Abstract
The hypothesis that environmental factors alter somatically heritable epigenetic marks and change long-term patterns of gene expression is an exciting possibility in human disease research. Because most common diseases, and many quantitative traits, are influenced by both genetic and environmental factors, environmentally induced changes in epigenetic structures can provide a mechanistic link between genes and environment. We believe that inter-individual differences in the epigenetic modification of genes will explain a much greater fraction of inter-individual phenotypic variation than differences in genotype, alone.
Keywords: inter-individual variation, gene expression, DNA methylation, DNA methylation
One of the long-awaited promises of human genome sequencing and whole genome association analysis was the identification of genes involved in common human diseases.1 In fact, human geneticists have been able to deliver on this promise in a number of cases, identifying genes involved in type 2 diabetes,2,3 asthma4,5 and many other diseases.6–8 In the minds of the public, the identification of disease genes was one step on the road to personalized medicine. Unfortunately, it has turned out to be a rather small step, in most instances.
The reason that most of us have not had our genomes sequenced9 (even if we could get our genomes sequenced for $1000) is that, in the case of type 2 diabetes, for example, the identification of “risk” alleles at the disease loci provides little predictive power; each genetic “risk” variant is associated with an average odds ratio of 1.18 for SNPs at ten loci with very strong association.10 Although that fact may not be very surprising to many geneticists, given the multifactorial, multigenic nature of common diseases, it begs the question of what type of information would add to the predictive power of genetic risk in determining phenotype. In this vein, there is potential for different measures of “epigenotype” (DNA methylation, histone methylation/acetylation/sumoylation, etc.), acting as “readouts” of the impact of the environment, to add significant predictive power to genetic information, alone.11 Given this excitement, it is fair to evaluate the magnitude of the problem and to ask whether epigenetics, writ large, is up to the task.
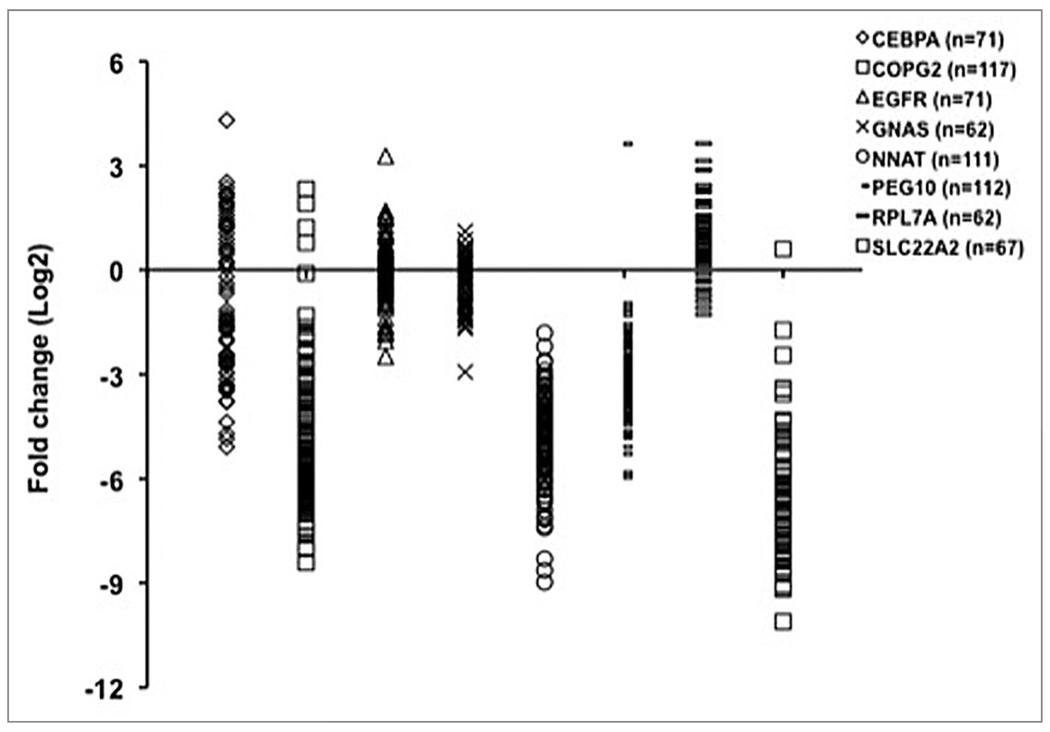
Inter-individual differences in phenotype, whether associated with disease or not, are generally assumed to reflect inter-individual differences in the expression of genes. Inter-individual differences in gene expression can be qualitative (functional product versus non-functional product, for example) or quantitative (amount of product or relative amounts of functional and non-functional products, etc.). In fact, one of the most surprising observations to emerge from human transcriptome profiling is the very high level of inter-individual variability found in steady state mRNA levels of many genes. The inter-individual differences (often an order of magnitude) do not appear to be a result of a technical artifact attributable to the arrays because similar large inter-individual differences for many genes have been validated by quantitative RT-PCR. Several examples from our own laboratory are shown in Figure 1 but additional examples may be found in reports from several groups.12–15
Figure 1.
Inter-individual variation in gene expression level. Steady-state mRNA levels were measured in placentas from unrelated individuals. Each symbol represents the mRNA level in one individual. Fold-changes were calculated relative to the housekeeping gene. mRNA levels of EGFR and GNAS are slightly less variable than the other six genes, which show extensive inter-individual variation.
Many inter-individual differences in mRNA level appear heritable and can be treated as quantitative traits (so-called “expression QTLs” or “eQTLs”), in much the same way as height or blood pressure.16,17 Over the past few years, several groups have used genome-wide association approaches to map the genetic determinants of inter-individual differences in the level of specific mRNAs. Both cis- and trans-acting genetic factors have been identified18–21 although cis-acting factors are more likely to be identified for a number of reasons.12,22
The good news to come out of such analyses is that many of the SNPs associated with eQTLs are very close to the genes, themselves, and can be expected to represent promoter “strength” alleles, binding sites for repressors, or other easily envisioned functional categories, although not proven to be in most cases.22 The bad news is that the associated SNPs explain only a small fraction of the variance observed in transcript level.14 So, we are left, once again, with the conundrum that a trait that shows moderate heritability (transcript level of a particular gene) and that can be mapped to specific sites in the genome, refuses to behave in a predictable fashion, at least on an individual basis. It is enough to drive any geneticist testifying before a congressional committee on personalized medicine to distraction.
It is fair to ask whether measures of epigenotype would perform any better in explaining the level of variance observed in transcript levels. In theory, at least, it is possible for epigenetic measures that can be continuous variables (0–100% methylation of a particular CpG site, for example) to predict widely varying transcript levels better than genetic measures that tend to be trichotomous (AA, AB, BB), at best.
We have examined inter-individual differences in DNA methylation and gene expression in children conceived in vivo or in vitro in a recent report.23 While the design and scale of our experiment did not permit us to map epigenetic determinants of inter-individual differences in transcript level, genome-wide, we did find that a fraction of genes that exhibited significant differences between groups in CpG site methylation also exhibited significant differences in transcript level.23 We hypothesize that many such correlations indicate cis-effects of DNA methylation on gene expression. Regression of CpG methylation level on transcript level in these cases can provide an estimate of the fraction of variance in transcript level that can be accounted for by cis-acting epigenetic factors.
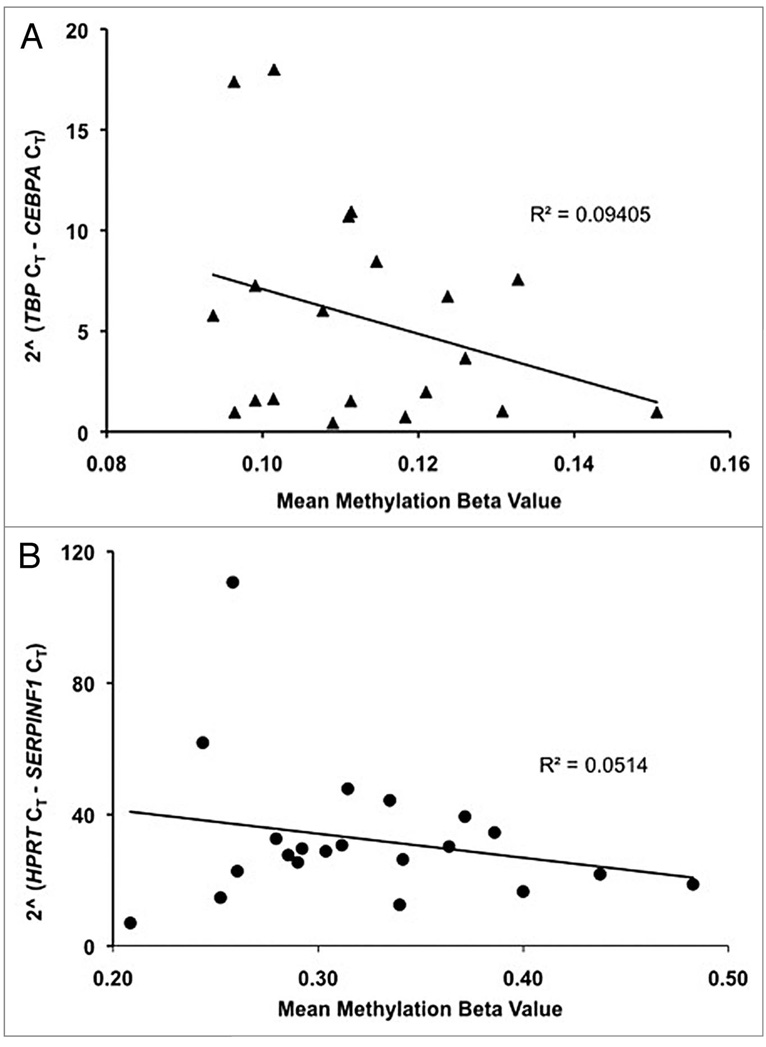
Two of the best examples derived from our study23 are shown in Figure 2. Methylation of a single CpG site adjacent to the CCAAT/enhancer binding protein alpha (CEBPA) locus accounts for approximately 10% of the inter-individual variance in cord blood transcript level, while the methylation of a single site adjacent to serpin peptidase inhibitor, clade F, member 1 (SERPINF1) accounts for approximately 5% of inter-individual differences in transcript level in placenta. While these numbers are nothing about which epigeneticists should puff up their chests, they compare favorably with estimates for the fraction of variation in transcript level explained by of cis-acting genetic factors (approximately 5%).20
Figure 2.
Inter-individual differences in DNA methylation and mRNA levels. CEBPA and SERPINF1 are two genes that exhibited significant differences in CpG site methylation and also exhibited significant differences in transcript levels between children conceived in vivo or in vitro (in cord blood and placenta, for CEBPA and SERPINF1, respectively). Sodium bisulfite modified DNA from cord blood and placenta of ten children conceived in vitro and thirteen children conceived in vivo were assayed, in duplicate, for the fraction of each CpG site methylated in each sample (the “mean methylation beta value”) using the “GoldenGate” hybridization/primer extension/ligation and amplification protocol.29,30 The extent of methylation at a given CpG site was determined by comparing the proportion of signal from methylated and unmethylated alleles the “beta value” in the DNA sample.29 (A) Methylation of a single CpG site (cg23071874, www.illumina.com) adjacent to the CEBPA locus accounts for ~10% of the inter-individual variance in cord blood transcript level, (B) methylation of a single site (cg27102649, www.illumina.com) adjacent to the SERPINF1 locus accounts for ~5% of transcript level differences in placenta.
Given that there are many epigenetic modifications that could be assayed in a similar fashion, the addition of epigenetic factors can be expected to explain a greater and greater proportion of inter-individual variance. Taking into account the fact that the regressions shown in Figure 2 do not control for any inter-individual genetic differences that might confound the effect of methylation on transcript level, it is heartening that the magnitude of the epigenetic effect seen is within the range of those discovered in whole-genome association searches for genetic factors affecting mRNA levels.12,20 If trans-acting epigenetic factors account for a similar fraction of inter-individual variance as trans-acting genetic factors (approximately 38%)20 the combination of the two types of information should be powerful, indeed.11 Even if genetic and epigenetic effects are only additive, rather than synergistic, the resulting increase in predictive power is likely to be large enough to fulfill the expectations of personalized medicine; i.e., patient specific information that provides relative risk estimates that are large enough to affect patient behavior and/or standards of care.
With respect to our studies of the possible effects of in vitro conception on epigenetic marks and patterns of gene expression,23 what could be the outcome of any validated effects? There is no question that the great majority of children conceived through the assisted reproductive technologies appear normal at birth.24 However, as a group, these children are of lower birth weight and are more often born prematurely (even when multiple pregnancies are taken into consideration).25,26 These issues put some of these children at risk for development of systemic diseases such as obesity, hypertension and/or cardiovascular disease in adulthood and possibly other ailments in later life.27,28 We should not forget that the oldest child conceived through in vitro fertilization is only 30 years old. Given all of the observations on the range of inter-individual variability observed, the challenge facing us is the identification of other “markers” that will accurately select which individuals among the whole cohort are at risk for the development of health problems later in life. Could specific epigenetic marks prove to be such “markers”? And, even better, could any marks altered by the process be re-altered, thus allowing for clinical intervention?
Acknowledgements
This work was supported by the National Institutes of Health (R01 HD048730 to C.S. and C.C.).
References
- 1.Manolio TA, Brooks LD, Collins FS. A HapMap harvest of insights into the genetics of common disease. J Clin Invest. 2008;118:1590–1605. doi: 10.1172/JCI34772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sladek R, Rocheleau G, Rung J, Dina C, Shen L, Serre D, et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445:828–830. doi: 10.1038/nature05616. [DOI] [PubMed] [Google Scholar]
- 3.Grant SF, Thorleifsson G, Reynisdottir I, Benediktsson R, Manolescu A, Sainz J, et al. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat Genet. 2006;38:320–323. doi: 10.1038/ng1732. [DOI] [PubMed] [Google Scholar]
- 4.Denham S, Koppelman GH, Blakey J, Wjst M, Ferreira MA, Hall IP, Sayers I. Meta-analysis of genome-wide linkage studies of asthma and related traits. Respir Res. 2008;9:38. doi: 10.1186/1465-9921-9-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dixon AL, Liang L, Moffatt MF, Chen W, Heath S, Wong KC, et al. A genome-wide association study of global gene expression. Nat Genet. 2007;39:1202–1207. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- 6.Barrett JC, Hansoul S, Nicolae DL, Cho JH, Duerr RH, Rioux JD, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat Genet. 2008;40:955–962. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Libioulle C, Louis E, Hansoul S, Sandor C, Farnir F, Franchimont D, et al. Novel Crohn disease locus identified by genome-wide association maps to a gene desert on 5p13.1 and modulates expression of PTGER4. PLoS Genet. 2007;3:58. doi: 10.1371/journal.pgen.0030058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang F, Sarginson J, Crombie C, Walker N, St. Clair D, Shaw D. Genetic association between schizophrenia and the DISC1 gene in the Scottish population. Am J Med Genet B Neuropsychiatr Genet. 2006;141:155–159. doi: 10.1002/ajmg.b.30274. [DOI] [PubMed] [Google Scholar]
- 9.“question of the year”, Nature Genetics. 2009 http://www.nature.com/ng/qoty/index.html.
- 10.Scott LJ, Mohlke KL, Bonnycastle LL, Willer CJ, Li Y, Duren WL, et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316:1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bjornsson TH, Fallin DM, Feinberg PA. An integrated epigenetic and genetic approach to common human disease. Trends in Genetics. 2004;20:350–358. doi: 10.1016/j.tig.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 12.Cheung VG, Spielman RS. Genetics of human gene expression: mapping DNA variants that influence gene expression. Nat Rev Genet. 2009;10:595–604. doi: 10.1038/nrg2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peters EH, Rojas-Caro S, Brigell MG, Zahorchak RJ, des Etages SA, Ruppel PL, et al. Quality-controlled measurement methods for quantification of variations in transcript abundance in whole blood samples from healthy volunteers. Clin Chem. 2007;53:1030–1037. doi: 10.1373/clinchem.2006.078154. [DOI] [PubMed] [Google Scholar]
- 14.Deutsch S, Lyle R, Dermitzakis ET, Attar H, Subrahmanyan L, Gehrig C, et al. Gene expression variation and expression quantitative trait mapping of human chromosome 21 genes. Hum Mol Genet. 2005;14:3741–3749. doi: 10.1093/hmg/ddi404. [DOI] [PubMed] [Google Scholar]
- 15.Whitney AR, Diehn M, Popper SJ, Alizadeh AA, Boldrick JC, Relman DA, Brown PO. Individuality and variation in gene expression patterns in human blood. Proc Natl Acad Sci USA. 2003;100:1896–1901. doi: 10.1073/pnas.252784499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Monks SA, Leonardson A, Zhu H, Cundiff P, Pietrusiak P, Edwards S, et al. Genetic inheritance of gene expression in human cell lines. Am J Hum Genet. 2004;75:1094–1105. doi: 10.1086/426461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morley M, Molony CM, Weber TM, Devlin JL, Ewens KG, Spielman RS, Cheung VG. Genetic analysis of genome-wide variation in human gene expression. Nature. 2004;430:733–734. doi: 10.1038/nature02797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smirnov DA, Morley M, Shin E, Spielman RS, Cheung VG. Genetic analysis of radiation-induced changes in human gene expression. Nature. 2009;459:587–591. doi: 10.1038/nature07940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gilad Y, Rifkin SA, Pritchard JK. Revealing the architecture of gene regulation: the promise of eQTL studies. Trends in genetics. 2008;24:408–415. doi: 10.1016/j.tig.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Price AL, Patterson N, Hancks DC, Myers S, Reich D, Cheung VG, Spielman RS. Effects of cis and trans genetic ancestry on gene expression in African Americans. PLoS Genet. 2008;4:1000294. doi: 10.1371/journal.pgen.1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stranger BE, Nica AC, Forrest MS, Dimas A, Bird CP, Beazley C, et al. Population genomics of human gene expression. Nat Genet. 2007;39:1217–1224. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rockman MV, Kruglyak L. Genetics of global gene expression. Nat Rev Genet. 2006;7:862–872. doi: 10.1038/nrg1964. [DOI] [PubMed] [Google Scholar]
- 23.Katari S, Turan N, Bibikova M, Erinle O, Chalian R, Foster M, et al. DNA methylation and gene expression differences in children conceived in vitro or in vivo. Hum Mol Genet. 2009;18:3769–3778. doi: 10.1093/hmg/ddp319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ludwig AK, Sutcliffe AG, Diedrich K, Ludwig M. Post-neonatal health and development of children born after assisted reproduction: a systematic review of controlled studies. Eur J Obstet Gynecol Reprod Biol. 2006;127:3–25. doi: 10.1016/j.ejogrb.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 25.Jackson R, Gibson K, Wu YW, Croughan MS. Perinatal outcomes in singletons following in vitro fertilization: a meta-analysis. Obstet & Gynecol. 2004;103:551–563. doi: 10.1097/01.AOG.0000114989.84822.51. [DOI] [PubMed] [Google Scholar]
- 26.Schieve LA, Meikle SF, Ferre C, Peterson H, Jeng G, Wilcox L. Low and very low birth weights in infants conceived with use of Assisted reproductive technology. NEJM. 2002;346:731–737. doi: 10.1056/NEJMoa010806. [DOI] [PubMed] [Google Scholar]
- 27.Grace KS, Sinclair KD. Assisted reproductive technology, epigenetics and long-term health: a developmental time bomb still ticking. Semin Reprod Med. 2009;27:409–416. doi: 10.1055/s-0029-1237429. [DOI] [PubMed] [Google Scholar]
- 28.Barker DJP. The developmental origins of adult disease. J Am Coll Nutr. 2004;23:588–595. doi: 10.1080/07315724.2004.10719428. [DOI] [PubMed] [Google Scholar]
- 29.Bibikova M, Lin Z, Zhou L, Chudin E, Garcia EW, Wu B, et al. High-throughput DNA methylation profiling using universal bead arrays. Genome Res. 2006;16:383–393. doi: 10.1101/gr.4410706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fan JB, Gunderson KL, Bibikova M, Yeakley JM, Chen J, Wickham Garcia E, et al. Illumina universal bead arrays. Methods Enzymol. 2006;410:57–73. doi: 10.1016/S0076-6879(06)10003-8. [DOI] [PubMed] [Google Scholar]