Abstract
A variety of experimental methods have been developed for the analysis of protein interactions. The majority of these methods either require disruption of the cells to detect molecular interactions or rely on indirect detection of the protein interaction. The bimolecular fluorescence complementation (BiFC) assay provides a direct approach for the visualization of molecular interactions in living cells and organisms. The BiFC approach is based on the facilitated association between two fragments of a fluorescent protein when the fragments are brought together by an interaction between proteins fused to the fragments. The BiFC approach has been used for visualization of interactions among a variety of structurally divers interaction partners in many different cell types and. It enables detection of transient complexes as well as complexes formed by a subpopulation of the interaction partners. It is essential to include negative controls in each experiment in which the interface between the interaction partners has been mutated or deleted. The BiFC assay has been adapted for simultaneous visualization of multiple protein complexes in the same cell and the competition for shared interaction partners. A ubiquitin-mediated fluorescence complementation (UbFC) assay has also been developed for visualization of the covalent modification of proteins by ubiquitin family peptides. These fluorescence complementation assays have a great potential to illuminate a variety of biological interactions in the future.
1. Introduction
Most cellular functions are regulated and executed by protein complexes. Extensive networks of protein interactions have been identified through both biochemical and genetic approaches. Visualization of protein complexes in living cells enables determination whether putative interactions occur in the normal cellular environment and identification of the subcellular locations of these complexes. The bimolecular fluorescence complementation (BiFC) approach enables visualization of protein complexes in living cells and organisms. It is based on the discoveries that the association between two nonfluorescent fragments of a fluorescent protein can produce a fluorescent complex and that this association can be facilitated by an interaction between proteins fused to the fragments. The BiFC assay has been used to visualize interactions between proteins from many different structural classes in a variety of cell types and species. The BiFC approach provides several unique advantages for the investigation of molecular complexes in living cells. Modified versions of the BiFC assay have been used to visualize the competition between alternative interaction partners and the covalent modification of proteins by ubiquitin family peptides. The BiFC approach is applicable for the visualization of a wide range of molecular interactions.
The following sections will briefly discuss the significance of protein interactions and the methods that are available for their study. The historical development of complementation strategies for the analysis of protein interactions is outlined briefly and the direct visualization of interactions using BiFC analysis is compared with a subset of other imaging approaches. The fundamental principles of BiFC analysis and the requirements for the application of this approach are reviewed. Instrumentation that can be used to conduct BiFC assays is described in general terms. A selection of interactions that have been visualized using the BiFC assay and the cell types and organisms in which this assay has been used are listed. Some of the limitations and assumptions of the BiFC approach are discussed. The extensions of BiFC analysis for the simultaneous visualization of multiple protein interactions and the visualization of ubiquitin conjugates are described. Finally, some future opportunities and challenges are presented.
1.1 Roles of protein interactions in regulatory complexity
The mammalian genomes that have been sequenced contain only slightly more annotated genes than the genomes of plants, insects and nematodes. Thus, the arguably more complex developmental programs and adaptive responses of mammals are not specified by a dramatically larger number of genes. The principal mechanism whereby mammals achieve a greater number of biological functions than the number of proteins encoded in their genomes is through combinatorial interactions among these proteins. Indeed, most regulatory mechanisms in mammalian cells rely on the activities of protein complexes rather than the actions of individual proteins. Through interactions with different partners under different conditions, each protein can perform different functions in different cell types and in response to different extracellular stimuli (see Box 1 on the complexity generated by combinatorial interactions). For instance, a transcription factor can bind to thousands of loci in a mammalian genome. However, in a particular cell type at any one time, the protein may regulate only a few genes. This combination of great versatility and context-dependent specificity is made possible by interactions that specify the functions of a protein in each cell.
Box 1. Combinatorial interactions can generate complexity.
The connectivity of a protein interaction network can be characterized by the average number of interaction partners for each protein. The distribution of the number of interaction partners about this mean varies for different types of networks (random, scale-free etc.). The number of binary complexes that can be formed among proteins encoded by a genome is a product of the number of proteins and the number of interaction partners for each protein. Thus, for interaction networks with the same connectivity, the number of complexes is proportional to the square of the number of proteins. The total number of protein complexes is also dramatically affected by the average number of proteins in a complex and the distribution about that average. An increase in the average number of components in each complex results in a dramatic increase in the total number of complexes. The combination of higher connectivity and a larger number of components in each complex can therefore produce a large increase in the number of protein complexes without a change in the size of the proteome.
2. Approaches for the investigation of protein interactions
Protein interactions have been investigated using many approaches under different experimental conditions. Most of the experimental approaches that enable direct detection of interactions such as biochemical co-purification require removal of the proteins from their normal environment. Conversely, most studies of protein interactions in their normal environment such as genetic analysis of the combined effects of mutations rely on indirect consequences of the interactions. The combined use of genetic and biochemical approaches has identified thousands of potential protein interactions. The largely non-overlapping interaction networks identified by high-throughput interaction screens at the proteomic level (Ito, et al., 2001, Gavin, et al., 2002, Giot, et al., 2003, Li, et al., 2004, Stelzl, et al., 2005) suggest that the number of potential protein complexes is orders of magnitude larger than the number of proteins encoded by the genome. The cell specificity and the subcellular localization of the vast majority of these complexes remain unknown. It is also possible that some of the interactions identified under non-native conditions do not occur in cells.
2.1 Studies of protein interactions using complementation assays
Complementation approaches enable investigation of protein interactions in their normal environment, and can allow direct detection of the interaction. Complementation between protein fragments was first observed between proteolytic fragments of ribonuclease (Richards, 1958) and has been subsequently described for many proteins. Importantly, complementation between some protein fragments is conditional, and can be facilitated by tethering the fragments in close proximity to each other (Table 1) (Kerppola, 2006). This enables the detection of protein interactions and other molecular processes in living cells (see Box 2 on the detection of protein interactions using complementation approaches). Complementation between fragments of a variety of different proteins enables detection of protein interactions in living cells. However, visualization of the subcellular localization of protein complexes requires that the function produced by complementation can be detected with high spatial resolution. This is possible by using fluorescent ligands that bind to the complementation complex or by using bimolecular fluorescence complementation, which produces an intrinsically fluorescent complex.
Table 1.
Comparison of complementation methods using fragments of different proteins.
| Protein | Detection | Spatial resolutiona |
Time resolutiona | Experimental systemsa | Reference |
|---|---|---|---|---|---|
| Ubiquitin | Ub-protease coupled reporters |
Cell population | Day | Yeast | (Rossi, et al., 1997) |
| β-galactosidase | FDG hydrolysis | Cellular | Hours | Cultured cells, D. melanogaster |
(Johnsson and Varshavsky, 1994) |
| Dihydrofolate reductase |
Fl-MTX binding | Sub-cellular | Minutes | Cultured cells, plants | (Pelletier, et al., 1998) |
| GFP variants | Intrinsic fluorescence | Sub-cellular | Minutes-Hours | Cultured cells, plants, fungi | (Hu, et al., 2002) (Ghosh, et al., 2000) |
|
SynechocystisdnaE intein |
Reporter ligation | Cell population | Hours | Cultured, implanted cells | (Ozawa, et al., 2001) |
| β-lactamase | CCF2/AM hydrolysis | Cellular | Minutes | Cultured cells, primary neurons |
(Galarneau, et al., 2002, Spotts, et al., 2002, Wehrman, et al., 2002) |
| Firefly luciferase | Luciferin hydrolysis | Cell population | Hours | Cultured, implanted cells | (Paulmurugan, et al., 2002) |
| Renilla luciferase | Coelenterazine luminescence |
Cell population | Minutes-Hours | Cultured, implanted cells | (Paulmurugan and Gambhir, 2003) |
| Gaussia luciferase | Coelenterazine luminescence |
Cell population | Minutes | Cultured cells | (Remy and Michnick, 2006) |
| TEV protease | Coupled reporters | Cellular | Minutes | Cultured cells | (Wehr, et al., 2006) |
The spatial and time resolution as well as the experimental systems used reflect those reported in publications using these approaches, and are not intended to represent the limits of performance of these methods.
Box 2. Complementation approaches for the detection of protein interactions.
Many proteins can be divided into fragments that can associate to produce a functional complex. This phenomenon is classically known as complementation by analogy with the ability of different mutations to complement each other to produce an organism with a normal phenotype. Complementation between protein fragments is due to the large favorable free energy of folding of most proteins and the fact that many proteins fold in multiple stages that involve initial interactions among neighboring amino acid residues and subsequent interactions between partially folded secondary structure elements. Nevertheless, the great majority of protein fragments are unable to produce a functional complex because of misfolding or the inability of partially folded fragments to associate with each other. Only a small subset of protein fragments has the potential to associate to form a functional complex. Experimental applications of protein fragment complementation are therefore critically dependent on the identification of fragments that can associate with each other under relevant conditions.
The correct folding of a protein occurs in competition with misfolding, which is often irreversible. The ability of protein fragments to associate with each other can depend on the frequency of their collisions, which depends on the effective local concentrations of the fragments. Thus, the association of some protein fragments can be facilitated by tethering them in proximity to each other. The association of such fragments can be conditional on their molecular proximity even if the fragments can associate independent of tethering when they are present at sufficiently high concentrations. The conditional association of protein fragments is a powerful reporter of molecular proximity, and can be used to investigate many biological processes that involve changes in molecular proximity, including protein interactions, nucleoprotein complex formation and covalent protein modifications (Hu, et al., 2002, Rackham and Brown, 2004, Fang and Kerppola, 2004, Stains, et al., 2005).
Fragments of many proteins have been identified that can complement each other under specific experimental conditions (see Table 1 for a small subset). The nature of the fragments that can undergo complementation varies between different proteins. In many cases the fragments are predicted to exist in a partially unfolded state due to the absence of interactions that are necessary for formation of some secondary structure elements within the individual fragments. Unfortunately, the structures of protein fragments that can undergo complementation as well as intermediates in their folding pathway have not been characterized. Thus, the characteristics of protein fragments required for complementation are unknown. The identification of protein fragments that can support complementation therefore remains a largely empirical process.
It is not clear why some protein fragments exhibit conditional complementation whereas others apparently associate with each other independent of mechanisms that would tether them in proximity to each other. It is possible that differences in the folding pathways of the fragments alter their susceptibility to misfolding, and affect the probability of productive association. Nevertheless, it is likely that all fragments that exhibit conditional complementation also have the potential to associate with each other independent of tethering under some conditions such as high protein concentrations.
2.2 Visualization of protein interactions in living cells
Direct visualization of protein interactions in living cells enables validation of complex formation in the normal environment and determination of their subcellular localization. Two principal methods have been used to visualize the localization of protein interactions in living cells. Fluorescence resonance energy transfer (FRET) analysis is based on changes in the fluorescence intensities and lifetimes of two fluorophores that are brought sufficiently close together. Bimolecular fluorescence complementation (BiFC) analysis is based on the formation of a fluorescent complex by fragments of fluorescent proteins whose association is facilitated by an interaction between proteins fused to the fragments. Other methods such as fluorescence correlation spectroscopy (FCS) (Brock and Jovin, 1998), image correlation spectroscopy (Petersen, et al., 1993) and complementation approaches using fragments of other proteins (Table 1) have also been used. Bioluminescence resonance energy transfer (BRET) analysis enables detection of protein interactions in live cells, but current methods do not enable visualization of the subcellular locations of the protein complexes (Pfleger and Eidne, 2006). Studies of protein interactions using the FRET assay have been described in several recent reviews (Zhang, et al., 2002, Jares-Erijman and Jovin, 2003, Miyawaki, 2003, Zal and Gascoigne, 2004, Bunt and Wouters, 2004, Galbraith, 2004, Schmid and Neumeier, 2005). This review focuses on critical principles and assumptions underlying the BiFC assay and on selected applications of this approach.
2.3 Challenges for the visualization of protein interactions
One major challenge for the direct detection of protein interactions is that most proteins have multiple partners in the cell, and only a small subpopulation interacts with a particular partner. In most experimental approaches, interactions between the proteins under study with other cellular proteins interfere with detection of the complex being investigated. This problem is often addressed by overexpression of the proteins under study to outcompete endogenous interaction partners and to produce a larger amount of complexes. This strategy carries the risk that protein overexpression produces non-native complexes or alters the characteristics of the complexes that are formed. One advantage of complementation approaches is that complexes that are formed with other partners are not detected, enabling selective observation of the complex under investigation. A fundamental difference between the FRET and BiFC approaches is that FRET analysis is based on measurement of the difference in fluorescence intensity or lifetime of one fluorophore in the presence and absence of a second fluorophore. In contrast BiFC analysis is based on the formation of a fluorescent complex from non-fluorescent constituents. This makes BiFC analysis potentially more sensitive and avoids interference from changes in fluorescence intensity or lifetime caused by cellular conditions unrelated to protein interactions. Conversely, FRET provides the potential for real-time observation of complex formation and dissociation, whereas BiFC analysis does not enable real-time detection of complex dynamics.
3. Bimolecular fluorescence complementation analysis
BiFC analysis is based on the formation of a fluorescent complex when two fragments of a fluorescent protein are brought together by an interaction between proteins fused to the fragments (see Fig. 1). In E. coli, fluorescence complementation was first detected using fragments of a green fluorescent protein (GFP) variant fused to artificial peptide sequences designed to form an anti-parallel coiled coil (Ghosh, et al., 2000). In mammalian cells, expression of YFP fragments fused to calmodulin and its target peptide was shown to produce fluorescence that was modulated by the Ca++ concentration (Nagai, et al., 2001). However, fluorescence complementation by these fragments was not shown to require calmodulin binding to M13 and the fragments have not been used for studies of other interactions. Conditional fluorescence complementation was demonstrated between fragments of the enhanced yellow fluorescent protein (YFP) fused to transcription factors containing the basic region – leucine zipper (bZIP) domain or the Rel domain (Hu, et al., 2002). Mutations that prevented the interactions reduced the fluorescence, demonstrating that the native protein interactions facilitated fluorescence complementation (Hu, et al., 2002). The same fragments were shown to support complementation when fused to other bZIP or Rel family proteins (Hu, et al., 2002). Subsequent studies have shown that these fragments can be used to study interactions among a variety of structurally unrelated proteins, validating the BiFC approach as a general strategy for the visualization of molecular interactions in living cells (Kerppola, 2006).
Figure 1. Principle of Bimolecular Fluorescence Complementation (BiFC).
BiFC analysis is based on the facilitated association between two fluorescent protein fragments when they are brought together by an interaction between proteins fused to the fragments. The individual fragments are non-fluorescent. Please see the text for factors that can influence bimolecular fluorescent complex formation. The image on the right shows BiFC analysis of complexes formed between Fos and Jun transcription factors. Image acquired by Changdeng Hu (Hu, et al., 2002).
3.1 Requirements for bimolecular fluorescence complementation analysis
BiFC analysis does not require information about the structures of the interaction partners or of their interaction interface. The association of the fluorescent protein fragments fused to the interaction partners does not require that the interaction partners position the fragments in a specific orientation or within a fixed distance from each other. Nevertheless, steric constrains can prevent association of the fragments within a complex. The fragments must have sufficient freedom of motion in the complex to collide with each other and to undergo the mutually induced folding required to form the β-barrel structure. Flexible linkers between the interaction partners and the fluorescent protein fragments can uncouple the motions of the fluorescent protein fragments from those of the interaction partners in the complex and may facilitate bimolecular fluorescent complex formation. The interaction partners do not need to form a complex with a long half-life since transient interactions can be trapped by the association of the fluorescent protein fragments (see Box 3 on the dynamics of BiFC complexes). Additionally, it is not necessary for a large fraction of the interaction partners to associate with each other in order to detect fluorescence complementation because cells have low background fluorescence and unassociated fragments do not interfere with detection of an interaction using the BiFC assay.
Box 3. Dynamics of bimolecular fluorescent complexes.
 The dynamics of bimolecular fluorescence complementation have been investigated in order to elucidate the pathway for fluorescent complex formation (Hu, et al., 2002). In vitro studies using purified proteins indicate that the initial association between the fusion proteins (complex I) is mediated by the interaction partners. This interaction occurs in competition with mutually exclusive interactions with alternative interaction partners (complexes II). The association between the fluorescent protein fragments is slower and produces an intermediate (complex III), that undergoes the slow chemical reactions (maturation) required to produce the peptide fluorophore (complex IV). This causes a delay in detection of bimolecular fluorescence complementation following formation of the protein complex (Hu, et al., 2002). The length of this delay depends on the intensity of the signal and the sensitivity and background of the detector. The latter two steps are irreversible under some conditions (Hu, et al., 2002, Magliery, et al., 2005). Some fluorescent protein fragments purified from E. coli as intein fusions can undergo at least partial maturation prior to association, resulting in rapid fluorescence complementation (Demidov, et al., 2006). The same study demonstrated a decrease in the fluorescence intensity when the conditions were adjusted to disfavor the interaction, suggesting that bimolecular fluorescent complex formation can be reversible (dashed arrow). Unassociated fluorescent protein fragments as well as fragments that are present in complexes that do not contain complementary fragments undergo irreversible misfolding in vitro (Hu, et al., 2002) (complexes V).
The dynamics of bimolecular fluorescence complementation have been investigated in order to elucidate the pathway for fluorescent complex formation (Hu, et al., 2002). In vitro studies using purified proteins indicate that the initial association between the fusion proteins (complex I) is mediated by the interaction partners. This interaction occurs in competition with mutually exclusive interactions with alternative interaction partners (complexes II). The association between the fluorescent protein fragments is slower and produces an intermediate (complex III), that undergoes the slow chemical reactions (maturation) required to produce the peptide fluorophore (complex IV). This causes a delay in detection of bimolecular fluorescence complementation following formation of the protein complex (Hu, et al., 2002). The length of this delay depends on the intensity of the signal and the sensitivity and background of the detector. The latter two steps are irreversible under some conditions (Hu, et al., 2002, Magliery, et al., 2005). Some fluorescent protein fragments purified from E. coli as intein fusions can undergo at least partial maturation prior to association, resulting in rapid fluorescence complementation (Demidov, et al., 2006). The same study demonstrated a decrease in the fluorescence intensity when the conditions were adjusted to disfavor the interaction, suggesting that bimolecular fluorescent complex formation can be reversible (dashed arrow). Unassociated fluorescent protein fragments as well as fragments that are present in complexes that do not contain complementary fragments undergo irreversible misfolding in vitro (Hu, et al., 2002) (complexes V).
The non-productive folding of the fluorescent protein fragments that are not in a complex containing a complementary fragment is critical for the specificity of the BiFC assay. Bimolecular fluorescent complex formation is likely to be energetically favorable even when the fluorescent protein fragments are not fused to proteins that interact with each other. However, the stimulation of fluorescence complementation by an interaction between proteins fused to the fragments is likely to be determined by kinetic rather than thermodynamic factors. The efficiency of bimolecular fluorescent complex formation is determined by the frequency of productive collisions between the fluorescent protein fragments relative to the rate of non-productive folding. Fusion of the fragments to interaction partners can increase the rate of potentially productive collisions relative to the rates of non-productive collisions with other proteins in the cell. The spectral characteristics of the bimolecular fluorescent complex and the intact fluorescent protein are indistinguishable, indicating that the β-barrel structure and the tripeptide fluorophore are likely to be identical in the bimolecular complex and the intact protein.
An essential requirement for fusion proteins to be used for BiFC analysis is that the fluorescent protein fragments do not associate with each other efficiently in the absence of an interaction between the proteins fused to the fragments. Spontaneous association between the fluorescent protein fragments can be affected by the characteristics of the proteins fused to the fragments. It is therefore essential to test the requirement for a specific interaction interface for complementation by each combination of interaction partners to be studied using the BiFC approach (Figure 2). Fragments of some fluorescent proteins can form fluorescent complexes in the absence of fusions to specific interaction partners (Cabantous, et al., 2005). Likewise, high-level expression of many fusion proteins containing the fragments used for BiFC analysis can result in fluorescent complex formation independent of specific interactions. The fluorescence produced by spontaneous association of the fragments used for BiFC analysis is frequently reduced when the fragments are fused to proteins that do not interact with each other. It is therefore essential to test the effects of mutations that eliminate the protein interaction on fluorescence complementation and to express the fusion proteins at levels comparable to their endogenous counterparts (Fig. 2).
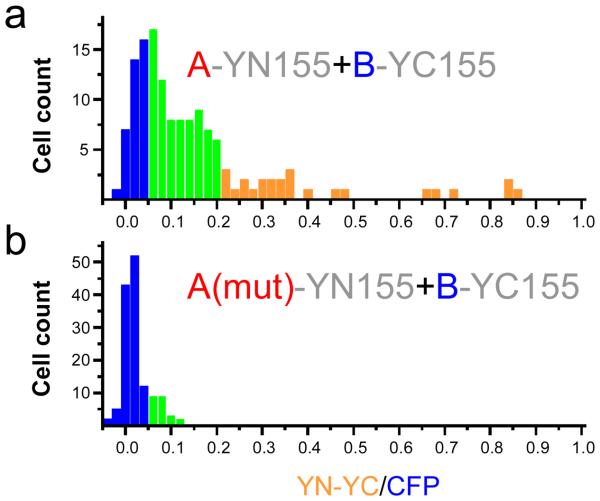
Figure 2. Determination of the specificity of bimolecular fluorescence complementation by mutational analysis.
The specificity of bimolecular fluorescence complementation should be tested by examining the effects of mutations that prevent the association of the interaction partners (data adapted from (Hu, et al., 2002)). The wild type (a) and mutated (b) interaction partners should be expressed at the same concentrations. The plasmids should be co-transfected together with an internal reference (i.e. CFP). The fluorescence intensities produced by bimolecular fluorescence complementation (YN-YC) and the internal reference (CFP) are measured in individual cells. The distribution of ratios between the fluorescence intensities in individual cells are plotted in each histogram.
3.2 Design of fusion proteins for bimolecular fluorescence complementation analysis
The association of the fluorescent protein fragments can be enhanced when they are tethered in the same macromolecular complex. However, the association of the fragments is not determined by molecular proximity alone. Steric constraints can influence the probability of association between the fragments when the fusion proteins form a complex. Fusion proteins that produce optimal signal must generally be identified empirically by testing several combinations of proteins with the fragments fused to different positions. The fluorescence signal produced by bimolecular fluorescence complementation varies widely for interactions between different partners and for different fusions to the same partners. For true interaction partners, it is virtually always possible to find a combination of fusion proteins that produces a detectable signal. Also, since autofluorescence in the visible range is extremely low in most cells, the signal from bimolecular fluorescence complementation is often orders of magnitude higher than cellular background fluorescence.
3.3 Instruments that can be used for bimolecular fluorescence complementation analysis
Fluorescence complementation can be detected using the same instruments that are used to detect intact fluorescent proteins. The fluorescence intensity of cells expressing fluorescent protein fragment fusions that can form bimolecular fluorescent complexes is generally less than 10% of the fluorescence intensity of cells expressing comparable levels of intact fluorescent proteins. It is likely that only a subset of the fragments associate with each other since the fluorescence intensity of BiFC complexes produced in vitro is comparable to that of intact fluorescent proteins (Hu, et al., 2002). It is important to optimize the instrument for detection of the bimolecular fluorescent complex(es) in order to maximize the signal to background ratio of the assay.
Bimolecular fluorescent complex formation in vitro can be monitored using a fluorometer. In such experiments, the signal produced through complex formation between specific interaction partners can be several orders of magnitude greater than the signal produced through complex formation between proteins in which the interaction interface has been mutated (Hu, et al., 2002, Hu and Kerppola, 2003). Bimolecular fluorescent complex formation in cell populations can also be monitored using a fluorometer, but this requires a strong signal and efficient rejection of signal resulting from scatter of the excitation beam.
Microscopy provides the greatest spatial resolution, and often the greatest sensitivity for detection of bimolecular fluorescent complexes. Virtually any fluorescence microscope can be used to determine the subcellular localization of BiFC complexes. In many cases, the signal in cells that express fusions to proteins that interact with each other is more than ten-fold higher than the signal in cells that express non-interacting fusions (Hu, et al., 2002). Microscopy also allows analysis of the variation in fluorescence intensities and subcellular distributions of complexes among different cells in the population. A large number of cells must be analyzed and strategies to avoid experimental bias must be implemented in order to obtain results that are representative for the cell population. It is generally straightforward to visualize multiple BiFC complexes with different spectral characteristics through the use of excitation and emission filters. This enables comparison of interactions among several proteins in the same cell using multicolor BiFC analysis (see section 9).
Flow cytometry enables determination of the fluorescence intensities of a large number of individual cells. An instrument with sensitive and stable detectors and accurate correction for scattered excitation light is necessary to obtain quantitative data. Flow analysis is less susceptible to experimental bias, but requires careful attention to identical treatment of samples in order to allow comparison of data from different cell populations. Multi-wavelength detector systems enable normalization of fluorescence intensities using internal controls and simultaneous analysis of multiple BiFC complexes in each cell.
3.4 Effects of fluorescent protein fragments on fusion protein properties
Similar to other approaches that make use of fusion proteins, it is necessary to examine the possibility that the fluorescent protein fragments fused to the interaction partners alter their functions. Ideally, the fusion proteins should be tested by substituting them for their endogenous counterparts. This is practical only in prokaryotes and yeast, so alternative assays for the functions of the fusion proteins must be used in other eukaryotes. It is also important to examine potential consequences of the stabilization of the interaction between the fusion proteins by bimolecular fluorescent complex formation (see Box 3 on the dynamics of BiFC complexes). Changes in the dynamics of the interaction caused by association of the fluorescent protein fragments can alter the properties of the complex.
4. Experimental strategies for BiFC analysis
The strategies for investigation of a specific protein interaction must be designed with the purpose of the experiment in mind. However, there are some general strategies that can be useful for the study of many interactions
4.1 Design of plasmid vectors for fusion protein expression
Plasmid vectors must be designed for fusion of the proteins of interest to the N- and C-terminal fragments of a fluorescent protein. In most cases, fusions to both the N- and C-terminal ends of the proteins of interest should be tested. Schematic diagrams of the different permutations of fusion proteins that can be examined are shown in Figure 3. The following general guidelines for the construction of expression vectors for BiFC analysis should be considered:
Figure 3. Recommended combinations of fusion proteins to be tested for bimolecular fluorescence complementation.
Multiple combinations of fusion proteins should be tested for bimolecular fluorescence complementation. Amino- and carboxyl-terminal fusions can be used to test eight distinct combinations (a through h). Although it may appear that combinations e-h would not be favorable for bimolecular complex formation, this will depend on the precise structures and flexibilities of the fusion proteins, which are difficult to predict.
4.1.1 Choice of fluorescent protein fragments for fusions
Several combinations of fluorescent protein fragments that support bimolecular fluorescence complementation have been identified (Hu, et al., 2002, Hu and Kerppola, 2003, Shyu, et al., 2006). Some combinations of fluorescent protein fragments recommended for BiFC analysis are listed Table 2. For most purposes, fragments of YFP truncated at residue 155 (YN155 – N-terminal residues 1-154; and YC155 – C-terminal residues 155-238) are recommended, as they exhibit a relatively high complementation efficiency when fused to many interaction partners, yet produce low fluorescence when fused to proteins that do not interact with each other. Fragments of YFP truncated at residue 173 (YN173 - N-terminal residues 1-172; and YC173 - C-terminal residues 172-238) can also be used. Complementation between fragments of Venus truncated at either residue 155 or 173 (VN155, VC155 or VN173, VC173) often produces brighter fluorescence, but can result in higher non-specific signal due to complementation between fragments fused to proteins that do not interact specifically in the absence of the fluorescent protein fusions.
Table 2.
Combinations of fluorescent protein fragments recommended for BiFC analysis.
| Fusionsa | Purpose | Excitation filter(s) | Emission filter(s) | Reference |
|---|---|---|---|---|
| A-YN155 B-YC155 |
A-B interaction | 500/20 nm | 535/30 nm | (Hu, et al., 2002) |
| A-YN173 B-YC173 |
A-B interaction | 500/20 nm | 535/30 nm | (Hu and Kerppola, 2003) |
| A-CN155 B-CC155 |
A-B interaction | 436/10 nm | 470/30 nm | (Hu and Kerppola, 2003) |
| A-YN155 B-CN155 Z-CC155 |
Competition between A and B for interaction with Z |
500/20 nm and 436/10 nm |
535/30 nm and 470/30 nm |
(Hu and Kerppola, 2003) |
YN155 corresponds to residues 1-154 of EYFP. YC155 corresponds to residues 155-238 of EYFP (Zhang, et al., 2002). YN173 corresponds to residues 1-172 of EYFP. YC173 corresponds to residues 173-238 of EYFP. VN155 corresponds to residues 1-154 of Venus (Nagai, et al., 2002). VC155 corresponds to residues 155-238 of Venus. VN173 corresponds to residues 1-172 of Venus. VC173 corresponds to residues 173-238 of Venus. CN155 corresponds to residues 1-154 of ECFP (Zhang, et al., 2002). CC155 corresponds to residues 155-238 of ECFP.
4.1.2 Choice of positions to which the fragments are fused
The fluorescent protein fragments can support fluorescence complementation when they are fused to either the N- or C-terminal ends of the proteins to be investigated. Ideally, all eight different combinations of fusion proteins should be tested for complementation (Figure 3). The positions of the fusions should be determined empirically based on the following criteria:
i) The fusions must allow the fragments of the fluorescent proteins to associate with each other if the fusion proteins interact. Information about the structure and location of the interaction interface may be useful since fusions near that interaction interface can in some cases produce more efficient complementation than fusions far from the interface. However, structural information is not necessary since multiple combinations of fusion proteins can be tested by screening for fluorescence complementation. A simple strategy for the identification of fusion proteins that support bimolecular fluorescence complementation is to fuse each of the fluorescent protein fragments to the N- and C-terminal end of each interaction partner, and to test all eight combinations of fusion proteins that contain both fragments for complementation (Figure 3).
ii) The fusions must not affect the subcellular distributions or the stabilities of the proteins. The localization and stability of each fusion protein should be compared with those of the endogenous or wild type proteins lacking the fusions.
iii) The fusions must not affect the functions of the proteins to be investigated. Ideally, the functions of the proteins should be tested using assays that evaluate all of the known functions of the endogenous proteins, and these assays should be performed under the conditions used to visualize the protein interactions.
iv) In general, the fragments should be fused to the full length proteins whose properties are to be investigated. In many cases, comparisons with truncated proteins will be important to test the roles of specific regions of the proteins in the interaction. In these cases, it is essential that the fluorescent proteins fragments are fused to the same positions of the full length and truncated proteins.
4.1.2 Choice of peptide linkers to connect the proteins to the fluorescent protein fragments
Peptide linkers are recommended to connect the fragments of the fluorescent proteins to the proteins of interest. These linkers should provide flexibility for independent motion of the fluorescent protein fragments and the interaction partners, allowing the fluorescent protein fragments to associate when the proteins interact. We have used the RSIAT and RPACKIPNDLKQKVMNH (single amino acid code) linker sequences in many fusion constructs used for BiFC analysis (Hu, et al., 2002, Hu and Kerppola, 2003). These linkers have been used for the visualization of interactions between many structurally unrelated proteins. Linker sequences encoding multiple copies of a GGGS sequence have also been successfully used in many BiFC experiments. Although these linker sequences worked well for the proteins we have examined, it is possible that other linkers are optimal for complementation between other proteins.
4.2 Strategies for fusion protein expression
The strategy for expression of the fusion proteins should be based on the purpose of the experiment. To determine whether a pair of proteins can interact in cells and to determine the subcellular location of the complex, a transient expression system can be used. However, the levels of protein expression in different cells in a transiently transfected population are likely to vary over a wide range. Protein overexpression can result in mis-localization of the proteins and formation of non-native complexes. This problem can be ameliorated by the use of plasmids with weak promoters and plasmid vectors that do not replicate in mammalian cells. Additionally, cells can be transfected using small amounts of plasmid DNA, and they can be observed soon after transfection, before the protein expression level becomes too high.
To ensure that the observed fluorescence signal reflects native interactions, the fusion proteins should be expressed at levels comparable to the endogenous proteins. The levels of protein expression can be controlled by using inducible expression vectors integrated into the genome of stable cell line. Such cell lines allow control of protein expression at relatively uniform levels in the cell population.
4.3 Controls required for interpretation of BiFC experiments
To interpret results from BiFC analysis, it is necessary to include negative controls in each experiment (Figure 2). This is essential since the fluorescent protein fragments are able to form fluorescent complexes with low efficiency in the absence of a specific interaction. This non-specific complementation is generally reduced when the fragments are fused to proteins that do not interact with each other. Thus, expression of the fluorescent protein fragments alone frequently produces more fluorescence than expression of fusions to proteins that do not specifically interact with each other. The validity of bimolecular fluorescence complementation results must be confirmed by testing complementation by fusion proteins in which the interaction interface has been mutated (Hu, et al., 2002, Hu and Kerppola, 2003, Grinberg, et al., 2004). The mutated proteins should be fused to the fluorescent protein fragments in the same way as the wild-type protein. The level of expression and the localization of the mutated proteins should be compared with the wild type protein by western blot and indirect immunofluorescence analyses. The efficiencies of fluorescence complementation by the wild type and mutant proteins should be quantified as described in section 4.4 below.
If the interaction interface has not been previously characterized, it is possible to screen for mutations that alter the efficiency of fluorescence complementation using the BiFC assay. If such mutations selectively alter complementation with a particular interaction partner, and they do not affect the levels of expression or subcellular locations of the proteins, it is likely that complementation by the wild type fusion proteins reflects a specific interaction. The BiFC assay can therefore be used to determine whether two proteins interact in cells without prior knowledge of the location or the structural nature of the interaction interface.
4.4. Quantification of the efficiency of bimolecular fluorescence complementation
The efficiency of fluorescence complementation is defined as the fluorescence intensity produced by bimolecular fluorescent complex formation, relative to the levels of fusion proteins present in the cell. The efficiencies of bimolecular fluorescence complementation by structurally unrelated fusion proteins cannot be used to compare the efficiencies of complex formation since the efficiency of bimolecular fluorescence complementation is influenced by many factors in addition to the efficiency of complex formation. For example, the levels of expression of the fusion proteins and the spatial arrangement of the fragments of the fluorescent protein affect the efficiency of fluorescence complementation. However, in situations where these factors are constant, differences in the efficiency of bimolecular fluorescence complementation can provide information about the relative efficiencies of complex formation. Thus, the effects of single amino acid substitutions on complex formation can be examined by testing their effects on complementation efficiency as long as the substitutions do not affect protein expression or localization (Hu, et al., 2002, Hu and Kerppola, 2003).
To quantify the efficiency of fluorescence complementation, it is necessary to include an internal control in the experiment to normalize for differences in transfection efficiency and the level of protein expression. For this purpose, cells are co-transfected with plasmids encoding the two fusion proteins (e.g. fused to YN155 and YC155), together with a plasmid encoding a full-length fluorescent protein with distinct spectral characteristics (e.g., CFP). The fluorescence intensities produced by both bimolecular fluorescence complementation (e.g., YN-YC) and the internal control (e.g., CFP) are measured in individual cells. The ratio of YN-YC to CFP emission is calculated after subtraction of background signal (Figure 2). The ratio of YN-YC to CFP fluorescence is a measure of the efficiency of bimolecular fluorescence complementation. The ratios for different structurally related fusion proteins can reflect the relative efficiencies of complex formation in living cells.
4.5. Interpretation of results from bimolecular fluorescence complementation analysis
-
(i) Fluorescence complementation detected
If fluorescence is detected when wild type proteins fused to the fluorescent protein fragments are expressed, and this signal is eliminated or significantly reduced by mutations that do not affect the expression or localization of the protein, it is likely that the fluorescence reflects a specific interaction between the proteins fused to the fluorescent protein fragments. If mutations that are known to eliminate the interaction of the wild type proteins do not eliminate the fluorescence, then the bimolecular complementation is due to non-specific interactions between the chimeric fusion proteins. Other combination of fusion proteins or linkers can be tested to determine if they produce a specific signal
-
(ii) No fluorescence complementation detected
The lack of fluorescence complementation does not prove the absence of an interaction, even if fluorescence complementation is observed when the same fusion protein is expressed with fusions to other interaction partners. The spatial arrangement of the fluorescent protein fragments can sterically prevent bimolecular complex formation. In addition, fusion of the fluorescent protein fragments can alter protein structure, which could selectively eliminate interactions with some proteins. Only in cases where fluorescence complementation can be induced by an external signal can the lack of fluorescence complementation in the absence of the signal be tentatively considered to reflect the absence of an interaction or a change in protein complex architecture. If no complementation is observed between fusions to proteins that are known to interact based on other assays, additional fusion proteins containing different linker sequences or fluorescent protein fragments should be tested.
5. Examples of proteins interactions that have been visualized using the BiFC assay
BiFC analysis has been used to visualize interactions among a wide range of proteins in many different cell types and organisms (Table 3). The results have validated interactions between many putative interaction partners and have identified several new complexes. Discovery of the subcellular locations of many protein complexes has provided new insights into their functions.
Table 3.
Examples of protein interactions that have been visualized using the BiFC assay
| Category | Class | Proteins1 | Organism | References |
|---|---|---|---|---|
| Peptides | Coiled coil | Anti-parallel NZ-CZ | E. coli | (Ghosh, et al., 2000) |
| Heat shock | Hsc70, Hsp90-TPR1, TPR2A, TPR2B | E. coli | (Magliery, et al., 2005) | |
| Nuclear proteins |
bZIP | Fos-Jun-ATF2; BATF-Jun; Maf-Sox | Mammalian cells | (Hu, et al., 2002, Deppmann, et al., 2003, Rajaram and Kerppola, 2004, Liu, et al., 2006) |
| Rel | p50,IkB-p65 | Mammalian cells | (Hu, et al., 2002) | |
| bHLHZIP | Myc, Mxi1, Mad3, Mad4-Max, Mist-Mist | Mammalian cells | (Grinberg, et al., 2004, Zhu, et al., 2004) | |
| Bromodomain | AcH4-Brd2; SPA-1, P-TEFb-Brd4 | Mammalian cells | (Kanno, et al., 2004, Farina, et al., 2004, Jang, et al., 2005) | |
| Smad | PKB/Akt, Smad4-Smad3 | Mammalian cells | (Remy, et al., 2004) | |
| IRF-Ets | IRF8-PU.1 | Mammalian cells | (Laricchia-Robbio, et al., 2005) | |
| Winged helix | AcFKH1-CPCR1 | A. chrysogenum | (Hoff and Kuck, 2005) | |
| Ubiquitination | E3 ligase-substrate | Skp2-Myc | Mammalian cells | (von der Lehr, et al., 2003) |
| Grr1-Hof1 EID1-ASK1,2,4,9,13,14,15,SSK1 AtCUL1-ASK1,EID1, P0CA-ASK1 |
S. cerevisiae S. alba, P. crispum |
(Blondel, et al., 2005) (Stolpe, et al., 2005, Marrocco, et al., 2006, Pazhouhandeh, et al., 2006) |
||
| Peptide conjugates | Jun-Ub, Jun-SUMO1 | Mammalian cells | (Fang and Kerppola, 2004) | |
| Plant | Type IV secretion | VirE2-VirD4 | A tumefaciens | (Atmakuri, et al., 2003, Cascales, et al., 2005) |
| pathogens | Host-pathogen | VirE2, VirF, H2A-AtVIP, VirE3 | N. tabacum | (Tzfira, et al., 2004, Loyter, et al., 2005, Lacroix, et al., 2005) |
| Signaling | MAP kinase network |
MEKK3-EKK3-IκBα; MEKK2-IκBβ; ERK1- p65; ERK2-p65 |
Mammalian cells | (Schmidt, et al., 2003) |
| PKB-PDK kinases | PKB/Akt, PDK1-hFt1 | Mammalian cells | (Remy and Michnick, 2004) | |
| Heterotrimeric G proteins |
Gβ1-Gγ7 |
D. discoideum, Mammalian cells |
(Hynes, et al., 2004, Hynes, et al., 2004) | |
| Phospholipases | PLCβ2-PLCδ1 | Mammalian cells | (Guo, et al., 2005) | |
| Apoptosis Photosensitivity |
Bif1-Bax, TRAF6-Src FpsA-FpsA |
Mammalian cells A. nidulans |
(Takahashi, et al., 2005, Wang, et al., 2006) (Blumenstein, et al., 2005) |
|
| Enzyme complexes |
ACCS | ACS1, ACS4 - ACS6, ACS7, ACS8 | E. coli | (Tsuchisaka and Theologis, 2004) |
| P450 | P450C2, P450E1-P450 reductase; P4502C2-BAP31 |
Mammalian cells | (Ozalp, et al., 2005, Szczesna-Skorupa and Kemper, 2006) |
|
| Membrane proteins |
Integrin signaling | Integrin αIIbβ3, Syk-Src | Mammalian cells | (de Virgilio, et al., 2004) |
| Arf GTPases | Arf1, Arf3, Arf4, Arf5-GBF1 | Mammalian cells | (Niu, et al., 2005) | |
| Lectin-glycoprotein Cytokine receptors APP processing |
MCFD2, Cathepsin-ERGIC53 gp130 – LIFR, gp130 APP-Notch2, APP |
Mammalian cells Mammalian cells Mammalian cells |
(Nyfeler, et al., 2005) (Giese, et al., 2005) (Chen, et al., 2006) |
|
| Nucleic acid binding |
RNA binding | IMP, FMRP, hStau1, IRP1, PTB1-RNA; Nef-Nef; NXF1-Y14 |
Mammalian cells | (Rackham and Brown, 2004),(Ye, et al., 2004, Schmidt, et al., 2006) |
| DNA binding | Zif268, PBSII-DNA | In vitro | (Stains, et al., 2005) | |
| Plant proteins |
Transcription factors |
FIE-MEA; bZIP63-bZIP63; LSD1-LSD1; bHLH1-OFP1; SAD, BPBF-GAMYB; LIP19-OBF19; GRP23-RBP36B |
N. benthamiana, N. tabacum, A. thaliana, Allium sp. |
(Bracha-Drori, et al., 2004, Walter, et al., 2004, Hackbusch, et al., 2005, Diaz, et al., 2005, Shimizu, et al., 2005, Ding, et al., 2006) |
| Protein modification |
PFTα-PFTβ; T143c-T143c |
N. benthamiana, A. thaliana |
(Bracha-Drori, et al., 2004, Walter, et al., 2004) |
|
| Flowering | FD-FT | N. benthamiana | (Abe, et al., 2005) | |
| Plastid division | MinD1-MinE1; FtsZ1,ARC6-FtsZ2 | N. tabacum | (Maple, et al., 2005) | |
| Enzyme complex | AtSufE-AtSufS, AtNifS | N. tabacum | (Xu and Moller, 2006) | |
Protein pairs that have been tested are separated by a dash. In cases where several protein pairs have been tested, the alternative partners are separated by a comma. Different combinations of proteins that have been tested are separated by semi-colons.
5.1 BiFC analysis of nuclear proteins
The BiFC assay was originally developed using transcription regulatory proteins as a model and has been used to visualize interactions among many different classes of transcription factors (Hu, et al., 2002, Hu and Kerppola, 2003, Deppmann, et al., 2003, Grinberg, et al., 2004, Rajaram and Kerppola, 2004, Zhu, et al., 2004, Kanno, et al., 2004, Farina, et al., 2004, Diaz, et al., 2005, Jang, et al., 2005, Laricchia-Robbio, et al., 2005). These studies have provided insights into the regulation of subcellular localization by protein interactions. In many cases, the localization of transcription factor complexes differs from those of the individual proteins (Hu, et al., 2002, Grinberg, et al., 2004, Rajaram and Kerppola, 2004, Shyu, et al., 2006). Interactions among transcription factors can therefore regulate their subcellular and subnuclear localization. The ATF2 transcription regulatory protein is localized to the cytoplasm when expressed alone, but is translocated into the nucleus upon dimerization with Jun family transcription factors (Hu, et al., 2002, Liu, et al., 2006). Complexes formed by Max with different Myc and Mad family members are localized to different subnuclear locations (Grinberg, et al., 2004). Complexes between the exon junction complex components Y14 and NXF1 are formed only during ongoing transcription and are localized to nuclear splicing speckles (Schmidt, et al., 2006). Further studies of the mechanisms regulating the localization of protein complexes will increase our understanding of the roles of compartmentalization in regulating protein function.
5.2 BiFC analysis of enzyme-substrate complexes
Interactions between several enzymes and their protein substrates have been visualized using the BiFC assay. Complementation between ubiquitin ligases, kinases and guanine nucleotide exchange factors and potential substrate proteins has been used to investigate substrate recognition in living cells (von der Lehr, et al., 2003, de Virgilio, et al., 2004, Blondel, et al., 2005, Niu, et al., 2005, Remy, et al., 2004, Stolpe, et al., 2005). Determination of the substrate specificities and subcellular sites of action of these enzymes in living cells has yielded new hypotheses for their functions. The In Saccharomyces cerevisiae ubiquitin E3 ligase Grr1 interacts with the Hof1 regulator of cytokinesis within the bud neck (Blondel, et al., 2005). This interaction is restricted to the M phase of the cell cycle, presumably because of rapid degradation of Hof1 following cytokinesis. The substrate specificities and sites of action of ubiquitin ligases in many other organisms have also been visualized using the BiFC assay (von der Lehr, et al., 2003, Blondel, et al., 2005, Stolpe, et al., 2005, Marrocco, et al., 2006).
5.3 BiFC analysis of signal transduction pathways
Interactions among many signaling proteins have been visualized using the BiFC assay. The study of membrane protein interactions presents unique challenges because of the role of the membrane environment in determination of interaction specificity. BiFC analysis has therefore been particularly valuable for the visualization of membrane protein interactions (de Virgilio, et al., 2004, Hynes, et al., 2004, Guo, et al., 2005, Ozalp, et al., 2005, Giese, et al., 2005). Contrary to expectation, the constrained mobilities of membrane proteins apparently neither prevent the association of the fluorescent protein fragments nor eliminate the requirement for specific protein interactions. The integrin αIIβ3 receptor exhibited a specific interaction with the Src kinase upon interaction with extracellular fibrin (de Virgilio, et al., 2004). These complexes were localized to membrane ruffles, focal adhesions and focal complexes. These results demonstrate spatially restricted assembly of signaling complexes. Further studies of interactions among additional signal transduction proteins will enable elucidation of the spatial organization of signal transduction networks.
5.4 BiFC analysis of complex relocalization
The BiFC assay is ideally suited for visualization of the subcellular localization of protein complexes. However, it is important to establish that the association of the fluorescent protein fragments does not result in mis-localization of the complex. In the case of the differences in subnuclear localization by BiFC complexes formed by Max with different Myc/Max/Mad family proteins, a similar re-localization of Max was observed in cells that overexpressed different Myc/Max/Mad family proteins in the absence of fluorescence complementation (Grinberg, et al., 2004). Likewise, ATF-2 was localized to different subcellular compartments in cells that expressed different levels of the Jun dimerization partner (Shyu, et al., 2006). Upon isopreterenol stimulation, the β and γ subunits of the heterotrimeric G-protein internalized as a complex separate from the β-adrenergic receptor (Hynes, et al., 2004, Hynes, et al., 2004). A complex formed by the guanine nucleotide exchange factors GBF1 and the small GTPase Arf1 was recruited to the golgi in cells treated with brefeldin A (Niu, et al., 2005). Similarly, a complex formed by the Bcl-2 family proteins Bif-1 and Bax was relocated to mitochondria in cells induced to undergo apoptosis (Takahashi, et al., 2005). Bimolecular fluorescence complementation enables visualization of complex localization in many different subcellular compartments and does not appear to interfere with the translocation of protein complexes between these compartments.
5.5 BiFC analysis of interactions induced by post-translational modifications
Many interactions are thought to require specific post-translational modifications. The requirement for these modifications in living cells can be tested using the BiFC assay. The bromodomain protein Brd2 binds selectively to acetylated histones. Complementation between Brd2 and H4 required both the bromodomain of Brd2 and the H4 tail that contains the acetylation site (Kanno, et al., 2004). Fluorescence complementation by the ERGIC53 receptor and cathepsins catZ and catC required the lectin-binding domain of ERGIC53, suggesting that these interactions require ligand glycosylation (Nyfeler, et al., 2005). BiFC can therefore be used to detect post-translational modifications that alter protein interactions in cells.
5.6 BiFC analysis of interactions on molecular scaffolds
The interaction that brings together the fluorescent protein fragments need not be direct. Fusion proteins that are brought together by assembly in a macromolecular complex can produce bimolecular fluorescent complexes in the absence of direct contact between the proteins that are fused to the fragments. Similarly, fusion proteins that bind to the same molecular scaffold can support fluorescence complementation even if the fusion partners do not contact each other directly. Using this principle, RNA binding by the human zipcode-binding protein 1 ortholog (IMP1), the iron regulatory protein (IRP1), the fragile X mental retardation protein (FMRP), and the human Staufen homologue (hStau1) was visualized in living cells (Rackham and Brown, 2004). Likewise, co-occupancy by zinc-finger DNA binding proteins on oligonucleotides has been detected in vitro (Stains, et al., 2005). Complexes between the exon junction complex components Y14 and NXF1 were also detected only in the presence of newly synthesized transcripts (Schmidt, et al., 2006). Therefore, although the association of fluorescent protein fragments in the BiFC assay is bimolecular, this assay is not limited to the visualization of binary interactions.
6 BiFC analysis of interactions in different organisms
The BiFC assay has been used to visualize interactions in a variety of species from many different phyla (see column 4 of Table 3). In E. coli, many endogenous and heterologous proteins as well as peptides have been shown to associate using BiFC analysis (Ghosh, et al., 2000, Hu, et al., 2002, Tsuchisaka and Theologis, 2004, Magliery, et al., 2005). In Agrobacterium tumefaciens interactions between components of the type IV secretion machinery have been visualized and the interaction between these components is blocked by inhibitors of transformation (Atmakuri, et al., 2003, Cascales, et al., 2005). In Saccharomyces cerevisiae interactions between the Grr1 ubiquitin E3 ligase and the Hof1 regulator of cytokinesis have been shown to be regulated during the cell cycle (Blondel, et al., 2005). In the slime mold Dictyostelium discoides, interactions between the β and γ subunits of heterotrimeric G proteins have been visualized (Hynes, et al., 2004). In the filamentous fungus Acremonium chrysogenum, interactions between transcription factors have been visualized in the nucleus (Hoff and Kuck, 2005). In tobacco, onion and Arabidopsis thaliana, interactions between many different types of proteins have been visualized by introducing expression vectors encoding the fusion proteins using Agrobacterium infiltration or particle bombardment (Tzfira, et al., 2004, Bracha-Drori, et al., 2004, Walter, et al., 2004, Abe, et al., 2005, Li, et al., 2005, Lacroix, et al., 2005, Loyter, et al., 2005, Diaz, et al., 2005, Hackbusch, et al., 2005, Maple, et al., 2005, Shimizu, et al., 2005). In C. elegans, BiFC has been used to mark cells in which specific promoters are transcribed, but the role of specific protein interactions was not examined (Zhang, et al., 2004). The BiFC assay is therefore likely to be generally applicable for visualization of protein interactions in virtually every cell type and organism that can be genetically modified to express proteins that are fused to the fluorescent protein fragments.
7 Screens using the BiFC approach
Complementation assays, and in particular the yeast two-hybrid transcription activation assay, have been used to identify new interaction partners for many proteins. The Ft1 protein was identified as an interaction partner of PKB/Akt using a BiFC-based library screening approach (Remy and Michnick, 2004). The advantage of BiFC-based screens is that the interactions can be detected within the cell, and the effects of stimuli on the interaction can be directly tested. One limitation of BiFC-based screens is that differences in protein expression levels are likely to influence the partners that can be identified. Nevertheless, BiFC analysis has the potential to identify partners that interact with a protein of interest under specific cellular conditions. BiFC analysis can also be used to identify synthetic molecules or cellular factors that can modulate protein interactions.
8 Analysis of complex dynamics using the BiFC approach
Interactions between many proteins are regulated in response to extracellular stimuli or developmental programs. Given the time required for fluorophore maturation and the stabilization of protein interactions by association of the fluorescent protein fragments (Hu, et al., 2002, Magliery, et al., 2005) (see Box 3), it is unclear how closely bimolecular fluorescent complex fluorescence reflects the dynamics of complex formation and dissociation. However, some fluorescent protein fragments purified as intein fusions from E. coli can undergo at least partial maturation prior to association (Demidov, et al., 2006). Moreover, the addition of high concentrations of protein competitors (Guo, et al., 2005) or adjusting conditions to destabilize complexes formed by nucleic acid hybridization (Demidov, et al., 2006) can reduce the fluorescence of bimolecular fluorescent complexes in vitro.
Fluorescence complementation by fusions to MEKK and IκB was induced within 2 minutes of TNFα treatment of HEK293 cells, and returned to basal level within 15 minutes of removal of the stimulus (Schmidt, et al., 2003). MEKK3 exhibited selective complementation with IκBα whereas MEKK2 exhibited selective fluorescence complementation with IκBβ. Similarly, insulin enhanced and TGF-β treatment reduced fluorescence complementation by PKB/Akt and Smad3 within 30 minutes of stimulation (Remy, et al., 2004). Complementation between fluorescent protein fragments fused to PLCβ2 and PLCδ1 was reduced by approximately 50% within 5 minutes of acetylcholine or carbachol treatment of HEK293 cells (Guo, et al., 2005). The fluorescence of bimolecular fluorescent complexes can therefore be dynamically modulated in response to regulatory signals. It is unclear if the rapid changes in fluorescence intensity observed in these experiments reflect the rates of formation or dissociation of protein complexes. It is important to consider the possibility that changes in fluorescence may reflect changes in protein synthesis or degradation.
9 Simultaneous visualization of several protein complexes
The discovery of the green fluorescent protein has transformed cell biology. This transformation has been accelerated by the development of numerous variants with altered spectral and photophysical characteristics (Zhang, et al., 2002). We have developed a multi-color BiFC assay that enables simultaneous visualization of multiple protein complexes in the same cell (Hu and Kerppola, 2003). The multicolor BiFC assay is based on complementation between fragments of different fluorescent proteins that produce bimolecular fluorescent complexes with distinct spectra (Fig. 4) (Hu and Kerppola, 2003). The fragments are fused to alternative interaction partners such that complexes formed between different partners can be visualized independently in the same cell, using different excitation and emission wavelengths.
Figure 4. Simultaneous visualization of multiple protein complexes in the same cell using multicolor BiFC analysis.
Multicolor BiFC analysis is based on the enhanced association of different fluorescent protein fragments through interactions between different proteins fused to the fragments. The bimolecular fluorescent complexes formed by fragments of different fluorescent proteins have distinct spectra and can be distinguished using interference filters. Since bimolecular fluorescent complex formation can stabilize protein interactions at least in vitro, the relative efficiencies of complex formation do not necessarily reflect the equilibrium binding affinities of the interaction partners in the cell. However, the rate of association between the fluorescent protein fragments in a complex (t½ ≈ 1 min) is slower than the rate of dissociation for the majority of protein interactions in the cell. It is therefore likely that the relative fluorescence intensities observed in the multicolor BiFC assay reflect the ratio of complexes formed with each interaction partner shortly after synthesis. Quantitative comparison of the efficiencies of complex formation between alternative interaction partners requires that the fluorescent protein fragments can associate with the same efficiency within each complex. This is likely to be true only when the structures of the alternative interaction partners are closely related and should be verified by using fusion proteins with different linker sequences. In cases where quantitative comparison of the efficiencies of complex formation is not possible using multicolor BiFC analysis, this assay can be used for qualitative comparison of the distributions of complexes formed with different interaction partners. The images below the diagrams display competition between full length Jun and the bZIP domain of Jun for dimerization with the bZIP domain of Fos (the fluorescence of these complexes are shown in green and red respectively). Image acquired by Changdeng Hu (Hu and Kerppola, 2003).
9.1 Comparison of the distributions of different complexes in the same cell
Direct comparison of the distributions of multiple complexes in the same cell eliminates the need to identify markers that co-localize with the complexes as is necessary for comparison of complex distributions between different cells. Comparison of complexes in the same cell also allows determination whether differences in complex localization reflect intrinsic differences in their localization signals. When the distributions of the complexes are compared between different cells, it is possible that indirect effects of the expression of different fusion proteins in different cells alter the localization of the complexes. Similarities in the distributions of two or more protein complexes suggest that the complexes have related functions, especially if their distributions are coordinately regulated.
9.2 Competition among mutually exclusive interaction partners for complex formation
Interactions with different structurally related interaction partners are often mediated by the same contact interface. For some intensely studied proteins, scores of putative interaction partners have been identified using in vitro assays and genetic screens in yeast. It is physically impossible for one protein to simultaneously associate with all of these partners. Interactions with many of these partners are therefore likely to be mutually exclusive. This results in competition for interactions among alternative partners in the cell. Competition among mutually exclusive interaction partners is likely to be an important determinant of the specificity of protein interactions in the cell.
Interactions with different partners can occur in distinct subcellular locations. The selectivity of protein interactions in the cell is determined by many factors including the relative binding affinities of alternative interaction partners and the local concentrations of each protein. It is difficult to predict the selectivity of protein interaction in cells based on in vitro studies, since many factors, including covalent modifications, differences in subcellular distributions and interactions with other cellular proteins can affect complex formation.
The multicolor BiFC assay can be used to investigate the competition between mutually exclusive interaction partners for complex formation with a common partner (Hu and Kerppola, 2003, Grinberg, et al., 2004). When two mutually exclusive interaction partners fused to fragments of different fluorescent proteins are expressed with a limiting amount of a shared interaction partner fused to a complementary fragment, the proportion of bimolecular fluorescent complexes formed with each interaction partner reflects the relative efficiencies of complex formation with each interaction partner in the cell. To use multicolor BiFC analysis for investigation of the relative efficiencies of complex formation, it is necessary to design fusion proteins that exhibit equal efficiencies of association between the fluorescent protein fragments upon formation of each of the complexes to be compared (Hu and Kerppola, 2003, Grinberg, et al., 2004). Differences in the fluorescence intensities of bimolecular fluorescent complexes formed by different combinations of fluorescent protein fragments and possible differences in the intrinsic efficiencies of association of fragments from different fluorescent proteins can be normalized by using proteins in which these fragments are fused to the same interaction partners as described (Hu and Kerppola, 2003, Grinberg, et al., 2004). Thus, multicolor BiFC analysis enables comparison of the relative efficiencies of complex formation by alternative interaction partners in the normal cellular environment.
10. Experimental strategies for multicolor BiFC analysis
The strategies for multicolor BiFC analysis of several complexes in the same cell are for the most part identical to those used for BiFC analysis of individual complexes described in section 4 above. In addition to these, it is necessary to consider the requirements for separation of the signals from two different BiFC complexes in the same cell.
10.1 Design of plasmid vectors for multicolor BiFC analysis
The basic principles for the design of plasmids for BiFC analysis (section 4.1) also apply to the design of plasmids for multicolor BiFC analysis. The main difference is that it is important to select fluorescent protein fragments that provide maximal spectral separation of the fluorescence signals from different bimolecular complexes. There are several combinations of fragments that can be used for multicolor BiFC analysis (Hu and Kerppola, 2003). Complex formation by CC155 (C-terminal fragment of CFP) with YN155 versus CN155 (N-terminal fragment of CFP) results in complexes with good spectral separation and high complementation efficiency (Table 2). These combinations are therefore appropriate for the simultaneous analysis of two protein interactions. For the simultaneous analysis of more than two interactions, more selective interference filters and more complex spectral separation algorithms are required.
If quantitative comparison of the efficiencies of complex formation is contemplated, the fluorescent protein fragments should be fused to the alternative interaction partners in the same manner. It is essential that steric constraints to the association between the fluorescent protein fragments are identical in each complex. One way to test this is to determine if the fragments fused to the alternative interaction partners associate with the same efficiency with the shared interaction partner. This can be accomplished by comparing the fluorescence intensities of cells expressing the proteins fused to the same fluorescent protein fragments in the absence of competitors.
10.2 Strategies for co-expression of proteins for multicolor BiFC analysis
To investigate the competition between two alternative interaction partners (e.g. A and B) for a shared partner (e.g. Z), the three proteins should be fused to fluorescent protein fragments that can form spectrally distinct bimolecular fluorescent complexes (i.e. A-YN155, B-CN155 and Z-CC155; Figure 5; for definitions of fusion proteins, see Table 2). Cell can be transiently co-transfected with plasmid vectors expressing each of the fusion proteins. Alternatively, cell lines that express the fusion proteins can be developed. Ideally, the proteins should be expressed at levels comparable to their endogenous counterparts. Because of the large differences in excitation and emission spectra, the fluorescence signals from the two complexes can be separated with less than 2% cross-talk.
Figure 5. Simultaneous visualization of multiple protein complexes using multicolor fluorescence complementation analysis.
(a) Two alternative interaction partners, A and B, are fused to fragments of different fluorescent protein fragments (YN155 and CN155 respectively). These fusions are co-expressed in cells with a shared interaction partner, Z, fused to a complementary fragment (CC155). Complexes formed by A-YN155 and Z-CC155 can be distinguished from complexes formed by B-CN155 and Z-CC155 based on their fluorescence spectra. (b) Schematic representation of the visualization of multiple protein complexes in the same cell (A-YN155-Z-CC155, cytoplasmic and perinuclear; B-CN155-Z-CC155, nuclear and perinuclear).
10.3 Quantitation of competition between alternative interaction partners using multicolor BiFC analysis
We have developed two methods for quantification of the efficiencies of complex formation using the multicolor BiFC assay (Hu and Kerppola, 2003, Grinberg, et al., 2004). Both methods can provide information about the relative efficiencies of complex formation by mutually exclusive interaction partners in living cells. The absolute fluorescence intensities of bimolecular fluorescent complexes formed by different interaction partners can not be used to compare their efficiencies of complex formation since many factors unrelated to the efficiency of complex formation affect the fluorescence intensities. However, the relative efficiencies of fluorescence complementation between different interaction partners can be compared in the same cell. As long as the steric constraints to fragment association and other factors affecting fluorescence complementation are equivalent for the alternative complexes, this strategy can be used to determine the relative efficiencies of complex formation by alternative interaction partners.
11 Limitations of the multicolor BiFC assay for analysis of the efficiencies of protein interactions in cells
The multicolor BiFC assay of the relative efficiencies of complex formation does not provide information about the binding affinities of the interaction partners, but about their relative efficiencies of complex formation. The efficiencies of complex formation do not necessarily reflect binding affinities since the complexes are not at thermodynamic equilibrium. Association of the fluorescent protein fragments is relatively slow (t½ ≈ 60s), and can stabilize the protein interactions at least in vitro (Hu, et al., 2002). Under these conditions, the relative efficiencies of complex formation reflect competition between alternative interaction partners prior to association of the fluorescent protein fragments. In this case, the multicolor BiFC assay is predicted to give a valid estimate of the relative efficiencies of complex formation for proteins with rapid exchange rates, but interaction partners with slow rates of association may not reach equilibrium prior to association of the fluorescent protein fragments.
Since the multicolor BiFC approach compares the relative amounts of different bimolecular fluorescent complexes, differences between the levels of expression and rates of degradation of the fusion proteins can affect the ratio of complexes that are formed. It is essential to compare the levels of expression of the alternative interaction partners and to take any differences in their expression levels into account when interpreting the data.
12 Interaction partners whose competition has been visualized using the multicolor BiFC assay
The multicolor BiFC assay has been used to visualize the relative efficiencies of dimerization among the bZIP domains of Fos, Jun and ATF2. The results show that the bZIP domains of Fos-Jun heterodimers form more efficiently than either Fos-ATF2 or Jun-ATF2 heterodimers in living cells (Hu and Kerppola, 2003). Jun-ATF2 heterodimers are thought to regulate the expression of many genes. Heterodimer formation may therefore be regulated by regions outside the bZIP domains of these proteins. Alternatively, Jun-ATF2 heterodimers may form only in cells or in subcellular locations where Jun is present in excess relative to the amount of Fos.
Multicolor BiFC analysis has also been used to compare the relative efficiencies of Max interactions with the bHLHZip domain of Myc versus Mad family proteins (Grinberg, et al., 2004). Max favored heterodimer formation with bMyc over homodimerization, consitent with the intrinsic binding affinities of these complexes measured in vitro. This preference was reversed by point mutations in the leucine zipper of Max. The dimerization proferences of these bHLHZIP proteins in living cells reflected their relative binding affinities in vitro.
13. Visualization of ubiquitin family peptide conjugates in cells
Fluorescence complementation assays have been adapted for the visualization of covalent protein modifications in living cells. Conjugation of ubiquitin-family peptides to many substrate proteins modulates their functions and stabilities. Ubiquitin conjugates have been visualized in cells using a ubiquitin mediated fluorescence complementation (UbFC) assay (Fang and Kerppola, 2004). In this approach, the fluorescent protein fragments used for BiFC analysis are fused to ubiquitin and to a putative substrate protein. The covalent attachment of ubiquitin to a substrate can facilitate association between the fragments, enabling selective visualization of the ubiquitin conjugate (Fig. 6). The UbFC assay shares many of the characteristics of the BiFC assay. Of particular significance is the ability to visualize a small subpopulation of modified proteins in the presence of an excess of unmodified proteins. This is critical for studies of ubiquitin-family peptide conjugation since the proportion of most proteins that is modified by a ubiquitin family peptide at any one time is small.
Figure 6. Visualization of ubiquitin-family peptide conjugates using the UbFC assay.
The ubiquitin-mediated fluorescence complementation (UbFC) assay is based on the association between non-fluorescent fragments of a fluorescent protein when they are brought together by covalent conjugation of ubiquitin fused to one fragment to a substrate fused to the complementary fragment. The fluorescent protein fragment must be fused to the N-terminal ends of ubiquitin-family peptides for them to retain their abilities to be conjugated to substrate proteins at their C-terminal ends. Fusions to the N-terminus of ubiquitin and SUMO1 do not interfere with their conjugation to many substrates (Fang and Kerppola, 2004). It is essential to determine if the conjugates containing the fluorescent protein fragments retain the biological functions of the unmodified conjugates. Since ubiquitin can be conjugated in different monomeric and polymeric configurations to substrates, it is important to establish that the stoichiometry and configuration of the ubiquitin conjugate are not altered by the fusions. The images below the diagrams display the total pool of Jun and the Jun-ubiquitin conjugate in blue and green respectively. Images acquired by Hiromi Ikeda and Deyu Fang (Fang and Kerppola, 2004).
13.1 Limitations of the UbFC assay for the detection of ubiquitin family peptide conjugates
One well-established function of ubiquitin conjugation is to induce degradation of the modified protein. It is therefore possible that some conjugates are not detected before they are degraded. It is also possible that only conjugates modified at specific lysine residues or conjugates with a specific linkage between the ubiquitin monomers are detected. Such effects of the steric arrangement of fluorescent protein fragments on fluorescence complementation can be detected by comparing ubiquitin mediated fluorescence complementation using substrates in which the fluorescent protein fragments are fused to different ends of the protein. If fusions to opposite ends of the substrate protein produce comparable results, it is unlikely that steric factors influence the detection of the conjugates.
14 Ubiquitin family peptide conjugates that have been visualized using the UbFC assay
The UbFC assay has been used to visualize conjugates formed by different ubiquitin-family peptides with the Jun protein. Surprisingly, ubiquitinated Jun is exported from the nucleus and is translocated to lysosomes for degradation. In contrast, SUMO1-modified Jun is localized to nuclear foci. Multicolor analysis of ubiquitination and SUMO1-conjugation confirmed that the conjugates are localized to different subcellular compartments in the same cell. Thus, different peptide modifications can induce translocation to different subcellular locations, with different consequences for protein function. Recent studies have revealed a broad range of functions for ubiquitin family peptide modifications, and more will undoubtedly be discovered. The direct visualization of these conjugates using the UbFC assay will be a valuable tool for the investigation of these modifications in living cells.
15 Comparison of BiFC analysis with other methods for the visualization of protein interactions in living cells
Several methods have been developed to study protein interactions in living cells. One of the most commonly employed methods is FRET (Zhang, et al., 2002, Jares-Erijman and Jovin, 2003, Miyawaki, 2003, Zal and Gascoigne, 2004, Bunt and Wouters, 2004, Galbraith, 2004, Schmid and Neumeier, 2005). To investigate protein interactions using FRET, two different fluorophores are either chemically linked or genetically fused to the two proteins whose interaction is to be examined. The interaction is detected based on a change in the intensity or lifetime of the donor fluorophore or the intensity of the acceptor. Compared to the BiFC assay, FRET analysis in living cells generally requires higher levels of protein expression for reliable detection of energy transfer. In addition, structural information, or a great deal of luck, is required to place the two fluorophores within 100 Å of each other. This is the maximum distance over which significant energy transfer between fluorescent proteins can be detected. The fraction of proteins that interact with each other must also be sufficiently high to produce a change in the donor and acceptor fluorescence intensities that is greater than changes caused by fluctuations in fluorescence intensity due to other effects. To exclude alternative interpretations of the results, numerous controls must be performed and the fluorescence intensities must be measured with high quantitative accuracy. Despite these limitations, FRET has been successfully used for the analysis of many protein interactions in living cells (Sorkin, et al., 2000, Li, et al., 2001, Majoul, et al., 2002, Hink, et al., 2003, Larson, et al., 2003, Miyawaki, 2003, Tsien, 2003). A great advantage of FRET relative to BiFC analysis is the real-time detection of complex formation and dissociation, which potentially allows analysis of the interaction under equilibrium conditions. FRET is therefore potentially superior to BiFC analysis in studies of the kinetics of protein association and dissociation.
Several characteristics of the BiFC assay make it useful for studying protein interactions. First, it enables direct visualization of protein interactions and does not rely on their secondary effects. Second, the interactions can be visualized in living cells, eliminating potential artifacts associated with cell lysis or fixation. Third, the proteins are expressed in a relevant biological context, ideally at levels comparable to their endogenous counterparts. Thus, they are predicted to reflect the properties of native proteins, including any post-translational modifications. Fourth, the BiFC assay does not require stoichiometric complex formation but can detect interactions between a subpopulation of each protein. Fifth, multicolor BiFC analysis allows simultaneous visualization of multiple protein complexes in the same cell and enables analysis of the competition between alternative interaction partners for complex formation. Finally, BiFC does not require specialized equipment, apart from an inverted fluorescence microscope equipped with objectives that allow imaging of fluorescence in cells. The simple detection of bimolecular complex formation requires no post-acquisition image processing for interpretation of the data. Consequently, BiFC is a powerful tool for scientists seeking to understand protein interactions in living cells.
16. Future opportunities and challenges
The visualization of molecular interactions in living cells and organisms has a great potential for elucidating fundamental biological mechansism. There do not appear to be any limitations on use of the BiFC approach in essentially any aerobic organism and cell type except for those that encode endogenous fluorescent or luminescent proteins. Likewise, the BiFC assay appears to be applicable for the visualization of interactions among a wide variety of structurally and functionally divers proteins. Future applications of the BiFC assay will likely include studies in transgenic animals and plants where the developmental and tissue-specific regulation of protein interactions can be investigated in the context of the living organism. Studies in cultured cells will likely make greater use of regulated expression systems to control fusion protein expression at more uniform levels in the cell population. The native transcription regulatory regions may be used to control expression of the fusion proteins in studies where the timing and the cell-type specificity of protein expression are critical but gene replacement through homologous recombination is impractical.
Multicolor BiFC analysis of the subcellular distributions of complexes formed with different partners is likely to provide insight into functional differences between complexes formed with different partners. The multi-color BiFC approach will also allow the incorporation of internal standards into experiments and elimination of many artefactual sources of variation in fluorescence intensity through ratiometric imaging. These advantages will prove particularly valuable in adaptation of the BiFC approach for high-throughput analysis and library screens. Many biological processes occur asynchronously in different cells in a population. Simultaneous comparison of several molecular interactions in the same cell can be used to detect heterogeneity in cell populations.
The BiFC approach can be used for many purposes in addition to the study of protein interactions. The wide range of spectral variants that are produced by different combinations of fluorescent protein fragments lends itself to combinatorial tagging of cells in studies of cell lineages and migration. The UbFC approach can be applied to the analysis of conjugates formed by other ubiquitin family peptides with a wide range of different substrates. These studies will lead to a better understanding of the relationships between different ubiquitin family peptide modifications of the same substrate.
There are several limitations of present versions of the BiFC approach that provide opportunities for future improvements. Chief among these limitations is the stabilization of the interaction by association of the fluorescent protein fragments under some conditions. A better understanding of the dynamics of bimolecular fluorescent complex formation in living cells and the development of complementing fragments that minimally perturb the dynamic exchange of interaction partners would be valuable contributions. Another limitation is the intrinsic ability of fluorescent protein fragments to associate independent of an interaction between proteins fused to the fragments. This major source of background signal in the BiFC assay varies depending on the fusion proteins and their levels of expression. Development of fluorescent protein fragments with a reduced tendency for intrinsic association, but without a reduction in their ability to associate when brought together by a protein interaction would make the assay less sensitive to the levels of protein expression. Finally, a variety of fluorescent protein variants with useful photophysical characteristics including photo-activation, photo-conversion and time-dependent spectral transformation have been identified (Terskikh, et al., 2000, Patterson and Lippincott-Schwartz, 2002, Ando, et al., 2002, Chudakov, et al., 2003). Fragments that form bimolecular fluorescent complexes with photophysical properties appropriate for specific experimental purposes would be a useful addition to the present selection of spectral variants. Screening of combinatorial libraries of fluorescent protein fragments also provides a strategy for the identification of novel characteristics that may not be found within the sequence space that can be explored using mutated variants of full length fluorescent proteins.
ACKNOWLEDGEMENTS
I thank C-D Hu for his participation in the design and implementation of the BiFC assay in mammalian cells and all members of the Kerppola laboratory for their contributions to the improvement and application of the BiFC approach.
References
- 1.Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, Ikeda Y, Ichinoki H, Notaguchi M, Goto K, Araki T. FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science. 2005;309:1052–1056. doi: 10.1126/science.1115983. [DOI] [PubMed] [Google Scholar]
- 2.Ando R, Hama H, Yamamoto-Hino M, Mizuno H, Miyawaki A. An optical marker based on the UV-induced green-to-red photoconversion of a fluorescent protein. Proc Natl Acad Sci U S A. 2002;99:12651–12656. doi: 10.1073/pnas.202320599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atmakuri K, Ding ZY, Christie PJ. VirE2, a type IV secretion substrate, interacts with the VirD4 transfer protein at cell poles of Agrobacterium tumefaciens. Molecular Microbiology. 2003;49:1699–1713. doi: 10.1046/j.1365-2958.2003.03669.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blondel M, Bach S, Bamps S, Dobbelaere J, Wiget P, Longaretti C, Barral Y, Meijer L, Peter M. Degradation of Hof1 by SCFGrr1 is important for actomyosin contraction during cytokinesis in yeast. Embo Journal. 2005;24:1440–1452. doi: 10.1038/sj.emboj.7600627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blumenstein A, Vienken K, Tasler R, Purschwitz J, Veith D, Frankenberg-Dinkel N, Fischer R. The Aspergillus nidulans phytochrome FphA represses sexual development in red light. Current Biology. 2005;15:1833–1838. doi: 10.1016/j.cub.2005.08.061. [DOI] [PubMed] [Google Scholar]
- 6.Bracha-Drori K, Shichrur K, Katz A, Oliva M, Angelovici R, Yalovsky S, Ohad N. Detection of protein-protein interactions in plants using bimolecular fluorescence complementation. Plant Journal. 2004;40:419–427. doi: 10.1111/j.1365-313X.2004.02206.x. [DOI] [PubMed] [Google Scholar]
- 7.Brock R, Jovin TM. Fluorescence correlation microscopy (FCM)-fluorescence correlation spectroscopy (FCS) taken into the cell. Cell Mol Biol (Noisy-le-grand) 1998;44:847–856. [PubMed] [Google Scholar]
- 8.Bunt G, Wouters FS. International Review of Cytology - a Survey of Cell Biology. 2004;237:205. doi: 10.1016/S0074-7696(04)37005-1. Vol. 237 (International Review of Cytology-a Survey of Cell Biology) + [DOI] [PubMed] [Google Scholar]
- 9.Cabantous S, Terwilliger TC, Waldo GS. Protein tagging and detection with engineered self-assembling fragments of green fluorescent protein. Nature Biotechnology. 2005;23:102–107. doi: 10.1038/nbt1044. [DOI] [PubMed] [Google Scholar]
- 10.Cascales E, Atmakuri K, Liu Z, Binns AN, Christie PJ. Agrobacterium tumefaciens oncogenic suppressors inhibit T-DNA and VirE2 protein substrate binding to the VirD4 coupling protein. Molecular Microbiology. 2005;58:565–579. doi: 10.1111/j.1365-2958.2005.04852.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen CD, Oh SY, Hinman JD, Abraham CR. Visualization of APP dimerization and APP-Notch2 heterodimerization in living cells using bimolecular fluorescence complementation. Journal of Neurochemistry. 2006;97:30–43. doi: 10.1111/j.1471-4159.2006.03705.x. [DOI] [PubMed] [Google Scholar]
- 12.Chudakov DM, Belousov VV, Zaraisky AG, Novoselov VV, Staroverov DB, Zorov DB, Lukyanov S, Lukyanov KA. Kindling fluorescent proteins for precise in vivo photolabeling. Nat Biotechnol. 2003;21:191–194. doi: 10.1038/nbt778. [DOI] [PubMed] [Google Scholar]
- 13.de Virgilio M, Kiosses WB, Shattil SJ. Proximal, selective, and dynamic interactions between integrin alpha II beta 3 and protein tyrosine kinases in living cells. Journal of Cell Biology. 2004;165:305–311. doi: 10.1083/jcb.200402064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Demidov VV, Dokholyan NV, Witte-Hoffmann C, Chalasani P, Yiu HW, Ding F, Yu Y, Cantor CR, Broude NE. Fast complementation of split fluorescent protein triggered by DNA hybridization. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:2052–2056. doi: 10.1073/pnas.0511078103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deppmann CD, Thornton TM, Utama FE, Taparowsky EJ. Phosphorylation of BATF regulates DNA binding: a novel mechanism for AP-1 (activator protein-1) regulation. Biochemical Journal. 2003;374:423–431. doi: 10.1042/BJ20030455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Diaz I, Martinez M, Isabel-LaMoneda I, Rubio-Somoza I, Carbonero P. The DOF protein, SAD, interacts with GAMYB in plant nuclei and activates transcription of endosperm-specific genes during barley seed development. Plant Journal. 2005;42:652–662. doi: 10.1111/j.1365-313X.2005.02402.x. [DOI] [PubMed] [Google Scholar]
- 17.Ding YH, Liu NY, Tang ZS, Liu J, Yang WC. Arabidopsis GLUTAMINE-RICH PROTEIN23 is essential for early embryogenesis and encodes a novel nuclear PPR motif protein that interacts with RNA polymerase II subunit III. Plant Cell. 2006;18:815–830. doi: 10.1105/tpc.105.039495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fang DY, Kerppola TK. Ubiquitin-mediated fluorescence complementation reveals that Jun ubiquitinated by Itch/AIP4 is localized to lysosomes. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:14782–14787. doi: 10.1073/pnas.0404445101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Farina A, Hattori M, Qin J, Nakatani Y, Minato N, Ozato K. Bromodomain protein Brd4 binds to GTPase-activating SPA-1, modulating its activity and subcellular localization. Molecular and Cellular Biology. 2004;24:9059–9069. doi: 10.1128/MCB.24.20.9059-9069.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Galarneau A, Primeau M, Trudeau LE, Michnick SW. Beta-lactamase protein fragment complementation assays as in vivo and in vitro sensors of protein protein interactions. Nat Biotechnol. 2002;20:619–622. doi: 10.1038/nbt0602-619. [DOI] [PubMed] [Google Scholar]
- 21.Galbraith DW. Cytometry, 4th Edition: New Developments. Methods in Cell Biology. 2004;75:153–169. doi: 10.1016/s0091-679x(04)75006-2. [DOI] [PubMed] [Google Scholar]
- 22.Gavin AC, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon AM, Cruciat CM, Remor M, Hofert C, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 23.Ghosh I, Hamilton AD, Regan L. Antiparallel leucine zipper-directed protein reassembly: application to the green fluorescent protein. J. Am. Chem. Soc. 2000;122:5658–5659. [Google Scholar]
- 24.Giese B, Roderburg C, Sommerauer M, Wortmann SB, Metz S, Heinrich PC, Muller-Newen G. Dimerization of the cytokine receptors gp130 and LIFR analysed in single cells. Journal of Cell Science. 2005;118:5129–5140. doi: 10.1242/jcs.02628. [DOI] [PubMed] [Google Scholar]
- 25.Giot L, Bader JS, Brouwer C, Chaudhuri A, Kuang B, Li Y, Hao YL, Ooi CE, Godwin B, Vitols E, Vijayadamodar G, Pochart P, et al. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- 26.Grinberg AV, Hu CD, Kerppola TK. Visualization of Myc/Max/Mad family dimers and the competition for dimerization in living cells. Molecular and Cellular Biology. 2004;24:4294–4308. doi: 10.1128/MCB.24.10.4294-4308.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo YJ, Rebecchi M, Scarlata S. Phospholipase C beta(2) binds to and inhibits phospholipase C delta(1) Journal of Biological Chemistry. 2005;280:1438–1447. doi: 10.1074/jbc.M407593200. [DOI] [PubMed] [Google Scholar]
- 28.Hackbusch J, Richter K, Muller J, Salamini F, Uhrig JF. A central role of Arabidopsis thaliana ovate family proteins in networking and subcellular localization of 3-aa loop extension homeodomain proteins. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:4908–4912. doi: 10.1073/pnas.0501181102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hink MA, Borst JW, Visser AJ. Fluorescence correlation spectroscopy of GFP fusion proteins in living plant cells. Methods Enzymol. 2003;361:93–112. doi: 10.1016/s0076-6879(03)61007-4. [DOI] [PubMed] [Google Scholar]
- 30.Hoff B, Kuck U. Use of bimolecular fluorescence complementation to demonstrate transcription factor interaction in nuclei of living cells from the filamentous fungus Acremonium chrysogenum. Current Genetics. 2005;47:132–138. doi: 10.1007/s00294-004-0546-0. [DOI] [PubMed] [Google Scholar]
- 31.Hu CD, Chinenov Y, Kerppola TK. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol Cell. 2002;9:789–798. doi: 10.1016/s1097-2765(02)00496-3. [DOI] [PubMed] [Google Scholar]
- 32.Hu CD, Kerppola TK. Simultaneous visualization of multiple protein interactions in living cells using multicolor fluorescence complementation analysis. Nat Biotechnol. 2003;21:539–545. doi: 10.1038/nbt816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hynes TR, Mervine SM, Yost EA, Sabo JL, Berlot CH. Live cell imaging of G(s) and the beta(2)-adrenergic receptor demonstrates that both alpha(s) and beta(1)gamma(7) internalize upon stimulation and exhibit similar trafficking patterns that differ from that of the beta(2)-adrenergic receptor. Journal of Biological Chemistry. 2004;279:44101–44112. doi: 10.1074/jbc.M405151200. [DOI] [PubMed] [Google Scholar]
- 34.Hynes TR, Tang LN, Mervine SM, Sabo JL, Yost EA, Devreotes PN, Berlot CH. Visualization of g protein beta gamma dimers using bimolecular fluorescence complementation demonstrates roles for both beta and gamma in subcellular targeting. Journal of Biological Chemistry. 2004;279:30279–30286. doi: 10.1074/jbc.M401432200. [DOI] [PubMed] [Google Scholar]
- 35.Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci U S A. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jang MK, Mochizuki K, Zhou MS, Jeong HS, Brady JN, Ozato K. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Molecular Cell. 2005;19:523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 37.Jares-Erijman EA, Jovin TM. FRET imaging. Nat Biotechnol. 2003;21:1387–1395. doi: 10.1038/nbt896. [DOI] [PubMed] [Google Scholar]
- 38.Johnsson N, Varshavsky A. Split ubiquitin as a sensor of protein interactions in vivo. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:10340–10344. doi: 10.1073/pnas.91.22.10340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kanno T, Kanno Y, Siegel RM, Jang MK, Lenardo MJ, Ozato K. Selective recognition of acetylated histones by bromodomain proteins visualized in living cells. Molecular Cell. 2004;13:33–43. doi: 10.1016/s1097-2765(03)00482-9. [DOI] [PubMed] [Google Scholar]
- 40.Kerppola TK. Complementary methods for studies of protein interactions in living cells. Nature Methods. 2006;3:969–971. doi: 10.1038/nmeth1206-969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kerppola TK. Visualization of molecular interactions by fluorescence complementation. Nature Reviews Molecular Cell Biology. 2006;7:449–456. doi: 10.1038/nrm1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lacroix B, Vaidya M, Tzfira T, Citovsky V. The VirE3 protein of Agrobacterium mimics a host cell function required for plant genetic transformation. Embo Journal. 2005;24:428–437. doi: 10.1038/sj.emboj.7600524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Laricchia-Robbio L, Tamura T, Karpova T, Sprague BL, McNally JG, Ozato K. Partner-regulated interaction of IFN regulatory factor 8 with chromatin visualized in live macrophages. PNAS. 2005;102:14368–14373. doi: 10.1073/pnas.0504014102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Larson DR, Ma YM, Vogt VM, Webb WW. Direct measurement of Gag-Gag interaction during retrovirus assembly with FRET and fluorescence correlation spectroscopy. J Cell Biol. 2003;162:1233–1244. doi: 10.1083/jcb.200303200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li HY, Ng EK, Lee SM, Kotaka M, Tsui SK, Lee CY, Fung KP, Waye MM. Protein-protein interaction of FHL3 with FHL2 and visualization of their interaction by green fluorescent proteins (GFP) two-fusion fluorescence resonance energy transfer (FRET) J Cell Biochem. 2001;80:293–303. [PubMed] [Google Scholar]
- 46.Li JX, Krichevsky A, Vaidya M, Tzfira T, Citovsky V. Uncoupling of the functions of the Arabidopsis VIN protein in transient and stable plant genetic transformation by Agrobacterium. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:5733–5738. doi: 10.1073/pnas.0404118102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li S, Armstrong CM, Bertin N, Ge H, Milstein S, Boxem M, Vidalain PO, Han JD, Chesneau A, Hao T, Goldberg DS, Li N, et al. A map of the interactome network of the metazoan C. elegans. Science. 2004;303:540–543. doi: 10.1126/science.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu H, Deng1 X, Shyu YJ, Li JJ, Taparowsky EJ, Hu C-D. Mutual regulation of c-Jun and ATF2 by transcriptional activation and subcellular localization. EMBO Journal. 2006;25:1058–1069. doi: 10.1038/sj.emboj.7601020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Loyter A, Rosenbluh J, Zakai N, Li JX, Kozlovsky SV, Tzfira T, Citovsky V. The plant VirE2 interacting protein 1. A molecular link between the Agrobacterium T-complex and the host cell chromatin? Plant Physiology. 2005;138:1318–1321. doi: 10.1104/pp.105.062547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Magliery TJ, Wilson CGM, Pan WL, Mishler D, Ghosh I, Hamilton AD, Regan L. Detecting protein-protein interactions with a green fluorescent protein fragment reassembly trap: Scope and mechanism. Journal of the American Chemical Society. 2005;127:146–157. doi: 10.1021/ja046699g. [DOI] [PubMed] [Google Scholar]
- 51.Majoul I, Straub M, Duden R, Hell SW, Soling HD. Fluorescence resonance energy transfer analysis of protein-protein interactions in single living cells by multifocal multiphoton microscopy. J Biotechnol. 2002;82:267–277. doi: 10.1016/s1389-0352(01)00042-3. [DOI] [PubMed] [Google Scholar]
- 52.Maple J, Aldridge C, Moller SG. Plastid division is mediated by combinatorial assembly of plastid division proteins. Plant Journal. 2005;43:811–823. doi: 10.1111/j.1365-313X.2005.02493.x. [DOI] [PubMed] [Google Scholar]
- 53.Marrocco K, Zhou YC, Bury E, Dieterle M, Funk M, Genschik P, Krenz M, Stolpe T, Kretsch T. Functional analysis of EID1, an F-box protein involved in phytochrome A-dependent light signal transduction. Plant Journal. 2006;45:423–438. doi: 10.1111/j.1365-313X.2005.02635.x. [DOI] [PubMed] [Google Scholar]
- 54.Miyawaki A. Visualization of the spatial and temporal dynamics of intracellular signaling. Dev Cell. 2003;4:295–305. doi: 10.1016/s1534-5807(03)00060-1. [DOI] [PubMed] [Google Scholar]
- 55.Nagai T, Ibata K, Park ES, Kubota M, Mikoshiba K, Miyawaki A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nature Biotechnology. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- 56.Nagai T, Sawano A, Park ES, Miyawaki A. Circularly permuted green fluorescent proteins engineered to sense Ca2+ Proc Natl Acad Sci U S A. 2001;98:3197–3202. doi: 10.1073/pnas.051636098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Niu TK, Pfeifer AC, Lippincott-Schwartz J, Jackson CL. Dynamics of GBF1, a brefeldin A-sensitive Arf1 exchange factor at the Golgi. Molecular Biology of the Cell. 2005;16:1213–1222. doi: 10.1091/mbc.E04-07-0599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nyfeler B, Michnick SW, Hauri HP. Capturing protein interactions in the secretory pathway of living cells. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:6350–6355. doi: 10.1073/pnas.0501976102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ozalp C, Szczesna-Skorupa E, Kemper B. Bimolecular fluorescence complementation analysis of cytochrome P4502C2, 2E1, and NADPH-cytochrome P450 reductase molecular interactions in living cells. Drug Metabolism and Disposition. 2005;33:1382–1390. doi: 10.1124/dmd.105.005538. [DOI] [PubMed] [Google Scholar]
- 60.Ozawa T, Kaihara A, Sato M, Tachihara K, Umezawa Y. Split Luciferase as an Optical Probe for Detecting Protein-Protein Interactions in Mammalian Cells Based on Protein Splicing. Anal. Chem. 2001;73:2516–2521. doi: 10.1021/ac0013296. [DOI] [PubMed] [Google Scholar]
- 61.Patterson GH, Lippincott-Schwartz J. A photoactivatable GFP for selective photolabeling of proteins and cells. Science. 2002;297:1873–1877. doi: 10.1126/science.1074952. [DOI] [PubMed] [Google Scholar]
- 62.Paulmurugan R, Gambhir SS. Monitoring protein-protein interactions using split synthetic renilla luciferase protein-fragment-assisted complementation. Anal Chem. 2003;75:1584–1589. doi: 10.1021/ac020731c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Paulmurugan R, Umezawa Y, Gambhir SS. Noninvasive imaging of protein-protein interactions in living subjects by using reporter protein complementation and reconstitution strategies. Proc Natl Acad Sci U S A. 2002;99:15608–15613. doi: 10.1073/pnas.242594299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pazhouhandeh M, Dieterle M, Marrocco K, Lechner E, Berry B, Brault V, Hemmer O, Kretsch T, Richards KE, Genschik P, Ziegler-Graff V. F-box-like domain in the polerovirus protein P0 is required for silencing suppressor function. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:1994–1999. doi: 10.1073/pnas.0510784103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pelletier JN, Campbell-Valois FX, Michnick SW. Oligomerization domain-directed reassembly of active dihydrofolate reductase from rationally designed fragments. Proc Natl Acad Sci U S A. 1998;95:12141–12146. doi: 10.1073/pnas.95.21.12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Petersen NO, Hoddelius PL, Wiseman PW, Seger O, Magnusson KE. Quantitation of membrane receptor distributions by image correlation spectroscopy: concept and application. Biophys. J. 1993;65:1135–1146. doi: 10.1016/S0006-3495(93)81173-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pfleger KDG, Eidne KA. Illuminating insights into protein-protein interactions using bioluminescence resonance energy transfer (BRET) Nature Methods. 2006;3:165. doi: 10.1038/nmeth841. [DOI] [PubMed] [Google Scholar]
- 68.Rackham O, Brown CM. Visualization of RNA-protein interactions in living cells: FMRP and IMP1 interact on mRNAs. Embo Journal. 2004;23:3346–3355. doi: 10.1038/sj.emboj.7600341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rajaram N, Kerppola TK. Synergistic transcription activation by Maf and Sox and their subnuclear localization are disrupted by a mutation in Maf that causes cataract. Molecular and Cellular Biology. 2004;24:5694–5709. doi: 10.1128/MCB.24.13.5694-5709.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Remy I, Michnick SW. A highly sensitive protein-protein interaction assay based on Gaussia Luciferase. Nature Methods. 2006;3:977–979. doi: 10.1038/nmeth979. [DOI] [PubMed] [Google Scholar]
- 71.Remy I, Michnick SW. Regulation of apoptosis by the Ft1 protein, a new modulator of protein kinase B/Akt. Molecular and Cellular Biology. 2004;24:1493–1504. doi: 10.1128/MCB.24.4.1493-1504.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Remy I, Montmarquette A, Michnick SW. PKB/Akt modulates TGF-beta signalling through a direct interaction with Smad3. Nature Cell Biology. 2004;6:358–365. doi: 10.1038/ncb1113. [DOI] [PubMed] [Google Scholar]
- 73.Richards FM. On the enzymic activity of subtilisin-modified ribonuclease. Proc. Natl. Acad. Sci. USA. 1958;44:162–166. doi: 10.1073/pnas.44.2.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rossi F, Charlton CA, Blau HM. Monitoring protein-protein interactions in intact eukaryotic cells by beta-galactosidase complementation. Proc Natl Acad Sci U S A. 1997;94:8405–8410. doi: 10.1073/pnas.94.16.8405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schmid JA, Neumeier H. Evolutions in science triggered by green fluorescent protein (GFP) Chembiochem. 2005;6:1149. doi: 10.1002/cbic.200500029. + [DOI] [PubMed] [Google Scholar]
- 76.Schmidt C, Peng BL, Li ZK, Sclabas GM, Fujioka S, Niu JG, Schmidt-Supprian M, Evans DB, Abbruzzese JL, Chiao PJ. Mechanisms of proinflammatory cytokine-induced biphasic NF-kappa B activation. Molecular Cell. 2003;12:1287–1300. doi: 10.1016/s1097-2765(03)00390-3. [DOI] [PubMed] [Google Scholar]
- 77.Schmidt U, Richter K, Berger AB, Lichter P. In vivo BiFC analysis of Y14 and NXF1 mRNA export complexes: preferential localization within and around SC35 domains. Journal of Cell Biology. 2006;172:373–381. doi: 10.1083/jcb.200503061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shimizu H, Sato K, Berberich T, Miyazaki A, Ozaki R, Imai R, Kusano T. LIP19, a basic region leucine zipper protein, is a fos-like molecular switch in the cold signaling of rice plants. Plant and Cell Physiology. 2005;46:1623–1634. doi: 10.1093/pcp/pci178. [DOI] [PubMed] [Google Scholar]
- 79.Shyu YJ, Liu H, Deng X, Hu CD. Identification of new fluorescent protein fragments for bimolecular fluorescence complementation analysis under physiological conditions. Biotechniques. 2006;40:61–66. doi: 10.2144/000112036. [DOI] [PubMed] [Google Scholar]
- 80.Sorkin A, McClure M, Huang F, Carter R. Interaction of EGF receptor and grb2 in living cells visualized by fluorescence resonance energy transfer (FRET) microscopy. Curr Biol. 2000;10:1395–1398. doi: 10.1016/s0960-9822(00)00785-5. [DOI] [PubMed] [Google Scholar]
- 81.Spotts JM, Dolmetsch RE, Greenberg ME. Time-lapse imaging of a dynamic phosphorylation-dependent protein-protein interaction in mammalian cells. PNAS. 2002;99:15142–15147. doi: 10.1073/pnas.232565699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Stains CI, Porter JR, Ooi AT, Segal DJ, Ghosh I. DNA sequence-enabled reassembly of the green fluorescent protein. Journal of the American Chemical Society. 2005;127:10782–10783. doi: 10.1021/ja051969w. [DOI] [PubMed] [Google Scholar]
- 83.Stelzl U, Worm U, Lalowski M, Haenig C, Brembeck FH, Goehler H, Stroedicke M, Zenkner M, Schoenherr A, Koeppen S, Timm J, Mintzlaff S, et al. A human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 84.Stolpe T, Süßlin C, Marrocco K, Nick P, Kretsch T, Kircher S. In planta analysis of protein-protein interactions related to light signaling by bimolecular fluorescence complementation. Protoplasma. 2005;226:137–146. doi: 10.1007/s00709-005-0122-6. [DOI] [PubMed] [Google Scholar]
- 85.Szczesna-Skorupa E, Kemper B. BAP31 is involved in the retention of cytochrome P4502C2 in the endoplasmic reticulum. Journal of Biological Chemistry. 2006;281:4142–4148. doi: 10.1074/jbc.M509522200. [DOI] [PubMed] [Google Scholar]
- 86.Takahashi Y, Karbowski M, Yamaguchi H, Kazi A, Wu J, Sebti SM, Youle RJ, Wang HG. Loss of Bif-1 suppresses Bax/Bak conformational change and mitochondrial apoptosis. Molecular and Cellular Biology. 2005;25:9369–9382. doi: 10.1128/MCB.25.21.9369-9382.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Terskikh A, Fradkov A, Ermakova G, Zaraisky A, Tan P, Kajava AV, Zhao X, Lukyanov S, Matz M, Kim S, Weissman I, Siebert P. “Fluorescent timer”: protein that changes color with time. Science. 2000;290:1585–1588. doi: 10.1126/science.290.5496.1585. [DOI] [PubMed] [Google Scholar]
- 88.Tsien RY. Imagining imaging's future. Nat Rev Mol Cell Biol. 2003;(Suppl):SS16–21. [PubMed] [Google Scholar]
- 89.Tsuchisaka A, Theologis A. Heterodimeric interactions among the 1-amino-cyclopropane-1-carboxylate synthase polypeptides encoded by the Arabidopsis gene family. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:2275–2280. doi: 10.1073/pnas.0308515101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tzfira T, Vaidya M, Citovsky V. Involvement of targeted proteolysis in plant genetic transformation by Agrobacterium. Nature. 2004;431:87–92. doi: 10.1038/nature02857. [DOI] [PubMed] [Google Scholar]
- 91.von der Lehr N, Johansson S, Wu SQ, Bahram F, Castell A, Cetinkaya C, Hydbring P, Weidung I, Nakayama K, Nakayama KI, Soderberg O, Kerppola TK, et al. The F-Box protein Skp2 participates in c-Myc proteosomal degradation and acts as a cofactor for c-Myc-regulated transcription. Molecular Cell. 2003;11:1189–1200. doi: 10.1016/s1097-2765(03)00193-x. [DOI] [PubMed] [Google Scholar]
- 92.Walter M, Chaban C, Schutze K, Batistic O, Weckermann K, Nake C, Blazevic D, Grefen C, Schumacher K, Oecking C, Harter K, Kudla J. Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant Journal. 2004;40:428–438. doi: 10.1111/j.1365-313X.2004.02219.x. [DOI] [PubMed] [Google Scholar]
- 93.Wang KZQ, Wara-Aswapati N, Boch JA, Yoshida Y, Hu CD, Galson DL, Auron PE. TRAF6 activation of PI 3-kinase-dependent cytoskeletal changes is cooperative with Ras and is mediated by an interaction with cytoplasmic Src. Journal of Cell Science. 2006;119:1579–1591. doi: 10.1242/jcs.02889. [DOI] [PubMed] [Google Scholar]
- 94.Wehr MC, Laage R, Bolz U, Fischer TM, Grünewald S, Scheek S, Bach A, Nave K-A, Rossner MJ. Monitoring Regulated Protein-Protein Interactions Using Split-TEV. Nature Methods. 2006 doi: 10.1038/nmeth967. this issue. [DOI] [PubMed] [Google Scholar]
- 95.Wehrman T, Kleaveland B, Her JH, Balint RF, Blau HM. Protein-protein interactions monitored in mammalian cells via complementation of beta -lactamase enzyme fragments. Proc Natl Acad Sci U S A. 2002;99:3469–3474. doi: 10.1073/pnas.062043699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Xu XM, Moller SG. AtSufE is an essential activator of plastidic and mitochondrial desulfurases in Arabidopsis. Embo Journal. 2006;25:900–909. doi: 10.1038/sj.emboj.7600968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ye HH, Choi HJ, Poe J, Smithgall TE. Oligomerization is required for HIV-1 nef-induced activation of the Src family protein-tyrosine kinase, Hck. Biochemistry. 2004;43:15775–15784. doi: 10.1021/bi048712f. [DOI] [PubMed] [Google Scholar]
- 98.Zal T, Gascoigne NR. Using live FRET imaging to reveal early protein-protein interactions during T cell activation. Current Opinion in Immunology. 2004;16:418–427. doi: 10.1016/j.coi.2004.05.019. [DOI] [PubMed] [Google Scholar]
- 99.Zhang J, Campbell RE, Ting AY, Tsien RY. Creating new fluorescent probes for cell biology. Nature Reviews Molecular Cell Biology. 2002;3:906–918. doi: 10.1038/nrm976. [DOI] [PubMed] [Google Scholar]
- 100.Zhang SF, Ma C, Chalfie M. Combinatorial marking of cells and organelles with reconstituted fluorescent proteins. Cell. 2004;119:137–144. doi: 10.1016/j.cell.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 101.Zhu LQ, Tran T, Rukstalis JM, Sun PC, Damsz B, Konieczny SF. Inhibition of Mist1 homodimer formation induces pancreatic acinar-to-ductal metaplasia. Molecular and Cellular Biology. 2004;24:2673–2681. doi: 10.1128/MCB.24.7.2673-2681.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]