Abstract
From a structural perspective, the factors controlling and the mechanisms underlying the toxic effects of ionizing radiation remain elusive. We have studied the consequences of superhelical/torsional stress on the magnitude and mechanism of DSBs induced by low-energy, short-range, high-LET Auger electrons emitted by 125I, targeted to plasmid DNA by m[125I]iodo-p-ethoxyHoechst 33342 (125IEH). DSB yields per 125I decay for torsionally relaxed nicked (relaxed circular) and linear DNA (1.74 ± 0.11 and 1.62 ± 0.07, respectively) are approximately threefold higher than that for torsionally strained supercoiled DNA (0.52 ± 0.02), despite the same affinity of all forms for 125IEH. In the presence of DMSO, the DSB yield for the supercoiled form remains unchanged, whereas that for nicked and linear forms decreases to 1.05 ± 0.07 and 0.76 ± 0.03 per 125I decay, respectively. DSBs in supercoiled DNA therefore result exclusively from direct mechanisms, and those in nicked and linear DNA, additionally, from hydroxyl radical-mediated indirect effects. Iodine-125 decays produce hydroxyl radicals along the tracks of Auger electrons in small isolated pockets around the decay site. We propose that relaxation of superhelical stress after radical attack could move a single-strand break lesion away from these pockets, thereby preventing further breaks in the complementary strand that could lead to DSBs.
INTRODUCTION
The genomic DNA of both prokaryotes and eukaryotes is negatively supercoiled and in a torsionally strained state (1). Prokaryotic chromosomal DNA is circular, compacted by supercoiling, and further condensed by topological constraints imposed by histone-like structural proteins (2), including the factor for inversion stimulation (Fis), the integration host factor (IHF), and the heat-stable nucleoid-structuring protein (H-NS). Eukaryotic DNA is organized on histones as nucleosomes, which in turn are compacted to form chromatin fibers.
Several studies have focused on understanding how torsional stress arising from supercoiling affects various fundamental genetic processes in the cell. Prokaryotic transcription is intimately linked to DNA supercoiling (3). Additionally, because it can bring two distal regulatory elements of DNA into proximity through DNA bending/looping (4), supercoiling is thought to play a key role in gene regulation (5) and recombination (6). Furthermore, it is crucial in accurate initiation of DNA duplication at replication origins (7). Although the influence of DNA super-coiling on basic biological processes has been investigated in some depth, its role in the susceptibility of DNA to damage by ionizing radiation remains poorly understood.
Supercoiling is thought to be among the determinants of DNA sensitivity to ionizing radiation. Miller et al. (8) have shown in plasmid DNA that the yield of single-strand breaks (SSBs) induced by external-beam ionizing radiation increases with negative supercoiling. However, Milligan et al. (9) have reported insignificant variations in SSB yield over a similar range of superhelical density and have concluded that, if subtle variation is not ignored, the trend would be slightly in the opposite direction. Ozols et al. (10), comparing the yield of double-strand breaks (DSBs) induced by pulsed electrons in supercoiled and relaxed DNA templates, have supported the observations of Milligan and coworkers. When ionization events throughout the medium are low in density and uniformly distributed, it seems, therefore, that supercoiling does not influence the susceptibility of DNA to strand breaks generated by either X- or γ-ray external-beam radiation.
Internal emitters that undergo radioactive decay by electron capture and/or internal conversion result in the emission of a surge of low-energy (<1 keV) Auger electrons (11–16). Since many of these electrons traverse a very short distance (few nm), the density of the hydroxyl radicals (•OH) generated will be very high around the decaying atom and will decrease drastically as a function of the length of their tortuous path. Even under circumstances in which the radionuclide atoms are uniformly distributed in medium, the decay of these radionuclides will result in the formation of randomly dispersed “hot spots” (volumes densely traversed by electrons and occupied by •OH and other radicals) and “cold spots” (volumes sparsely traversed by electrons and deficient in •OH and other radicals). Therefore, it is inappropriate to extrapolate from the dependence of X- or γ-ray-induced DSBs on supercoiling to conclusions concerning short-range, Auger electron radiation or localized ionizing radiation from internal emitters.
Among the Auger electron emitters under consideration for therapeutic applications, 125I has been most studied (16). The decay of this radionuclide produces a cascade of extremely low-energy electrons, ~21 on average; of these, ~18 electrons have a range <40 nm and ~13 electrons travel <5 nm (16). Dosimetric calculations indicate that (1) most of the energy released is deposited in the immediate vicinity of the decay site, (2) there is a sharp and significant drop in the energy deposited (from ~109 cGy to ~106 cGy) as a function of increasing distance (few nm) from the decaying 125I atom (12, 14–16), and (3) the DSB yield per decay drops precipitously as the 125I atom is displaced a few angstroms from the central axis of DNA (17). Consistent with these theoretical expectations, studies in which the 125I atom has been positioned in proximity to the DNA molecule, by either DNA incorporation of an 125I-labeled nucleoside analog or binding of an 125I-labeled DNA groove binder or intercalator, show that the decay of this Auger electron emitter leads to the efficient induction of SSBs and DSBs in naked DNA (e.g. synthetic oligonucleotides, bacterial DNA, plasmid DNA) and in eukaryotic chromatin [reviewed in ref. (16)], whereas studies in which the 125I atom has been placed at a distance from the DNA molecule (e.g. 125I-antipyrine) indicate that its decay is relatively ineffective in the induction of DSBs (18, 19). In the former case, the experimental results have also supported the mechanistic notions that (1) the bulk of SSBs formed are principally consequent to indirect, •OH-mediated ionizations (19, 20) and (2) in naked DNA molecules, DSBs (~0.5–1.0 per decaying 125I atom) are induced mainly by direct ionizations (19–22). In the latter case, the findings indicate that DSB formation in eukaryotic chromatin is caused predominantly (~90%) by indirect, •OH-mediated ionizations (23–25). Finally, in addition to damage caused by direct (unscavengable) and indirect (scavengable) ionizations of the DNA molecule, the neutralization of the highly positively charged metastable 125mTe atom (daughter of 125I) has been postulated to be responsible for up to 50% of the damage seen in DNA (13, 26–28).
Many questions concerning the biophysical mechanisms of DSB production by the decay of Auger electron-emitting 125I remain, including the role of DNA supercoiling (bending/compaction). Previously, we had proposed that chromatin structure (highly compacted DNA) provides conditions conducive to the formation of >1 DSB per 125I decay by indirect mechanism(s) since a cluster of •OH produced by decay of 125I may attack a DNA site that is hundreds of nucleotides away from the decaying atom but placed in proximity to it by the supercoiling of DNA (23). Herein, using a naked plasmid DNA model, we address the role of DNA compaction resulting from negative supercoiling in the production of radiation-induced DSBs. We have exposed all three topological forms of pUC19 DNA (supercoiled, nicked and linear) to the DNA minor-groove-binder m-iodo-p-ethoxyHoechst 33342 labeled with 125I (125IEH). Surprisingly, our studies have shown that the DSB yield consequent to the decay of 125I is markedly reduced by supercoiling of plasmid DNA, contrary to the expectations based on experiments in mammalian cells (23).
MATERIALS AND METHODS
Synthesis of 125I-Labeled m-Iodo-p-ethoxyHoechst 33342 (125IEH)
The radioiodinated analog of the DNA minor-groove-binding drug m-iodo-p-ethoxyHoechst 33342 (125IEH) was synthesized as described previously (18, 29). In essence, lactoperoxidase (0.02 units/4 μl) and aqueous hydrogen peroxide (10 μl of 0.24%) were combined with a mixture of Na125I (37–74 MBq/10–20 μl 0.1 M NaOH) and trimethylstannyl-Hoechst 33342 (30 μg/20 μl DMSO) in acetate buffer (40 μl, 0.2 M, pH 4.9). After a 15-min incubation at room temperature, the reaction was stopped with tri-n-butylamine (2%) in methanol (100 μl). The radioiodinated product was purified on a SepPak® Plus C18 cartridge (Waters) to remove all ionic impurities including free radioiodide. The radiochemical purity of 125IEH was greater than 98%, and its specific activity was ~81.4 TBq/mmol. Aliquots of 125IEH (~5.2 MBq) were dispensed in 1.5-ml microfuge tubes and blown dry with argon.
Preparation of 3HT-pUC19 Plasmid DNA
Stocks of E. coli DH5α bacterial cultures were grown with pUC19 in Luria broth for 16 h at 37°C in the presence of ampicillin (50 μg/ml) and 3H-thymidine ([3H]dThd, 37 MBq). The plasmids (3HT-pUC19) were isolated using the Qiagen Maxi preparation kit and dissolved in PBS (pH 7.4). The concentration determined at A260 was 0.748 μg/μl. The plasmid DNA was stored at −20°C.
Preparation of Nicked-Circular 3HT-pUC19 Plasmid DNA
Restriction endonuclease EcoR1 (New England Biolabs), being a homodimer, under normal conditions of digestion cuts both strands in the DNA between the bases G and A in the recognition sequence 5′ GAATTC 3′, leading to a DSB. However, when supercoiled DNA is intercalated with ethidium bromide, EcoR1 severs only one of the two DNA strands in its recognition sequence, resulting in relaxation of supercoiling and production of a nicked-circular molecule (30). Since our model plasmid DNA contains an EcoR1 site, we digested supercoiled 3HT-pUC19 plasmid DNA (100 μg in 134 μl 1× PBS) with EcoR1 (800 units, 40 μl) in the presence of ethidium bromide (0.4 mg/ml) and EcoR1 buffer (1×, 400 μl, New England Biolabs) for 24 h at 37°C. Aliquots were assessed on agarose gel for nicked DNA formation, another aliquot of nicking reaction cocktail was added, and the incubation was continued for 16 h. The reaction was arrested by the addition of EDTA to a final concentration of 10 mM. Agarose gel analysis confirmed that very little supercoiled DNA was left undigested. The reaction mixture was extracted with phenol (pH >7.5) and chloroform:isoamyl alcohol (24:1, v/v), followed by five n-butanol extractions to remove ethidium bromide completely. DNA was ethanol-precipitated and redissolved in 1× PBS. DNA concentration was 0.71 μg/μl. This nicked DNA preparation was found to have traces of linear DNA, which was removed by digesting it with λ exonuclease and RecJf for 24 h at 37°C. Agarose gel analysis indicated that linear DNA was completely digested into mononucleotides. The λ exonuclease was inactivated thermally at 65°C for 10 min, and the reaction mixture was extracted with phenol and chloroform:amyl alcohol (24:1, v/v). Homogeneous nicked-circular 3HT-pUC19 plasmid DNA was ethanol-precipitated and dissolved in 1× PBS. The DNA concentration was 0.303 μg/μl.
Preparation of Linear 3HT-pUC19 Plasmid DNA
Supercoiled 3HT-pUC19 plasmid DNA (100 μg in 134 μl 1× PBS) was digested with EcoR1 (1000 units, 50 μl) in EcoR1 buffer (1×, 500 μl, New England Biolabs) for 16 h at 37°C. The reaction mixture was assessed for linear DNA formation on 1% agarose gel, another 100 units of EcoR1 was added, and the incubation was continued for another 12 h at 37°C. Subsequent agarose gel analysis indicated 100% linearization and no residual supercoiled DNA left undigested. The reaction mixture was extracted with phenol (pH >7.5) and chloroform:isoamyl alcohol (24:1, v/v) to remove EcoR1. Linear 3HT-pUC19 DNA was precipitated with ethanol and redissolved in 1× PBS (pH 7.4). The DNA concentration was 0.375 μg/μl, with a total yield of ~75 μg.
Incubation of Supercoiled, Nicked and Linear Forms of 3HT-pUC19 Plasmid DNA with m-[125I]Iodo-p-ethoxyHoechst 33342 (125IEH)
The incubation mixtures consisted of 3HT-pUC19 plasmid DNA (9 μg, 5.3 pmol) in one of three topological states (supercoiled, nicked or linear) and 125IEH (~1.48 MBq) in 1× PBS (100 μl, pH 7.4) without or with DMSO (0.2 M). DNA solutions were added to microfuge tubes containing dry 125IEH, mixed vigorously for 5 min, and incubated at 4°C. The 125IEH: DNA molar ratio was in the range of 2:1 to 3:1. Fluorescence titration analysis using IEH had indicated previously that under these conditions all dye molecules in solution are bound to DNA (29). Each incubation was performed in duplicate. At various intervals (0, 5, 12, 19, 36, 44, 52, 65 and 72 days), aliquots (6 μl) were removed from the incubation mixture, combined with loading-dye–glycerol mixture (3 μl) and PBS (3 μl), and loaded onto 1% agarose gels in TAE buffer (0.5×) with ethidium bromide (0.5 μg/ml). We estimate the random pipetting error in filling the agarose gels to be 3–7% of the total volume of the sample (12 μl). The gels were run at 200 V (~7 V/cm) for 1 h and then photographed on a transilluminator (long wave) attached to a CCD camera. The DNA bands corresponding to the supercoiled, nicked and linear forms were excised from the gel. For 125IEH–nicked DNA incubations, bands corresponding to linear DNA produced by DSBs and intact nicked DNA were obtained, and for 125IEH–linear DNA incubations, bands corresponding to DNA fragments and intact linear DNA were acquired. The DNA in each of these bands was quantified by dissolving the gel pieces in Opti-Fluor scintillation fluid (10 ml) and assaying the associated tritium in a liquid scintillation counter. Since the DNA bands also contained undecayed 125IEH bound to DNA, correction was made for spillover of 125I into the tritium energy window using a 125I standard. Control incubations without 125IEH were set up in parallel for each of these topological forms.
Calculations show that 540 ng (0.318 pmol) control DNA loaded on agarose gel carried 2.475 pmol 3H (specific activity of [3H]dThd, molecular weight 242.2, is 740 GBq/mmol). Each molecule of pUC19 was associated with approximately 7.78 3H atoms on day zero of the incubation. Given the 12.32-year half-life of tritium, even after 60 days the number of 3H atoms decayed per plasmid molecule is insignificant (~0.08 = 1% 3H present on day 0). Therefore, we did not see any detectable damage (SSBs) to DNA in the control samples over time. This reassured us that, during the entire incubation, there was no change in the concentration of DNA unless the incubation mixture contained 125IEH and that the strand breaks therefore resulted entirely from 125I decays.
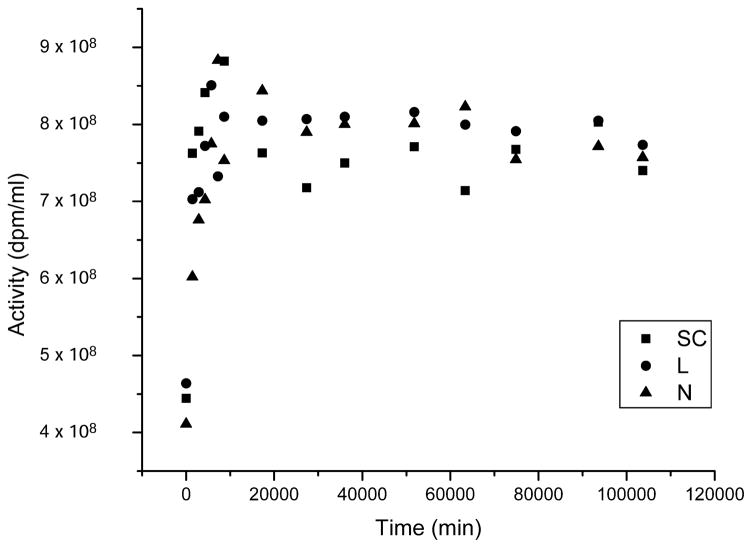
The 125I activity of the incubation medium was established in parallel with the gel analysis as described previously (20). Aliquots of incubation mixture were centrifuged at 13,000 rpm for 3 min, and the radioactivity in the supernatant (2 μl) was assayed in a γ-ray counter and calculated as dpm/ml (this procedure was carried out daily during the first week and thereafter only when gel analysis was performed). These determinations indicate that 125IEH was not completely solubilized immediately after the addition of DNA to the tube: 125I activity in solution increased steeply for the first 6 days before reaching a plateau (Fig. 1). The amount of 125IEH solubilized was independent of the topological form and was consistent with our previous findings for supercoiled DNA (20). The number of decays accumulated in any given time is calculated by quantifying the area under the curve for activity (dpm/ml) as a function of time (min). This is obtained by integrating the function within the limits of the accumulation time using Origin (OriginLab Corporation, Northampton, MA).
FIG. 1.
Dissolved 125I activity present in incubations of supercoiled (SC, ■), linear (L, ●) and nicked (N, ▲) forms of 3HT-pUC19 DNA with 125IEH as a function of time.
Determination of Association Constants of Hoechst 33342 with Various Forms of DNA
Stock solutions of the three forms of pUC19 DNA (200 μM) were diluted with PBS to give a series of DNA solutions (3 ml each) with concentrations in nucleotide phosphate ranging from 0 μM to 75 μM. A stock solution of Hoechst 33342 in water (5.84 mM) was diluted to 20.45 μM before use. For fluorescence titrations, DNA samples (3 ml) were mixed with Hoechst 33342 (25 μl) to give a final Hoechst 33342 concentration of 0.169 μM. Fluorescence spectra were recorded at 25°C in a PerkinElmer LS50B luminescence spectrometer (slit width 5 nm, excitation wavelength 335 nm, emission wavelength 450 nm). The fluorescence intensity of Hoechst 33342 at 450 nm was plotted as a function of DNA concentration (in nucleotides). Fluorescence intensity in the presence of excess DNA was assumed to reflect complete binding of Hoechst 33342 to DNA, with no unbound Hoechst 33342 in solution. Concentrations of bound and unbound Hoechst 33342 at each DNA concentration were calculated from the titration profiles, and the association constant was obtained from a Scatchard plot.
Calculation of 125IEH-Induced Double-Strand Breaks in Supercoiled, Nicked and Linear 3HT-pUC19 Plasmid DNA
The fraction of DNA (supercoiled, nicked or linear form) remaining intact at each time was plotted as a function of the number of decays/ml accumulated during that period. Since at zero decay, the fraction of intact DNA equals unity, the regression line was forced through 1. Statistically, this is permissible because, in the absence of forcing through 1, the y intercept is not significantly different. The DSB calculations were based on the assumption that the binding of 125IEH to DNA and thus the strand breaks follow a Poisson distribution.
Supercoiled DNA
The mean number of DSBs (XDSB) per DNA molecule is calculated from the fraction of linear DNA formed at each time after exposure of supercoiled DNA to a given number of 125I disintegrations as described previously (20):
where FL is the fraction of linear DNA formed as a result of 125I decays.
Nicked DNA
DSBs are solely responsible for the disappearance of intact nicked DNA and the formation of linear DNA. The mean number of DSBs (XDSB) per DNA molecule at any given dose is determined from the fractions of intact nicked DNA remaining at that time and linear DNA formed as a result of exposure to 125I:
where FL is the fraction of linear and fragmented DNA resulting from DSBs due to 125I decays (1 − FN(i)) and FN(i) is the fraction of intact nicked DNA at any given time (ratio of 3H dpm in intact nicked DNA remaining after 125I exposure to that of unirradiated control nicked DNA).
Linear DNA
Because DSBs are the only cause of reduction in intact linear DNA, the rate of formation of DSBs must be equal to the rate of disappearance of linear DNA. The mean number of DSBs (XDSB) per DNA molecule is calculated from the experimentally observed fractions of fragmented DNA formed and intact linear DNA remaining after exposure to a given number of 125I disintegrations:
where Ff is the fraction of fragmented DNA occurring as a result of DSBs due to 125I decays (1 − FI(i)) and FI(i) is the fraction of intact linear DNA remaining at any given time (ratio of 3H dpm in intact linear DNA remaining after 125I exposure to that of unirradiated control linear DNA).
For all three forms of DNA, the rate of formation of DSBs per DNA molecule per decay of 125I is obtained by plotting XDSB as a function of the number of accumulated 125I decays per ml. Since the fraction of break-down products resulting from DSBs in nicked and linear DNA is determined from two different gel lanes, namely, unirradiated control and 125I-exposed DNA, the random 3–7% pipetting error mentioned above is carried over into the XDSB values calculated and is manifest in the scatter in the linear regressions used to calculate the DSB yield. To overcome these sample-loading shortcomings, we have carried out duplicate experiments and plotted them together in such a way as to nullify random errors associated with pipetting, loading and recovery of samples. The standard errors for the slopes of the linear regressions reflect these random errors. The straight lines are forced through zero, because at zero decay there would be no DSBs. The slopes of these linear regressions reflect the DSB yield (YDSB) expressed as the number of DSBs generated in one DNA molecule per decay of 125I per ml, and the reciprocal of the slopes represents D0, the number of decays per ml required to form one DSB in one DNA molecule. When multiplied by the total number of plasmid DNA molecules per ml, the slope gives the yield of DSB per decay of 125I:
where the concentration of DNA ([DNA]) = 3.06 × 1013 molecules/ml.
Statistical Analysis
To compare the slopes of linear regressions, statistical analysis was carried out with Graph Pad Prism 4 (Graph Pad Software) as described previously (20). The adherence of the x and y coordinates of two or more linear regressions to the null hypothesis is indicated by a global P value for the comparison of slopes. If P > 0.05 for two or more separate linear fits, each representing that of two sets of data points, the null hypothesis cannot be rejected, and the slopes are not significantly different. If P < 0.05, the slopes are statistically significantly different.
RESULTS
Detection of 125IEH-Induced Double-Strand Breaks in Supercoiled, Nicked and Linear 3HT-pUC19 Plasmid DNA
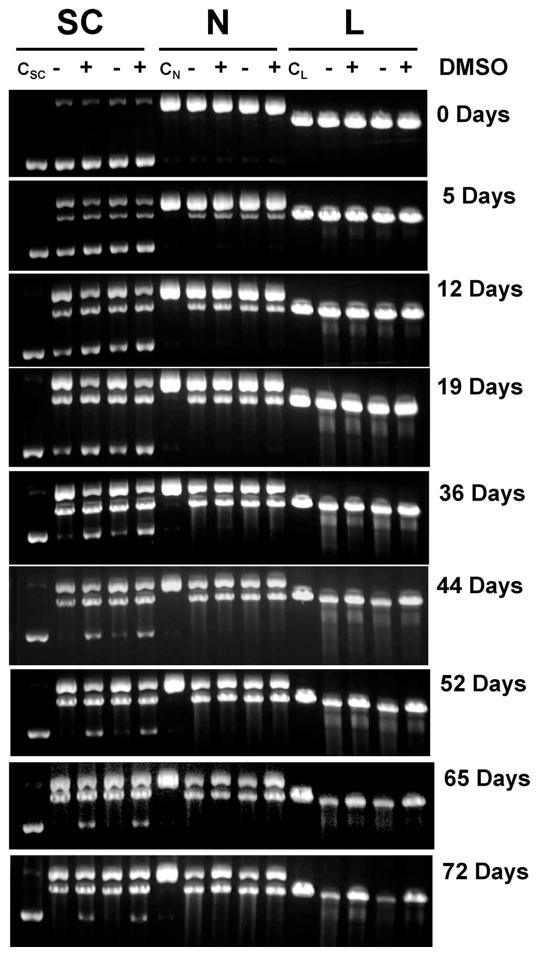
Tritium-labeled DNA enables us to quantify the amount of DNA in each band accurately and precludes the need for taking into account the differential staining of supercoiled, nicked and linear DNA by ethidium bromide (31). Figure 2 shows the analysis of DNA from a representative experiment at various times after accumulation of 125I decays. Controls of supercoiled, nicked and linear pUC19 DNA without 125IEH remain intact over the entire period of incubation at 4°C, as seen from lanes CSC, CN and CL, respectively. In the presence of 125IEH, all three forms of DNA vanish gradually with time as a function of accumulated 125I decays, manifested by reduction in the band intensities of intact DNA. The disappearance of supercoiled DNA occurs through both SSBs and DSBs, resulting in nicked and linear DNA, respectively, which have electrophoretic mobilities that are different from the parent supercoiled DNA (Fig. 2). At the 125IEH to DNA ratio of ~3:1 in the current study, the probability of formation of linear from nicked DNA is low, and the majority of linear DNA is formed directly from supercoiled DNA through DSBs (20). The electrophoretic migration of nicked and linear DNA is unaffected by SSBs; therefore, any reduction in the intensity of intact nicked or linear DNA as a function of time is due to DSBs. In the linear form (2686 bp), DSBs produce shorter DNA fragments (<2686 bp) that can be seen as a smear or a ladder in gels run at or after 19 days. In the nicked form, a single DSB produces a linear DNA fragment, whereas multiple DSBs on the same nicked DNA molecule lead to shorter fragments.
FIG. 2.
Agarose gel analysis of supercoiled (SC), nicked (N) and linear (L) forms of 3HT-pUC19 plasmid DNA incubated with 125IEH at 4°C in PBS (pH 7.4): lanes CSC, CN and CL represent control incubations of supercoiled, nicked and linear DNA without 125IEH; − and + indicate absence and presence of DMSO (0.2 M), respectively. Gels containing ethidium bromide were visualized using ultraviolet (320 nm) transillumination.
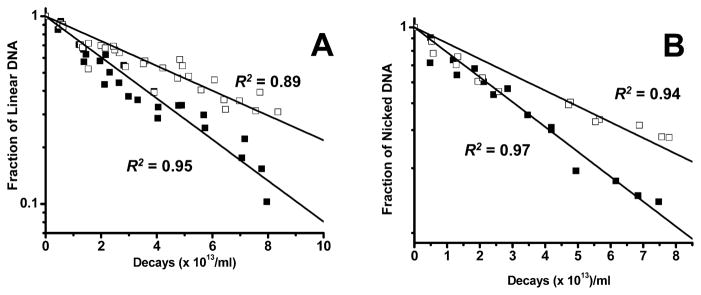
A comparison of −DMSO and +DMSO lanes for linear DNA incubations from 44 days to 72 days shows that in the absence of DMSO more than half of the linear DNA has broken down into smaller fragments, and in its presence a much larger fraction remains intact. A similar trend occurs for nicked DNA incubations. The rate of disappearance for linear and nicked DNA under −DMSO and +DMSO conditions, denoted by the fraction of intact DNA remaining (ratio of 3H dpm in 125I-irradiated DNA to that in un-irradiated control as function of 125I decays), is higher in the absence of DMSO (Fig. 3), suggesting that the number of DSBs for a given number of decays is greater under nonscavenging conditions, because the 125I activity in all four incubation mixtures is similar. Thus, in the absence of DMSO, 125IEH-induced DSBs in nicked or linear DNA are caused by both direct and indirect mechanisms, whereas in its presence, DSBs occur predominantly by direct ionization of DNA.
FIG. 3.
Quantitative analysis of agarose gel electrophoresis for disappearance of linear 3HT-pUC19 plasmid DNA (panel A) and nicked 3HT-pUC19 plasmid DNA (panel B) as a function of 125I decays accumulated in the absence (■) and presence (□) of DMSO.
In the nicked plasmid, a single DSB leads to a linear plasmid (2686 bp). The intensities of linear DNA bands resulting from DSBs in −DMSO and +DMSO lanes appear to be the same, despite the higher rate of disappearance in the −DMSO lane, suggesting that a small fraction of plasmid molecules undergo more than one DSB, producing shorter fragments (<2686 bp). Therefore, the DSB yield determined by this method is an underestimate. The agarose gel data indicate that DSBs in nicked and linear DNA are caused by both direct and indirect effects of Auger electron radiation in contrast to those in supercoiled DNA, which are caused only by direct action of decaying 125IEH (20).
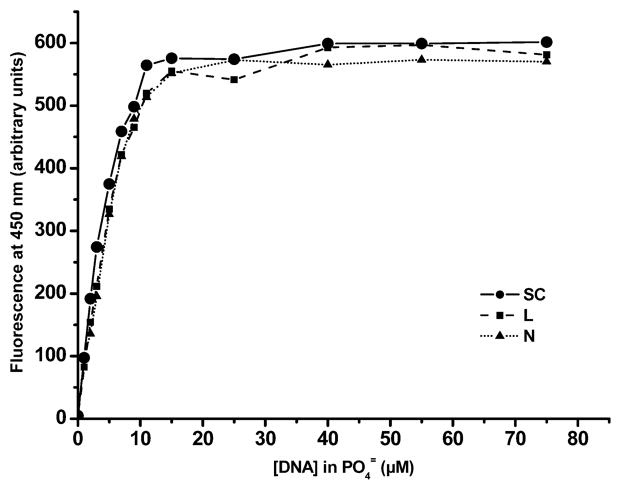
To verify whether all 125IEH present in solution is bound to DNA and to rule out the possibility that the variation in the number of DSBs with superhelical stress is due to differences in binding affinities of supercoiled, nicked and linear forms for the carrier ligand Hoechst 33342, we studied the binding equilibrium. Our attempts to directly measure the binding parameters for 125IEH with DNA failed because (1) the 125IEH–DNA complex dissociates during agarose gel electrophoresis (0.5× TAE, pH 8.3, 0.5 μg/ml ethidium bromide) and (2) 125IEH is highly soluble in reagents such as ethanol and isopropanol, which are used to precipitate the 125IEH–DNA complex to determine the bound fraction of 125IEH. Therefore, we monitored the binding by fluorescence spectroscopy using the same conditions as in the 125IEH–DNA incubations. The fluorescence titration profiles of the supercoiled, nicked and linear forms of pUC19 plasmid DNA with Hoechst 33342, in the concentration range used in the 125IEH–DNA incubations, are very similar (Fig. 4). All three topological forms of pUC19 plasmid DNA are in B conformation and have the same minor groove width and length. Consequently, the binding of 125IEH to DNA is not affected by DNA supercoiling and the number of 125IEH molecules bound to the supercoiled, nicked and linear forms must be about the same at any given ligand and DNA concentration. In contrast, the affinity of intercalators such as ethidium to the DNA varies among the supercoiled, nicked and linear forms, because the intercalation of DNA by each molecule of ethidium results in the unwinding of the base pairs by 26° (32). Since both strands in supercoiled DNA are covalently closed, the ability of this form to unwind upon intercalation is topologically constrained compared with the nicked and linear forms. Consequently, the number of ethidium molecules bound to the supercoiled form is less than that bound to the nicked and linear forms. Fluorescence of Hoechst 33342 reaches a plateau at ~20 μM DNA nucleotide concentration for all three forms, indicating that all the ligand molecules present in solution are bound at this concentration. Furthermore, since the actual concentration of DNA in the 125IEH–DNA incubations is 284 μM (⋙20 μM), very few unbound 125IEH molecules could exist in the incubation mixtures. The slopes of Scatchard plots for Hoechst 33342 binding to all three forms of pUC19 DNA are not significantly different from each other (P = 0.31, null hypothesis), and a single fit of the three data sets gives an association constant (Ka) of 3.42 ± 0.81 × 107 M−1, a value in good agreement with the Ka reported previously for calf thymus DNA (29).
FIG. 4.
Fluorescence titration curves of supercoiled (SC), linear (L) and nicked (N) forms of pUC19 DNA with Hoechst 33342 (0.169 μM) in PBS (pH 7.4). Fluorescence maxima for all three forms of pUC19 DNA occur at approximately the same DNA concentration (20 μM), indicating saturation of ligand binding to DNA. In 125IEH–DNA incubations, the concentration of 125IEH is 284 μM, ensuring that there is little unbound 125IEH in mixtures.
Calculation of 125IEH-Induced Double-Strand Break Yields
The method and formulas for calculating DSB yields in all three topological forms are based on the assumption that ligand binding to DNA and hence subsequent strand breaks upon 125I decay follow a Poisson distribution (33). In supercoiled DNA, the DSB yield is calculated from the appearance of linear DNA. For nicked and linear DNA, the DSB yield is assessed by the disappearance of intact nicked and linear molecules. This seems ideal, because a nicked or linear DNA molecule can vanish from an intact DNA band only when the latter acquires at least one DSB. If it receives multiple DSBs, it will also be gone, so the DSB values we report may be slightly underestimated. In the case of nicked DNA, since >1 DSB results in a product with fragments shorter than linear DNA (<2686 bp), the 3H dpm values for linear DNA (2686 bp) produced by DSBs and intact nicked DNA remaining may not add up to match that of control unirradiated DNA. For both nicked and linear DNA in the absence of DMSO, the DSB yield is >1, and the resulting product cannot be recovered fully from the gel and quantified accurately. This is the main reason for calculating DSB yield from the disappearance of intact nicked or linear DNA. Under gel electrophoresis conditions, intact linear DNA (2686 bp) can be separated from fragments shorter than linear DNA by 100 bp. The number of preferred Hoechst-binding tetranucleotide sites (AATT, TAAT, ATAT, TATA and TTAA) in pUC19 is 39, and there are only three such sites within 100 bp from each end of the linear pUC19 molecule, indicating that the probability of DSBs leading to fragments in the size range 2586–2686 bp is very low. These facts suggest that estimation from the agarose gels (Fig. 2) of the fraction of intact linear DNA remaining after each dose must be fairly accurate.
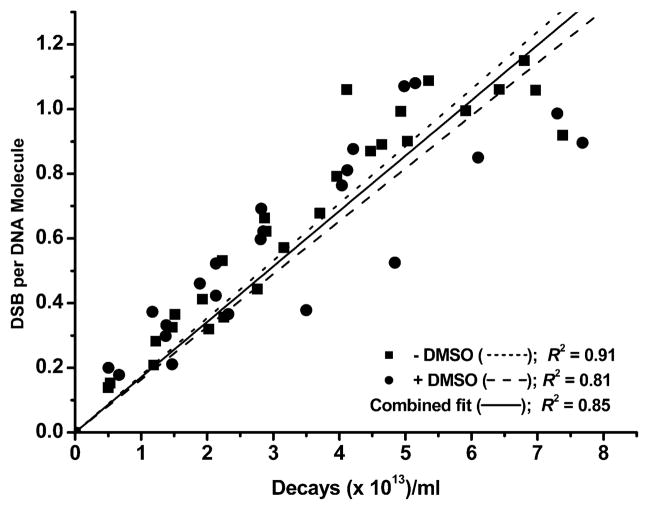
The variation of mean DSB yield per DNA molecule with 125IEH decay in the absence and presence of DMSO for all three forms of plasmid DNA is shown in Figs. 5 and 6. From the linear regressions for supercoiled DNA (Fig. 5), it can be seen that DMSO has no significant effect on the rate of DSB formation, and DSBs in the supercoiled form are predominantly the result of direct effects. Accordingly, D0(DSB), the number of decays required to produce a single DSB in the absence of DMSO [(5.65 ± 0.19) × 1013 decays per ml], is about the same as that in the presence of DMSO [(6.12 ± 0.34) × 1013 decays per ml], leading to a dose modification factor (DMF) of 1.08 ± 0.07 (Table 1). As expected, DSB yields are close, 0.54 ± 0.02 without DMSO and 0.50 ± 0.03 with DMSO, and a P value (null hypothesis) of 0.2 (P > 0.05) suggests that the slopes of these linear regressions (broken lines in Fig. 5) are not significantly different. Therefore, a single fit of the −DMSO and +DMSO data sets (solid line in Fig. 5) gives a DSB yield of 0.52 ± 0.02 per decay for scavenging and nonscavenging conditions. This value is in agreement with that reported previously (20), thereby increasing the confidence in DSB yields determined for the nicked and linear forms in parallel.
FIG. 5.
Mean DSB yield per DNA molecule for supercoiled (SC) pUC19 plasmid DNA without or with DMSO, calculated from the appearance of linear (L) DNA as a function of accumulated 125I decays. Linear regressions were carried out and R2 was obtained using Origin. Regression lines were forced through zero.
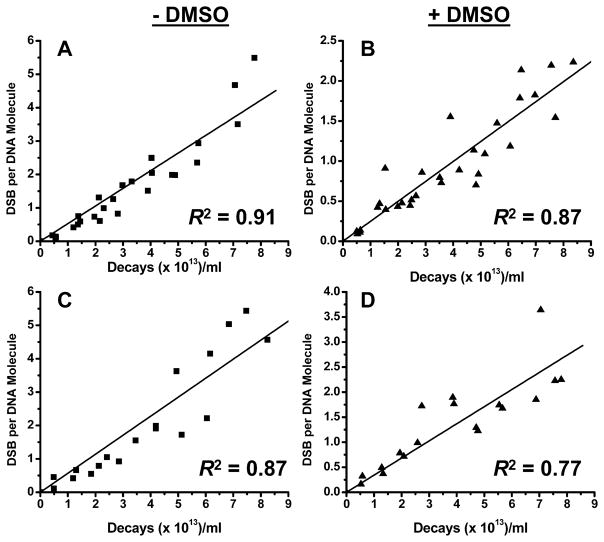
FIG. 6.
Mean DSB yield per DNA molecule for linear and nicked DNA: linear form −DMSO (panel A); linear form +DMSO (panel B); nicked form −DMSO (panel C); nicked form +DMSO (panel D). DSB yields were calculated from the rate of disappearance of intact linear or nicked DNA as a function of accumulated 125I decays. Linear regressions were carried out and R2 was obtained using Origin. Regression lines were forced through zero.
TABLE 1.
Comparison of DSB Yields among Different Forms of Plasmid DNA: Dependence on DNA Topology
| D0DSB × 1013 (decays/ml) |
DSBs/decay |
||||
|---|---|---|---|---|---|
| Topological state of plasmid DNA | −DMSO | +DMSO | DMFD0DSB | −DMSO | +DMSO |
| 0.54 ± 0.02* | 0.50 ± 0.03* | ||||
| Supercoiled | 5.65 ± 0.19 | 6.12 ± 0.34 | 1.08 ± 0.07 | 0.52 ± 0.02* | |
| Linear | 1.89 ± 0.08 | 4.02 ± 0.16 | 2.13 ± 0.12 | 1.62 ± 0.07 | 0.76 ± 0.03 |
| Nicked circular | 1.76 ± 0.11 | 2.93 ± 0.20 | 1.66 ± 0.16 | 1.74 ± 0.11 | 1.05 ± 0.07 |
P = 0.2.
It has been observed that 125I-iodoantipyrene, an agent that does not bind to DNA and is freely available in solution, causes DSBs in plasmid supercoiled DNA by indirect mechanisms only [no DSBs are formed in supercoiled DNA in the presence of DMSO (19)]. This suggests that the decay of non-DNA-bound 125IEH, for example, the insoluble fraction bound to the wall of the tubes, could lead to DSBs only by indirect mechanisms. However, our current (Fig. 5) and previous (20) data show that in supercoiled DNA ~100% of DSBs are caused by direct ionizations, indicating that insoluble reaction-tube-bound 125IEH is ineffective and that all of the DSBs in DNA are caused by solubilized DNA-bound 125IEH.
For the linear form [Fig. 6A (−DMSO) and B (+DMSO)], D0DSB values of (1.89 ± 0.08) × 1013 and (4.02 ± 0.16) × 1013 decays/ml without and with DMSO, respectively, are observed with a DMF of 2.13 ± 0.12 (Table 1). Thus, for a given number of 125I decays, DSBs are approximately twofold more likely to occur in the absence of DMSO in linear plasmid DNA, emphasizing the contribution of scavengable, •OH-mediated, indirect mechanisms to DSB formation. A DSB yield of 1.62 ± 0.07 per decay, which is approximately threefold higher than that of the supercoiled form (0.52 ± 0.02 per decay), decreases to 0.76 ± 0.03 in the presence of DMSO (Table 1), indicating that in the torsionally relaxed linear form, DSBs occur through both the direct action of Auger electrons with bond breakage in the complementary DNA strands (unscavengable by DMSO) and indirect mechanisms mediated by •OH (scavengable by DMSO). We attribute the difference in the magnitude and mechanism of Auger electron-induced DSBs between the supercoiled and linear forms to torsional stress.
This conclusion is strengthened by the DSB yields obtained for nicked plasmid DNA, which also lacks torsional stress [Fig. 6C (−DMSO) and D (+DMSO)]. D0DSB values of (1.76 ± 0.11) × 1013 and (2.93 ± 0.20) × 1013 decays/ml in the absence and presence of DMSO, respectively, with a DMF of 1.66 ± 0.16 (Table 1) denote an approximately 1.7-fold higher DSB yield in the absence of DMSO than in its presence, again pointing to the contribution of indirect mechanisms. A DSB yield of 1.74 ± 0.11 per decay, which is about three times higher than that of the supercoiled form, decreases to 1.05 ± 0.07 in the presence of DMSO (Table 1), suggesting that, in the torsionally relaxed nicked form, DSBs occur through both direct action and indirect mechanisms mediated by •OH.
DISCUSSION
We have been examining the biophysical mechanisms underlying DSB production by 125I. Herein we show that DNA topology plays a major role in modulating the mode and efficiency of DSB production in naked DNA.
Analysis of 125IEH-Induced Double-Strand Breaks in Supercoiled, Nicked and Linear DNA
In our current study, the DSB yields from 125IEH bound to the minor groove of supercoiled pUC19 DNA are in excellent agreement with those that we reported recently (20). Since the P value (null hypothesis) of 0.2 (P > 0.05) for the ±DMSO data sets suggests that these DSB yields are not significantly different, we deduce that DSBs per decay produced in the supercoiled form are derived only from the direct action of Auger electrons and have an average value of 0.52 ± 0.02 (Table 1). Based on our previous findings (19) that no DSBs are formed in supercoiled DNA in the presence of DMSO after the decay of 125I-antipyrine (an agent with no affinity to DNA; average distance between 125I atoms and supercoiled DNA ~42 nm), we also conclude that all DSBs detected occur only in the supercoiled DNA molecules to which the radioiodinated Hoechst 33342 molecule is bound (internal DSBs).
For the linear form of pUC19 DNA, the DSB yields per decay of 125IEH in the absence and presence of DMSO are approximately three- and ~1.5-fold higher, respectively, than those for supercoiled DNA (Table 1). Because these two DSB yields in linear DNA are significantly different from each other (P <0.0001), it appears that (1) indirect mechanisms are operative in linear DNA, (2) the contribution from •OH-mediated indirect mechanisms is 0.86 ± 0.08 per 125IEH decay (1.62 ± 0.07 − 0.76 ± 0.03), and (3) given that all 125IEH molecules are DNA-bound under the experimental conditions, the 0.76 ± 0.03 DSB (~50%) induced by direct mechanism(s) occurs within the DNA molecule to which the 125IEH molecule is bound (internal DSB). A DSB yield of 1.62 ± 0.07 per 125I decay in the absence of DMSO also implies that at least ~0.62 DSB must take place in neighboring linear DNA molecules (external DSB), within a radius of 500 nm [maximum range of 98% of Auger electrons emitted (16)] from the decaying 125I. Under the experimental conditions, we calculate that about seven DNA molecules on average are present in a sphere of 480-nm radius.
A detailed analysis was carried out to dissect various sources contributing to the DSBs from 125IEH decays in linear DNA (Table 2). In principle, DSBs due to direct ionization could be present within either the DNA molecule where 125I decayed (*LD) or neighboring DNA molecules (LD), i.e., *LD + LD = ~0.8 per decay of 125IEH. Because the likelihood of DSBs due to direct ionizations in neighboring DNA molecules approaches zero under the current experimental conditions in which (1) the average center-to-center distance between DNA molecule and 125IEH is ~320 nm [based on the number of DNA molecules in one cm3 (1 ml) being 3.06 × 1013, with a single DNA molecule therefore occupying 0.0327 × 109 nm3, and each DNA molecule occupying a cube of the above volume with the length of each side of the cube (320 nm) taken as the center-to-center distance between two DNA molecules], (2) the range of most of the electrons is <40 nm, and (3) all 125IEH molecules are bound to DNA, it can be assumed that LD = 0 and therefore *LD = ~0.8. These assignments are further justified by the following observations: (1) none of the DSBs formed by the decay of 125I free in solution (unbound to DNA) are caused by direct ionization even when the average distance of 125I to pUC19 DNA molecules is 42 nm (18, 19); (2) the probability of SSBs and DSBs in prokaryotic DNA molecules within which a DNA-incorporated 125I atom decays is very high (16, 34, 35) as a consequence of the high ionization density in the immediate vicinity around the decaying atom and the much lower density in neighboring DNA molecules (18, 19); and (3) in supercoiled DNA, 125IEH-induced DSBs arise only from direct effects and are confined to the DNA molecule within which the 125I decays (19, 20). The DSB yield per 10 decays would lead to the disappearance of 16 linear DNA molecules, eight by indirect ionizations and eight by direct mechanisms. Since each DSB in the latter case must have been generated by the decay of one 125I atom, the decay of the remaining two 125I atoms must have caused one DSB in each of two linear DNA molecules (to which they were bound) by •OH-mediated DSB (*LID). These results (Table 2) would indicate that (1) 80% of DSBs in the DNA molecules within which a decay occurs are caused by direct effects [(0.8 *LD/0.8 *LD + 0.2 *LID) × 100] and (2) 20% of DSBs are caused by indirect effects [(0.2 *LID/0.8 *LD + 0.2 *LID) × 100] and (2) 100% of DSBs generated in neighboring molecules are mediated by indirect effects [(0.6 LID/0.0 LD + 0.6 LID) × 100].
TABLE 2.
Analysis of DSB Yields in Linear pUC19 DNA per 125IEH Decay
| Source of DSB | DSB yield per 125I decay | Percentage | ||
|---|---|---|---|---|
| * LD | ~0.8 |
|
~50% |
|
| LD | 0 | 0% | ||
| * LID | ~0.2 |
|
~10% |
|
| LID | 0.6 | ~40% | ||
Notes. D: Direct ionization(s). ID: •OH-mediated indirect ionization(s).
L: Linear DNA molecule whose DSB is consequent to decay of 125IEH that is bound to it (internal DSB). L: Linear DNA molecule whose DSB is consequent to decay of 125IEH bound to neighboring molecule(s) (external DSB).
For the torsionally relaxed nicked form of pUC19 DNA, the mechanism of DSB production is reminiscent of that in the linear form, as might be expected with the absence of torsional stress. In the absence of DMSO, both direct (1.05 ± 0.07) and indirect (0.69 ± 0.13) effects contribute to a total DSB yield per decay of 1.74 ± 0.11 (Table 1). The DSB yield without DMSO is approximately three times higher than that in the supercoiled form, and the yield with DMSO is twice that in supercoiled DNA, whereas the yield from direct effects in the nicked form (1.05 ± 0.07) is somewhat higher than that in the linear form (0.76 ± 0.03). These differences may be attributed to the circularity of the nicked form, which brings a DNA strand of the same molecule closer to the decay site. An analysis similar to that for the linear form (Table 3) discloses that the DSB yield due to direct ionization of DNA by bound 125IEH decays (*ND) is unity, and that from direct ionization in neighboring DNA molecules (ND) is zero. DSBs mediated by indirect effects (~0.7 per decay) are generated only by 125IEH decays occurring on neighboring DNA molecules (NID). Therefore, for the nicked form of plasmid DNA, it appears that (1) ~60% of DSBs are produced by direct effects and ~40% by indirect effects, (2) all DSBs caused by direct effects occur on the DNA molecule to which decaying 125IEH is bound, and (3) all DSBs mediated by indirect effects take place on DNA molecules surrounding the one to which the decaying 125IEH is bound.
TABLE 3.
Analysis of DSB Yields in Nicked/Relaxed pUC19 DNA per 125IEH Decay
| Source of DSB | DSB yield per 125I decay | Percentage | ||
|---|---|---|---|---|
| * ND | ~1.0 |
|
~59% |
|
| ND | 0 | 0% | ||
| * NID | 0 |
|
0% |
|
| NID | ~0.7 | ~41% | ||
Notes. D: Direct ionization(s). ID: •OH-mediated indirect ionization(s).
N: Nicked DNA molecule whose DSB is consequent to decay of 125IEH that is bound to it (internal DSB). N: Nicked DNA molecule whose DSB is consequent to decay of 125IEH bound to neighboring molecule(s) (external DSB).
It is clear that the mode of DSB production in linear and nicked plasmid DNA is different from that in their supercoiled counterpart. From our data (Table 1), the following statements can be made: (1) In linear and nicked DNA, DSBs are caused by both direct and indirect mechanisms, whereas in supercoiled DNA only direct effects operate; (2) even the unscavengable DSB yields of 0.76 ± 0.03 and 1.05 ± 0.07 obtained with DMSO for linear and nicked DNA, respectively, due to the direct action of Auger electrons, are greater than the DSB yield of 0.52 ± 0.02 measured for supercoiled DNA (P < 0.0001). Because the direct action of Auger electrons is confined to a highly localized decay site, the higher values may be attributed to local conformational differences at or around the decay site/ligand-binding site of the various DNA forms: DNA ± 10 bp from the decay site is rather straight for linear and nicked pUC19 DNA but bent and conformationally different for supercoiled DNA, although globally all three topological forms adopt B DNA structure.
Mechanisms of Double-Strand Break Production in Supercoiled, Nicked and Linear DNA
Ionizing radiation (external beam such as X rays, γ rays, pulsed electrons) in general has been shown to produce DSBs in naked DNA predominantly through •OH-mediated indirect effects. The radiosensitivity of supercoiled DNA to pulsed electrons (400 keV) is almost the same as that of relaxed plasmid DNA, ruling out a role for supercoiling (torsional stress/compaction/conformation) in the efficiency of DSB production (10). This conclusion is convincing because (1) external-beam radiation (both high-energy photons and electrons) generates DSBs predominantly through •OH-mediated indirect mechanisms, (2) the distribution of •OH formed during irradiation is uniform throughout the DNA solution, (3) the chances of a DNA strand encountering an •OH are the same throughout the solution at any given dose regardless of whether the DNA is or is not supercoiled, and (4) the radicals are very small in size and can diffuse to the sugar-phosphate backbone of naked DNA whether it is compacted and bent due to supercoiling or has an extended shape in the relaxed circular or linear state.
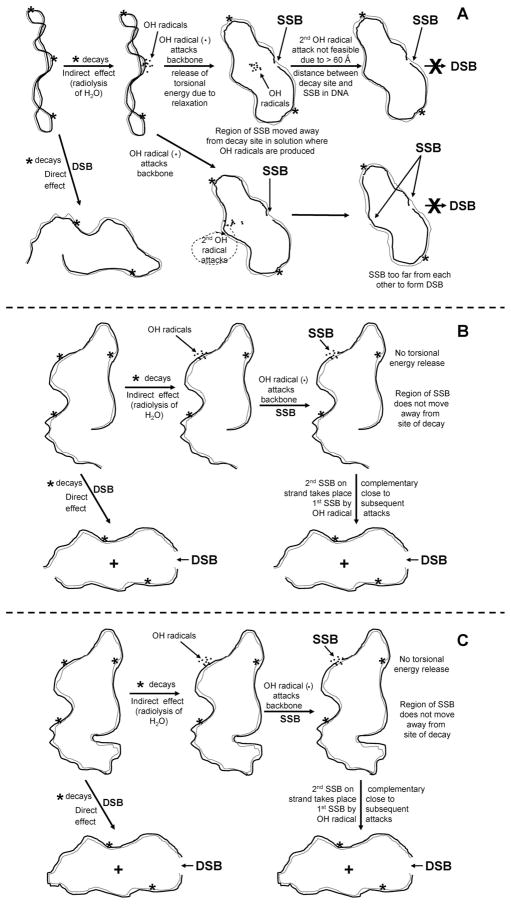
For DNA-bound internal emitters such as the low-energy Auger electron emitter 125I, covalently attached to a minor-groove-binding Hoechst ligand, DNA supercoiling plays a major role in determining the efficiency of DSB production (Table 1). To account for the occurrence of both direct and indirect mechanisms in the absence of DNA supercoiling, we provide a qualitative explanation based on the dynamics of the transition, upon nicking, from supercoiled to relaxed state. An explanation for the absence of indirect-effect-mediated DSBs in supercoiled DNA, based on the dynamics of relaxation of supercoiled DNA, is shown schematically in Fig. 7A. When an 125I atom within an 125IEH molecule bound to supercoiled DNA decays, the following events lead to the appearance of either nicked or linear DNA molecule(s): (1) Simultaneous strand scissions in the complementary strands separated by ~10 bp or less produce a DSB in supercoiled DNA, which we have previously shown to be caused solely by direct ionization of the DNA backbone (19, 20), and (2) scission in one of the two complementary strands results in relaxation of supercoiled DNA and formation of nicked DNA, with ~90% of the relaxation induced through indirect •OH-mediated action and 10% by direct ionization of the DNA backbone of one of the complementary DNA strands (18–20). We propose that when an •OH produced by the radiolysis of water molecules around the decaying 125I atom reacts with one of the two DNA backbones and nicks a supercoiled DNA molecule, the release of torsional energy by the latter (process of relaxation) displaces the nicked region away from the cluster of •OH around the decaying atom by more than 60 Å, the range of •OH (36), so that either the damaged area becomes totally inaccessible to another •OH attack or, alternatively, the additional SSBs formed are too distant from the original SSB to generate a DSB (Fig. 7A). This dynamic model explains why contributions from indirect effects are absent in supercoiled DNA. For γ rays, where the density of •OH in solution is uniform, the formation of DSBs would not be affected by such a mechanism since the process of relaxation after an •OH attack and the formation of an SSB would not spare the complementary DNA strand from further •OH attacks proximal to the first SSB. On the other hand, the nicking of one of the strands in a nicked or linear DNA molecule by an •OH does not induce dynamic changes as would occur in the case of relaxation of a supercoiled DNA molecule. In the absence of supercoiling, the region on the DNA strand with the first nick caused by •OH attack remains at the site of decay since there are no dynamic changes in topology, thus making it vulnerable to subsequent •OH attacks that can cut the complementary strand within 10 bp from the nick, leading to a DSB (Fig. 7B and C). The behavior of nicked relaxed DNA fits well within the framework of our model with lack of dynamic changes resulting from the relaxation of torsional stress. The half-life of hydroxyl radicals in the aqueous environment inside cells is 10−9 s (37). Recent work by Crut et al. (38) using magnetic tweezers as a probe to measure the kinetics of plectoneme removal suggests that the complete relaxation of the supercoiled form of pUC19 DNA would take 8 × 10−3 s, a time scale much greater than the half-life of hydroxyl radicals, indicating that our model is less likely to be true. However, supercoiling simulated in these experiments is unnatural. Furthermore, the dependence of the relaxation kinetics on the strength of the magnetic force and the unidirectional movement of the beads used to detect the relaxation of supercoils upon nicking make this an artificial system. Therefore, it is not possible to rule out or to negate our dynamic model on the basis of these findings. Although the time of relaxation of a covalently closed, circular, supercoiled DNA upon nicking has not been measured directly, theoretical calculations based on the hydrodynamics of DNA movement in solution suggest that it may take up to a few hundred nanoseconds for a supercoiled DNA of size similar to that of pUC19 to relax completely (39). However, for our model (Fig. 7) to be valid, it is actually sufficient if the segmental dynamics of relaxation at the site of nicking (±10 bp) occur within the time frame of the T1/2 of hydroxyl radicals.
FIG. 7.
Proposed mechanisms for DSB formation in pUC19 DNA caused by 125I decays from minor-groove-bound 125IEH. Supercoiled DNA (panel A), linear DNA (panel B), nicked DNA (panel C).
We have proposed this mechanism as a possibility in the absence of any other reasonable explanation. Our model (Fig. 7) is highly speculative and hypothetical. Nevertheless, we have shown that the magnitudes and mechanisms of DSBs produced by 125IEH for two extreme topologies, namely, DNA with physiological supercoiling and DNA without supercoiling, are radically different. Further experiments with DNA topoisomers are under way to explore the relationship between linking number and 125IEH-induced DSBs. On the basis of our current work, we would expect a gradual increase in indirect effect-mediated DSBs in topoisomers with a decrease in the number of supercoils. Although currently we do not have experimental evidence to support our dynamic model, we believe it reasonably explains our observations.
Conclusion
Our data indicate that the DSB yield in plasmid DNA irradiated by low-energy electrons emitted after 125I decay varies by a factor of three depending on the DNA form: 0.52 ± 0.02, 1.74 ± 0.11 and 1.62 ± 0.07 per 125IEH decay for supercoiled, nicked and linear DNA, respectively. Furthermore, whereas the DSBs in the supercoiled form are caused solely by direct ionizations, those induced in nicked and linear forms are consequent to direct ionizations and •OH-mediated indirect ionizations involving neighboring DNA molecules. We hypothesize that the dynamics of relaxation of supercoiled plasmid DNA immediately after the first nick in one of the two strands of double-stranded DNA is responsible for the differences in DSB production efficiencies and their underlying mechanisms.
Acknowledgments
This work was supported in part by NIH 5 R01 CA015523 (A. I. Kassis). P. Balagurumoorthy was a National Research Service Awardee under NIH 5 T32 CA009078 (B. F. Demple).
References
- 1.Leonard MW, Patient RK. Evidence for torsional stress in transcriptionally activated chromatin. Mol Cell Biol. 1991;11:6128–6138. doi: 10.1128/mcb.11.12.6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thanbichler M, Wang SC, Shapiro L. The bacterial nucleoid: A highly organized and dynamic structure. J Cell Biochem. 2005;96:506–521. doi: 10.1002/jcb.20519. [DOI] [PubMed] [Google Scholar]
- 3.Wang JC, Lynch AS. Transcription and DNA supercoiling. Curr Opin Genet Dev. 1993;3:764–768. doi: 10.1016/s0959-437x(05)80096-6. [DOI] [PubMed] [Google Scholar]
- 4.Adhya S. Multipartite genetic control elements: Communication by DNA loop. Annu Rev Genet. 1989;23:227–250. doi: 10.1146/annurev.ge.23.120189.001303. [DOI] [PubMed] [Google Scholar]
- 5.Sarkar PS, Bagga R, Balagurumoorthy P, Brahmachari SK. A novel approach to design of cis-acting DNA structural elements for regulation of gene expression. in vivo Curr Sci. 1991;60:586–591. [PubMed] [Google Scholar]
- 6.Sumners DW, Ernst C, Spengler SJ, Cozzarelli NR. Analysis of the mechanism of DNA recombination using tangles. Q Rev Biophys. 1995;28:253–313. doi: 10.1017/s0033583500003498. [DOI] [PubMed] [Google Scholar]
- 7.Kornberg A, Baker TA. DNA Replication. 2. W. H. Freeman; New York: 1992. [Google Scholar]
- 8.Miller JH, Nelson JM, Ye M, Swenberg CE, Speicher JM, Benham CJ. Negative supercoiling increases the sensitivity of plasmid DNA to single-strand break induction by X-rays. Int J Radiat Biol. 1991;59:941–949. doi: 10.1080/09553009114550831. [DOI] [PubMed] [Google Scholar]
- 9.Milligan JR, Arnold AD, Ward JF. The effect of superhelical density on the yield of single-strand breaks in γ-irradiated plasmid DNA. Radiat Res. 1992;132:69–73. [PubMed] [Google Scholar]
- 10.Ozols A, Prise KM, Michael BD. A comparison of the radiosensitivity of relaxed and supercoiled plasmid DNA. Int J Radiat Biol. 1999;75:83–90. doi: 10.1080/095530099140834. [DOI] [PubMed] [Google Scholar]
- 11.Auger P. Sur les rayons β secondaires produits dans un gaz par des rayons X. C R Hebd Seances Acad Sci. 1925;180:65–68. [Google Scholar]
- 12.Sastry KSR, Rao DV. Dosimetry of low energy electrons. In: Rao DV, Chandra R, Graham MC, editors. Physics of Nuclear Medicine: Recent Advances. American Institute of Physics; Woodbury, NY: 1984. pp. 169–208. [Google Scholar]
- 13.Charlton DE, Pomplun E, Booz J. Some consequences of the Auger effect: Fluorescence yield, charge potential, and energy imparted. Radiat Res. 1987;111:553–564. [PubMed] [Google Scholar]
- 14.Kassis AI, Sastry KSR, Adelstein SJ. Kinetics of uptake, retention, and radiotoxicity of 125IUdR in mammalian cells: Implications of localized energy deposition by Auger processes. Radiat Res. 1987;109:78–89. [PubMed] [Google Scholar]
- 15.Pomplun E, Booz J, Charlton DE. A Monte Carlo simulation of Auger cascades. Radiat Res. 1987;111:533–552. [PubMed] [Google Scholar]
- 16.Kassis AI. The amazing world of Auger electrons. Int J Radiat Biol. 2004;80:789–803. doi: 10.1080/09553000400017663. [DOI] [PubMed] [Google Scholar]
- 17.Charlton DE, Humm JL. A method of calculating initial DNA strand breakage following the decay of incorporated 125I. Int J Radiat Biol. 1988;53:353–365. doi: 10.1080/09553008814552501. [DOI] [PubMed] [Google Scholar]
- 18.Kassis AI, Harapanhalli RS, Adelstein SJ. Comparison of strand breaks in plasmid DNA after positional changes of Auger electron-emitting iodine-125. Radiat Res. 1999;151:167–176. [PubMed] [Google Scholar]
- 19.Kassis AI, Harapanhalli RS, Adelstein SJ. Strand breaks in plasmid DNA after positional changes of Auger electron-emitting iodine-125: Direct compared to indirect effects. Radiat Res. 1999;152:530–538. [PubMed] [Google Scholar]
- 20.Balagurumoorthy P, Chen K, Bash RC, Adelstein SJ, Kassis AI. Mechanisms underlying production of double-strand breaks in plasmid DNA after decay of 125I-Hoechst. Radiat Res. 2006;166:333–344. doi: 10.1667/RR3591.1. [DOI] [PubMed] [Google Scholar]
- 21.Kandaiya S, Lobachevsky PN, D’Cunha G, Martin RF. DNA strand breakage by 125I-decay in a synthetic oligodeoxynucleotide: Fragment distribution and evaluation of DMSO protection effect. Acta Oncol. 1996;35:803–808. doi: 10.3109/02841869609104030. [DOI] [PubMed] [Google Scholar]
- 22.Panyutin IG, Neumann RD. Radioprobing of DNA: Distribution of DNA breaks produced by decay of 125I incorporated into a triplex-forming oligonucleotide correlates with geometry of the triplex. Nucleic Acids Res. 1997;25:883–887. doi: 10.1093/nar/25.4.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walicka MA, Adelstein SJ, Kassis AI. Indirect mechanisms contribute to biological effects produced by decay of DNA-incorporated iodine-125 in mammalian cells in vitro: Double-strand breaks. Radiat Res. 1998;149:134–141. [PubMed] [Google Scholar]
- 24.Kassis AI, Walicka MA, Adelstein SJ. Double-strand break yield following 125I decay: Effects of DNA conformation. Acta Oncol. 2000;39:721–726. doi: 10.1080/028418600750063785. [DOI] [PubMed] [Google Scholar]
- 25.Elmroth K, Stenerlöw B. DNA-incorporated 125I induces more than one double-strand break per decay in mammalian cells. Radiat Res. 2005;163:369–373. doi: 10.1667/rr3339. [DOI] [PubMed] [Google Scholar]
- 26.Booz J, Paretzke HG, Pomplun E, Olko P. Auger-electron cascades, charge potential and microdosimetry of iodine-125. Radiat Environ Biophys. 1987;26:151–162. doi: 10.1007/BF01211409. [DOI] [PubMed] [Google Scholar]
- 27.Lobachevsky PN, Martin RF. Iodine-125 decay in a synthetic oligodeoxynucleotide. II. The role of Auger electron irradiation compared to charge neutralization in DNA breakage. Radiat Res. 2000;153:271–278. doi: 10.1667/0033-7587(2000)153[0271:idiaso]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 28.Li WB, Friedland W, Jacob P, Panyutin IG, Paretzke HG. Simulation of 125I decay in a synthetic oligodeoxynucleotide with normal and distorted geometry and the role of radiation and non-radiation actions. Radiat Environ Biophys. 2004;43:23–33. doi: 10.1007/s00411-004-0231-1. [DOI] [PubMed] [Google Scholar]
- 29.Harapanhalli RS, McLaughlin LW, Howell RW, Rao DV, Adelstein SJ, Kassis AI. [125I/127I]iodoHoechst 33342: Synthesis, DNA binding, and biodistribution. J Med Chem. 1996;39:4804–4809. doi: 10.1021/jm9602672. [DOI] [PubMed] [Google Scholar]
- 30.Brahmachari SK, Shouche YS, Cantor CR, McClelland M. Sequences that adopt non-B-DNA conformation in form V DNA as probed by enzymic methylation. J Mol Biol. 1987;193:201–211. doi: 10.1016/0022-2836(87)90637-1. [DOI] [PubMed] [Google Scholar]
- 31.Panyutin IV, Luu AN, Panyutin IG, Neumann RD. Strand breaks in whole plasmid DNA produced by the decay of 125I in a triplex-forming oligonucleotide. Radiat Res. 2001;156:158–166. doi: 10.1667/0033-7587(2001)156[0158:sbiwpd]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 32.Saenger W. Principles of Nucleic Acid Structure. Springer-Verlag; New York: 1984. [Google Scholar]
- 33.Cowan R, Collis CM, Grigg GW. Breakage of double-stranded DNA due to single-stranded nicking. J Theor Biol. 1987;127:229–245. doi: 10.1016/s0022-5193(87)80133-9. [DOI] [PubMed] [Google Scholar]
- 34.Krisch RE, Ley RD. Induction of lethality and DNA breakage by the decay of iodine-125 in bacteriophage T4. Int J Radiat Biol. 1974;25:21–30. doi: 10.1080/09553007414550021. [DOI] [PubMed] [Google Scholar]
- 35.Linz U, Stöcklin G. Chemical and biological consequences of the radioactive decay of iodine-125 in plasmid DNA. Radiat Res. 1985;101:262–278. [PubMed] [Google Scholar]
- 36.Roots R, Okada S. Estimation of life times and diffusion distances of radicals involved in X-ray-induced DNA strand breaks or killing of mammalian cells. Radiat Res. 1975;64:306–320. [PubMed] [Google Scholar]
- 37.Hall EJ. Radiobiology for the Radiologist. 4. J. B. Lippincott; Philadelphia: 1994. [Google Scholar]
- 38.Crut A, Koster DA, Seidel R, Wiggins CH, Dekker NH. Fast dynamics of supercoiled DNA revealed by single-molecule experiments. Proc Natl Acad Sci USA. 2007;104:11957–11962. doi: 10.1073/pnas.0700333104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marko JF. DNA under high tension: Overstretching, undertwisting, and relaxation dynamics. Phys Rev E. 1998;57:2134–2149. [Google Scholar]