Abstract
Although the classification of cell types often relies on the identification of cell surface proteins as differentiation markers, flow cytometry requires suitable antibodies and currently permits detection of only up to a dozen differentiation markers in a single measurement. We use multiplexed mass-spectrometric identification of several hundred N-linked glycosylation sites specifically from cell surface–exposed glycoproteins to phenotype cells without antibodies in an unbiased fashion and without a priori knowledge. Our cell surface–capturing (CSC) technology, which covalently labels extracellular glycan moieties on live cells, enables the detection and relative quantitative comparison of the cell surface N-glycoproteomes of T and B cells, as well as monitoring changes in the abundance of cell surface N-glycoprotein markers during T-cell activation and the controlled differentiation of embryonic stem cells into the neural lineage. A snapshot view of the cell surface N-glycoprotein will enable detection of panels of N-glycoproteins as potential differentiation markers that are currently not accessible by other means.
The molecular composition of the plasma membrane and its dynamic changes determine how a cell can interact with its environment. Proteins embedded in the membrane that have exposed, extracellular domains are crucial for cell-cell communication, interaction with pathogens, binding of chemical messengers and responses to environmental perturbations1,2. As cell surface proteins confer specific cellular functions and are easily accessible, they are often used as markers to classify cell types3 and as drug targets4. By using available antibodies against cell surface proteins, cells are thus often classified or immunophenotyped according to their cell-surface-protein expression profile5. This approach has been used to immunophenotype cells of the immune system, and for the development of the cluster of differentiation (CD) nomenclature for antibodies against cell surface molecules. The latter has been used to classify the ~220 currently known cell types6.
Despite these successes, most cell surface proteins remain undetectable due to a lack of suitable antibodies7. A comparison of the relatively small number of available CD cell surface protein markers (~320) with the number of predicted human transmembrane proteins (~13,000) or the 3,094 membrane glycoproteins currently annotated in the UniProt database illustrates the gap between the number of potential cell surface markers and those currently used8,9. Furthermore, owing to a lack of enabling technology, cell surface protein analysis has been limited to measuring ~12 CD molecules in parallel. Finally, the development of a new CD assay is time consuming and expensive: in each case, an optimal antibody needs to be selected, and cell surface expression for the target protein needs to be tested and validated by flow cytometry or immunohistochemistry10. Therefore, in spite of its successes, the current gold-standard approach for classifying cells by CD profiling has substantial limitations.
Currently, it is not known how many different proteins are expressed on the surface of any particular cell, and at what levels each is expressed11. In cases in which copy numbers of proteins are published, these values are often not measured from the same cell type, and are imprecise due to variations in antibody affinities. Similarly, inferring quantitative cell surface protein data from gene transcript measurements is also problematic. Transcript microarrays cannot accurately predict the quantity, nor the specific cellular location of the corresponding proteins11. Likewise, although quantitative PCR of selected mRNAs from suspected or bona fide cell surface–expressed proteins can reveal up- or downregulation of specific transcripts, it provides no information about their translation or the cellular location of the final products. Thus, new experimental approaches are required to more fully and reliably characterize cell types by means of their cell surface proteome.
Mass spectrometry (MS)–based proteomics permits sensitive, parallel identification and quantification of substantial numbers of peptides or proteins12,13. To date, the identification of the cell surface proteome by quantitative MS has been hampered by the difficulty in obtaining homogenous and highly enriched plasma membrane protein isolates, the limited relative abundance of surface membrane proteins compared with cytoskeletal or cytosolic components, and the difficulty in resolving and identifying hydrophobic proteins and peptides14,15. Commonly, analyses of the cell surface proteome are attempted by first enriching plasma membrane proteins by subcellular fractionation, with subsequent mass spectrometric analysis of these isolates16–18. Such studies typically identify a small percentage of bona fide cell surface proteins that are largely contaminated with other proteins, specifically those from intracellular membranes. The direct experimental identification of membrane surface proteins by mass spectrometry therefore remains a considerable challenge.
In an attempt to overcome the specificity problem for membrane surface protein analysis, chemical tagging strategies have been used in conjunction with subcellular fractionation. Biotinylation of lysine residues of the extracellular domains of plasma membrane proteins has been a popular choice to identify cell surface proteins19–21. Other approaches have included monitoring of selected peptides22, lectin-based methods23,24, cell surface shaving25, two phase separation26 and antibody-mediated membrane enrichment27 strategies. Although all of these methods are able to identify membrane proteins to some extent, they still lack the specificity and selectivity needed for conclusive and comprehensive analysis of the surface membrane proteome.
In 2003, we developed a novel approach for the selective isolation of N-linked glycosylation sites28 from glycoproteins in blood serum and from cellular samples. The method proved to be very specific, and the reduction of sample complexity increased sensitivity in glycoprotein identifications in complex samples. As most cell surface proteins and secreted proteins are known or predicted to be glycosylated, we sought to adapt this technology to selective identification of cell surface glycoproteins29. The cell surface capturing (CSC) technology described here enables the comprehensive and quantitative analysis of the cell surface glycoprotein landscape at very high specificity. We hope that this technology will be useful for identifying new cell surface protein targets for drug development, improving classification of cell types and enhancing our understanding of the cell surface proteome and its function.
RESULTS
Selective identification of cell surface N-glycoproteins
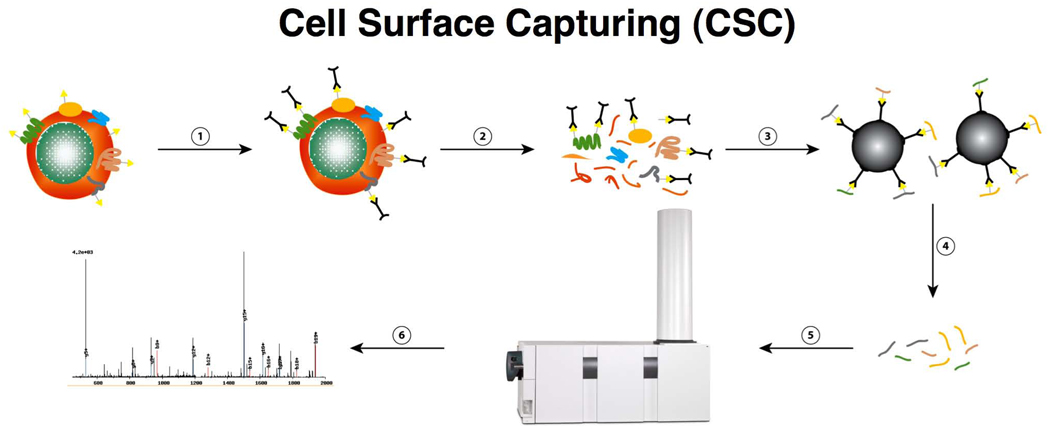
Our strategy for selective chemical tagging of cell surface glycoproteins on the intact, living cell, followed by high-affinity enrichment and gel-free LC-MS/MS analysis of peptides derived from the tagged proteins is diagrammed in Figure 1. The specific steps of this tandem affinity-labeling strategy involve (i) gentle, covalent chemical labeling of oxidized carbohydrate-containing proteins on live cells using the bi-functional linker molecule, biocytin hydrazide (BH)30,31, (ii) affinity enrichment of BH-labeled peptides, (iii) specific enzymatic peptide release that permits systematic and selective identification of N-linked glycosylation sites from the surface glycoproteins and (iv) subsequent peptide and protein identification by means of reversed phase capillary liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS). The key difference when compared with other published approaches, and which leads to the superior selectivity of the present method for cell surface proteins, is the oxidation of cell surface polysaccharides on living cells, combined with subsequent BH labeling. CSC technology is geared toward the identification of glycoproteins through the detection of tryptic peptides containing N-linked glycosylation sites from the extracellular domains of glycoproteins. The reaction conditions were carefully optimized using dose-response and time-curve experiments to both maximize cell viability, as judged by the integrity of the cells in the forward/sideward scatter in combination with propidium iodide uptake, and prevent side reactions involving the uncatalyzed oxidation agent. Harsher oxidation conditions to label specifically cell surface glycoproteins using higher concentrations of sodium periodate (>2.5 mM) led to increased cell death. Although the sensitivity to sodium periodate depends on the cell type, cell lysis prevents labeling from being restricted to the cell surface, as judged by fluorescence-activated cell sorting (FACS), immunohistochemistry and MS analysis (data not shown).
Figure 1.
CSC uses a multistep tandem affinity labeling strategy to confer the desired specificity for the glycoproteins on the cell surface. These steps involve (1) tagging reactive groups from plasma-membrane proteins (yellow triangles, glycans; black, bi-functional linker molecules), (2) cell homogenization, (3) protein digestion and affinity purification, (4) peptide release, (5) peptide analysis by LC-MS/MS and (6) peptide or protein identification.
Efficient and selective CSC labeling of glycoproteins
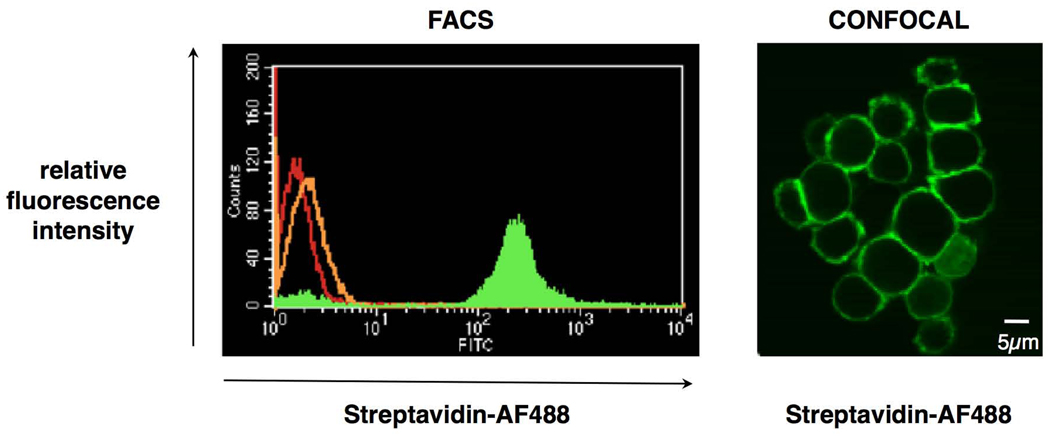
We first examined the labeling efficiency of the CSC technology using the Jurkat human T-lymphocyte cell line as a substrate. Cells were oxidized, treated with BH, and subsequently labeled with streptavidin-AF488 to visualize the degree of biotinylation and the localization of the biotin groups added. Results (Fig. 2, FACS) show homogenous fluorescent labeling. In contrast, only a slight increase in fluorescence intensity could be observed after BH treatment in the absence of oxidation of the cells, indicating very few BH-available aldehyde or keto groups on the cell surface before oxidation treatment, compared with untreated cells. The CSC labeling conditions were optimized for efficient labeling, while maintaining maximum viability of the cells.
Figure 2.
The CSC reaction is selective for glycans on the cell surface. (a) Ungated FACS histogram showing three populations of Jurkat T-lymphocytes: control cells (red), cells treated with biocytin hydrazide only (orange) and cells treated with sodium periodate and biocytin hydrazide (green). The streptavidin-AF488 fluorophore was used for detection. (b) Confocal image of Jurkat T-lymphocytes upon visualization of the tagged cell-surface polysaccharides. Cells were oxidized, labeled with biotin hydrazide and incubated with streptavidin-AF488 (green) after permeabilization.
We next investigated the specificity of the cell surface labeling. Jurkat cells were again oxidized and labeled with BH on ice to minimize endocytosis and exocytosis, then additionally permeabilized before treatment with streptavidin-AF488. Cells were then visualized using a confocal microscope (Fig. 2). The distribution of the streptavidin fluorescent dye in the central cellular stack section of labeled cells confirmed that the CSC labeling was specific for the cell surface, and that the BH reagent was not able to penetrate the cell membranes. This indicates that the CSC approach can label the cell surface of a live cell population both selectively and homogenously.
Selective identification of cell surface N-glycoproteins
We next performed a proof-of-principle CSC experiment where we labeled Jurkat cells as described above, and analyzed the recovered N-linked glycosylation sites by LC-MS/MS. The measured MS/MS spectra were searched against a human protein sequence database using SEQUEST32, and the search results were validated using the TransProteomicPipeline33. The results were then imported into a cell surface protein atlas. In this way, 110 proteins were identified from ~5 × 107 Jurkat T-lymphocytes, at a probability of ≥0.9 using ProteinProphet software34 (Supplementary Table 1 online), which, for this data set, represented a false-discovery rate of <1%. Of these, 104 proteins (95%) were CSC-labeled cell surface glycoproteins, and included 43 CD-annotated proteins, providing a broad view of the specific Jurkat T lymphocyte surface glycoprotein landscape and the corresponding N-linked glycosylation sites (visualized in Supplementary Fig. 1 online via CYTOSCAPE35). Jurkat T-lymphocytes treated with neuramidase (to eliminate terminal sialic acid residues) yielded a similar number of glycoprotein identifications as those processed using the CSC technology (Supplementary Fig. 2 and Supplementary Table 2 online). Furthermore, a control peptide N-glycosidase A (PNGaseA) treatment of bead-bound glycopeptides from human Jurkat T cells failed to identify any N-linked glycosylation sites, indicating either that PNGaseA-cleavable 3-linked fucose structures do not exist on Jurkat T cells or are rare, or that potential N-linked glycosylation sites are below the MS detection limit. The CSC technology was further applied to Drosophila melanogaster Kc167 cells, where no sialic acid can be detected36. The identification of 91 cell surface glycoproteins through 210 N-linked glycosylation sites (92% specificity for glycosylation sites) indicates that the CSC technology does not depend on sialic acid residues. (Supplementary Figs. 3 and 4 online).
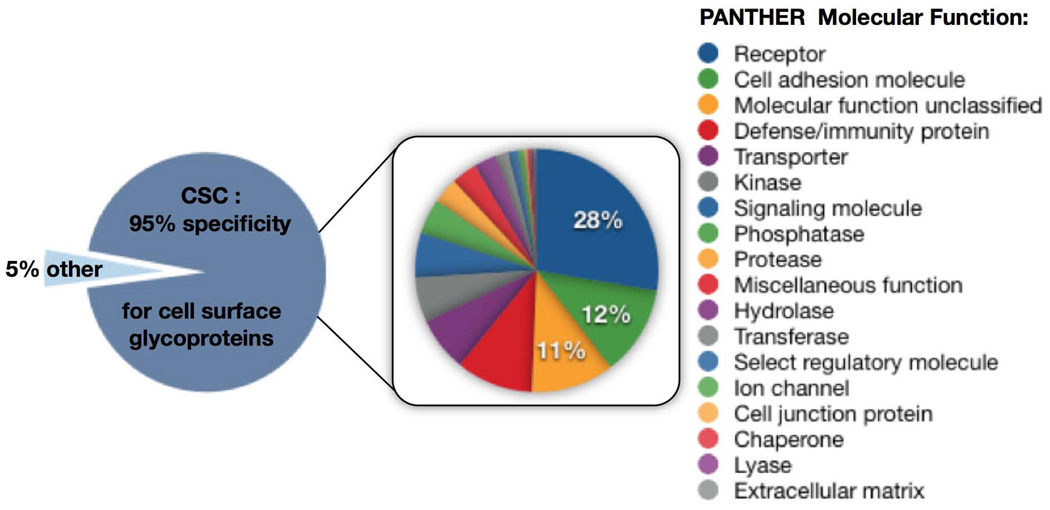
The CSC strategy proved to be very robust, and substantially more selective for MS-based identification of cell surface glycoproteins than previously published methods. The 104 CSC-identified proteins in Jurkat T cells covered a wide range of molecular functions, pathways and biological processes found in the PANTHER database37. This indicates that the CSC technology could, in a single analysis, perform a more comprehensive view of the cell surface proteome than previously possible in multiple experiments (Fig. 3, Supplementary Table 1 and Supplementary Table 3a–c online). In this data set, for example, proteins with presumably higher abundance, such as CD98, as well as lower abundance receptors, such as the G-protein–coupled receptor P2Y purinoceptor 8, were identified with confidence. Also, among the CSC-identified proteins, were receptor phosphatases such as CD45, and receptor tyrosine kinases such as TYRO3 which are important for phosphorylation-dependent signal transduction. The CSC technology also identified single-transmembrane and multitransmembrane proteins. Indeed, glycoproteins containing cell surface transmembrane domains, identified from the Jurkat T-lymphocytes, had between 1 and 25 transmembrane domains predicted by the transmembrane hidden Markov model (TMHMM) algorithm38 (Supplementary Table 1 online). We note that many of the proteins in Supplementary Table 1 would not be readily detected by immunology-based methods, because of a lack of specific antibodies.
Figure 3.
Specificity of the CSC technology and molecular functions of the proteins identified. 110 proteins were identified in a CSC experiment, including 43 CD proteins from Jurkat T-lymphocytes with a specificty of 95% for cell surface glycoproteins. 5% were identified nonspecifically. The PANTHER molecular function analysis of the 104 cell surface glycoproteins is displayed on the right.
These results showed an unprecedented degree of specificity for the detection of cell surface proteins, with <5% of identified proteins resulting from co-isolation of intracellular and nonglycosylated proteins. Indeed, five of the six contaminating proteins were identified by only a single unique peptide (Supplementary Table 1 online), and thus represent lower confidence and potential false-positive identifications. As a repository of cell surface–expressed glycoproteins does not exist for Jurkat T-lymphocytes or any other cell type, and available antibodies against cell surface proteins have never been tested on these particular cells, many of the proteins identified in this single CSC experiment represent new cell surface glycoprotein identifications for this particular cell type. Finally, as these identified glycoproteins effectively represent a ‘snapshot’, analogous to a cell surface ‘barcode’ of the Jurkat cell surface proteome under normal growth conditions, these results also highlighted the potential of the CSC technology for selective and multiplexed MS analysis of cell surface proteins under changing growth conditions, following stimulation or in various disease states.
CSC reveals cell surface topology of membrane N-glycoproteins
The CSC technology is not restricted to the identification of cell surface proteins; it also identifies the specific glycosylation sites on cell surface proteins. The 110 proteins identified in the experiment described above were inferred from 313 identified peptides (Supplementary Table 5 online). Of these, 289 (92%) were N-linked glycosylation sites containing the consensus N-glycosylation NXS/T motif (where X is any amino acid, except proline). N-linked glycosylation sites also showed a predictable fixed mass shift (of 0.984 Da) in the MS and MS/MS spectra as a result of the conversion of glycosylated asparagine to aspartic acid, by means of the enzymatic deglycosylation step. This mass shift can be observed and confirmed in a mass spectrometer with sufficient mass accuracy, and in so doing, positively identifies the N-glycosylation site(s) in the original protein (Supplementary Fig. 4). As the identified glycosylation site is expected to be on the outside of the plasma membrane, the identification of N-linked glycosylation sites can facilitate the prediction and confirmation of the transmembrane topology of cell surface proteins (data not shown). Data obtained from all our CSC experiments combined have shown that 93.7% of identified glycopeptides contain one NXS/T motif, 6.0% have two motifs and 0.3% have three. We have not identified peptides with four or more glycosylation sites. Sixty percent of identified N-linked glycosylation sites are at an NXT motif, with the remaining 40% at an NXS motif. In turn, the glycoproteins identified by CSC have between one and eight unique N-linked glycosylation sites. From these data, we conclude that CSC-identified peptides carrying the mass shift signature within the N-glycosylation motif, represent experimentally confirmed cell surface protein N-glycosylation sites.
CSC technology is applicable to primary cells, tissues and organs
As the initial evaluation of the CSC technology was with cultured cell lines, we next set out to assess the performance of the CSC technology with primary cells and tissues. A prerequisite for the CSC method is that the critical oxidation and cell surface labeling must be done with live cells in solution. Thus successful application to tissues and primary cells would first require getting viable cells into solution. As a proof-of-principle experiment toward this goal, we applied the CSC technology to the identification of cell surface proteins from mouse splenic cells; protocols for making single-cell suspensions of splenocytes from splenic tissues are well established. Upon applying the CSC method to tissue-derived primary mouse splenocytes, we identified 87 proteins at a ProteinProphet protein probability ≥0.9 (Supplementary Table 6 online) and at a false-discovery rate of <1%. Of these, 82 proteins (94%) were CSC-labeled cell surface proteins, including 38 CD-annotated proteins. These 87 proteins were identified by means of 282 peptides, of which 248 (88%) contained at least one NXS/T motif (Supplementary Table 7 online). As expected, we observed cell surface proteins that were annotated as cell-type specific. These include known T-lymphocyte (e.g., CD3δ and CD8α) and B-lymphocyte (e.g., CD22 and CD72) markers, indicating that multiple cell types were present in the primary splenocyte population. Most of the identified cell surface glycoproteins, however, could not be attributed to a specific cell type. In summary, these data show that the CSC technology is equally applicable to cultured and primary cells, as well as tissues in cases where live single-cell suspensions can be generated, such as liver or brain.
CSC technology for quantitative cell surface protein scanning
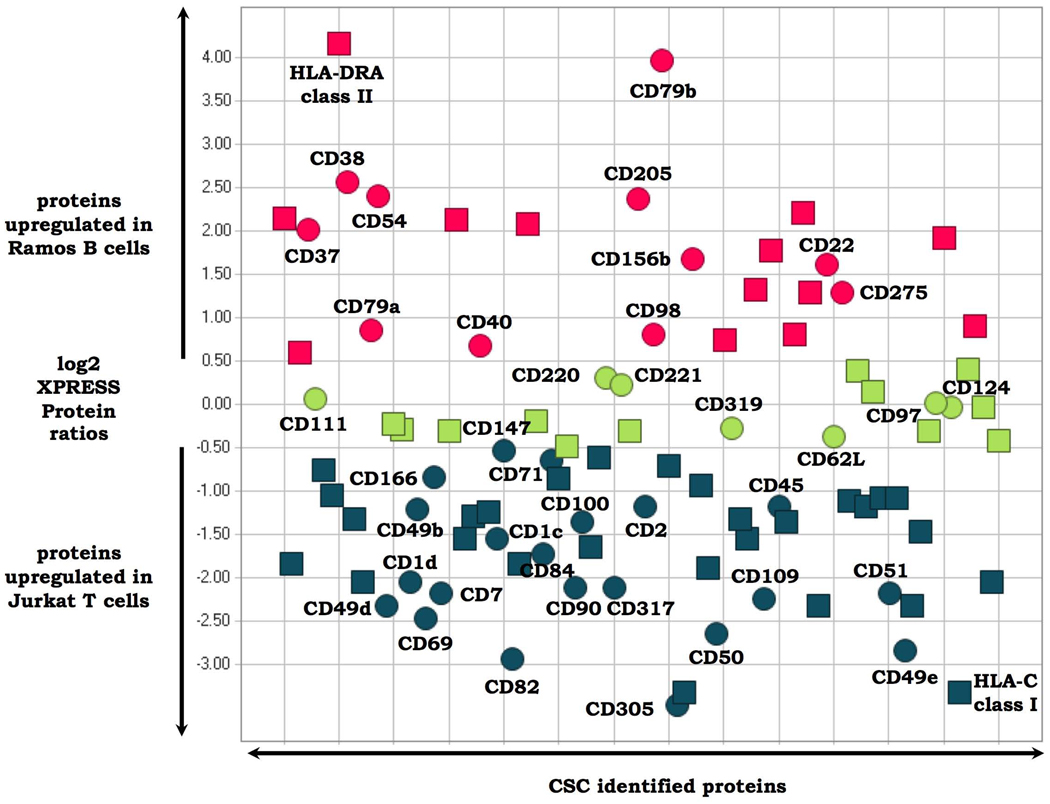
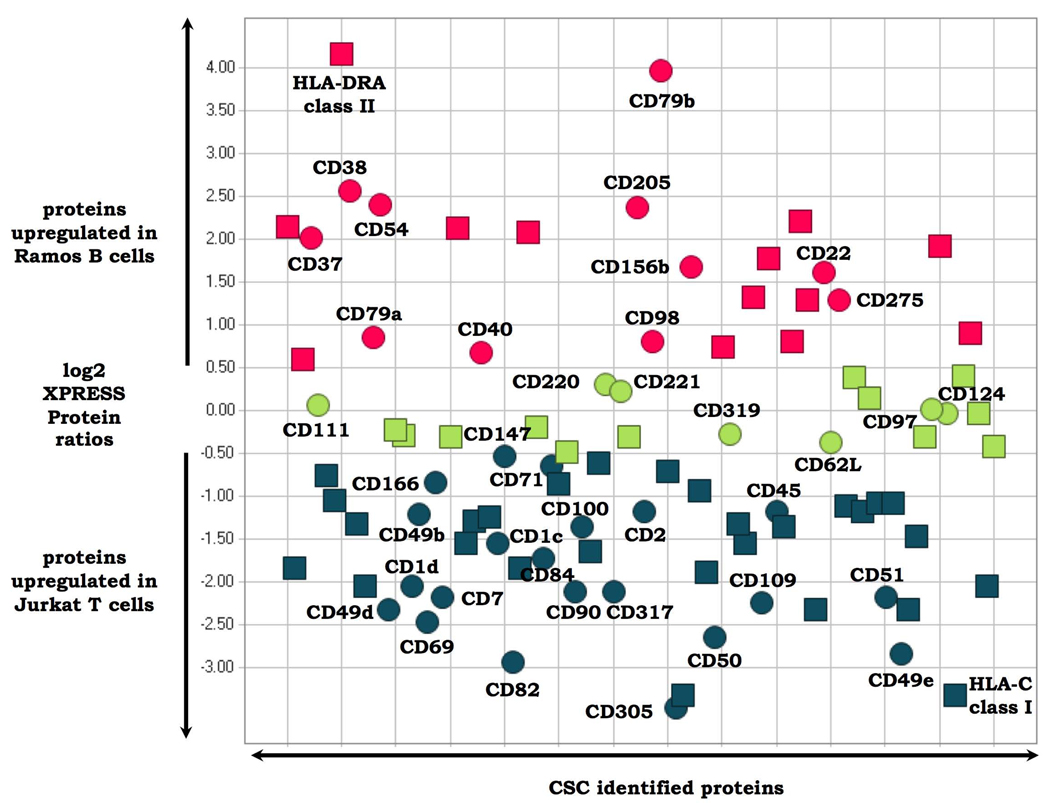
We next investigated the integration of the CSC technology with quantitative proteomic workflows to detect differences in the cell surface glycoproteome between related cell types, and upon perturbation of a specific cell type. In the first set of experiments, cultured Ramos B and human Jurkat T human lymphocytes were labeled with isotopically light and heavy SILAC reagents, respectively, and the cell surface glycoproteins were analyzed as previously described. We identified 96 proteins at a ProteinProphet protein probability of ≥0.9 and quantified (Supplementary Table 8 online). Of these, 93 (97%) were CSC-labeled cell surface glycoproteins, including 40 CD-annotated proteins. The 96 proteins were identified by means of 281 peptides, with 274 peptides (97%) containing one or more NXS/T motif (Supplementary Table 9 online). Quantitative analysis of the signal intensities of the respective isotopically labeled heavy and light peptides also indicated which glycoproteins were expressed on the surface of both cell types in similar amounts, which proteins were overrepresented on Ramos B, compared with Jurkat T-lymphocytes, and which proteins were overrepresented on the T, compared with B lymphocytes (Fig. 4). Selected CSC protein identifications are also indicated in Figure 4 to illustrate the capacity of the quantitative CSC technology to detect relative cell surface protein expression differences for known (and unknown) B- and T-cell markers. For example, CD79b is a component of the B-cell receptor that is expressed in B cells, but not in T cells39. Similarly, CD69 is a well-known T-cell activation marker40. Apart from these known markers, a number of other glycoproteins—CD and non-CD annotated—showed differential cell surface expression, indicating the different functional capacities of these two cell types (Supplementary Table 8 online). FACS experiments with a panel of selected antibodies confirmed the CSC-detected regulation of glycoproteins at the single-cell level (data not shown). The phenotype, and therefore the identity of the B- and T-cell type under investigation, can be deduced from the generated CSC data in a single experiment, without using antibodies and at a far greater level of complexity than that possible using antibodies. The data thus show that quantitative CSC technology has the capacity to classify cell types with greater detail, both in the number of markers and the range of marker proteins, than is possible by the conventional use of antibodies.
Figure 4.
Cell surface glycoproteins differentially expressed on Jurkat T versus Ramos B lymphocytes. Glycoproteins present on the cell surfaces of both cell types in similar amounts are depicted in green. Cell surface proteins which are upregulated on T cells versus B cells are depicted in blue. Cell surface proteins which are downregulated in B cells versus T cells are depicted in red. Circles indicate proteins which are CD annotated. Squares represent proteins without CD annotation. Selected CSC-identified proteins are labeled. Glycopeptides used to quantify all proteins are listed in Supplementary Table 8 and Supplementary Table 9 online.
In a second set of quantitative CSC SILAC experiments, we investigated alterations in the T-cell surface glycoproteome induced by T-cell activation41. Jurkat T-lymphocytes were subjected to co-stimulation with anti-CD3 and anti-CD28 antibodies for 24 h, the timing based on control FACS experiments showing that the expected upregulation of the T-cell activation marker CD69 was clearly evident at this time point (data not shown). In this experiment, 119 proteins were identified and quantified at a ProteinProphet protein probability of ≥0.9 (Supplementary Table 10 online). Of these, 112 proteins (94%) were CSC labeled cell surface proteins, including 47 CD-annotated proteins. The 119 proteins were identified by means of 1,035 peptide identifications, with 920 peptides (88%) containing the NXS/T motif (Supplementary Table 11 online). These results showed, for example, that the CSC SILAC experiment confirmed the strong upregulation of CD69, and downregulation of T-cell receptor (TCR) cell surface expression upon CD3/CD28 co-stimulation, as would be expected (Fig. 5)41. The data also showed additional up- and downregulation of a variety of cell surface glycoproteins (some expected and others not previously known) in response to this mode of stimulation, all in a single experiment (Supplementary Table 10 online). Indeed, the relative expression changes of 47 different CD proteins were measured simultaneously in this MS experiment. CSC experiments with Drosophila cells to assess the accuracy and reproducibility of quantifying cell surface proteome changes detectable by the CSC technology showed that 76% of the quantified features had a coefficient of variation (CV) ≤30% and the average coefficient of variance of all 1,210 aligned features was 24%. CVs between 10% and 13% for technical replicates were reported42. Together, these data demonstrated that quantitative CSC technology allows for the profiling of changes in cell surface marker proteins, upon perturbation of the cellular system, at a much more detailed level than previously possible by conventional methods for cellular phenotyping.
Figure 5.
Up- and downregulation of Jurkat T-cell proteins after costimulation with CD3/CD28 co-stimulation for 24h. Glycoproteins which are present on the cell surface before and after stimulation in similar amounts are depicted in green. Cell surface glycoproteins which are upregulated upon T-cell stimulation are depicted in red. Cell surface glycoproteins which are downregulated upon T-cell stimulation are depicted in blue. Circles indicate proteins which are CD annotated. Squares represent proteins with out Cd annotation. Selected CSC-identified proteins are labeled. Glycopeptides used to quantify allglycol proteins are listed in Supplementary Table 10 and Supplementary Table 11 online.
CSC detects cell surface changes during ES cells differentiation
In a third set of quantitative CSC experiments, we used an established mouse embryonic stem (ES) cell model system for lineage-specific differentiation43,44 to investigate alterations in the mouse ES cell-surface glycoproteome during controlled differentiation into the neural lineage. Using CSC technology in combination with label-free proteotypic peptide quantification, we followed changes in the cell surface glycoprotein landscape from the ES cell stage without feeder layers through the embryoid body stage at day 4 to the neural progenitor cell stadium at day 8 (Fig. 6)45. These experiments identified and quantified 341 cell surface glycoproteins, including 53 CD-annotated proteins, at a ProteinProphet protein probability of ≥0.9 (Supplementary Table 12 online). The glycoproteins were identified by means of 971 total glycopeptide identifications (Supplementary Table 13 online).
Figure 6.
Up- and downregulation of glycoproteins on the surfaces of ES cells during their controlled differentiation into neural progenitors. Glycoproteins which are present on the cell surface at time point 1 (ES, ES cells), time point 2 (EB, embryoid bodies) and time point 3 (NP, neural progenitors) and their relative changes upon differentiation are depicted. The color code in the boxes adjacent to the Gene IDs indicates proteotypic peptide MS1 intensities calculated by SUPERHIRN as a percentage of the most intense peptide in shades of yellow. Fading yellow color in each row indicates less surface-expression and vice versa. Identified proteins cannot be directly compared with each other in terms of intensities within columns due to the MS approach used. CD-annotated proteins are shown in bold. Selected CSC-identified proteins mentioned in the text are labeled red. Full lists of all identified and quantified glycoproteins and glycopeptides are provided in Supplementary Table 12 and Supplementary Table 13 online, respectively.
The result shows, for example, that such antibody-independent CSC experiments can both confirm the expected downregulation of the LIF receptor (CD118), and the upregulation of FGF receptor 2 (CD332) during differentiation into the neural lineage, similar to standard hypothesis-driven FACS experiments (data not shown). However, the multiplexed CSC data also reveal for the first time relative quantitative up- and downregulation of sets of cell surface glycoproteins as a systems response to differentiation on the protein level, all in a single discovery-driven CSC experiment (Supplementary Table 12 online). Indeed, the relative expression changes of 53 different CD proteins were measured at the same time in one MS experiment. Cell surface glycoproteins in all PANTHER functional categories were affected during the guided differentiation process, indicating a dramatic remodeling of the cell surface subproteome indicating new cellular functions in changing microenvironments. Together, these data demonstrated that quantitative CSC technology allows for the profiling of changes in cell surface marker proteins, upon perturbation of the cellular system, at a much more detailed level than previously possible by conventional methods for cellular phenotyping.
DISCUSSION
For most cell types, it is not known which proteins are expressed at the cell surface, and how these protein expression patterns change quantitatively upon perturbation or differentiation. For the most part, the paucity of this highly relevant information stems from a lack of specific antibodies that support the immunodetection of cell surface proteins. Such methods, including flow cytometry and immunohistochemistry, have been the mainstay of cell-surface-protein analysis to date. The generation of antibodies with validated specificity remains a major hurdle for such studies, and several factors complicate the establishment of multiplexed assays for identifying sets of cell surface proteins in a single measurement. These include variations in the affinities of available antibodies and the limited range of suitable fluorophores available for multicolor flow cytometry. Thus, it is not currently feasible to test all available antibodies against suspected cell surface proteins on one particular cell type to generate a more global view of the cell surface protein landscape and the development of immunoassays for additional proteins is slow and tedious.
We have developed a robust MS-based technology for the multiplexed identification and quantification of cell surface glycoproteins and their N-linked glycosylation sites. The CSC method provides a more comprehensive and more specific view of the cell surface protein landscape compared to prior methods, and to a large extent, circumvents the issues and limitations mentioned above. It takes advantage of the fact that glycosylation is a common distinguishing feature of most cell surface proteins. By selectively tagging cell surface polysaccharides as they are presented on the surface of live cells, we have been able to exclusively target the pool of cell surface-expressed glycoproteins for subsequent in-depth analysis, using conventional quantitative proteomic technologies.
To achieve the desired high specificity for cell surface glycopeptides, we used a gentle, multistep chemical tagging approach for covalently labeling glycoproteins by means of their extracellular glycan moieties. This chemistry works for essentially all modes of protein glycosylation. This is in contrast to the use of lectins, which isolate subsets of the glycoproteome based on their respective substrate specificity. The CSC data generated from insect cells (Supplementary Table 3 online), where sialic acid residues cannot be detected, as well as the neuraminidase experiment (Supplementary Table 2 online) confirm that the sodium periodate concentrations used within the CSC experiments enabled glycoprotein labeling and that CSC technology is not dependent on terminal silac acid residues for glycoprotein identification36,46–48. However, in the neuraminidase control experiment we could observe a slight decrease in redundant peptide observations due to less total ion current (TIC) signal intensity from the N-linked glycosylation sites. This is to be expected, as we loose the TIC signal that would result from the additional capturing and release from formerly sialylated glycopeptides. The insect cell data also show that the CSC technology enables the identification of the cell surface glycoproteome in an organism where almost no antibodies exist to detect these cell surface glycoproteins. Moreover, although affinity-based or subcellular fractionation–based methods for isolating cell surface proteins are confounded by co-isolation on noncell surface (membrane) proteins, the high specificity and selectivity obtained by CSC, through the combination of live cell labeling, avidin-affinity peptide enrichment and PNGaseF cleavage of N-glyosylation modifications, allow us to confidently assert that the identified proteins are bona fide cell surface proteins. Additionally, by using spectrometers with sufficient mass resolution to clearly distinguish between asparagine and deamidated asparagine (±0.984 Da), we could directly confirm the physiological sites of N-glycosylation on the identified peptides/proteins, also with high confidence. The CSC technology does not, however, allow us to make statements about potential N-linked glycosylation sites that were not detected.
We have also discovered some additional benefits from the high confidence we can ascribe to both the N-glycosylation sites identified and confirmation of their extracellular orientation, when using CSC. First, confirmed extracellular protein N-glycosylation sites from our CSC studies are now being tested, in combination with protein transmembrane prediction algorithms, as restrictive parameters for greatly improved prediction of the orientation of glycoproteins in the plasma membrane. Second, in silico protein-folding algorithms such as Robetta49 can similarly use information about confirmed N-linked glycosylation sites as restrictive parameters for modeling protein structure, as the N-linked glycosylation sites must be on the surface of the computed three-dimensional protein model. Lastly, identified cell surface glycopeptides can be used as target sequences for antibody generation50 again because the identified glycopeptide sequences are necessarily on the surface of the native glycoprotein, and are therefore likely accessible to soluble antibodies.
The CSC technology allows, for the first time, a more comprehensive view of the cell surface protein landscape in qualitative and quantitative terms than has previously been possible. Restricting proteomic analyses solely to the cell membrane enables us to map surface glycoproteins in an unprecedented and unbiased way. Therefore, CSC technology enables broad-based phenotyping of cells without antibodies. The glycoproteins identified represent a wide range of protein classes, which can now be observed in a rapid and multiplexed fashion, with or without perturbation or other alteration of the cell system under investigation. Without a priori knowledge, the CSC technology thus enables the identification of new and unknown proteins relevant to the system under study, as well as the identification of known proteins whose signaling involvement may not have been predicted from existing data. As shown above (Fig. 6 and (Supplementary Table 12 online).), 53 CD proteins could be monitored during ES cell differentiation into the neural lineage in one such CSC experiment, revealing a cellular systems response at the protein level during differentiation and also enabling evaluation of the pluripotent quality of the ES cells. A quantitative understanding of the proteins at the cell surface, especially receptors, is a pre-requisite for modeling the cues needed for the guided differentiation of stem cells or self-renewal.
Although these proteins could also potentially be observed with antibodies, it would take several separate analyses to screen cells with all available antibodies. In addition, the non-CD proteins also identified in the same CSC experiment may represent new cell surface protein identifications for the particular ES cell differentiation states, many of which likely cannot be otherwise measured because there are no suitable antibody reagents for cell screening. Thus, due to the limited bandwidth provided by antibody-based characterization of cell types, most such characterizations are made on the basis of only a few known cell surface markers. We think, therefore, that CSC technology will allow the identification of a wide range of cell surface proteins, representing a unique pattern for each cell type, analogous to a broader cell surface protein ‘barcode’. This barcode, which could be established from homogenous cell populations, would contain information about the predominant cell surface–expressed proteins, as well as their relative abundance. CSC technology could thus be used to compare various cell types and states, based on their unique cell surface barcodes, and thus phenotype them without the use of or initial need for antibodies. We expect that phenotyping cells in this way could be an especially useful approach in discovery-driven clinical experiments for the detection of cell surface, disease-related, differentiation markers or potential therapeutic targets.
The CSC method is a robust, enabling technology, which can be integrated and combined with various experimental workflows. It is ideally suited for multiplexed, discovery-driven identification and quantification of cell surface glycoproteins. The dynamic range of the CSC technology and of glycoproteins that can be measured in a single experiment is restricted by the currently available MS instrumentation. A limitation for the current implementation of CSC is the relatively high number of cells (~5 × 107) needed to detect the wide range of cell surface glycoproteins simultaneously by MS. Another limitation is that CSC necessarily reveals an average view of the cell surface glycoproteome over the measured cells. In contrast, antibody-based methods, such as flow cytometry or immunohistochemistry, have the sensitivity to analyze single cells. We see immunohistochemistry, FACS and CSC technology as complementary technologies on different levels which can profit from each other to reveal cell surface protein signaling landscapes and networks. With continued optimization of CSC technology, in combination with the predicted development of MS hardware and single-reaction monitoring strategies, we can expect CSC to become even more useful for smaller and smaller cell populations, though single-cell proteomics still seems rather unlikely in the near future.
Like many other new technologies, CSC opens up new questions and avenues of investigation for study. For example, how many proteins are actually expressed at the cell surface? Which of these are potential drug targets? How many different cell types, differentiable by molecular patterns, actually exist, and what new information can we glean by applying CSC technology to poorly studied cell systems? In a new avenue of investigation, cell surface glycoprotein barcodes could be measured at different time points, for example during cellular activation and differentiation, to establish quantitative data points for the modeling and prediction of cellular processes, perhaps in combination with other ‘omics technologies such as genomics and transcriptomics.
Finally, we see cell surface protein barcodes as an essential prerequisite for systems biology research. Currently, we are using CSC to establish a quantitative cell surface protein atlas. Such an atlas would represent a new systems biology resource for targeted CSC multiple-reaction monitoring experiments. We therefore hope that new CSC users will be willing to share their data, as part of a common and mutually beneficial effort to unravel cell surface signaling networks on a global scale.
METHODS
Harvesting of cells and oxidation of cell surface glycoproteins
5 × 107 cells (Jurkat T, Ramos B Drosophila Kc167, or ES cells) were collected in a 50 ml tube and washed two times with labeling buffer (PBS pH6.5, 0.1%FBS). Subsequently, cells were oxidized for ten minutes in the dark at 4 °C with 1.6 mM sodium-meta-periodate (Piercenet.com). The cell pellet was washed two times with 50 ml labeling buffer to remove residual sodium-meta-periodate and to deplete dead cells/fragments. In control experiments, cells were treated with 100 mU of Neuraminidase (Sigma, N2876) in PBS at 37 °C for 30 min to remove terminal sialic acid residues before CSC labeling.
Cell surface labeling
The cell pellet was resuspended in 10 ml labeling buffer containing 5 mM Biocytin hydrazide (Biotium.com) for 60 min at 4 °C on a rotator on slow speed. Upon labeling, the cell pellet was washed two times with 50 ml labeling buffer to remove unreacted Biocytin hydrazide and to deplete dead cells/fragments.
Cell lysis and membrane preparation
The cell pellet was resuspended in 12 ml detergent free, ice-cold, hypotonic lysis buffer (10 mM Tris pH 7.5, 0.5mM MgCl2). After ten minutes on ice, cells were homogenized with forty strokes using a Dounce homogenizer (Wheaton, USA, 15 ml Dounce Tissue Grinder). Upon homogenization, an equal volume of membrane prep buffer (280 mM sucrose, 50 mM MES pH 6, 450 mM NaCl, 10 mM MgCl2) was added to the lysate. After ten minutes the lysate was centrifuged at 2500g at 4 °C for 10 min. The homogenization procedure was repeated one more time with the pellet. The combined supernatants (the membrane fraction) was centrifuged in an Ultracentrifuge (Beckmann Ultacentrifuge L8-M Ultra; Beckmann SW41 swing rotor, 6 × 12 ml tubes; #344059; Beckmann Ultra Clear Centrifuge tubes; 12 ml) at 35,000 r.p.m. (~150000g) for 1 h at 4 °C. Upon centrifugation the pellet was incubated with 0.025M Na2CO3, (pH 11) for 30 min on ice and the ultracentrifugation step was repeated once.
Digestion of labeled membrane preparation
The pellet was resuspended in 500µl 50 mM ammonium bicarbonate (Sigma) containing 0.05% of the acid-labile surfactant RapiGest (Waters.com). The membrane preparation was indirectly sonicated in continuous mode at 100% for 10 min in a VialTweeter (hielscher.com) to obtain a translucent solution. The proteins were digested for 4 h with LysC (Calbiochem) and subsequently overnight with trypsin (Promega). Upon digestion, the peptide mixture was boiled for ten minutes to inactivate the proteases and protease inhibitors were added (Roche Complete tabs).
Glycopeptide capture
1 ml of UltraLink Streptavidin Plus beads (Piercenet.com) were washed twice with 50 mM ammonium bicarbonate in Mobicols (Bocascientific.com). The peptide mixture was added to the beads and incubated for 1 h in a MACSMix head over head shaker (Miltenyi Biotec). The captured glycopeptides were washed intensively with 0.5 Triton X-100 (Sigma) in 50 mM Ammoniumbicarbonate; followed by 10 ml 5M sodium chloride, followed by 10 ml 100 mM sodium carbonate pH11, followed by 10 ml 100 mM Ammonium bicarbonate. Washing was performed in Mobicols connected to a Vac-ManLaboratory Vacuum Manifold (Promega).
Release of N-linked glycopeptides
Washed beads were incubated in 600 µl Ammonium-bicarbonate containing PNGaseF (NEB) overnight in a head-over-head shaker at 37 °C. Upon incubation, the beads were washed once with 500 µl 50 mM Ammoniumbicarbonat and once with 20% Acetonitrile. Eluates were combined and dried in a speedvac for subsequent LC-MS/MS analysis. In control experiments, PNGaseA (SIGMA, G0535) was used according to the manufactures instructions.
LC-MS and database analysis
Peptides were re-solubilized in 0.1% formic acid and analyzed by nanospray liquid chromatography tandem mass spectrometry (LC-MS/MS). CHIP-QTOF 6530 (Agilent), LTQ and LTQ-FT mass spectrometers (Thermoelectron) were used with HP 1100/1200 solvent delivery systems (Agilent). Peptides were pressure-loaded or loaded by an autosampler on a capillary reverse-phase C18 column (75 µm i.d. and 12 cm of bed length; 200 A, 5-µm C18 beads, Michrom BioResources). Peptides were eluted from the capillary column at a flow rate of 180 nl/min to the mass spectrometer through an integrated nanospray emitter tip. Needle voltage was set to 2.0 kV. The mobile phase used for a linear gradient elution of 15–35% B in 45 min followed by 35–70% B in 15 min consisted of (A) 0.1% formic acid (B) 100% acetonitrile. Both MS and MS/MS spectra were acquired with the instrument operating in the data-dependent mode of one MS scan followed by three MS/MS scans. Ion signals above a predetermined threshold automatically triggered the instrument to switch from MS to MS/MS mode for generating collision-induced dissociation (CID) spectra. All MS/MS spectra were searched against the International Protein Index (IPI) database (Version 3.26) using the SEQUEST algorithm. Statistical analysis of the data was performed by using a combination of ISB open source software tools (PeptideProphet, ProteinProphet (http://tools.proteomecenter.org/software.php). A ProteinProphet protein probability score of at least 0.9 was used to filter the data, followed by manual validation. Quantitative data analysis was performed using the XPRESS18 and SUPERHIRN18 software18. Data were imported, stored, annotated and validated within the SISYPHUS database software. Identified proteins within the CSC data set were validated and “contaminations” were singled out in a bioinformatic process, yielding 100% bona fide cell surface labeled glycoproteins. This bioinformatic process was developed and validates the CSC protein identifications by using four criteria. [1] A ProteinProphet Protein Probability cutoff is set depending on the individual data quality of the data set, limiting the false positive protein identification rate to below 1% [2] All identified proteins are analyzed with two transmembrane prediction algorithms (SOSUI, TMHMM) indicating hydrophobic protein sequence regions. CSC proteins must have one or more transmembrane segments (exception: GPI linkage). [3] All proteins must be identified with at least one peptide containing the consensus NXS/T glycosylation motif. [4] Every identified glycopeptide must have an asparagine to aspartic acid deamidation site with a MS measured mass difference of 0.986 Da, indicating the cell surface labeling and the enzymatic deamidation during the CSC workflow. If all four criteria match we can be confident that the CSC MS identified protein was at the cell surface at the time of the CSC labeling. Please note that this statement can be made solely on the basis of the generated data in combination with bioinformatic tools without consulting public protein databases and their annotations.
FACS
Flow cytometric analysis was performed on CSC labeled lymphocytes. Cells labeled with Biocytin hydrazide were incubated with Streptavidin Alexa Fluor 488 (#S-11223, Molecular Probes) at 4 °C for 20 min. Labeled cells were washed twice and analyzed using a FACScan (BD Biosciences) in combination with CellQuest software.
Confocal imaging
To detect labeled cell surface glycoproteins, we stained cells for 20min with Streptavidin Alexa Fluor 488 at 1:100 dilution in blocking solution and washed with PBS. Upon washing, cells were transferred onto cover slides and fixed with 4% paraformaldehyde for 10 min and subsequently washed with PBS. The nucleus was counterstained for five minutes with 1 µg/ml DAPI (Merck) and washed with PBS. To detect labeled cell surface glycoproteins and potentially labeled intracellular glycoproteins, cells were permeabilized for 5 min with 0.1% saponin. Permeabilized cells were stained for 20 min with Streptavidin Alexa Fluor 488 at 1:100 dilution in blocking solution and washed with PBS. In control experiments, Neuraminidase treated and untreated cells were stained with peanut agglutinin-Rhodamine (Axxora) according to the manufacturers instructions. Cover slides were mounted with Fluoromount-G (Southern Biotechnology). Confocal laser scan microscopy images were taken with a 64 × 1.4 oil objective using a Leica TCS SP2 AOBS and filters as follows: AF488 emission 501–554nm, excitation 490nm, DAPI emission 409–478nm, excitation 365nm. Images were merged with Photoshop 6.0 and LCS Lite (Leica) software.
Cell culture
Human Ramos B cells and Jurkat T cells were cultivated in RPMI1640 supplemented with 10%FCS, 100Units of Pen/Strep and 0.1 mM β-mercaptoethanol (Invitrogen.com). SILAC labeling of cells was performed according to the manufacturer’s instruction (Invitrogen.com). CD3/CD28 co-stimulation was performed as previously described41. ES cells were cultured and differentiated into neural progenitor cells, as described43,44.
Supplementary Material
ACKNOWLEDGMENTS
This work has been funded in part with federal funds from the National Heart, Lung, and Blood Institute, National Institutes of Health (NIH), under contract no. N01-HV-28179 (to R.A.), from NIH RO1-AI51344-01 (to J.D.W.), from NIH N01-HV-28179-22 (to B.W.), from National Center of Competence in Research (NCCR) Neural Plasticity and Repair (to B.W.). Thanks to Anne-Claude Gingras and Peter Zandstra for critical reading of the manuscript; to Andreas Hofmann and Thomas Bock for supplying information and graphics support; to Alexander Schmidt for LTQ-FT performance; to Jimmy Eng, Andy Keller, Alexey Nesvizhskii, David Shteynberg, Luis Mendoza, Josh Tasman, James Eddes, Andreas Panagiotidis and Patrick Pedrioli for bioinformatic support.
Footnotes
Accession numbers. The described MS data can be downloaded in the open source mzXML format (http://www.peptideatlas.org/repository/) and was integrated into the publicly accessible PeptideAtlas database (http://www.peptideatlas.org).
AUTHOR CONTRIBUTIONS
B.W., R.A., and J.D.W. planned the project. B.W., C.H., D.B.-F. M.B. and R.O. carried out experimental work. R.S. carried out the Drosophila experiments. B.W., D.B.-F. analyzed the data. B.W., R.A. and J.D.W. wrote the paper. All authors discussed the results and commented on the manuscript.
References
- 1.von Heijne G. The membrane protein universe: what's out there and why bother? J. Intern. Med. 2007;261:543–557. doi: 10.1111/j.1365-2796.2007.01792.x. [DOI] [PubMed] [Google Scholar]
- 2.Wollscheid B, et al. Lipid raft proteins and their identification in T lymphocytes. Subcell. Biochem. 2004;37:121–152. doi: 10.1007/978-1-4757-5806-1_3. [DOI] [PubMed] [Google Scholar]
- 3.Hosen N, et al. CD96 is a leukemic stem cell-specific marker in human acute myeloid leukemia. Proc. Natl. Acad. Sci. USA. 2007;104:11008–11013. doi: 10.1073/pnas.0704271104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hopkins AL, Groom CR. The druggable genome. Nat. Rev. Drug Discov. 2002;1:727–730. doi: 10.1038/nrd892. [DOI] [PubMed] [Google Scholar]
- 5.Stewart CC, Nicholson JKA. Immunophenotyping. Wiley-Liss: New York; 2000. [Google Scholar]
- 6.Zola H, et al. CD molecules 2006–human cell differentiation molecules. J. Immunol. Methods. 2007;319:1–5. doi: 10.1016/j.jim.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 7.Zola H. Medical applications of leukocyte surface molecules–the CD molecules. Mol. Med. 2006;12:312–316. doi: 10.2119/2006-00081.Zola. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ahram M, Litou ZI, Fang R, Al-Tawallbeh G. Estimation of membrane proteins in the human proteome. Silico Biol. (Gedrukt) 2006;6:379–386. [PubMed] [Google Scholar]
- 9.Nicholson IC, Ayhan M, Hoogenraad NJ, Zola H. In silico evaluation of two mass spectrometry-based approaches for the identification of novel human leukocyte cell-surface proteins. J. Leukoc. Biol. 2005;77:190–198. doi: 10.1189/jlb.0804450. [DOI] [PubMed] [Google Scholar]
- 10.Craig FE, Foon KA. Flow cytometric immunophenotyping for hematologic neoplasms. Blood. 2008;111:3941–3967. doi: 10.1182/blood-2007-11-120535. [DOI] [PubMed] [Google Scholar]
- 11.Evans EJ, et al. The T cell surface–how well do we know it? Immunity. 2003;19:213–223. doi: 10.1016/s1074-7613(03)00198-5. [DOI] [PubMed] [Google Scholar]
- 12.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 13.Bantscheff M, Schirle M, Sweetman G, Rick J, Kuster B. Quantitative mass spectrometry in proteomics: a critical review. Anal. Bioanal. Chem. 2007;389:1017–1031. doi: 10.1007/s00216-007-1486-6. [DOI] [PubMed] [Google Scholar]
- 14.Macher BA, Yen TY. Proteins at membrane surfaces-a review of approaches. Mol. Biosyst. 2007;3:705–713. doi: 10.1039/b708581h. [DOI] [PubMed] [Google Scholar]
- 15.Josic D, Clifton JG. Mammalian plasma membrane proteomics. Proteomics. 2007;7:3010–3029. doi: 10.1002/pmic.200700139. [DOI] [PubMed] [Google Scholar]
- 16.Pasini EM, et al. In-depth analysis of the membrane and cytosolic proteome of red blood cells. Blood. 2006;108:791–801. doi: 10.1182/blood-2005-11-007799. [DOI] [PubMed] [Google Scholar]
- 17.Andersen JS, Mann M. Organellar proteomics: turning inventories into insights. EMBO Rep. 2006;7:874–879. doi: 10.1038/sj.embor.7400780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han DK, Eng JK, Zhou H, Aebersold R. Quantitative profiling of differentiation-induced microsomal proteins using isotope-coded affinity tags and mass spectrometry. Nat. Biotechnol. 2001;19:946–951. doi: 10.1038/nbt1001-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nunomura K, et al. Cell surface labeling and mass spectrometry reveal diversity of cell surface markers and signaling molecules expressed in undifferentiated mouse embryonic stem cells. Mol. Cell. Proteomics. 2005;4:1968–1976. doi: 10.1074/mcp.M500216-MCP200. [DOI] [PubMed] [Google Scholar]
- 20.Rybak JN, et al. In vivo protein biotinylation for identification of organ-specific antigens accessible from the vasculature. Nat. Methods. 2005;2:291–298. doi: 10.1038/nmeth745. [DOI] [PubMed] [Google Scholar]
- 21.Zhang W, Zhou G, Zhao Y, White MA, Zhao Y. Affinity enrichment of plasma membrane for proteomics analysis. Electrophoresis. 2003;24:2855–2863. doi: 10.1002/elps.200305569. [DOI] [PubMed] [Google Scholar]
- 22.Arnott D, et al. Selective detection of membrane proteins without antibodies: a mass spectrometric version of the Western blot. Mol. Cell. Proteomics. 2002;1:148–156. doi: 10.1074/mcp.m100027-mcp200. [DOI] [PubMed] [Google Scholar]
- 23.Lewandrowski U, Moebius J, Walter U, Sickmann A. Elucidation of N-glycosylation sites on human platelet proteins: a glycoproteomic approach. Mol. Cell. Proteomics. 2006;5:226–233. doi: 10.1074/mcp.M500324-MCP200. Medline CrossRef. [DOI] [PubMed] [Google Scholar]
- 24.Kaji H, Yamauchi Y, Takahashi N, Isobe T. Mass spectrometric identification of N-linked glycopeptides using lectin-mediated affinity capture and glycosylation site-specific stable isotope tagging. Nat. Protocols. 2006;1:3019–3027. doi: 10.1038/nprot.2006.444. [DOI] [PubMed] [Google Scholar]
- 25.Wu CC, MacCoss MJ, Howell KE, Yates JR. A method for the comprehensive proteomic analysis of membrane proteins. Nat. Biotechnol. 2003;21:532–538. doi: 10.1038/nbt819. [DOI] [PubMed] [Google Scholar]
- 26.Elortza F, et al. Proteomic analysis of glycosylphosphatidylinositol-anchored membrane proteins. Mol. Cell. Proteomics. 2003;2:1261–1270. doi: 10.1074/mcp.M300079-MCP200. [DOI] [PubMed] [Google Scholar]
- 27.Watarai H, et al. Plasma membrane-focused proteomics: dramatic changes in surface expression during the maturation of human dendritic cells. Proteomics. 2005;5:4001–4011. doi: 10.1002/pmic.200401258. [DOI] [PubMed] [Google Scholar]
- 28.Zhang H, Li XJ, Martin DB, Aebersold R. Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nat. Biotechnol. 2003;21:660–666. doi: 10.1038/nbt827. [DOI] [PubMed] [Google Scholar]
- 29.Varki A, Cummings R, Esko J. Essentials of Glycobiology. Cold Spring Harbor Press, NY: Cold Spring Harbor Laboratory Press; 2002. [PubMed] [Google Scholar]
- 30.Bayer EA, Ben-Hur H, Wilchek M. Biocytin hydrazide-a selective label for sialic acids, galactose, and other sugars in glycoconjugates using avidin-biotin technology. Anal. Biochem. 1988;170:271–281. doi: 10.1016/0003-2697(88)90631-8. [DOI] [PubMed] [Google Scholar]
- 31.Gahmberg CG, Andersson LC. Selective radioactive labeling of cell surface sialoglycoproteins by periodate-tritiated borohydride. J. Biol. Chem. 1977;252:5888–5894. [PubMed] [Google Scholar]
- 32.Yates JR, Eng JK, McCormack AL. Mining genomes: correlating tandem mass spectra of modified and unmodified peptides to sequences in nucleotide databases. Anal. Chem. 1995;67:3202–3210. doi: 10.1021/ac00114a016. [DOI] [PubMed] [Google Scholar]
- 33.Keller A, Eng J, Zhang N, Li XJ, Aebersold R. A uniform proteomics MS/MS analysis platform utilizing open XML file formats. Mol. Syst. Biol. 2005;1 doi: 10.1038/msb4100024. 2005.0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 35.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.North SJ, et al. Glycomic studies of Drosophila melanogaster embryos. Glycoconj. J. 2006;23:345–354. doi: 10.1007/s10719-006-6693-4. [DOI] [PubMed] [Google Scholar]
- 37.Thomas PD, et al. PANTHER: a browsable database of gene products organized by biological function, using curated protein family and subfamily classification. Nucleic Acids Res. 2003;31:334–341. doi: 10.1093/nar/gkg115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 39.Schamel WW, Reth M. Monomeric and oligomeric complexes of the B cell antigen receptor. Immunity. 2000;13:5–14. doi: 10.1016/s1074-7613(00)00003-0. [DOI] [PubMed] [Google Scholar]
- 40.Sancho D, Gomez M, Sanchez-Madrid F. CD69 is an immunoregulatory molecule induced following activation. Trends Immunol. 2005;26:136–140. doi: 10.1016/j.it.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 41.Kim JE, White FM. Quantitative analysis of phosphotyrosine signaling networks triggered by CD3 and CD28 costimulation in Jurkat cells. J. Immunol. 2006;176:2833–2843. doi: 10.4049/jimmunol.176.5.2833. [DOI] [PubMed] [Google Scholar]
- 42.Schiess R, et al. Analysis of cell surface proteome changes via label-free, quantitative mass spectrometry. Mol. Cell Proteomics. doi: 10.1074/mcp.M800172-MCP200. (2008 MCP published November 25, 2008, 10.1074/mcp.M800172-MCP 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bibel M, et al. Differentiation of mouse embryonic stem cells into a defined neuronal lineage. Nat. Neurosci. 2004;7:1003–1009. doi: 10.1038/nn1301. [DOI] [PubMed] [Google Scholar]
- 44.Bibel M, Richter J, Lacroix E, Barde Y. Generation of a defined and uniform population of CNS progenitors and neurons from mouse embryonic stem cells. Nat. Protocols. 2007;2:1034–1043. doi: 10.1038/nprot.2007.147. [DOI] [PubMed] [Google Scholar]
- 45.Mueller LN, et al. SuperHirn - a novel tool for high resolution LC-MS-based peptide/protein profiling. Proteomics. 2007;7:3470–3480. doi: 10.1002/pmic.200700057. [DOI] [PubMed] [Google Scholar]
- 46.Jarvis DL. Developing baculovirus-insect cell expression systems for humanized recombinant glycoprotein production. Virology. 2003;310(1–7) doi: 10.1016/s0042-6822(03)00120-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aumiller JJ, Hollister JR, Jarvis DL. A transgenic insect cell line engineered to produce CMP-sialic acid and sialylated glycoproteins. Glycobiology. 2003;13:497–507. doi: 10.1093/glycob/cwg051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Viswanathan K, et al. Engineering sialic acid synthetic ability into insect cells: identifying metabolic bottlenecks and devising strategies to overcome them. Biochemistry. 2003;42:15215–15225. doi: 10.1021/bi034994s. [DOI] [PubMed] [Google Scholar]
- 49.Chivian D, et al. Automated prediction of CASP-5 structures using the Robetta server. Proteins. 2003;53 Suppl 6:524–533. doi: 10.1002/prot.10529. [DOI] [PubMed] [Google Scholar]
- 50.Martin D, Wollscheid B, Aebersold R, Watts J. Methods for Characterizing Glycoproteins and Generating Antibodies for Same. 066661-0148. US Patent. 2008
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.