differentiated PD59 cybrid under basal conditions and 10 weeks after a single treatment with MTD–TFAM or MTD–TFAM complexed with human mtDNA. Shown are velocities of individual mitochondria that moved. Data were analyzed by one-way, nonparametric ANOVA with post-hoc Dunn's testing. Mitochondrial movement measured in five differentiated CTL cybrid lines was 0.232 ± 0.017 (SEM) μm/sec. Mitochondrial movement velocity in differentiated SH-SY5Y was 0.21 μm/sec. The calculated velocity for baseline PD59 was 0.178 μm/sec. *p < 0.05 compared with basal movement; #p < 0.01 compared with MTD–TFAM treatment. Color images available online at www.liebertonline.com/hum.
Studies of mitochondrial respiration at room air oxygen levels may miss abnormalities that are present when respiration is studied at the lower oxygen levels typically found in tissues (Gnaiger, 2003). This has been shown in the case of respiration in cells from Leigh syndrome patients carrying SURF1 mutations that interfere with the assembly of cytochrome oxidase (Pecina et al., 2004). Using whole cell respiration, we examined the normoxic–anoxic transition to estimate the Km of the respiratory chain for oxygen (Pecina et al., 2004). We found that single treatments of both CTL and PD cybrids with MTD–TFAM ± mtDNA yielded trends toward increased apparent oxygen affinity (lowered Km values) of mitochondrial respiration (Fig. 3b).
Mitochondria are transported antero- and retrogradely in neuronal processes, and mitochondrial transport velocities are reduced in differentiated PD cybrids (P.A. Trimmer, unpublished data). We differentiated PD59 cybrid lines into nondividing neurons (Borland et al., 2008) and found that mitochondrial transport velocities in processes were reduced in the untreated and MTD–TFAM-treated cells but were restored to normal in the MTD–TFAM + mtDNA treated cybrid cells (Fig. 3c).
We sought preliminary evidence for whether the mitochondrial genome structure was altered after cells had been treated with MTD–TFAM ± mtDNA. We amplified the mtDNA coding regions, using overlapping primer pairs (pairs A–H; Bannwarth et al., 2005, 2006); subjected the resulting ≈2-kb amplicons to cleavage by Surveyor nuclease, a plant-derived, mismatch-cleaving endonuclease; and then separated the products by automated electrophoresis (Experion; Bio-Rad). In our hands this approach can detect ≈2% heteroplasmy and provides an estimate of numbers and relative abundances of individual heteroplasmic fragments. We grouped detected heteroplasmies into bins 100 bp in size and determined the percentage distribution of heteroplasmies in each of the eight ≈2-kb amplicons. These distributions for cybrid PD59 mtDNA in the basal state, and after treatments with MTD–TFAM or MTD–TFAM + mtDNA, are shown in Supplementary Fig. 6 (see www.liebertonline.com/hum), along with the distribution of heteroplasmies found in the mtDNA generated from pooled human genomic DNA that was introduced into PD59 and the other cybrids.
We found low-abundance heteroplasmies throughout the PD59 mitochondrial genome that were overall lower in the source mtDNA used. Treatment with MTD–TFAM shifted the greatest number of detectable heteroplasmies to the central ≈50% of the genome, whereas treatment with MTD–TFAM + mtDNA shifted the greatest number of detectable heteroplasmies to the distal ≈40% of the genome. This finding suggests that the substantial increase in PD59 mtDNA copy number after MTD–TFAM treatment did not result from uniform replication of all available mtDNAs.
The Surveyor nuclease approach does not identify mutations causing heteroplasmies, and thus the identities of the specific heteroplasmies shown in Supplementary Fig. 6 and their biological significance for mitochondrial function are not known. We also observed that treatment with Surveyor nuclease of all eight ≈2-kb amplicons from the three PD59 cybrid groups, but not from the source mtDNA, resulted in severalfold losses of total amplicon intensities (data not shown). This suggests that all the cybrid amplicons contain many heteroplasmies at levels lower (2%) than are detectable by our gel system. Overall, our initial exploration of low-abundance mtDNA mutations in this PD cybrid line before and after restored respiration revealed a complex result that is not easily interpreted in terms of mtDNA genotype–phenotype relationships. The properties of mtDNA used to create this cell line that led to such marked mtDNA depletion are not known. Our findings also suggest that PD mtDNA can exert an influence over its own abundance, potentially leading to situations of mtDNA depletion and respiratory impairment like that observed in some of our PD cybrid lines.
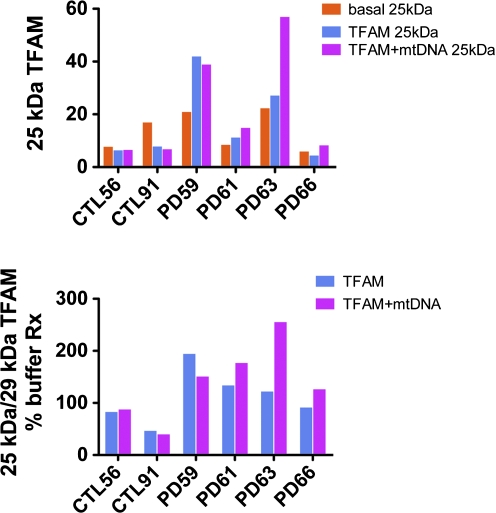
Human TFAM with its mitochondrial localization signal is either 29 or 28.4 kDa in size, depending on which methionine is used for initiation of translation (Pastukh et al., 2007). After mitochondrial importation and processing it is converted to a 204-amino acid, 24.4-kDa mature form (Pastukh et al., 2007). By Western blot analysis we assayed levels of TFAM before (≈29 kDa) and after (≈25 kDa) mitochondrial importation/processing into cybrid cells 9–11 weeks after treatment. We found in all four PD cybrids, but not in either of the two CTL cybrid lines, that MTD–TFAM and MTD–TFAM +mtDNA treatments increased the amounts of 25-kDa TFAM and the 25-kDa:29-kDa TFAM ratios (Fig. 4). This finding suggests that MTD–TFAM ± mtDNA treatments activate a mitochondrial biogenesis program that increases endogenous TFAM, mtDNA gene copy number, mtDNA gene expression, and levels of ETC proteins encoded by the nuclear genome (Scarpulla, 2006, 2008).
FIG. 4.
Effects of treatment with MTD–TFAM alone or complexed with mtDNA on cybrid cell TFAM levels. Total protein extracts from CTL and PD cybrids before and 9–11 weeks after a single treatment with MTD–TFAM or MTD–TFAM complexed with mtDNA were analyzed by Western blot for TFAM levels. Top: Levels of ≈25-kDa TFAM normalized to levels of β-actin. Bottom: 25-kDa:29-kDa TFAM ratios expressed as a percentage of baseline. Color images available online at www.liebertonline.com/hum.
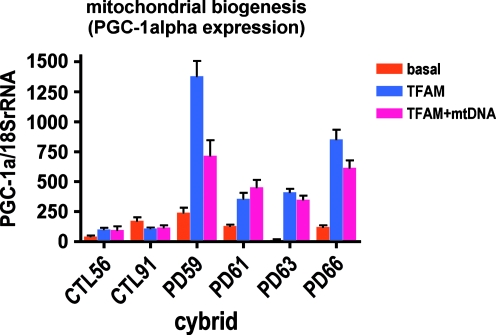
To explore further the possibility that ProtoFection technology increases mitochondrial biogenesis, we examined by qPCR the expression of PGC-1α (PPAR-γ-related cofactor-1α), a major regulator of mitochondrial biogenesis (Scarpulla, 2008). Figure 5 shows that treatment of PD but not CTL cybrids with either MTD–TFAM or MTD–TFAM complexed with mtDNA led to markedly increased PGC-1α expression 9 to 11 weeks after treatment.
FIG. 5.
Effects of treatment with MTD–TFAM alone or complexed with mtDNA on cybrid cell PGC (peroxisome proliferator-activated receptor [PPAR]-γ-related cofactor)-1α expression. cDNAs from CTL and PD cybrids before and 9–11 weeks after a single treatment with MTD–TFAM or MTD–TFAM complexed with mtDNA were analyzed by RT-qPCR for PGC-1α and results are normalized to 18S rRNA levels. Color images available online at www.liebertonline.com/hum.
Discussion
In a proof-of-principle approach, we have applied a novel protein-mediated transfection technology (“ProtoFection”) to augment mitochondrial physiology in the cybrid model of sporadic PD. We were surprised that a single incubation of cybrid cells for a few hours with MTD–TFAM protein or MTD–TFAM protein complexed with human mtDNA produced such robust long-term changes in mitochondrial physiology, including increases in mitofilin, TFAM, and multiple nuclear genome-encoded ETC proteins. Because treatment with MTD–TFAM alone or complexed with mtDNA also increased long-term PGC-1α expression, mitochondrial biogenesis appears to be activated through as yet undefined mechanisms.
Our experiments were not designed to increase knowledge of Parkinson's pathobiology, particularly regarding mtDNA. The causal involvement of mtDNA in the pathogenesis of sporadic PD at any stage remains controversial and will not be confirmed until abnormalities in mtDNAs from PD brains or neurons are shown to reproduce essential PD pathogenic events after expression in cells or animals. Unique mtDNA deletions in individual nigral neurons (Bender et al., 2006, 2008; Krishnan et al., 2008; Reeve et al., 2008) or clustered heteroplasmic mutations in PD frontal cortex homogenates (Smigrodzki et al., 2004; Parker and Parks, 2005; Smigrodzki and Khan, 2005), although suggestive of mtDNA involvement in disease pathogenesis, are restricted logically to being compelling correlations. Although our findings demonstrate that mtDNA-derived biogenic deficiencies of cybrid cells expressing platelet mtDNAs of PD patients can be substantially improved, they do not demonstrate mtDNA causality for PD pathogenesis. Our results should be viewed as correcting a bioenergetic deficiency that may or may not be involved in PD pathogenesis but likely is functionally important to the synaptic life of neurons.
There are many unanswered questions regarding mechanisms of action of MTD–TFAM + mtDNA and its potential clinical utility in PD and related diseases. These include lack of knowledge about the intramitochondrial or long-term fate of exogenous MTD–TFAM + mtDNA complexes in terms of their interactions with endogenous TFAM mtDNA in nucleoids (Wang and Bogenhagen, 2006; Holt et al., 2007; Kang et al., 2007; Kucej and Butow, 2007; Bogenhagen et al., 2008). We do not understand how imported mitochondrial (25-kDa) TFAM levels are increased for such a long time after a single treatment, nor do we understand how mitochondrially localized MTD protein activates PGC-1α expression. It seems likely that the observed long-term increases in mtDNA gene expression and resulting respiration changes derive in part from increased 25-kDa TFAM, but that remains to be demonstrated. We found variations in mitochondrial responses to MTD–TFAM ± mtDNA treatments. In general, cybrids with normal baseline respiration (referenced to parental SH-SY5Y) had no substantive alterations in respiration and minimal changes, if any, in mtDNA gene copy numbers, gene expression, or ETC complex subunit protein levels. Cybrids with substantial basal loss of mitochondrial function, such as PD59, showed robust improvements. These early findings suggest that our approach to mitochondrial gene therapy may exert some selectivity toward improving bioenergetics of impaired cells and not altering the mitochondrial physiology of more normal cells.
Finally, there are no animal models of sporadic PD that produce specific alterations of mtDNA beyond the TFAM conditional knockout model (“MitoPark”) that produces selective mtDNA depletion in and progressive death of dopaminergic nigral neurons through bioenergetic deficiency (Ekstrand et al., 2007). Prevention or reversal of degeneration of nigral neurons in the MitoPark mouse could serve as a therapeutic test of the in vivo activity of our MTD–TFAM, alone or complexed with mouse mtDNA.
We also have not shown that introduced mtDNA is specifically amplified in the cybrid cells. Because our human mtDNA was generated from buffy coat genomic DNA derived from 80 to 100 donors, tracking of haplotypes or forensic sequencing of D-loops was not feasible. Therapeutic use in humans could conceivably use mtDNA generated from commercially prepared gDNA, similar to the preparation use in our studies. Single-donor gDNA with mtDNA sequenced to eliminate mutations or deletions could serve as a therapeutic and could be ethnically matched to the recipient. It is not yet clear whether this approach will be more efficacious compared with mtDNAs derived from pooled gDNA.
In addition, our MTD–TFAM + mtDNA treatments included an ≈2-fold excess of unbound MTD–TFAM based on EMSA. Thus, results of MTD–TFAM + mtDNA treatment include some effects of MTD–TFAM treatment alone. In the case of PD59, the substantial increase in mtDNA-encoded CO subunit 2 protein after treatment with MTD–TFAM +mtDNA, not seen with MTD–TFAM treatment alone, provides compelling circumstantial evidence for expression of the introduced mtDNA.
In spite of the preceding limitations our approach offers the potential to restore mitochondrial bioenergetics to severely impaired nigral neurons in PD brain. Neurons with absent CO activity are likely not able to engage in normal, energy-requiring synaptic activities and are also vulnerable to death. In addition, MTD–TFAM appears to be a unique tool with which to explore how brief introduction of a mitochondrial transcription factor produces long-term stimulation of mitochondrial bioenergetic physiology in impaired cells but not in cells with more normal function. This observation suggests that the MTD–TFAM approach possesses therapeutic selectivity for bioenergetically impaired cells. In normal adult male mice, weekly MTD–TFAM intravenous injections increase motor endurance and respiration in mitochondrial preparations from brain, heart, and skeletal muscle (R.R. Thomas and J.P. Bennett, unpublished data), providing encouraging preliminary evidence for the functional in vivo activity of this approach.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (NS39788 [J.P.B., P.A.T.], AG023443 [J.P.B., S.M.K.], and K18DC009121 [J.P.B.]) and by the D. Loy Stewart Research Fund. The authors thank Donna Bennett for providing the illustration of MTD–TFAM binding to mtDNA (Fig. 1c). P.M.K., C.K.Q., L.D.D., C.M.P., S.I., R.R.T., K.M.S., and K.E.B. carried out experiments; P.A.T. and J.P.B. designed and carried out experiments; S.I. developed exonuclease production of human mtDNA; S.M.K. invented ProtoFection; S.M.K. and F.R.P. developed and provided plasmids for recombinant MTD–TFAM production; F.R.P. developed and assisted with MTD–TFAM synthesis and purification procedures; all authors analyzed data; J.P.B. wrote the paper.
Author Disclosure Statement
S.M.K. and F.R.P. are officers in Gencia and have a financial interest in the development of ProtoFection. The other authors state that they have no conflicts of interest.
References
- Bannwarth S. Procaccio V. Paquis-Flucklinger V. Surveyor nuclease: A new strategy for a rapid identification of heteroplasmic mitochondrial DNA mutations in patients with respiratory chain defects. Hum. Mutat. 2005;25:575–582. doi: 10.1002/humu.20177. [DOI] [PubMed] [Google Scholar]
- Bannwarth S. Procaccio V. Paquis-Flucklinger V. Rapid identification of unknown heteroplasmic mutations across the entire human mitochondrial genome with mismatch-specific Surveyor nuclease. Nat. Protoc. 2006;1:2037–2047. doi: 10.1038/nprot.2006.318. [DOI] [PubMed] [Google Scholar]
- Bender A. Krishnan K.J. Morris C.M. Taylor G.A. Reeve A.K. Perry R.H. Jaros E. Hersheson J.S. Betts J. Klopstock T. Taylor R.W. Turnbull D.M. High levels of mitochondrial DNA deletions in substantia nigra neurons in aging and Parkinson disease. Nat. Genet. 2006;38:515–517. doi: 10.1038/ng1769. [DOI] [PubMed] [Google Scholar]
- Bender A. Schwarzkopf R.M. McMillan A. Krishnan K.J. Rieder G. Neumann M. Elstner M. Turnbull D.M. Klopstock T. Dopaminergic midbrain neurons are the prime target for mitochondrial DNA deletions. J. Neurol. 2008;255:1231–1235. doi: 10.1007/s00415-008-0892-9. [DOI] [PubMed] [Google Scholar]
- Bogenhagen D.F. Rousseau D. Burke S. The layered structure of human mitochondrial DNA nucleoids. J. Biol. Chem. 2008;283:3665–3675. doi: 10.1074/jbc.M708444200. [DOI] [PubMed] [Google Scholar]
- Borland M.K. Trimmer P.A. Rubinstein J.D. Keeney P.M. Mohanakumar K.P. Liu L. Bennett J.P., Jr. Chronic, low-dose rotenone reproduces Lewy neurites found in early stages of Parkinson's disease, reduces mitochondrial movement and slowly kills differentiated SH-SY5Y neural cells. Mol. Neurodegener. 2008;3:21. doi: 10.1186/1750-1326-3-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borland M.K. Mohanakumar K.P. Rubinstein J.D. Keeney P.M. Xie J. Capaldi R. Dunham L.D. Trimmer P.A. Bennett J.P., Jr. Relationships among molecular genetic and respiratory properties of Parkinson's disease cybrid cells show similarities to Parkinson's brain tissues. Biochim. Biophys. Acta. 2009;1792:68–74. doi: 10.1016/j.bbadis.2008.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassarino D.S. Fall C.P. Swerdlow R.H. Smith T.S. Halvorsen E.M. Miller S.W. Parks J.P. Parker W.D., Jr. Bennett J.P., Jr. Elevated reactive oxygen species and antioxidant enzyme activities in animal and cellular models of Parkinson's disease. Biochim. Biophys. Acta. 1997;1362:77–86. doi: 10.1016/s0925-4439(97)00070-7. [DOI] [PubMed] [Google Scholar]
- Dimauro S. Schon E.A. Mitochondrial disorders in the nervous system. Annu. Rev. Neurosci. 2008;31:91–123. doi: 10.1146/annurev.neuro.30.051606.094302. [DOI] [PubMed] [Google Scholar]
- Ekstrand M.I. Falkenberg M. Rantanen A. Park C.B. Gaspari M. Hultenby K. Rustin P. Gustafsson C.M. Larsson N.G. Mitochondrial transcription factor A regulates mtDNA copy number in mammals. Hum. Mol. Genet. 2004;13:935–944. doi: 10.1093/hmg/ddh109. [DOI] [PubMed] [Google Scholar]
- Ekstrand M.I. Terzioglu M. Galter D. Zhu S. Hofstetter C. Lindqvist E. Thams S. Bergstrand A. Hansson F.S. Trifunovic A. Hoffer B. Cullheim S. Mohammed A.H. Olson L. Larsson N.G. Progressive parkinsonism in mice with respiratory-chain-deficient dopamine neurons. Proc. Natl. Acad. Sci. U.S.A. 2007;104:1325–1330. doi: 10.1073/pnas.0605208103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garstka H.L. Schmitt W.E. Schultz J. Sogl B. Silakowski B. Perez-Martos A. Montoya J. Wiesner R.J. Import of mitochondrial transcription factor A (TFAM) into rat liver mitochondria stimulates transcription of mitochondrial DNA. Nucleic Acids Res. 2003;31:5039–5047. doi: 10.1093/nar/gkg717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S.S. Swerdlow R.H. Miller S.W. Sheeman B. Parker W.D., Jr. Davis R.E. Use of cytoplasmic hybrid cell lines for elucidating the role of mitochondrial dysfunction in Alzheimer's disease and Parkinson's disease. Ann. N. Y. Acad. Sci. 1999;893:176–191. doi: 10.1111/j.1749-6632.1999.tb07825.x. [DOI] [PubMed] [Google Scholar]
- Gnaiger E. Oxygen conformance of cellular respiration: A perspective of mitochondrial physiology. Adv. Exp. Med. Biol. 2003;543:39–55. doi: 10.1007/978-1-4419-8997-0_4. [DOI] [PubMed] [Google Scholar]
- Gu M. Cooper J.M. Taanman J.W. Schapira A.H. Mitochondrial DNA transmission of the mitochondrial defect in Parkinson's disease. Ann. Neurol. 1998;44:177–186. doi: 10.1002/ana.410440207. [DOI] [PubMed] [Google Scholar]
- Holt I.J. He J. Mao C.C. Boyd-Kirkup J.D. Martinsson P. Sembongi H. Reyes A. Spelbrink J.N. Mammalian mitochondrial nucleoids: Organizing an independently minded genome. Mitochondrion. 2007;7:311–321. doi: 10.1016/j.mito.2007.06.004. [DOI] [PubMed] [Google Scholar]
- Hutter E. Unterluggauer H. Garedew A. Jansen-Durr P. Gnaiger E. High-resolution respirometry: A modern tool in aging research. Exp. Gerontol. 2006;41:103–109. doi: 10.1016/j.exger.2005.09.011. [DOI] [PubMed] [Google Scholar]
- Iyer S. Thomas R.R. Portell F.R. Dunham L.D. Quigley C.K. Bennett J.P., Jr. Recombinant mitochondrial transcription factor A with N-terminal mitochondrial transduction domain increases respiration and mitochondrial gene expression. Mitochondrion (in press) 2009 doi: 10.1016/j.mito.2009.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang D. Kim S.H. Hamasaki N. Mitochondrial transcription factor A (TFAM): Roles in maintenance of mtDNA and cellular functions. Mitochondrion. 2007;7:39–44. doi: 10.1016/j.mito.2006.11.017. [DOI] [PubMed] [Google Scholar]
- Khan S.M. Bennett J.P., Jr. Development of mitochondrial gene replacement therapy. J. Bioenerg. Biomembr. 2004;36:387–393. doi: 10.1023/B:JOBB.0000041773.20072.9e. [DOI] [PubMed] [Google Scholar]
- Kosel S. Hofhaus G. Maassen A. Vieregge P. Graeber M.B. Role of mitochondria in Parkinson disease. Biol. Chem. 1999;380:865–870. doi: 10.1515/BC.1999.106. [DOI] [PubMed] [Google Scholar]
- Kraytsberg Y. Kudryavtseva E. McKee A.C. Geula C. Kowall N.W. Khrapko K. Mitochondrial DNA deletions are abundant and cause functional impairment in aged human substantia nigra neurons. Nat. Genet. 2006;38:518–520. doi: 10.1038/ng1778. [DOI] [PubMed] [Google Scholar]
- Krishnan K.J. Reeve A.K. Samuels D.C. Chinnery P.F. Blackwood J.K. Taylor R.W. Wanrooij S. Spelbrink J.N. Lightowlers R.N. Turnbull D.M. What causes mitochondrial DNA deletions in human cells? Nat. Genet. 2008;40:275–279. doi: 10.1038/ng.f.94. [DOI] [PubMed] [Google Scholar]
- Kucej M. Butow R.A. Evolutionary tinkering with mitochondrial nucleoids. Trends Cell Biol. 2007;17:586–592. doi: 10.1016/j.tcb.2007.08.007. [DOI] [PubMed] [Google Scholar]
- Mukai T. Matsubara K. Takagi Y. Isolation of circular DNA molecules from whole cellular DNA by use of ATP-dependent deoxyribonuclease. Proc. Natl. Acad. Sci. U.S.A. 1973;70:2884–2887. doi: 10.1073/pnas.70.10.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker W.D., Jr. Parks J.K. Mitochondrial ND5 mutations in idiopathic Parkinson's disease. Biochem. Biophys. Res. Commun. 2005;326:667–669. doi: 10.1016/j.bbrc.2004.11.093. [DOI] [PubMed] [Google Scholar]
- Pastukh V. Shokolenko I. Wang B. Wilson G. Alexeyev M. Human mitochondrial transcription factor A possesses multiple subcellular targeting signals. FEBS J. 2007;274:6488–6499. doi: 10.1111/j.1742-4658.2007.06167.x. [DOI] [PubMed] [Google Scholar]
- Pecina P. Gnaiger E. Zeman J. Pronicka E. Houstek J. Decreased affinity for oxygen of cytochrome-c oxidase in Leigh syndrome caused by SURF1 mutations. Am. J. Physiol. Cell. Physiol. 2004;287:C1384–C1388. doi: 10.1152/ajpcell.00286.2004. [DOI] [PubMed] [Google Scholar]
- Reeve A.K. Krishnan K.J. Elson J.L. Morris C.M. Bender A. Lightowlers R.N. Turnbull D.M. Nature of mitochondrial DNA deletions in substantia nigra neurons. Am. J. Hum. Genet. 2008;82:228–235. doi: 10.1016/j.ajhg.2007.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpulla R.C. Nuclear control of respiratory gene expression in mammalian cells. J. Cell. Biochem. 2006;97:673–683. doi: 10.1002/jcb.20743. [DOI] [PubMed] [Google Scholar]
- Scarpulla R.C. Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol. Rev. 2008;88:611–638. doi: 10.1152/physrev.00025.2007. [DOI] [PubMed] [Google Scholar]
- Schapira A.H. Mitochondrial dysfunction in neurodegenerative disorders. Biochim. Biophys. Acta. 1998;1366:225–233. doi: 10.1016/s0005-2728(98)00115-7. [DOI] [PubMed] [Google Scholar]
- Schapira A.H. Gu M. Taanman J.W. Tabrizi S.J. Seaton T. Cleeter M. Cooper J.M. Mitochondria in the etiology and pathogenesis of Parkinson's disease. Ann. Neurol. 1998;44:S89–S98. doi: 10.1002/ana.410440714. [DOI] [PubMed] [Google Scholar]
- Smigrodzki R. Parks J. Parker W.D. High frequency of mitochondrial complex I mutations in Parkinson's disease and aging. Neurobiol. Aging. 2004;25:1273–1281. doi: 10.1016/j.neurobiolaging.2004.02.020. [DOI] [PubMed] [Google Scholar]
- Smigrodzki R.M. Khan S.M. Mitochondrial microheteroplasmy and a theory of aging and age-related disease. Rejuvenation Res. 2005;8:172–198. doi: 10.1089/rej.2005.8.172. [DOI] [PubMed] [Google Scholar]
- Swerdlow R.H. Mitochondria in cybrids containing mtDNA from persons with mitochondriopathies. J. Neurosci. Res. 2007;85:3416–3428. doi: 10.1002/jnr.21167. [DOI] [PubMed] [Google Scholar]
- Swerdlow R.H. Parks J.K. Miller S.W. Tuttle J.B. Trimmer P.A. Sheehan J.P. Bennett J.P., Jr. Davis R.E. Parker W.D., Jr. Origin and functional consequences of the complex I defect in Parkinson's disease. Ann. Neurol. 1996;40:663–671. doi: 10.1002/ana.410400417. [DOI] [PubMed] [Google Scholar]
- Trimmer P.A. Borland M.K. Differentiated Alzheimer's disease transmitochondrial cybrid cell lines exhibit reduced organelle movement. Antioxid. Redox Signal. 2005;7:1101–1109. doi: 10.1089/ars.2005.7.1101. [DOI] [PubMed] [Google Scholar]
- Trimmer P.A. Borland M.K. Keeney P.M. Bennett J.P., Jr. Parker W.D., Jr. Parkinson's disease transgenic mitochondrial cybrids generate Lewy inclusion bodies. J. Neurochem. 2004;88:800–812. doi: 10.1046/j.1471-4159.2003.02168.x. [DOI] [PubMed] [Google Scholar]
- Wallace D.C. Animal models for mitochondrial disease. Methods Mol. Biol. 2002;197:3–54. doi: 10.1385/1-59259-284-8:003. [DOI] [PubMed] [Google Scholar]
- Wallace D.C. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: A dawn for evolutionary medicine. Annu. Rev. Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. Bogenhagen D.F. Human mitochondrial DNA nucleoids are linked to protein folding machinery and metabolic enzymes at the mitochondrial inner membrane. J. Biol. Chem. 2006;281:25791–25802. doi: 10.1074/jbc.M604501200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.