Abstract
Plasmodium falciparum, the causative agent of human malaria, invades host erythrocytes using several proteins on the surface of the invasive merozoite, which have been proposed as potential vaccine candidates. Members of the multi-gene PfRh family are surface antigens that have been shown to play a central role in directing merozoites to alternative erythrocyte receptors for invasion. Recently, we identified a large structural polymorphism, a 0.58 Kb deletion, in the C-terminal region of the PfRh2b gene, present at a high frequency in parasite populations from Senegal. We hypothesize that this region is a target of humoral immunity. Here, by analyzing 371 P. falciparum isolates we show that this major allele is present at varying frequencies in different populations within Senegal, Africa, and throughout the world. For allelic dimorphisms in the asexual stage antigens, Msp-2 and EBA-175, we find minimal geographic differentiation among parasite populations from Senegal and other African localities, suggesting extensive gene flow among these populations and/or immune-mediated frequency dependent balancing selection. In contrast, we observe a higher level of inter-population divergence (as measured by Fst) for the PfRh2b deletion, similar to that observed for SNPs from the sexual stage Pfs45/48 loci, which is postulated to be under directional selection. We confirm that the region containing the PfRh2b polymorphism is a target of humoral immune responses by demonstrating antibody reactivity of endemic sera. Our analysis of inter-population divergence suggests that in contrast to the large allelic dimorphisms in EBA-175 and Msp-2, the presence or absence of the large PfRh2b deletion may not elicit frequency-dependent immune selection, but may be under positive immune selection, having important implications for the development of these proteins as vaccine candidates.
Keywords: Plasmodium falciparum, Fst, selection, immunity, PfRh2b, antigen
INTRODUCTION
The battle against malaria, a major global health problem, has been complicated by the ongoing emergence of drug-resistant parasites and insecticide-resistant mosquitoes. Inadequate health systems and the lack of sustainable infrastructure exacerbate the problem in the countries most affected by the disease. Among the high priority potential control measures is the development of a vaccine against malaria. The design of such a vaccine is hampered by extensive genetic polymorphism in Plasmodium falciparum proteins; in particular, those expressed on the parasite’s surface that are considered ideal targets for the immune response and the development of highly specific vaccines (McCutchan 1988; Saul 1994; Conway 1997). Antigens exposed at the surface of the invasive merozoite form of the parasite, such as the EBA-175 and Msp-2 proteins, are targets of naturally acquired immunity (al-Yaman 1994), inducing antibodies that inhibit parasite growth in vitro that are frequently detected in sera from individuals living endemic areas (Epping 1988; Ramasamy 1990; Sim 1990; Thomas 1990; Ranford-Cartwright 1996; Daugherty 1997; Taylor 1998; Okenu 2000).
Many sequence polymorphisms have been described in the genes encoding asexual stage antigens. Identifying allele frequencies in antigen genes from different populations has revealed considerable polymorphism in parasite populations worldwide, which is an important consideration for vaccine development. Moreover, population genetic analysis of different malaria vaccine antigens in different populations has shown that Wright’s fixation index (Fst) is sensitive to the different selection strategies that act on different antigens (Conway 1997; Escalante 1998; Conway 2000; Silva 2000; Tanabe 2000; Binks 2001; Hoffmann 2001; Escalante 2002; Polley 2003). Some polymorphic loci, such as the sexual stage antigen Pfs48/45, have a high Fst value for mean heterogeneity in allele frequencies among populations, indicating a high degree of population divergence and possible directional selection (Conway 2000; Escalante 2002). Loci like EBA-175 and Msp-2 that are subject to immune-mediated balancing selection, however, have been shown to exhibit reduced divergence (low Fst) (Conway 1997; Binks 2001; A A Abdel-Muhsin 2003; Verra 2006). The Fst statistic, therefore, may be used as a proxy to estimate the degree to which loci are subject to balancing selection as a result of immune selection.
In addition, dimorphic allelic classes including relatively large stretches of amino acid sequence have been observed in several antigens, including EBA-175 (F-seg vs. C-seg) and Msp-2 (IC vs. FC), where the alleles within a class are much less divergent from each other than from alleles of the other class (Snewin 1991; Ware 1993), although recombination can occur between the two classes (Roy 2008).
New proteins that are currently being assessed as invasion-blocking vaccine candidates include members of the PfRh (Plasmodium falciparum reticulocyte binding protein homolog) family (Rayner 2005). PfRh family members are localized to the apical organelles of the invasive merozoite and are believed to play a role in the recognition of the erythrocyte and tight junction formation, the irreversible step in erythrocyte invasion. A recent study assessing variant expression and polymorphism in the PfRh proteins in Senegal identified a 0.58Kb deletion in the unique region of PfRh2b, the region which is predicted to confer binding to the erythrocyte and subsequent invasion (Jennings 2007).
In the present study, we analyze the distribution and immunogenicity of the large sequence deletion in the PfRh2b gene, in comparison with large allelic dimorphisms in the Msp-2 and EBA-175 genes, and SNPs (single nucleotide polymorphisms) in the Pfs45/48 gene, to investigate the geographic variation in the frequencies of these polymorphisms using population genetic analyses and to infer the existence of natural selection. Such knowledge will inform the rational prioritization of vaccine candidates.
MATERIALS AND METHODS
Plasmodium falciparum isolates and collection sites
Peripheral blood samples were collected from 371 P. falciparum-infected individuals in diverse geographical locations for genotyping of parasite antigen polymorphisms. Samples were collected from 4 different sites in Senegal: 63 isolates from Pikine (approximately 15 km from Dakar), 74 isolates from Thies (approximately 70 km from Dakar), 62 isolates from Passy (central Senegal), and 35 isolates from Velingara (southern Senegal). In addition, samples were collected in two other African countries: 36 isolates from Malawi and 25 from Tanzania. Samples from Asia included 18 samples from Thailand and 43 from Malaysia in addition to 15 from Brazil. These samples come from regions of the world with dramatically different entomological inoculation rates (EIR) that reflect local transmission rates: Pikine (EIR=1), Thies (EIR=10), Passy (EIR>100), Velingara (EIR>100), Malawi (EIR>200), Tanzania (EIR>200), Brazil (EIR = 2–20) Thailand and Malaysia (EIR undetermined). All samples were obtained with informed consent from patients and guardians and the studies received ethical clearance from ethical committees of all collaborating institutes and governments.
DNA extraction and genotyping
Parasite DNA was extracted from blood or from filter paper using QIAmp DNA Minikit (Qiagen, Valencia, CA, USA). The EBA-175 F-seg/C-seg genotype was determined by nested polymerase chain reaction (PCR) as described elsewhere (Toure 2001). The Pfs48/45 polymorphisms were amplified by PCR followed by restriction fragment length polymorphism (RFLP) with NlaIII and HpaI as previously described (Drakeley 1996). PCR-based Msp-2 genotyping was performed as previously described (Snounou 1999).
For the PfRh2b deletion, the following primers were used to amplify a gene fragment including the deletion using a hemi-nested PCR strategy, (forward primer: 5′ TAA TGA TAT AAA GGA TCT TGG TGA 3′, reverse primer: 5′ AGG AAA TCA TCC ATT TTG TTA TGG T 3′), PCR was carried out using the following conditions: initial 2-min denaturation at 94°C followed by 25 cycles with 20-s at 94°C, 20-s at 55°C and 2-min at 72°C and 2-min final extension at 72°C. For the second PCR, (forward primer: 5′ GGA TAA AAT ACT AGA AGG AAG TGA 3′, reverse primer: 5′ AGG AAA TCA TCC ATT TTG TTA TGG T 3′), PCR was carried out using the following conditions: initial 2-min denaturation at 94°C followed by 30 cycles with 2-s at 95°C, 2-s at 55°C and 30-s at 72°C and 2-min final extension at 72°C. All amplicons were separated on 1% agarose gel, stained with ethidium bromide and visualized using UV-trans illumination.
Population genetic analysis
To determine the extent of geographic variation in the frequencies of alleles, we employed Wright’s fixation index (Fst). Calculations were made as previously described (Drakeley 1996). Because mixed infections are not uncommon in highly endemic populations, we calculated the allele frequencies for bi-allelic populations using the following formula: for allele A = (no. of A isolates + (0.5 * no. of AB isolates)/(total no. of isolates) and allele B = (no. of B isolates + (0.5 * no. of AB isolates)/(total no. of isolates). Similarly for the tri-allelic locus, the following formula was used: for A= (no. of A isolates + (0.5 * no. of AB isolates) + (0.5* no. of AC isolates)/(total no. of isolates), for B = (no. of B isolates + (0.5 * no. of AB isolates) + (0.5* no. of BC isolates)/(total no. of isolates), and for C = (no. of C isolates + (0.5 * no. of AC isolates) + (0.5* no. of BC isolates)/(total no. of isolates). To explore associations in allele frequencies between different genes for different regions in Senegal, Fisher’s exact test was applied. This method for estimating allele frequencies is inherently biased due to the multiplicity of infection (multiclonality) found in many isolates of P. falciparum. The tendency of the bias is to force allele frequencies closer to 0.5 (50%) in samples composed of many mixed infections of high MOI. As a result, this approach will tend to underestimate the degree of divergence between populations due to heterogeneity in allele frequency between populations being smoothed away. This will create the illusion that loci not subject to balancing selection are actually maintained at intermediate allele frequencies in multiple populations, thereby decreasing the contrast in an Fst analysis. If a contrast in Fst between different loci is still observed despite this bias, the inference of immune-mediated balancing selection is conservative. In addition, though the estimates of allele frequency (and consequently Fst) are incorrect in absolute terms, they are comparable among loci due to similar sample sets used for each locus.
Antigens
The PfRh2b unique domain was amplified from 3D7 genomic DNA, cloned into pGEX-2T vector (Promega) and expressed as a GST-fusion protein in BL21 DE3 Rosetta pLysS cells (Novagen). Protein was purified with Glutathione sepharose columns, concentrated and stored in PBS. GST protein (pGEX-2T without insert) was expressed, purified, and used to determine non-specific background reactivity.
Endemic Sera
Plasma was obtained from patients in Senegal and Tanzania with uncomplicated malaria during the transmission season after informed consent was obtained. Patient plasma was collected in a highly endemic region of Senegal (n=67) and a highly endemic region of Tanzania (n=36), both having EIR>100. Seventy-two unexposed sera, obtained from Brigham and Women’s Hospital, Boston, MA, were used as controls.
Enzyme-linked immunosorbent assay
To measure IgG response against PfRh2b, microtiter plates were coated with 300ng of GST-PfRh2b in PBS at 4°C overnight. Plates were blocked with 1% milk in PBST (1X PBS + 0.05% Tween-20) for 1 hour at room temperature and washed 3 times with PBST. Individual plasma were added in duplicate at 1/800 dilution and incubated for 2 hours at room temperature. After washing 3 times with PBST, goat anti-human total IgG HRP-conjugated secondary antibodies (Southern Biotech) were added at 1/8000 dilution and incubated at room temperature for 2 hours. Plates were washed 5 times with PBST, developed using Sureblue TMB one-component substrate (KPL 52-00-02), and the reaction was stopped with 1N HCl. ELISA plates were read at 450nm. The positive cut-offs for PfRh2b was determined as an OD 3 times the standard deviation of the mean of the seventy-two unexposed Boston plasma.
RESULTS
Polymorphisms in PfRh2b, Msp-2, EBA-175 and Pfs48/45 are present at high frequency in Senegal and in populations throughout the world
The goal of this study was to determine the frequency of a novel polymorphism, the PfRh2b deletion, in the context of other well described polymorphisms in current vaccine candidate antigens (Figure 1). The PfRh2b deletion was present at a high frequency, ranging between 0.63 and 0.72, in all four sites in Senegal (Pikine, Thies, Velingara and Passy) (Table 1). The high frequency of this recently described polymorphism was intriguing and prompted our analysis in other countries within Africa and worldwide. The PfRh2b deletion was found at similar high frequencies in Tanzania, Malaysia and Malawi (0.87, 0.84 and 0.58 respectively), but in Thailand and Brazil the frequency was dramatically lower (0.10 and 0.08 respectively).
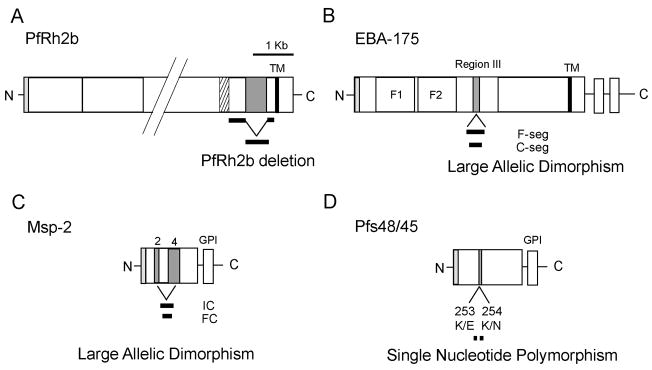
Figure 1. Polymorphic alleles of vaccine candidate antigens.
Schematic of the polymorphic alleles of genes in this study. (A) PfRh2b with large C-terminal deletion, (B) EBA-175 with F-segment and C-segment large allelic dimorphisms, (C) Msp-2 with IC and FC large allelic dimorphisms, and (D) Pfs48/45 single nucleotide polymorphisms with define the IIa1, IIa2, and IIc alleles.
Table 1.
Allele Frequencies of four P. falciparum genes in geographically distinct populations
| Senegal |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Allele | Pikine | Thies | Passy | Velingara | Tanzania | Malawi | Thailand | brazil | Malaysia |
| PfRh2b | Deletion | 0.68 (43/63) | 0.72 (51/71) | 0.70 (45/64) | 0.63 (29/46) | 0.87 (21/24) | 0.58 (21/36) | 0.10 (2/21) | 0.08 (1/13) | 0.84 (34 5/41) |
| Full length | 0.32 (20/63) | 0.28 (20/71) | 0.30 (19/64) | 0.37 (17/46) | 0.13 (3/24) | 0.42 (15/36) | 0.90 (19/21) | 0.92 (12/13) | 0.16 (6.5/41) | |
| EBA-175 | F-fragment | 0.62 (39/63) | 0.76 (56/74) | 0.47 (29/62) | 0.49 (17/35) | 0.44 (11/25) | 0.44 (16/36) | 0.71 (15/21) | 0.15 (2/13) | 0.50 (15/30) |
| C-fragment | 0.38 (24/63) | 0.24 (18/74) | 0.53 (33/62) | 0.51 (18/35) | 0.56 (14/25) | 0.56 (20/36) | 0.29 (6/21) | 0.85 (11/13) | 0.50 (15/30) | |
| MSP-2 | IC | 0.56 (32/57) | 0.62 (41/66) | 0.68 (38/56) | 0.67 (24/36) | 0.46 (13/28) | 0.57 (21/37) | 0.67 (14/21) | 0.86 (13/15) | 0.28 (9/32) |
| FC27 | 0.44 (25/57) | 0.38 (25/66) | 0.32 (18/56) | 0.33 (12/36) | 0.54 (15/28) | 0.43 (16/37) | 0.33 (7/21) | 0.14 (2/15) | 0.72 (23/32) | |
| pfs48/45 | IIa1 | 0.04 (1.5/35) | 0.08 (3.5/46) | 0.06 (3/48) | 0.09 (3/33) | 0.13 (3/22) | 0.27 (7/26) | 0.65 (13/20) | 0.00 (0/15) | 1.00 (38/38) |
| IIa2 | 0.01 (0.5/35) | 0.06 (3/46) | 0.05 (2.5/48) | 0.00 (0/33) | 0.10 (2/22) | 0.17 (4.5/26) | 0.00(0/20) | 1.00 (15/15) | 0.00 (0/38) | |
| IIc | 0.95 (33/35) | 0.86 (39.5/46) | 0.00 (42.5/48) | 0.91 (30/33) | 0.77 (17/22) | 0.56 (14.5/26) | 0.35 (7/20) | 0.00 (0/15) | 0.00 (0/38) | |
We next sought to compare the frequencies of the PfRh2b deletion with those of polymorphisms in other known malaria vaccine candidate antigens. Analysis of the EBA-175 dimorphic alleles from different geographical regions in Senegal revealed that in Thies and Pikine the predominant allele was F-segment (0.62 and 0.76 respectively), while in Passy and Velingara, the frequency of C-segment was predominant (0.53 and 0.51 respectively). The C-segment was found at the same frequency in Malawi and Tanzania and Malaysia (0.56) and at a high frequency in Brazil. However, in Thailand, the C-segment was observed at a low frequency (0.29) (Table 1).
For the Msp-2 locus, the IC dimorphic allele was predominant in all sites in Senegal, and in Brazil, Thailand and Malawi (Table. 1); whereas, in Tanzania and Malaysia the frequencies were lower at 0.46 and 0.28 respectively.
The results of Pfs48/45 analysis show that the predominant allele in the four Senegalese locations was type IIc, with a frequency between 0.86 and 0.95. The other two alleles were present at lower frequencies and type IIa2 was not found in Velingara. The type IIc was predominant in Tanzania and Malawi; however, IIa1 was predominant in Thailand and Malaysia, and IIa2 in Brazil (Table 1).
Applying Fisher’s exact test indicates that there was no association between allelic forms of PfRh2b, EBA-175, Pfs48/45 or Msp-2 in the Senegalese or in the global isolates (data not shown). Using the Mann-Whitney U test, we found no association between age or seasonality and the presence of deletion in Senegal (data not shown).
Geographical differentiation of dimorphisms between populations within Senegal, Africa and worldwide
Fst was calculated from allele frequencies to measure the genetic differentiation between the different populations. For the PfRh2b polymorphism, the inter-population Fst value was low between the Senegalese populations, and remained low when we included other African countries in the analysis (Table 2). By comparison, the Fst indices for the EBA-175 and the Msp-2 dimorphisms were also low within Senegal, and between the African countries. Within Senegal, the Fst index for Pfs48/45 was low (0.020); however, when we calculated the Fst including African countries the value was higher (0.070).
Table 2.
Fst indices of inter-population variance for four P. falciparum genes
| Polymorphism/Selection | Senegal | Africa | Africa & SE Asia | Africa, SE Asia & Brazil | |
|---|---|---|---|---|---|
| PfRh2b | Deletion | 0.0092 | 0.070 | 0.42 | 0.49 |
| EBA-175 | Large allelic dimorphism/Balancing | 0.057 | 0.020 | 0.040 | 0.16 |
| Msp-2 | Large alleic dimorphism/Balancing | 0.012 | 0.022 | 0.11 | 0.18 |
| Pfs48/45 | SNP/Directional | 0.022 | 0.075 | 0.43 | 0.70 |
The combined data from all global populations considered in this study—African populations, South East Asian and Latin American populations—demonstrate a higher Fst value for the PfRh2b polymorphism, when compared to the African populations alone: the Fst value of combined African populations and South East Asian populations was six times greater than the value for African populations. The increased Fst values when moving from African to global isolates was similar to that seen for the Pfs48/45 polymorphisms. Although higher Fst values are also observed for EBA-175 and Msp-2 dimorphisms compared with African populations alone, these values are significantly lower than those observed for the PfRh2b and Pfs48/45 polymorphisms, suggesting less genetic differentiation of these alleles (Table 2). Similar trends are observed when only monoclonal infections are used to estimate allele frequencies (Supplementary Figure 1), although the numbers of samples to estimate frequency in each population is much reduced.
Recognition of PfRh2b by human antibodies and influence of age on antibody response
The presence of the PfRh2b deletion worldwide prompted us to determine whether antibodies against the C-terminal region of PfRh2b exist, and could act as a selective pressure on this locus. We measured IgG responses against a unique region of PfRh2b and found that positive antibody responses exist in both Senegal and Tanzania (Figure 3A) and are acquired in an age-dependent manner, although not statistically significant by Kruskal-Wallis (p=0.17) (Figure 3B). This result implies that the humoral immune response could potentially select parasites harboring the PfRh2b deletion if such parasites invoked a reduced immune response relative to parasites with intact PfRh2b proteins.
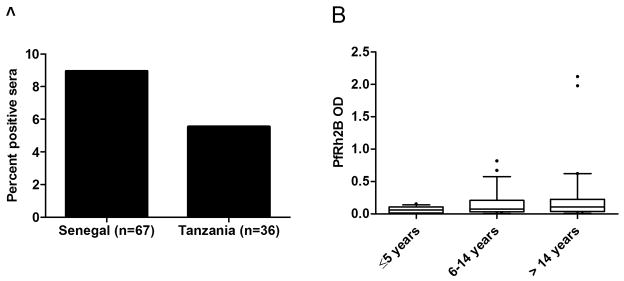
Figure 3. Immune responses against PfRh2b exist in two geographically distinct populations.
(A) Percent positive responses (defined as OD responses greater than 3 times the standard deviation of the mean of 72 unexposed Boston sera) for Senegal (n=67) and Tanzania (n=36). In Senegal, 8.96% of sera had positive IgG titers against PfRh2b; in Tanzania, 5.55% percent of sera were positive. (B) IgG against PfRh2b is acquired in an age-dependent manner; however mean differences were not statistically significant by Kruskal-Wallis (p=0.17)
DISCUSSION
It has become very clear that substantial genetic differentiation exists between parasite populations in Asian and African isolates (Volkman 2007; Neafsey 2008). Here we report the assessment of a new structural polymorphism, a large deletion in the P. falciparum erythrocyte invasion ligand PfRh2b, a dominant invasion ligand involved in sialic-acid independent invasion of erythrocytes. In addition to showing that it is widespread throughout the world, we further provide evidence that it elicits a humoral response which could be the selective pressure driving its frequency.
Using a population genetic analysis, we show that although the PfRh2b deletion is present worldwide, it appears to vary in frequency significantly in different populations. Our results show that the frequency of the deletion is high in all study sites with the exception of Thailand and Brazil. When calculating Fst for the PfRh2b deletion globally, the Fst value was very comparable to that observed for polymorphisms in Pfs48/45, which have been previously shown to be under directional selection (Conway 2000; Escalante 2002). There was no evidence from the inter-population variance that the PfRh2b polymorphism is under balancing selection globally. However, Fst calculation using only Senegalese sites revealed a value very close to zero (0.009), a similar result is found when only African sites are analyzed. This low Fst indicates little genetic differentiation, or divergence among African populations. Indeed, similar values were seen for all of the allelic polymorphisms considered in our study, suggesting the presence of significant gene-flow within the continent, similar to previous observations (Awadalla 2001).
The dramatic difference in the PfRh2b deletion frequencies in Brazil and Thailand population compared with the other populations invites the speculation the genetic background of the host in these populations may influence the allele frequencies, due to the presence of different erythrocyte receptor polymorphisms. Alternatively, the PfRh2b deletion may have originated in Africa, and could be at low frequency in Asian and American populations because it is has only recently arrived in those locations. We have some evidence that the PfRh2b deletion is associated with the use of protease-sensitive erythrocyte receptors for invasion (Lantos 2009). Previous studies have shown that parasite lines from different geographic areas express PfRh proteins variantly (Taylor 2002; Duraisingh 2003; Nery 2006; Bei 2007). This variation in expression may also possibly contribute to the fact that the PfRh2b polymorphism does not appear to be under balancing selection since alternative ligands can compensate for polymorphism in PfRh2b.
The PfRh2b deletion was previously described in only one lab strain, T9/96, from Thailand, and more recently in Senegalese isolates (Jennings 2007). The high prevalence of the deletion in many global populations may suggest that appearance of the deletion in the field is not recent; however, to test this hypothesis we need to measure the distribution of the PfRh2b deletion using matched archival and recent samples from the same region, or follow the prevalence in specific sites over time. The absence of the deletion in other lab strains, most of which were established in the 1980s, may on the other hand suggest that the deletion has spread recently.
We investigated if specific immune responses exist against the PfRh2b C-terminal region that harbors the PfRh2b deletion polymorphism. We hypothesize that the parasite has the ability to utilize two potential mechanisms for evading the immune response: variant expression and polymorphism. To test these hypotheses, we measured the IgG response to PfRh2b using sera from Senegal and Tanzania. The presence of an antibody response against the invasion ligand PfRh2b may be significant as these antibodies may be inhibitory to erythrocyte invasion. We found that antibodies against PfRh2b exist in Senegalese and Tanzania sera. The percentage of positive antibody responses by cut-off is similar in both sites, and the acquisition of antibody is correlated with increasing patient age. Such findings imply that this locus is recognized by the humoral immune response, which may serve as a selective force driving the frequency of the PfRh2b deletion if the deletion allele invokes a weaker response than the wild-type allele. Allele specificity of the immune reactivity, measured by competitive ELISA using the PfRh2b wildtype and deletion polymorphism, would further strengthen the hypothesis of immune selection. It has yet to be determined whether immune responses to the PfRh2b unique domain are inhibitory and whether the deletion contributes to immune evasion.
Our results show global variation in the genetic distribution of the PfRh2b deletion and Pfs48/45 polymorphisms, while the frequency distribution of the deletion seems similar in Africa, it is quite divergent worldwide relative to polymorphisms in antigens known to be subject to balancing selection, implying that this polymorphism may not be subject to strong balancing selection, or may be exposed to variable selection pressures across sites. Alternatively, the PfRh2b deletion may exhibit higher divergence among worldwide populations than the Msp-2 and EBA-175 polymorphisms because it is a younger allele, and has not had as much time to spread throughout the range of the parasite populations. We consider this unlikely due to the observation that the PfRh2b deletion is found at frequencies comparable to the Msp-2 and EBA-175 polymorphisms in African and Malaysian populations, whereas a younger allele would more likely be present at lower frequencies. Our findings imply that inclusion of antigens with such profiles may be efficacious in some, but not all countries, resulting in limited vaccine utility. In contrast, Msp-2 and EBA-175 are also involved in erythrocyte invasion, yet in our study, the EBA-175 and Msp-2 dimorphisms exhibit minimal genetic differentiation globally. This strongly suggests that they are under balancing selection, a criteria for inclusion in a universally applicable vaccine.
This population genetic study demonstrates that focusing on large sequence dimorphisms and deletions as vaccine candidates may prove to be an effective strategy for vaccine design. These large regions may inherently define discrete domains that can elicit protective antibody responses (Conway 1997). Using such molecular population genetic analyses to predict a region of selection, and validating this region using molecular and immunological tools, is an effective approach to identifying novel antigenic regions. This could be exploited in vaccine development, particularly by including both dimorphic regions in combination.
In our study we focus on a comparison of different antigens that are presumably all under selection either by humoral immunity or by red blood cell polymorphisms. We have chosen to focus on antigen polymorphisms for which data previously exists as controls for the measurement of genetic differentiation, known to be either under balancing selection (Msp-2) or directional selection (Pfs48/45). We realize that an expanded study comparing these antigen loci with SNPs in putative neutral loci or in drug-resistance loci may be informative (Anderson 2005).
In conclusion, it has been suggested that a good vaccine candidate would incorporate a region of a protein with very few allelic forms under strong balancing selection (Conway 1997). Our results suggest that in contrast to the EBA-175 and Msp-2 dimorphisms, the PfRh2b deletion may not be under balancing selection globally. However, the C-terminal region of PfRh2b harboring the PfRh2b deletion does elicit immune responses that may exert directional selection on the polymorphism. Further studies will determine whether naturally acquired antibodies to PfRh2b in endemic populations can functionally inhibit invasion. To our knowledge, large deletions on the order of 500bp have not been previously identified in other Plasmodium antigens; however small deletions on the order of 39bp are present in Msp-3 (McColl 1997). It will be of interest in the future to determine whether these deleted regions in other antigens are also more genetically differentiated between populations, suggesting that in general, large sequence deletions arise as a mechanism to avoid frequency-dependent immune selection.
Supplementary Material
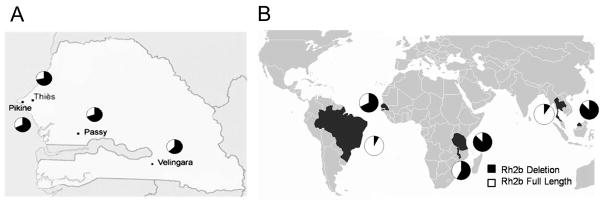
Figure 2. Allele frequencies in Senegal and worldwide.
Minimal variation in the frequency of a novel polymorphism in PfRh2b is observed in various regions of Senegal (A), whereas striking variation is observed worldwide (B). PCR typing of genomic DNA from global parasite isolates was used to determine the allele frequencies. This result represents the first comprehensive description of PfRh2b deletion frequency in field-isolated parasite populations.
Acknowledgments
We thank Terrie Taylor, Balbir Singh, Cameron Jennings, Daouda Ndiaye, Younous Diedhiou, Amadou Makhtar Mbaye, Omar Ly, Lamine Ndiaye, and Dior Diop for collecting samples, as well as all the patients who participated in the studies.
FUNDING STATEMENT
AA is Fogarty trainee supported by the National Institute of Health grant 5D43TW001503-09 to Dr. Dyann Wirth, AB is supported by a Harvard Institute for Global Health fellowship. MTD is supported by NIH RO1AI057919 and this work is supported by 1R03TW008053.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abdel-Muhsin AA, MJM, Awadalla P, Ali E, Suleiman S, Ahmed S, Walliker D, Babiker HA. Local differentiation in Plasmodium falciparum drug resistance genes in Sudan. Parasitology. 2003;126(5):391–400. doi: 10.1017/s0031182003003020. [DOI] [PubMed] [Google Scholar]
- al-Yaman F, Genton B, Anders RF, Falk M, Triglia T, Lewis D, Hii J, Beck HP, Alpers MP. Relationship between humoral response to Plasmodium falciparum merozoite surface antigen-2 and malaria morbidity in a highly endemic area of Papua New Guinea. Am J Trop Med Hyg. 1994;51(5):593–602. doi: 10.4269/ajtmh.1994.51.593. [DOI] [PubMed] [Google Scholar]
- Anderson TJ, Nair S, Sudimack D, Williams JT, Mayxay M, Newton PN, Guthmann JP, Smithuis FM, Tran TH, van den Broek IV, White NJ, Nosten F. Geographical distribution of selected and putatively neutral SNPs in Southeast Asian malaria parasites. Mol Biol Evol. 2005;22(12):2362–74. doi: 10.1093/molbev/msi235. [DOI] [PubMed] [Google Scholar]
- Awadalla P, Walliker D, Babiker H, Mackinnon M. The question of Plasmodium falciparum population structure. Trends Parasitol. 2001;17(8):351–3. doi: 10.1016/s1471-4922(01)02034-7. [DOI] [PubMed] [Google Scholar]
- Bei AK, Membi CD, Rayner JC, Mubi M, Ngasala B, Sultan AA, Premji Z, Duraisingh MT. Variant merozoite protein expression is associated with erythrocyte invasion phenotypes in Plasmodium falciparum isolates from Tanzania. Mol Biochem Parasitol. 2007;153(1):66–71. doi: 10.1016/j.molbiopara.2007.01.007. [DOI] [PubMed] [Google Scholar]
- Binks RH, Baum J, Oduola AM, Arnot DE, Babiker HA, Kremsner PG, Roper C, Greenwood BM, Conway DJ. Population genetic analysis of the Plasmodium falciparum erythrocyte binding antigen-175 (eba-175) gene. Mol Biochem Parasitol. 2001;114(1):63–70. doi: 10.1016/s0166-6851(01)00240-7. [DOI] [PubMed] [Google Scholar]
- Conway DJ. Natural selection on polymorphic malaria antigens and the search for a vaccine. Parasitol Today. 1997;13(1):26–9. doi: 10.1016/s0169-4758(96)10077-6. [DOI] [PubMed] [Google Scholar]
- Conway DJ, Cavanagh DR, Tanabe K, Roper C, Mikes ZS, Sakihama N, Bojang KA, Oduola AM, Kremsner PG, Arnot DE, Greenwood BM, McBride JS. A principal target of human immunity to malaria identified by molecular population genetic and immunological analyses. Nat Med. 2000;6(6):689–92. doi: 10.1038/76272. [DOI] [PubMed] [Google Scholar]
- Daugherty JR, Murphy CI, Doros-Richert LA, Barbosa A, Kashala LO, Ballou WR, Snellings NJ, Ockenhouse CF, Lanar DE. Baculovirus-mediated expression of Plasmodium falciparum erythrocyte binding antigen 175 polypeptides and their recognition by human antibodies. Infect Immun. 1997;65(9):3631–7. doi: 10.1128/iai.65.9.3631-3637.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drakeley CJ, Duraisingh MT, Povoa M, Conway DJ, Targett GA, Baker DA. Geographical distribution of a variant epitope of Pfs48/45, a Plasmodium falciparum transmission-blocking vaccine candidate. Mol Biochem Parasitol. 1996;81(2):253–7. doi: 10.1016/0166-6851(96)02718-1. [DOI] [PubMed] [Google Scholar]
- Duraisingh MT, Triglia T, Ralph SA, Rayner JC, Barnwell JW, McFadden GI, Cowman AF. Phenotypic variation of Plasmodium falciparum merozoite proteins directs receptor targeting for invasion of human erythrocytes. Embo J. 2003;22(5):1047–57. doi: 10.1093/emboj/cdg096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epping RJ, Goldstone SD, Ingram LT, Upcroft JA, Ramasamy R, Cooper JA, Bushell GR, Geysen HM. An epitope recognised by inhibitory monoclonal antibodies that react with a 51 kilodalton merozoite surface antigen in Plasmodium falciparum. Mol Biochem Parasitol. 1988;28(1):1–10. doi: 10.1016/0166-6851(88)90173-9. [DOI] [PubMed] [Google Scholar]
- Escalante AA, Grebert HM, Chaiyaroj SC, Riggione F, Biswas S, Nahlen BL, Lal AA. Polymorphism in the gene encoding the Pfs48/45 antigen of Plasmodium falciparum. XI. Asembo Bay Cohort Project. Mol Biochem Parasitol. 2002;119(1):17–22. doi: 10.1016/s0166-6851(01)00386-3. [DOI] [PubMed] [Google Scholar]
- Escalante AA, Lal AA, et al. Genetic polymorphism and natural selection in the malaria parasite Plasmodium falciparum. Genetics. 1998;149(1):189–202. doi: 10.1093/genetics/149.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann EH, da Silveira LA, Tonhosolo R, Pereira FJ, Ribeiro WL, Tonon AP, Kawamoto F, Ferreira MU. Geographical patterns of allelic diversity in the Plasmodium falciparum malaria-vaccine candidate, merozoite surface protein-2. Ann Trop Med Parasitol. 2001;95(2):117–32. doi: 10.1080/00034980120045833. [DOI] [PubMed] [Google Scholar]
- Jennings CV, Ahouidi AD, Zilversmit M, Bei AK, Rayner J, Sarr O, Ndir O, Wirth DF, Mboup S, Duraisingh MT. Molecular analysis of erythrocyte invasion in Plasmodium falciparum isolates from Senegal. Infect Immun. 2007;75(7):3531–8. doi: 10.1128/IAI.00122-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lantos PM, Ahouidi AD, Bei AK, Jennings CV, Sarr O, Ndir O, Wirth DF, Mboup S, Duraisingh MT. Erythrocyte invasion profiles are associated with a common invasion ligand polymorphism in Senegalese isolates of Plasmodium falciparum. Parasitology. 2009;136(1):1–9. doi: 10.1017/S0031182008005167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McColl DJ, Anders RF. Conservation of structural motifs and antigenic diversity in the Plasmodium falciparum merozoite surface protein-3 (MSP-3) Mol Biochem Parasitol. 1997;90(1):21–31. doi: 10.1016/s0166-6851(97)00130-8. [DOI] [PubMed] [Google Scholar]
- McCutchan TF, de la Cruz VF, Good MF, Wellems TE. Antigenic diversity in Plasmodium falciparum. Prog Allergy. 1988;41:173–92. doi: 10.1159/000415223. [DOI] [PubMed] [Google Scholar]
- Neafsey DE, Schaffner SF, Volkman SK, Park D, Montgomery P, Milner DA, Jr, Lukens A, Rosen D, Daniels R, Houde N, Cortese JF, Tyndall E, Gates C, Stange-Thomann N, Sarr O, Ndiaye D, Ndir O, Mboup S, Ferreira MU, Moraes SD, Dash AP, Chitnis CE, Wiegand RC, Hartl DL, Birren BW, Lander ES, Sabeti PC, Wirth DF. Genome-wide SNP genotyping highlights the role of natural selection in Plasmodium falciparum population divergence. Genome Biology. 2008;9(12):R171. doi: 10.1186/gb-2008-9-12-r171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nery S, Deans AM, Mosobo M, Marsh K, Rowe JA, Conway DJ. Expression of Plasmodium falciparum genes involved in erythrocyte invasion varies among isolates cultured directly from patients. Mol Biochem Parasitol. 2006;149(2):208–15. doi: 10.1016/j.molbiopara.2006.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okenu DM, Riley EM, Bickle QD, Agomo PU, Barbosa A, Daugherty JR, Lanar DE, Conway DJ. Analysis of human antibodies to erythrocyte binding antigen 175 of Plasmodium falciparum. Infect Immun. 2000;68(10):5559–66. doi: 10.1128/iai.68.10.5559-5566.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polley SD, Chokejindachai W, Conway DJ. Allele frequency-based analyses robustly map sequence sites under balancing selection in a malaria vaccine candidate antigen. Genetics. 2003;165(2):555–61. doi: 10.1093/genetics/165.2.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramasamy R, Jones G, Lord R. Characterisation of an inhibitory monoclonal antibody-defined epitope on a malaria vaccine candidate antigen. Immunol Lett. 1990;23(4):305–9. doi: 10.1016/0165-2478(90)90077-4. [DOI] [PubMed] [Google Scholar]
- Ranford-Cartwright LC, Taylor RR, Asgari-Jirhandeh N, Smith DB, Roberts PE, Robinson VI, Babiker HA, Riley EM, Walliker D, McBride JS. Differential antibody recognition of FC27-like Plasmodium falciparum merozoite surface protein MSP2 antigens which lack 12 amino acid repeats. Parasite Immunol. 1996;18(8):411–20. doi: 10.1046/j.1365-3024.1996.d01-137.x. [DOI] [PubMed] [Google Scholar]
- Rayner JC, Tran TM, Corredor V, Huber CS, Barnwell JW, Galinski MR. Dramatic difference in diversity between Plasmodium falciparum and Plasmodium vivax reticulocyte binding-like genes. Am J Trop Med Hyg. 2005;72(6):666–74. [PubMed] [Google Scholar]
- Roy SW, Ferreira MU, et al. Evolution of allelic dimorphism in malarial surface antigens. Heredity. 2008;100(2):103–10. doi: 10.1038/sj.hdy.6800887. [DOI] [PubMed] [Google Scholar]
- Saul MFGaAJ. Molecular Immunological Considerations in Malaria Vaccine Development. Boca Raton, FL: CRC Press; 1994. [Google Scholar]
- Silva NS, Silveira LA, Machado RL, Povoa MM, Ferreira MU. Temporal and spatial distribution of the variants of merozoite surface protein-1 (MSP-1) in Plasmodium falciparum populations in Brazil. Ann Trop Med Parasitol. 2000;94(7):675–88. doi: 10.1080/00034983.2000.11813591. [DOI] [PubMed] [Google Scholar]
- Sim BK, Orlandi PA, Haynes JD, Klotz FW, Carter JM, Camus D, Zegans ME, Chulay JD. Primary structure of the 175K Plasmodium falciparum erythrocyte binding antigen and identification of a peptide which elicits antibodies that inhibit malaria merozoite invasion. J Cell Biol. 1990;111(5 Pt 1):1877–84. doi: 10.1083/jcb.111.5.1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snewin VA, Herrera M, Sanchez G, Scherf A, Langsley G, Herrera S. Polymorphism of the alleles of the merozoite surface antigens MSA1 and MSA2 in Plasmodium falciparum wild isolates from Colombia. Mol Biochem Parasitol. 1991;49(2):265–75. doi: 10.1016/0166-6851(91)90070-m. [DOI] [PubMed] [Google Scholar]
- Snounou G, Zhu X, Siripoon N, Jarra W, Thaithong S, Brown KN, Viriyakosol S. Biased distribution of msp1 and msp2 allelic variants in Plasmodium falciparum populations in Thailand. Trans R Soc Trop Med Hyg. 1999;93(4):369–74. doi: 10.1016/s0035-9203(99)90120-7. [DOI] [PubMed] [Google Scholar]
- Tanabe K, Sakihama N, Nakamura Y, Kaneko O, Kimura M, Ferreira MU, Hirayama K. Selection and genetic drift of polymorphisms within the merozoite surface protein-1 gene of Plasmodium falciparum. Gene. 2000;241(2):325–31. doi: 10.1016/s0378-1119(99)00472-2. [DOI] [PubMed] [Google Scholar]
- Taylor HM, Grainger M, Holder AA. Variation in the expression of a Plasmodium falciparum protein family implicated in erythrocyte invasion. Infect Immun. 2002;70(10):5779–89. doi: 10.1128/IAI.70.10.5779-5789.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor RR, Allen SJ, Greenwood BM, Riley EM. IgG3 antibodies to Plasmodium falciparum merozoite surface protein 2 (MSP2): increasing prevalence with age and association with clinical immunity to malaria. Am J Trop Med Hyg. 1998;58(4):406–13. doi: 10.4269/ajtmh.1998.58.406. [DOI] [PubMed] [Google Scholar]
- Thomas AW, Carr DA, Carter JM, Lyon JA. Sequence comparison of allelic forms of the Plasmodium falciparum merozoite surface antigen MSA2. Mol Biochem Parasitol. 1990;43(2):211–20. doi: 10.1016/0166-6851(90)90146-d. [DOI] [PubMed] [Google Scholar]
- Toure FS, Mavoungou E, Ndong JM, Tshipamba P, Deloron P. Erythrocyte binding antigen (EBA-175) of Plasmodium falciparum: improved genotype determination by nested polymerase chain reaction. Trop Med Int Health. 2001;6(10):767–9. doi: 10.1046/j.1365-3156.2001.00789.x. [DOI] [PubMed] [Google Scholar]
- Verra F, Chokejindachai W, Weedall GD, Polley SD, Mwangi TW, Marsh K, Conway DJ. Contrasting signatures of selection on the Plasmodium falciparum erythrocyte binding antigen gene family. Mol Biochem Parasitol. 2006;149(2):182–90. doi: 10.1016/j.molbiopara.2006.05.010. [DOI] [PubMed] [Google Scholar]
- Volkman SK, Sabeti PC, DeCaprio D, Neafsey DE, Schaffner SF, Milner DA, Jr, Daily JP, Sarr O, Ndiaye D, Ndir O, Mboup S, Duraisingh MT, Lukens A, Derr A, Stange-Thomann N, Waggoner S, Onofrio R, Ziaugra L, Mauceli E, Gnerre S, Jaffe DB, Zainoun J, Wiegand RC, Birren BW, Hartl DL, Galagan JE, Lander ES, Wirth DF. A genome-wide map of diversity in Plasmodium falciparum. Nat Genet. 2007;39(1):113–9. doi: 10.1038/ng1930. [DOI] [PubMed] [Google Scholar]
- Ware LA, Kain KC, Lee Sim BK, Haynes JD, Baird JK, Lanar DE. Two alleles of the 175-kilodalton Plasmodium falciparum erythrocyte binding antigen. Mol Biochem Parasitol. 1993;60(1):105–9. doi: 10.1016/0166-6851(93)90033-t. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.