Abstract
Currently the ε4 allele of the apolipoprotein E gene (APOE) is the strongest genetic risk factor for late onset Alzheimer's disease (AD). However, inheritance of the APOE ε 4 allele is not necessary or sufficient for the development of AD. Genetic evidence suggests that multiple loci in a 70 kb region surrounding APOE are associated with AD risk. Even though these loci could represent surrogate markers in linkage disequilibrium with APOE ε4 allele, they could also contribute biological effects independent of the APOE ε4 allele. Our previous study identified multiple SNPs upstream from APOE that are associated with cerebrospinal fluid apoE levels, suggesting that a haplotype structure proximal to APOE can influence apoE expression. In this study, we examined apoE expression in human post-mortem brain (PMB), and constructed chromosome-phase-separated haplotypes of the APOE proximal region to evaluate their effect on PMB apoE expression. ApoE protein expression was found to differ among AD brain regions and to differ between AD and control hippocampus. In addition, an extended APOE proximal haplotype structure, spanning from the TOMM40 gene to the APOE promoter, may modulate apoE expression in a brain region-specific manner and may influence AD disease status. In conclusion, this haplotype-phenotype analysis of apoE expression in PMB suggests that either; (1) the cis-regulation of APOE expression levels extends far upstream of the APOE promoter or (2) an APOE ε4 allele independent mechanism involving the TOMM40 gene plays a role in the risk of AD.
Keywords: Alzheimer's disease, APOE, post-mortem brain, TOMM40
Introduction
The ε4 allele of the apolipoprotein E gene (APOE) is the strongest known genetic risk factor for late onset Alzheimer's disease (AD) (Coon et al. 2007; Mahley et al. 2006;Yu et al. 2007). However, inheritance of the ε4 allele of APOE is neither necessary nor sufficient for AD (Huang et al. 2004; Hyman et al. 1996). Other genetic factors, such as APOE promoter polymorphisms have been reported to be associated with AD, including −491, −427, −219 (Th1/E47cs), and +113 (Artiga et al. 1998; Bullido et al. 1998; Parker et al. 2005; Ramos et al. 2005; Town et al. 1998). In addition to the APOE core promoter, other cis-regulatory elements of APOE have been characterized, which include multienhancer 1 and 2 (ME1 and ME2), that influence APOE regulation in macrophages and adipocytes (Mak et al. 2002; Shih et al. 2000), hepatic control regions, HCR1 and HCR2, that play a role in APOE regulation in the liver (Allan et al. 1995; Simonet et al. 1993), and a brain control region (BCR) that can modulate APOE expression in neurons and microglial (Zheng et al. 2004).
ApoE plasma levels have been reported to be lower in AD compared to the mildly cognitively impaired (Fukumoto et al. 2003). In addition, APOE 3/ 4 heterozygotes have a higher proportion of apoE E3 than the E4 isoform in plasma whereas the opposite proportion is present in cerebrospinal fluid (CSF) (Fukumoto et al. 2003). Other evidence suggests that plasma apoE is produced mostly by hepatocytes while CSF apoE is secreted primarily by brain glial cells (Linton et al. 1991; Pitas et al. 1987). Thus apoE and its isoforms are likely to be metabolized or regulated differently in the liver compared to the central nervous system (CNS). In order to better understand the role of apoE in AD risk, it is crucial to study APOE expression levels in the central compartments such as CSF and post-mortem brain (PMB).
APOE expression studies, haplotype analysis of APOE promoter polymorphisms and apoE E4 protein isoform studies have been reported with varying results between studies (Beffert et al. 1999; Bertrand et al. 1995; Bray et al. 2004; Lambert et al. 1997, 1998, 2002; Matsui et al. 2007; Yamagata et al. 2001). Several studies have measured either APOE mRNA or apoE protein levels in PMB, however, there are contrasting reports on whether APOE mRNA or apoE protein levels are increased or decreased in autopsy samples from AD patients compared to controls (Beffert et al. 1999; Bertrand et al. 1995; Lambert et al. 1998, 2002; Matsui et al. 2007,Yamagata et al. 2001). It may be that the measurement of different brain regions in these studies played a role in the contrasting results because APOE expression levels appear to vary by brain region in both humans (Beffert et al. 1999; Bertrand et al. 1995; Lambert et al. 1998, 2002; Matsui et al. 2007; Yamagata et al. 2001) and mice (Levin-Allerhand et al. 2001; Xu et al. 1998). Identification of a brain region specific haplotype that influences APOE expression may help explain some of this variability between studies and lead to better understanding of AD risk.
Recently, our group has reported that multiple SNPs within the TOMM40 gene and the APOE promoter are associated with CSF apoE levels (Bekris et al. 2008). To our knowledge, associations between a distant proximal APOE haplotype and multiple brain region APOE PMB mRNA and apoE protein levels has not been reported. Thus, in this investigation we hypothesized that haplotypes that span from TOMM40 to the APOE promoter influence PMB apoE expression. APOE mRNA and protein levels in four human brain regions, from AD patients and controls, were quantified; and chromosome-phase-separated haplotypes consisting of a proximal APOE gene region were analyzed for their association with apoE expression levels.
Methods
Population Description
Four regions of human brain (cerebellum; CB, frontal lobe; FL, hippocampus; HP, temporal lobe; TL) were obtained from the Neuropathology Core of the Alzheimer's Disease Research Center (ADRC) at the University of Washington. All tissue was obtained following informed consent, flash frozen at time of autopsy, and stored at −80°C. Patients with late-onset Alzheimer’s disease AD (n=8) were volunteers in the UW ADRC, where they were diagnosed during life with probable AD and confirmed by post-mortem neuropathologic examination to have AD. Control individuals (n=8) were volunteers in the UW ADRC, were never diagnosed with a central nervous system disorder and post-mortem examination showed age-related changes only; that included sparse to moderate amyloid plaque and Braak stage of IV or less. Use of human tissue was approved by the University of Washington Institutional Review Board. Sample size was constrained by limits on post-mortem brain interval (PMI), Braak stage and amyloid plaque score. Sample criteria for inclusion in this investigation were: 1) a low PMI less than or equal to 8 hours to limit mRNA and protein degradation of the brain samples and 2) diagnosis in life of AD as well as AD pathology according to Braak stage and amyloid plaque score. The population PMI, Braak stage, and amyloid score are described in Table 1.
Table 1.
Population, Haplotype and Diplotype Description.
| 1st Chromosome | 2nd Chromosome | Diplotype | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Disease Status |
Age |
Gender |
Braak Stage |
Amyloid Plaque |
Post-Mortem Interval |
APOE |
rs11556505 |
rs59007384 |
rs157584 |
rs741780 |
rs449647 |
Haplotype |
rs11556505 |
rs59007384 |
rs157584 |
rs741780 |
rs449647 |
Haplotype |
H1 + or − |
| AD | 82 | Male | IV | Frequent | 8:00 | 4/4 | C | G | C | T | A | H6 | T | T | T | T | A | H2 | H1− |
| AD | 84 | Female | VI | Sparse | 5:00 | 3/4 | C | T | T | T | A | H3 | T | T | T | T | T | H5 | H1− |
| AD | 76 | male | VI | Frequent | 4:30 | 4/4 | C | T | T | T | A | H3 | T | T | T | T | A | H2 | H1− |
| AD | 78 | male | VI | Frequent | 7:15 | 4/4 | C | T | T | T | A | H3 | T | T | T | T | A | H2 | H1− |
| AD | 87 | Female | VI | Moderate | 5:00 | 3/3 | C | G | C | C | A | H1 | C | G | T | T | A | H4 | H1+ |
| AD | 78 | male | VI | Moderate | 4:15 | 3/4 | C | G | C | C | A | H1 | C | T | T | T | A | H3 | H1+ |
| AD | 93 | Female | VI | Sparse | 6:30 | 3/4 | C | G | C | C | A | H1 | T | T | T | T | A | H2 | H1+ |
| AD | 60 | Female | IV | Moderate | 7:30 | 3/3 | C | G | C | C | A | H1 | C | G | C | C | A | H1 | H1+ |
| Control | 93 | Male | IV | Sparse-Moderate | 4:00 | 3/3 | C | G | C | C | A | H1 | C | G | T | T | T | H7 | H1+ |
| Control | 89 | Female | III | Sparse | 5:30 | 3/3 | C | G | C | C | A | H1 | T | T | T | T | T | H5 | H1+ |
| Control | 94 | Female | III | Sparse | 6:00 | 3/3 | C | G | C | C | A | H1 | T | T | T | T | T | H5 | H1+ |
| Control | 86 | Male | II | > Sparse | 2:30 | 3/3 | C | G | C | C | A | H1 | C | G | T | T | A | H4 | H1+ |
| Control | 86 | Male | II | > Sparse | 2:30 | 3/3 | C | G | C | C | A | H1 | C | G | T | T | A | H4 | H1+ |
| Control | 79 | Female | II | Moderate | 3:00 | 3/4 | C | G | C | C | A | H1 | T | T | T | T | A | H2 | H1+ |
| Control | 87 | Female | II | None | 3:45 | 3/3 | C | G | C | C | A | H1 | C | G | C | C | A | H1 | H1+ |
| Control | 94 | Male | IV | Sparse | 6:00 | 3/3 | C | G | C | C | A | H1 | C | G | C | C | A | H1 | H1+ |
The halotype anlysed includes 5 SNPs: rs 11556505, rs59007384, rs157584, rs741780, rs449647. There are a total of 7 different haplotypes. Haplotype number was assigned according to its ranking in frequency. The most common H1 haplotype is associated with increased AD risk (OR: 3.00, 95% Cl: 1.35 – 6.68).
DNA, RNA, and Protein Extraction and Quantification
DNA, RNA, and protein were extracted from between 1 and 3 mg of post-mortem brain tissue using the Qiagen Allprep DNA/RNA MiniKit (Qiagen, Valencia, CA). Protein was isolated from the column flow through (after the ethanol step) using an acetone precipitation step according to Qiagen protocol (Qiagen) which precipitates soluble and insoluble protein. The protein sample was then resuspended in immunoprecipitation buffer (150mM NaCl, 50mM Tris-HCL (pH6.8), 0.5% NP-40, 0.5% Na-deoxycholate, 5 mM EDTA, 50 µg/ml pepstatin, 50 µg/ml leupeptin, 10 µg/ml aprotinin, 0.25mM PMSF). APOE mRNA was measured twice using duplicates within each quantitative real time RT-PCR (QRT-PCR) (Applied Biosystems, Foster City, CA). Beta-actin was used as a loading control. Minus RT samples were included and any samples that had a positive minus RT were repeated. All QRT-PCR results are presented as a ratio of APOE/Actin. ApoE protein levels were measured twice by Western blot using a goat polyclonal anti-human apoE antibody conjugated to HRP (AbCam, Cambridge, MA). Mouse anti-human beta-actin antibody (Sigma, St. Louis, MO) was used to measure beta-actin and utilized as a loading control. All results are presented as a ratio of ApoE/Actin. Western blot band intensity was quantified using ImageJ software (U. S. National Institutes of Health, http://rsb.info.nih.gov.offcampus.lib.washington.edu/ij/, 1997–2007). Results above the 97th percentile or below the 3rd percentile were considered as outliers and removed from the database (n=4). Data are presented as APOE/Actin (mRNA), ApoE/Actin (protein), or as a ratio of APOE/Actin (mRNA/protein).
SNPs and Genotyping
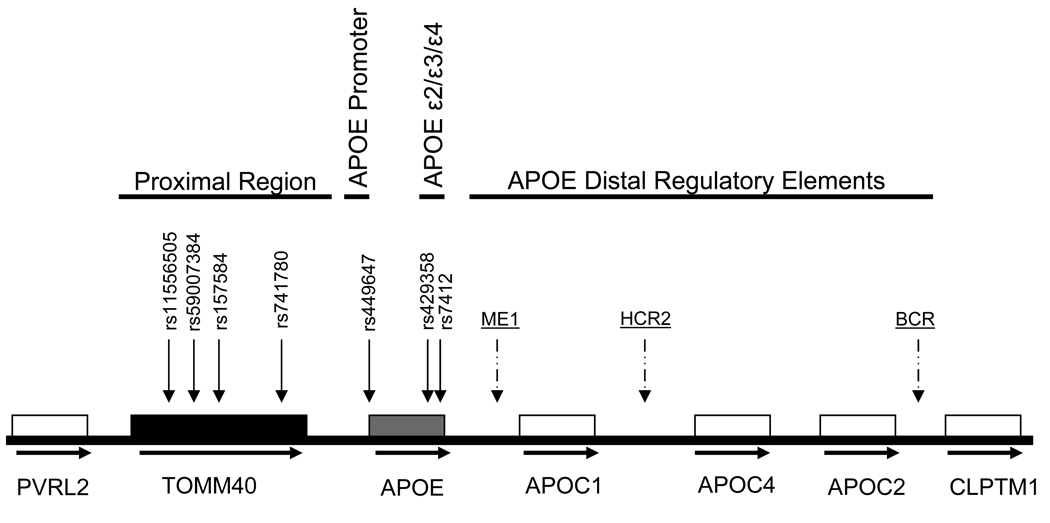
Five APOE proximal region SNPs were chosen according to the following criteria: 1) reported to be associated with AD or 2) reported to be associated with CSF apoE protein levels. Altogether seven SNPs were genotyped, including the rs429358 and rs7412 that define the APOE ε2/ε3/ε4 system plus APOE proximal SNPs (within the TOMM40 gene; rs11556505, rs59007384, rs157584, rs741780), and an APOE promoter SNP (5’ UTR of APOE, rs449647) (Figure 1). Genotypes were determined as previously described (Bekris et al. 2008). Briefly, TaqMan allelic discrimination detection using 96-well plates was utilized (Applied Biosystems). PCR reactions were carried out using a 9700 Gene Amp PCR System (Applied Biosystems). Plates were subjected to end-point read in a 7500 Real-Time PCR System (Applied Biosystems). All SNPs are in Hardy Weinberg equilibrium.
Figure 1.
APOE Gene Region Description. Horizontal arrows represent transcriptional orientation for each gene. Vertical arrows represent SNP location. Scale is approximate.
Molecular Haplotyping
Molecular haplotypes, also known as phase-separated haplotypes, were determined for five SNPs (rs11556505, rs59007384, rs157584, rs741780, and rs449647, Table 1 and Fig. 1) using the Allele Discriminating Long and Accurate PCR Haplotyping (ADLAPH) method (Yu et al. 2004). Briefly, ADLAPH combines allele-discriminating primers and long-range PCR amplification to amplify long genomic fragments from only one of the two chromosome homologues of a particular subject. The phase-separated long-rang PCR product is then genotyped by standard methods to yield one haplotype. The contrast with the original diploid genotypes is then carried out to provide the complementary haplotype. Primers sequences and PCR conditions of ADLAPH procedure will be provided upon request.
Statistical Analysis
We examined the relationship between apoE expression levels and APOE proximal region diplotypes by treating each sample from each brain region as an independent sample and using an independent t-test to determine if there was a difference in apoE expression levels between brain regions. No correction for multiple comparisons was used. Statistical analyses were performed using SPSS software (Version 14) and Prism (Version 3.03)
Results
APOE Expression Levels in PMB
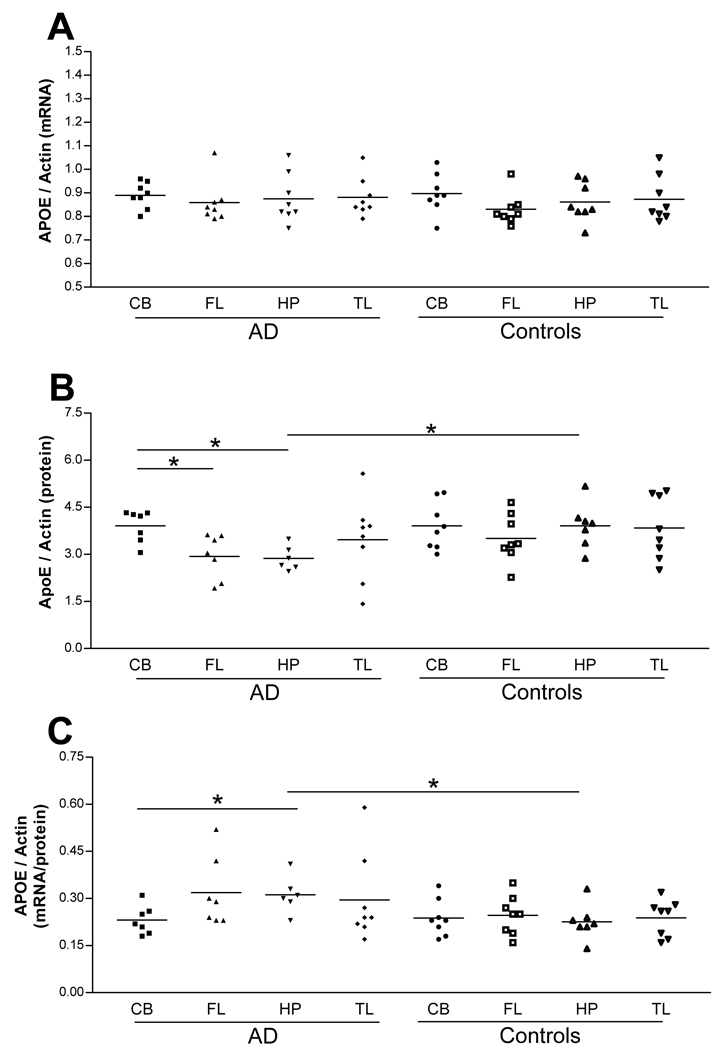
To determine if APOE mRNA and protein levels differ according to brain region in AD patients compared to controls, quantitative real time RT-PCR (qRT-PCR) and Western blot analyses were performed on PMB from eight AD patients and eight cognitively normal control subjects. Four regions from PMB (CB, FL, HP, TL) were used. Independent t-tests (without a correction for multiple comparisons) were used to determine if there is a difference in APOE mRNA, protein or mRNA/protein ratio levels between brain regions. AD patients and control subjects did not show a significant difference in APOE mRNA levels between brain regions (Fig. 2, panel A). CB apoE protein levels were found to be significantly higher than FL (p-value, 0.012) or HP (p-value, 0.002) in AD (Fig. 2, panel B). AD HP apoE protein levels were significantly lower than control HP (p-value, 0.009) (Fig. 2, panel B). To determine if there was a direct relationship between mRNA levels relative to protein for each sample, a ratio of APOE mRNA/apoE protein was calculated. This ratio is significantly higher in AD HP when compared to AD CB (p-value, 0.018) and control HP (p-value, 0.021) (Fig. 2, panel C).
Figure 2.
Expression levels of APOE mRNA, protein or mRNA/protein ratio for each brain region in AD patients and controls. APOE mRNA levels do not differ between brain regions (panel A). CB apoE protein levels are higher than FL (p-value, 0.012) or HP (p-value, 0.002) in AD (panel B). AD HP apoE protein levels are lower than control HP (p-value, 0.009) (panel B). AD HP apoE mRNA/protein are significantly higher than control HP (p-value, 0.021) or control CB (p-value, 0.018) (panel C). (cerebellum; CB, frontal lobe; FL, hippocampus; HP, temporal lobe; TL).
Haplotype and Diplotype Analyses
We next addressed the question of whether specific APOE proximal haplotypes, which were constructed using regional SNPs previously found to correlate with apoE CSF levels (Bekris et al. 2008), are associated with PMB APOE expression levels. The ADLAPH method was applied to construct the chromosome-phase-separated haplotypes in this region of interest; that spans 12.5 kb from TOMM40 exon 4 to APOE promoter and includes five SNPs (rs11556505, rs59007384, rs157584, rs741780, and rs449647). A total of seven haplotypes were identified from our 16 PMB study samples and were defined as H1–H7 (Table 1). The H1 haplotype (CGCCA) is the predominant haplotype and is present in 46.9% of all the samples including 31.3% and 62.5% of AD and controls, respectively. Because the final expression levels of a gene are largely determined by combined expression of two copies of the gene in the same genome, we further assessed the effect of the combined haplotypes (or diplotypes) of each subject in apoE expression. Such a diplotype setting represents the actual genomic environment in each individual. When the two phase-separated haplotypes from a subject were reconstructed to form a single diplotype, there were nine different diplotypes present in our study subjects. To simplify the analysis, we collapsed the diplotypes into two groups, either with (H1+) or without (H1−) the predominant haplotype H1. The H1- diplotype carriers were found to be present only in the AD group (50%) and thus highly associated with AD risk (OR: 3.0; 95% CI: 1.35–6.68) (Table 1).
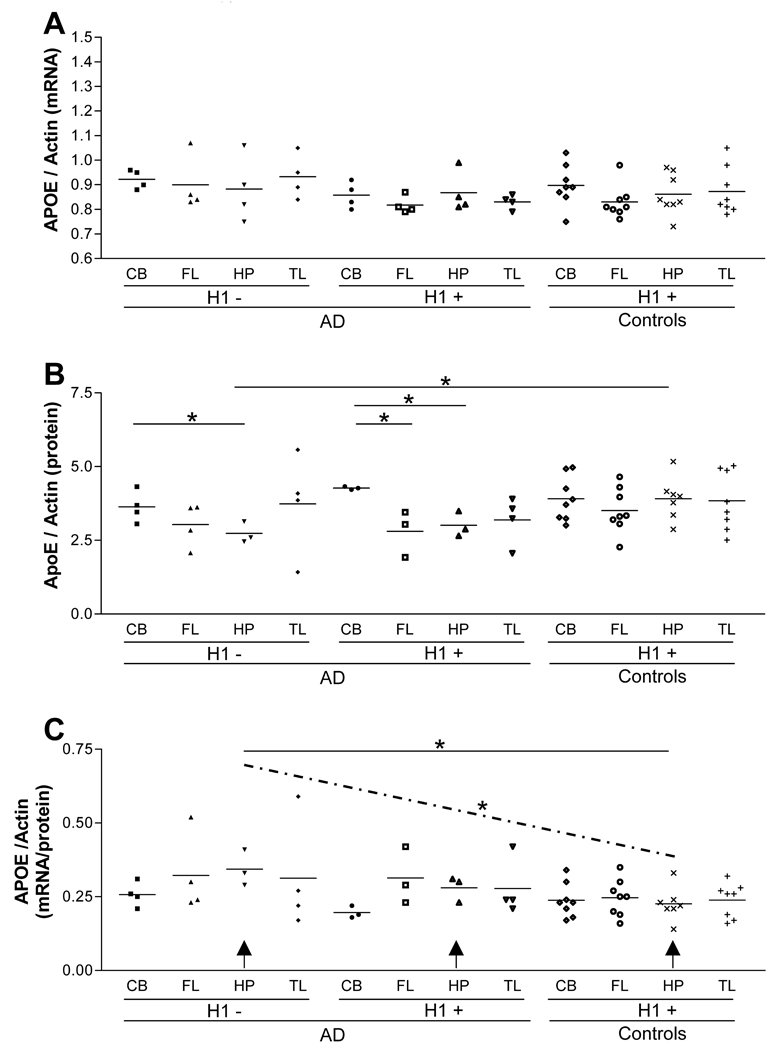
We then tested the dichotomized (H1+ vs. H1−) diplotypes for their association with APOE expression between brain regions using an independent sample t-test for each comparison. There was not a significant difference in APOE mRNA levels between brain regions according to the H1+/− dichotomized diplotypes (Fig. 3, panel A). There was a difference in apoE protein levels between the AD CB and the AD HP within the H1− group (p-value, 0.053). There was a significant difference in apoE protein levels between the AD CB and the AD FL (p-value, 0.003) and between the AD CB and the AD HP (p-value, 0.007) within the H1+ group (Fig. 3, panel B). AD HP APOE mRNA/protein ratio was significantly higher in the H1− diplotype than the control HP with H1+ diplotype (p-value, 0.018) (Fig. 3, panel C). Furthermore, there is a significant decreasing trend when APOE mRNA/protein ratios of HP were compared among three groups; AD H1−, AD H1+, and control H1+ (p-value, 0.011) (Fig. 3, panel C, dotted line). These results suggest an extended haplotype and diplotype that spans from TOMM40 to the APOE promoter region are likely to be associated with APOE expression in a brain region-specific manner; that could carry biological consequence for the risk of AD.
Figure 3.
Diplotype analysis of APOE expression in PMB. No subjects in the control group carry the H1−(panel A). The H1− diplotype AD apoE protein levels are significantly lower than H1 + displotype control HP (p-value, 0.030) (panel B). The H1 + AD CB has higher apoE protein levels than both the AD FL (p-value, 0.003) and the HP (p-value, 0.007) (panel B). The H1 − AD CB has higher apoE protein levels than H1−AD HP(p-value,0.053) (panel B). The H1 − diplotype AD HP APOE mRNA/protein ratio is significantly higher than H1+ diplotype control HP (p-value, 0.018) (panel C). There is a significant decreasing trend between the H1− AD HP, H1+ AD HP and H1+ control HP (indicated by arrows and dotted line) (p-value. 0.011) (panel C).
Discussion
APOE ε4 is an important genetic risk factor for AD (Coon et al. 2007; Mahley et al. 2006; Yu et al. 2007). APOE expression appears to differ between AD and controls according to brain region (Beffert et al. 1999; Bertrand et al. 1995; Lambert et al. 1998, 2002; Matsui et al. 2007; Yamagata et al. 2001) and the hippocampus plays a key role in AD pathogenesis (Allen et al. 2007; Csernansky et al. 2004; Hampel et al. 2002; Sluimer et al. 2008). Furthermore, it has been demonstrated in animal models that APOE distal regulatory elements have tissue specific activity (Allan et al. 1995; Mak et al. 2002; Shih et al. 2000; Simonet et al. 1993; Zheng et al. 2004). Therefore, besides the effect of APOE ε4, brain region specific expression levels of apoE, may also contribute to the risk of AD. In our previous investigations, SNPs proximal to APOE were found to be associated with AD risk (Yu et al. 2007) and these SNPs were also found to be associated with CSF apoE levels in cognitively normal subjects (Bekris et al. 2008). Thus, the present study is an extension of our previous study, and aims to test the hypothesis that PMB APOE mRNA or apoE protein levels vary according to brain region, haplotype structure, and AD disease status.
APOE expression from 16 PMB samples (8 AD and 8 controls) and 4 brain regions (CB, FL, HP, and TL) were measured. It was determined that brain apoE protein levels but not APOE mRNA levels differ between brain regions whereby AD HP apoE protein levels are significantly lower than the AD CB and control HP (Fig. 2, panel B), consistent with the findings of some (Beffert et al. 1999; Bertrand et al. 1995) but not others (Laws et al. 2002). Our finding that APOE mRNA levels were not different between AD patient and control brain region is inconsistent with previous reports where PMB APOE mRNA levels were either higher (Lambert et al. 2002; Matsui et al. 2007; Yamagata et al. 2001) or lower (Xu et al. 1999) in AD compared to controls. One potential explanation for the discrepant expression data between apoE mRNA and protein is sequestration of apoE protein in amyloid plaque in diseased regions of AD brain. However, a t-test comparing AD patients with frequent amyloid plaques and all the other amyloid plaque groups (sparse to moderate) did not show a significant difference in APOE mRNA, apoE protein, or APOE mRNA/protein (data not shown). Another explanation for this discrepancy is the degradation of PMB mRNA due to post-mortem interval (PMI). However, mRNA degradation was not found in our sample upon gel electrophoresis (data not shown) suggesting that APOE mRNA levels in brain may be highly variable between samples. To resolve such discrepancy, a larger sample size will be required in future studies.
Because our study sample size is small (n=16), and our SNPs of interest are not in strong linkage disequilibrium, as measured by R2 (data not shown), we constructed the chromosome-phase-separated haplotypes to eliminate estimation and uncertainty associated with inferred haplotyping. These haplotypes consist of five SNPs and extend from TOMM40 exon 4 to the APOE promoter. Within our study sample, seven haplotypes (H1–H7) were identified. Because each subject carries two haplotypes (or a single diplotype) in their genome, we further reconstructed this diplotype status for each subject. The H1 haplotype predominated in the controls (62.5%) and the H1− diplotype was only observed in the AD group (Table 1). This H1− diplotype was associated with AD risk (OR: 3.0; 95% CI: 1.35–6.68) (Table 1).
A direct relationship between mRNA levels relative to protein levels was evaluated by calculating an APOE mRNA/protein ratio. Higher APOE mRNA/protein ratios were found in: (1) AD HP when compared to AD CB and control HP (Fig. 2, panel C), (2) AD HP H1− diplotype group when compared to control HP H1+ diplotype group (Fig. 3, panel C). Additionally, there is a significant decreasing trend in this APOE mRNA/protein ratios when HP regions were compared among AD/H1−, AD/H1+, and control/H1+ groups (P-value, 0.011) (Fig. 3, panel C, dotted line). Thus, this H1 haplotype may influence APOE mRNA relative to apoE protein expression particularly in the HP. These results suggest that there may be an uncharacterized regulatory cis-element distantly proximal to APOE that is particularly active in the HP. It also supports the notion that an extended haplotype structure, spanning from the APOE promoter to a region proximal to the APOE gene, within the TOMM40 gene, may modulate apoE expression as well as AD risk.
Since this haplotype extends into the TOMM40 gene, an alternative explanation is that changes in apoE expression associated with AD may be merely a secondary consequence, and TOMM40 variants that alter mitochondrial function are the actual primary effecters for AD risk. Thus, it is important to note that recently, mitochondrial dysfunction and mitochondrial targeting of amyloid precursor protein (APP) has been reported in AD brain whereby APP appears to associate with the mitochondrial protein translocator, tom40, which is encoded by the TOMM40 gene (Anandatheerthavarada et al. 2003; Devi et al. 2006). Accumulation of Aβ within mitochondria has been reported to lead to modulation of mitochondrial function (Busciglio et al. 2002; Cardoso et al. 2001; Casley et al. 2002; Manczak et al. 2006). Therefore, it is possible that variants of tom40 differentially impact APP and Aβ translocation, mitochondrial energy metabolism or the oxidative stress response, and contribute a biological effect in AD.
A limitation of this investigation is the inclusion of both; AD patients with low Braak stage and sparse amyloid plaque as well as controls with moderately high Braak stage and moderate amyloid plaque score (Table 1). However, studies have shown that normal age-related changes in PMB can include an increase in neurofibrillary tangles and amyloid plaque as well as a high variability in Braak stage and amyloid plaque levels (Hof et al. 1996). Since amyloid plaque levels could be inversely associated with apoE protein levels in PMB due to sequestration (Glockner and Ohm, 2003), apoE protein levels from AD patients with frequent plaques were compared to apoE protein levels from AD patients with sparse to moderate plaques, and no significant difference was found. Another limitation of this study is the small sample size which may have lead to false negative results. Indeed, power calculations using α=5% for APOE mRNA data show a lack of power (i.e., AD HP compared to control HP: 1-β=5%) whereas the protein data does not show a lack of power (AD HP compared to control HP: 1-β=91.4%). Therefore, the results of this investigation must be approached with caution because sample sizes are small and multiple comparisons were not taken into account in any of these analyses. However, it is important to note that most of the interesting findings are derived, in part or fully, from the protein data, which has adequate power.
This exploratory investigation is novel and important because it is the only study, to our knowledge, that includes both APOE mRNA and apoE protein measurements within the same study sample, for multiple brain regions, thus allowing for the direct examination of APOE mRNA relative to apoE protein levels. Interestingly, high APOE mRNA levels relative to protein levels, as indicated by the mRNA/protein ratio, is present in the HP compared to other regions, and may implicate brain region specific post-transcriptional regulation such as protein degradation or translational inhibition. Recently important post-transcriptional regulators of temporal and spatial protein expression, such as microRNAs, have been characterized in the nervous system (Filipowicz et al. 2008; Mehler and Mattick, 2006). Other post-transcriptional mechanisms such as sequestration of protein into amyloid plaques could also play role. However, even though, individuals with frequent plaques were more likely to be on the H1− diplotype background, no significant difference was found in the levels of APOE expression according to amyloid plaque score when comparing frequent plaques to sparse or moderate plaques, suggesting that sequestration does not play a role. Alternatively, translational inhibition could play a role. For example, alternatively expressed large 5’ UTRs and regulation by miRNA expression under stress conditions have been described (Lytle et al. 2007). A recent report suggests that microRNA can modulate AD CSF and PMB APOE expression (Cogswell et al. 2008). APOE regulation has been reported to be influenced by distant regulatory regions that have tissue specific activity, such as ME1, ME2, HCR1, HCR2 and the BCR as far downstream as 41.9 kb, suggesting that regulation of APOE expression levels is complex and involves multiple distant cis-elements (Allan et al. 1995; Mak et al. 2002; Shih et al. 2000; Simonet et al. 1993; Zheng et al. 2004). Therefore, it may be proposed that an uncharacterized distant regulatory element exists in the APOE gene region that may contribute to a complex APOE regulatory system. In addition, specific haplotypes may contribute to high mRNA levels relative to protein levels, particularly in the AD HP, implicating an influence by genetic variability in the APOE proximal region on protein degradation or translational inhibition.
In summary, results from this exploratory investigation suggest that a haplotype structure that spans from the APOE promoter 12.5 kb upstream to the TOMM40 gene may influence APOE expression in a brain region and AD specific manner.
Acknowledgements
This work was funded by Funded by Veterans Affairs Biomedical Laboratory Research Development Merit Review and NIH Grant P50 AG00536.
References
- Allan CM, Walker D, Taylor JM. Evolutionary duplication of a hepatic control region in the human apolipoprotein E gene locus. Identification of a second region that confers high level and liver-specific expression of the human apolipoprotein E gene in transgenic mice. J Biol Chem. 1995;270:26278–26281. doi: 10.1074/jbc.270.44.26278. [DOI] [PubMed] [Google Scholar]
- Allen G, Barnard H, McColl R, Hester AL, Fields JA, Weiner MF, Ringe WK, Lipton AM, Brooker M, McDonald E, Rubin CD, Cullum CM. Reduced hippocampal functional connectivity in Alzheimer disease. Arch Neurol. 2007;64:1482–1487. doi: 10.1001/archneur.64.10.1482. [DOI] [PubMed] [Google Scholar]
- Anandatheerthavarada HK, Biswas G, Robin MA, Avadhani NG. Mitochondrial targeting and a novel transmembrane arrest of Alzheimer's amyloid precursor protein impairs mitochondrial function in neuronal cells. J Cell Biol. 2003;161:41–54. doi: 10.1083/jcb.200207030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artiga MJ, Bullido MJ, Sastre I, Recuero M, Garcia MA, Aldudo J, Vazquez J, Valdivieso F. Allelic polymorphisms in the transcriptional regulatory region of apolipoprotein E gene. FEBS Lett. 1998;421:105–108. doi: 10.1016/s0014-5793(97)01543-3. [DOI] [PubMed] [Google Scholar]
- Beffert U, Cohn JS, Petit-Turcotte C, Tremblay M, Aumont N, Ramassamy C, Davignon J, Poirier J. Apolipoprotein E and beta-amyloid levels in the hippocampus and frontal cortex of Alzheimer's disease subjects are disease-related and apolipoprotein E genotype dependent. Brain Res. 1999;843:87–94. doi: 10.1016/s0006-8993(99)01894-6. [DOI] [PubMed] [Google Scholar]
- Bekris LM, Millard SP, Galloway NM, Vuletic S, Albers JJ, Li G, Galasko DR, DeCarli C, Farlow MR, Clark CM, Quinn JF, Kaye JA, Schellenberg GD, Tsuang D, Peskind ER, Yu CE. Multiple SNPs within and surrounding the apolipoprotein E gene influence cerebrospinal fluid apolipoprotein E protein levels. J Alzheimers Dis. 2008;13:255–266. doi: 10.3233/jad-2008-13303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand P, Poirier J, Oda T, Finch CE, Pasinetti GM. Association of apolipoprotein E genotype with brain levels of apolipoprotein E and apolipoprotein J (clusterin) in Alzheimer disease. Brain Res Mol Brain Res. 1995;33:174–178. doi: 10.1016/0169-328x(95)00097-c. [DOI] [PubMed] [Google Scholar]
- Bray NJ, Jehu L, Moskvina V, Buxbaum JD, Dracheva S, Haroutunian V, Williams J, Buckland PR, Owen MJ, O'Donovan MC. Allelic expression of APOE in human brain: Effects of epsilon status and promoter haplotypes. Hum Mol Genet. 2004;13:2885–2892. doi: 10.1093/hmg/ddh299. [DOI] [PubMed] [Google Scholar]
- Bullido MJ, Artiga MJ, Recuero M, Sastre I, Garcia MA, Aldudo J, Lendon C, Han SW, Morris JC, Frank A, Vazquez J, Goate A, Valdivieso F. A polymorphism in the regulatory region of APOE associated with risk for Alzheimer's dementia. Nat Genet. 1998;18:69–71. doi: 10.1038/ng0198-69. [DOI] [PubMed] [Google Scholar]
- Busciglio J, Pelsman A, Wong C, Pigino G, Yuan M, Mori H, Yankner BA. Altered metabolism of the amyloid beta precursor protein is associated with mitochondrial dysfunction in Down's syndrome. Neuron. 2002;33:677–688. doi: 10.1016/s0896-6273(02)00604-9. [DOI] [PubMed] [Google Scholar]
- Cardoso SM, Santos S, Swerdlow RH, Oliveira CR. Functional mitochondria are required for amyloid beta-mediated neurotoxicity. FASEB J. 2001;15:1439–1441. doi: 10.1096/fj.00-0561fje. [DOI] [PubMed] [Google Scholar]
- Casley CS, Canevari L, Land JM, Clark JB, Sharpe MA. Beta-amyloid inhibits integrated mitochondrial respiration and key enzyme activities. J Neurochem. 2002;80:91–100. doi: 10.1046/j.0022-3042.2001.00681.x. [DOI] [PubMed] [Google Scholar]
- Cogswell JP, Ward J, Taylor IA, Waters M, Shi Y, Cannon B, Kelnar K, Kemppainen J, Brown D, Chen C, Prinjha RK, Richardson JC, Saunders AM, Roses AD, Richards CA. Identification of miRNA changes in Alzheimer's disease brain and CSF yields putative biomarkers and insights into disease pathways. J Alzheimers Dis. 2008;14:27–41. doi: 10.3233/jad-2008-14103. [DOI] [PubMed] [Google Scholar]
- Coon KD, Myers AJ, Craig DW, Webster JA, Pearson JV, Lince DH, Zismann VL, Beach TG, Leung D, Bryden L, Halperin RF, Marlowe L, Kaleem M, Walker DG, Ravid R, Heward CB, Rogers J, Papassotiropoulos A, Reiman EM, Hardy J, Stephan DA. A high-density whole-genome association study reveals that APOE is the major susceptibility gene for sporadic late-onset Alzheimer's disease. J Clin Psychiatry. 2007;68:613–618. doi: 10.4088/jcp.v68n0419. [DOI] [PubMed] [Google Scholar]
- Csernansky JG, Wang L, Joshi SC, Ratnanather JT, Miller MI. Computational anatomy and neuropsychiatric disease: Probabilistic assessment of variation and statistical inference of group difference, hemispheric asymmetry, and time-dependent change. Neuroimage. 2004;23:S56–S68. doi: 10.1016/j.neuroimage.2004.07.025. [DOI] [PubMed] [Google Scholar]
- Devi L, Prabhu BM, Galati DF, Avadhani NG, Anandatheerthavarada HK. Accumulation of amyloid precursor protein in the mitochondrial import channels of human Alzheimer's disease brain is associated with mitochondrial dysfunction. J Neurosci. 2006;26:9057–9068. doi: 10.1523/JNEUROSCI.1469-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Fukumoto H, Ingelsson M, Garevik N, Wahlund LO, Nukina N, Yaguchi Y, Shibata M, Hyman BT, Rebeck GW, Irizarry MC. APOE epsilon 3/epsilon 4 heterozygotes have an elevated proportion of apolipoprotein E4 in cerebrospinal fluid relative to plasma, independent of Alzheimer's disease diagnosis. Exp Neurol. 2003;183:249–253. doi: 10.1016/s0014-4886(03)00088-8. [DOI] [PubMed] [Google Scholar]
- Glockner F, Ohm TG. Hippocampal apolipoprotein D level depends on Braak stage and APOE genotype. Neuroscience. 2003;122:103–110. doi: 10.1016/s0306-4522(03)00529-3. [DOI] [PubMed] [Google Scholar]
- Hampel H, Teipel SJ, Alexander GE, Pogarell O, Rapoport SI, Moller HJ. In vivo imaging of region and cell type specific neocortical neurodegeneration in Alzheimer's disease. Perspectives of MRI derived corpus callosum measurement for mapping disease progression and effects of therapy. Evidence from studies with MRI, EEG and PET. J Neural Transm. 2002;109:837–855. doi: 10.1007/s007020200069. [DOI] [PubMed] [Google Scholar]
- Hof PR, Glannakopoulos P, Bouras C. The neuropathological changes associated with normal brain aging. Histol Histopathol. 1996;11:1075–1088. [PubMed] [Google Scholar]
- Huang W, Qiu C, von Strauss E, Winblad B, Fratiglioni L. APOE genotype, family history of dementia, and Alzheimer disease risk: A 6-year follow-up study. Arch Neurol. 2004;61:1930–1934. doi: 10.1001/archneur.61.12.1930. [DOI] [PubMed] [Google Scholar]
- Hyman BT, Gomez-Isla T, Rebeck GW, Briggs M, Chung H, West HL, Greenberg S, Mui S, Nichols S, Wallace R, Growdon JH. Epidemiological, clinical, and neuropathological study of apolipoprotein E genotype in Alzheimer's disease. Ann NY Acad Sci. 1996;802:1–5. doi: 10.1111/j.1749-6632.1996.tb32592.x. [DOI] [PubMed] [Google Scholar]
- Lambert JC, Araria-Goumidi L, Myllykangas L, Ellis C, Wang JC, Bullido MJ, Harris JM, Artiga MJ, Hernandez D, Kwon JM, Frigard B, Petersen RC, Cumming AM, Pasquier F, Sastre I, Tienari PJ, Frank A, Sulkava R, Morris JC, St Clair D, Mann DM, Wavrant-DeVrieze F, Ezquerra-Trabalon M, Amouyel P, Hardy J, Haltia M, Valdivieso F, Goate AM, Perez-Tur J, Lendon CL, Chartier-Harlin MC. Contribution of APOE promoter polymorphisms to Alzheimer's disease risk. Neurology. 2002;59:59–66. doi: 10.1212/wnl.59.1.59. [DOI] [PubMed] [Google Scholar]
- Lambert JC, Berr C, Pasquier F, Delacourte A, Frigard B, Cottel D, Perez-Tur J, Mouroux V, Mohr M, Cecyre D, Galasko D, Lendon C, Poirier J, Hardy J, Mann D, Amouyel P, Chartier-Harlin MC. Pronounced impact of Th1/E47cs mutation compared with -491 AT mutation on neural APOE gene expression and risk of developing Alzheimer's disease. Hum Mol Genet. 1998;7:1511–1516. doi: 10.1093/hmg/7.9.1511. [DOI] [PubMed] [Google Scholar]
- Lambert JC, Perez-Tur J, Dupire MJ, Galasko D, Mann D, Amouyel P, Hardy J, Delacourte A, Chartier-Harlin MC. Distortion of allelic expression of apolipoprotein E in Alzheimer's disease. Hum Mol Genet. 1997;6:2151–2154. doi: 10.1093/hmg/6.12.2151. [DOI] [PubMed] [Google Scholar]
- Laws SM, Hone E, Taddei K, Harper C, Dean B, McClean C, Masters C, Lautenschlager N, Gandy SE, Martins RN. Variation at the APOE-491 promoter locus is associated with altered brain levels of apolipoprotein E. Mol Psychiatry. 2002;7:886–890. doi: 10.1038/sj.mp.4001097. [DOI] [PubMed] [Google Scholar]
- Levin-Allerhand J, McEwen BS, Lominska CE, Lubahn DB, Korach KS, Smith JD. Brain region-specific up-regulation of mouse apolipoprotein E by pharmacological estrogen treatments. J Neurochem. 2001;79:796–803. doi: 10.1046/j.1471-4159.2001.00627.x. [DOI] [PubMed] [Google Scholar]
- Linton MF, Gish R, Hubl ST, Butler E, Esquivel C, Bry WI, Boyles JK, Wardell MR, Young SG. Phenotypes of apolipoprotein B and apolipoprotein E after liver transplantation. J Clin Invest. 1991;88:270–281. doi: 10.1172/JCI115288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lytle JR, Yario TA, Steitz JA. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5 UTR as in the 3 UTR. Proc Natl Acad Sci USA. 2007;104:9667–9672. doi: 10.1073/pnas.0703820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahley RW, Weisgraber KH, Huang Y. Apolipoprotein E4: A causative factor and therapeutic target in neuropathology, including Alzheimer's disease. Proc Natl Acad Sci USA. 2006;103:5644–5651. doi: 10.1073/pnas.0600549103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak PA, Laffitte BA, Desrumaux C, Joseph SB, Curtiss LK, Mangelsdorf DJ, Tontonoz P, Edwards PA. Regulated expression of the apolipoprotein E/C-I/C-IV/C-II gene cluster in murine and human macrophages. A critical role for nuclear liver X receptors alpha and beta. J Biol Chem. 2002;277:31900–31908. doi: 10.1074/jbc.M202993200. [DOI] [PubMed] [Google Scholar]
- Manczak M, Anekonda TS, Henson E, Park BS, Quinn J, Reddy PH. Mitochondria are a direct site of A beta accumulation in Alzheimer's disease neurons: Implications for free radical generation and oxidative damage in disease progression. Hum Mol Genet. 2006;15:1437–1449. doi: 10.1093/hmg/ddl066. [DOI] [PubMed] [Google Scholar]
- Matsui T, Ingelsson M, Fukumoto H, Ramasamy K, Kowa H, Frosch MP, Irizarry MC, Hyman BT. Expression of APP pathway mRNAs and proteins in Alzheimer's disease. Brain Res. 2007;1161:116–123. doi: 10.1016/j.brainres.2007.05.050. [DOI] [PubMed] [Google Scholar]
- Mehler MF, Mattick JS. Non-coding RNAs in the nervous system. J Physiol. 2006;575:333–341. doi: 10.1113/jphysiol.2006.113191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker GR, Cathcart HM, Huang R, Lanham IS, Corder EH, Poduslo SE. Apolipoprotein gene E4 allele promoter polymorphisms as risk factors for Alzheimer's disease. Psychiatr Genet. 2005;15:271–275. doi: 10.1097/00041444-200512000-00009. [DOI] [PubMed] [Google Scholar]
- Pitas RE, Boyles JK, Lee SH, Foss D, Mahley RW. Astrocytes synthesize apolipoprotein E and metabolize apolipoprotein E-containing lipoproteins. Biochim Biophys Acta. 1987;917:148–161. doi: 10.1016/0005-2760(87)90295-5. [DOI] [PubMed] [Google Scholar]
- Ramos MC, Matias S, Artiga MJ, Pozueta J, Sastre I, Valdivieso F, Bullido MJ. Neuronal specific regulatory elements in apolipoprotein E gene proximal promoter. Neuroreport. 2005;16:1027–1030. doi: 10.1097/00001756-200506210-00029. [DOI] [PubMed] [Google Scholar]
- Shih SJ, Allan C, Grehan S, Tse E, Moran C, Taylor JM. Duplicated downstream enhancers control expression of the human apolipoprotein E gene in macrophages and adipose tissue. J Biol Chem. 2000;275:31567–31572. doi: 10.1074/jbc.M005468200. [DOI] [PubMed] [Google Scholar]
- Simonet WS, Bucay N, Lauer SJ, Taylor JM. A far-downstream hepatocyte-specific control region directs expression of the linked human apolipoprotein E and C-I genes in transgenic mice. J Biol Chem. 1993;268:8221–8229. [PubMed] [Google Scholar]
- Sluimer JD, Vrenken H, Blankenstein MA, Fox NC, Scheltens P, Barkhof F, van der Flier WM. Whole-brain atrophy rate in Alzheimer disease: Identifying fast progressors. Neurology. 2008;70:1836–1841. doi: 10.1212/01.wnl.0000311446.61861.e3. [DOI] [PubMed] [Google Scholar]
- Town T, Paris D, Fallin D, Duara R, Barker W, Gold M, Crawford F, Mullan M. The −491A/T apolipoprotein E promoter polymorphism association with Alzheimer's disease: Independent risk and linkage disequilibrium with the known APOE polymorphism. Neurosci Lett. 1998;252:95–98. doi: 10.1016/s0304-3940(98)00567-9. [DOI] [PubMed] [Google Scholar]
- Xu PT, Gilbert JR, Qiu HL, Ervin J, Rothrock-Christian TR, Hulette C, Schmechel DE. Specific regional transcription of apolipoprotein E in human brain neurons. Am J Pathol. 1999;154:601–611. doi: 10.1016/S0002-9440(10)65305-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu PT, Gilbert JR, Qiu HL, Rothrock-Christian T, Settles DL, Roses AD, Schmechel DE. Regionally specific neuronal expression of human APOE gene in transgenic mice. Neurosci Lett. 1998;246:65–68. doi: 10.1016/s0304-3940(98)00247-x. [DOI] [PubMed] [Google Scholar]
- Yamagata K, Urakami K, Ikeda K, Ji Y, Adachi Y, Arai H, Sasaki H, Sato K, Nakashima K. High expression of apolipoprotein E mRNA in the brains with sporadic Alzheimer's disease. Dement Geriatr Cogn Disord. 2001;12:57–62. doi: 10.1159/000051236. [DOI] [PubMed] [Google Scholar]
- Yu CE, Devlin B, Galloway N, Loomis E, Schellenberg GD. ADLAPH: A molecular haplotyping method based on allele-discriminating long-range PCR. Genomics. 2004;84:600–612. doi: 10.1016/j.ygeno.2004.06.003. [DOI] [PubMed] [Google Scholar]
- Yu CE, Seltman H, Peskind ER, Galloway N, Zhou PX, Rosenthal E, Wijsman EM, Tsuang DW, Devlin B, Schellenberg GD. Comprehensive analysis of APOE and selected proximate markers for late-onset Alzheimer's disease: Patterns of linkage disequilibrium and disease/marker association. Genomics. 2007;89:655–665. doi: 10.1016/j.ygeno.2007.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng P, Pennacchio LA, Le Goff W, Rubin EM, Smith JD. Identification of a novel enhancer of brain expression near the apoE gene cluster by comparative genomics. Biochim Biophys Acta. 2004;1676:41–50. doi: 10.1016/j.bbaexp.2003.10.007. [DOI] [PubMed] [Google Scholar]