Abstract
A 34-residue α/β peptide, [IG(28-61)], derived from the C-terminal part of the B3 domain of the immunoglobulin binding protein G from Streptoccocus was studied using CD and NMR spectroscopy at various temperatures, and by differential scanning calorimetry. It was found that the C-terminal part (a 16-residue-long fragment) of this peptide, which corresponds to the sequence of the β-hairpin in the native structure, forms structure similar to the β-hairpin only at T = 313 K, and the structure is stabilized by non-native long-range hydrophobic interactions (Val47 – Val59). On the other hand, the N-terminal part of IG(28-61), which corresponds to the middle α-helix in the native structure, is unstructured at low temperature (283 K), and forms an α-helix-like structure at 305 K and only one helical turn is observed at 313 K. At all temperatures at which NMR experiments were performed (283, 305 and 313 K), we do not observe any long-range connectivities which would have supported packing between the C-terminal (β-hairpin) and the N-terminal (α-helix) parts of the sequence. Such interactions are absent, in contrast to the folding pathway of the B domain of protein G, proposed recently by Kmiecik and Koliński [Kmiecik, S.; Kolinski, A. Biophys J 2008, 94, 726-736], based on Monte Carlo dynamics studies. Alternative folding mechanisms are proposed and discussed.
Keywords: peptide structure, β-hairpin, α-helix, B3 domain of protein G, NMR, CD
INTRODUCTION
An important challenge in molecular biology is to understand how a nascent polypeptide chain acquires its three-dimensional and functional structure. The question of the mechanism of protein folding has intrigued scientists for many decades. Starting in the 1960’s, the determination of the three-dimensional structure of proteins by X-ray diffraction (to be joined in the 1980’s by NMR spectroscopy) provided a new basis for structure analysis and for studying the folding process. Significant progress started to be made when Anfinsen succeeded in refolding denatured and reduced ribonuclease into a fully active enzyme.1 Since then, different models of protein folding arising either from theoretical consideration,2-8 simulations,9-15 or experimental observations16-19 have been proposed.
Another method, which helps to deduce the folding mechanism, involves a conformational study of protein fragments corresponding to regular secondary structure. Studies of short fragments of proteins provide information about local interactions isolated from the protein context and, therefore, indicate the importance of these interactions in determining the secondary structure elements of proteins. It has been shown,20-22 that some short protein fragments can fold in aqueous solution into native-like conformations when no tertiary interactions are present. Thus, these fragments may play an important role as nucleation centers in initiating protein folding through local interactions,2,4,18,23 and provide knowledge about the earliest events of protein folding.
Many studies about water-soluble protein fragments corresponding to α-helix24-31 and β-hairpin20,32 have been carried out so far. However, none of the studies mentioned above considers an important question as to whether there is any dependence of peptide conformation on its length. Usually peptide fragment length was determined by the length of the corresponding regular secondary structure elements present in the structure of complete proteins.20,24-32 In our previous studies, we addressed this problem by studying peptides of different lengths from 6 up to 20 residues corresponding to the C-terminal β-hairpin from the B3 domain of protein G.22,33-35 We found that, even for peptides of different length, which share a common 6-residue fragment, the following conformational properties remain unchanged: (i) we found that the 6-residue sequence corresponding to the turn region in the structure of the β-hairpin always possesses a structure very similar to that observed in the structure of the native protein, regardless of the peptide length used in our study. Moreover, we performed our study over a relatively wide range of temperatures (283 - 323 K), and found that the conformational properties of the turn region are not temperature dependent;22,33-35 (ii) our previous study22,33-35 also showed that, regardless of the peptide length, the general shape of the investigated peptide resembles a β-hairpin-like structure, which is stabilized by long-range hydrophobic interactions between nonpolar residues. We did not find any evidence that hydrogen bonds participate in the structure stabilization (some hydrogen bonds are observed within the turn region, but they have only very local character) as was suggested in earlier studies.32
As described above, the sequence of the C-terminal β-hairpin can form a stable structure in solution without the presence of tertiary contacts in the remaining part of the protein. Based on this information, as well as on experimental and theoretical studies of the whole protein,36,37 it is generally accepted that, in the case of the immunoglobulin binding protein G, the first step in its folding pathway is the formation of the C-terminal β-hairpin.20,22,36,37 On the other hand, it is known that peptides corresponding to the middle α-helix fragment of protein G do not form regular secondary structure.38,39 Because the isolated middle fragment (corresponding to the α-helix in the native protein) does not form an α-helix, whereas the C-terminal β-hairpin has a strong tendency to form a native secondary structure, the study of the peptide which corresponds to both of these elements (α-helix and β-hairpin) could answer the question as to whether one element will induce native structure of the other.
In this work, we study a 34-residue peptide fragment (the whole protein being composed of 61 residues)40 corresponding to the C-terminal part of the B3 domain of the immunoglobulin binding protein G from Streptococcus. Structurally, the investigated fragment corresponds to the 16-residue C-terminal β-hairpin, 4-residue loop connecting the β-hairpin with the central α-helix, and the 14-residue α-helix fragment of the whole protein (see Figure 1). Recently, based on Monte-Carlo dynamics simulations, Kmiecik and Kolinski37 proposed that, in the immunoglobulin binding protein G family, the folding pathway can be described by three consecutive events: (i) formation of the C-terminal β-hairpin, (ii) formation of the middle α-helix structure packed to the C-terminal β-hairpin, and (iii) formation of the N-terminal β-hairpin packed to the C-terminal β-hairpin and the middle α-helix (see Figure 1). The results of these theoretical calculations are supported by deuterium exchange experiments performed by Kuszewski and coworkers.36 The aim of our present work was to answer the question as to whether the folding pathway of this family of proteins follows the mechanism proposed in earlier studies.36,37
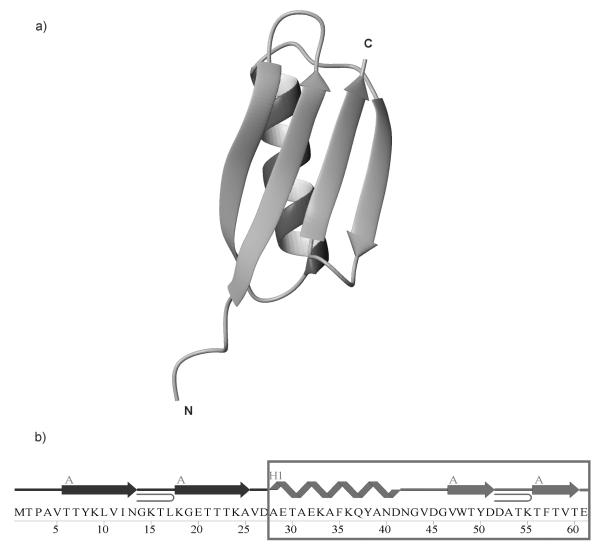
Figure 1.
(a) The X-ray structure of the B3 domain of protein G (1IGD).40 (b) The amino acid sequence of 1IGD. In (b), A denotes β-strands and H1 the α-helix. The boxed part of the sequence, IG(28-61), was studied in this work.
RESULTS
Differential Scanning Calorimetry (DSC)
As in our previous studies22,33-35 we used the DSC method to determine thermal stability41 and possible tendency to aggregation of the peptide.42 The DSC experiment can detect oligomerization/aggregation processes in a wide range of temperatures, using a very small amount of an investigated compound, and can show if the process of aggregation/oligomerization is reversible with changes of temperature. Additionally, a wide range of heating/cooling speed can be used to detect oligomerization/aggregation processes if the only interest is detection of such processes but not their thermodynamic/kinetic effects related to it.42
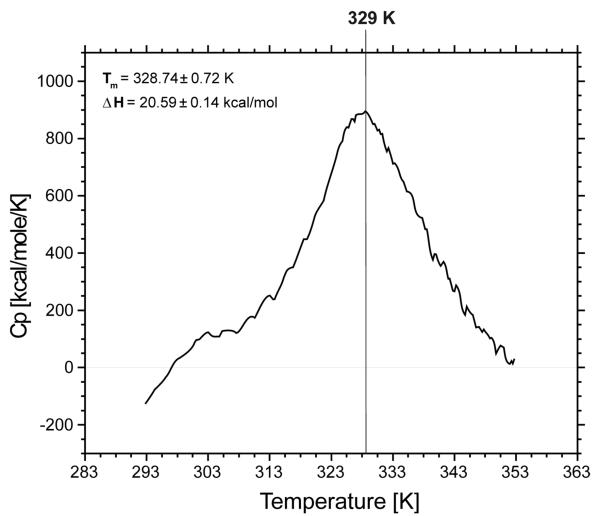
The heat capacity curve for IG(28-61) is presented in Figure 2. One relatively sharp peak, which corresponds to a folding/unfolding transition, is observed on the heat capacity curve. Based on this curve, and the use of a two-state model, the transition temperature 328.74±0.72 K was calculated; the enthalpy change related to this transition is ΔH = 20.59±0.14 kcal/mol. The transition temperature as well as enthalpy change observed for the IG(28-61) peptide is very similar to those reported in our previous studies22,33-35 for shorter peptides. Our study shows that changes in melting temperature and enthalpy of folding are not correlated with peptide length, and the range of changes obtained in the region of the melting point and in the enthalpy of folding in our study are similar to those obtained by other authors who used similar peptides but studied mostly single or multiple mutations with constant peptide length.43-45
Figure 2.
Heat capacity curve for IG(28-61) recorded in water at pH = 6.02.
CD measurements
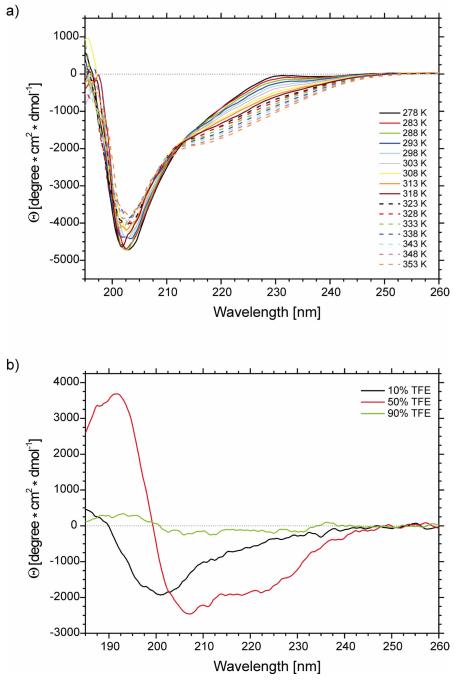
The CD spectra were recorded in pure water (pH = 6.02) (a) at 16 different temperatures, i.e., at 5 deg intervals between 278 and 353 K (Figure 3a) and (b) in 10%, 50% and 90% TFE/H2O mixture at 305 K (Figure 3b).
Figure 3.
CD spectra of IG(28-61) in (a) water at 16 different temperatures (pH = 6.02) and (b) water solutions of trifluoroethanol (10%, 50% and 90% of TFE).
As shown in Figure 3, and in Figure 2 of supplemental material, the molar ellipticity varies with temperature. The molar ellipticity for the IG(28-61) compound at γ = 201 nm becomes less negative, whereas at γ = 220 and γ = 230 nm it becomes more negative (Figure 3, and Figure 2 of supplemental material), with increasing temperature, which suggests creation of a more ordered hairpin/turn/helix structure at the cost of a random coil conformation.46,47 The calculated percentage of secondary structure elements, by using the SP37A CONTINLL method,48 is listed in Table IV of supplemental material. The changes in ellipticity at selected diagnostic wavelengths shown in Figure 2 of supplemental material are very small. Variation of the percentage of secondary structure elements, as calculated from CD spectra for IG(28-61), also change very little with temperature (less than 2%) (see Table IV of supplemental material), which prevents us from drawing any conclusions from these data. In our previous studies of shorter analogous peptides, we also observed that CD spectroscopy is not well suited to investigate temperature-induced conformational changes of short peptides22,33-35
NMR measurements
A more detailed structural analysis was carried out with NMR spectroscopy. 2D 1H-NMR spectra of IG(28-61) were recorded in water at pH = 6.02 at three different temperatures 283, 305 and 313 K, to examine the influence of temperature on the structure. The chemical shifts of the proton resonances for this peptide at these three temperatures are listed in Tables I – III of the supplemental material.
The chemical shifts of the amide protons of IG(28-61) at three different temperatures (283, 305 and 313 K) are plotted as a function of residue number in Figure 3 of supplemental material. Except for Gly43, Asp51, Thr54, Lys55 and Thr56, whose amide chemical shifts do not change significantly, the chemical shifts of all other amino acid residues show a tendency to move upfield with increasing temperature (Figure 3 of supplemental material). It should also be noted that the chemical shifts at 305 and 313 K change very little for almost all the amide protons in IG(28-61) peptide. The temperature coefficients for Gly43, Asp51, Thr54, Lys55 and Thr56 are Δδ/ΔT = −4.3, −3.1, 4, −3.5, and 1.2 ppb/K, respectively (see Table V of supplemental material). Thus, the amide protons of these residues could either be involved in a hydrogen bond or be buried in a hydrophobic region of the peptide (Δδ/ΔT < ∣4.5∣ ppb/K is the threshold below which an amide proton can be considered to be screened from the solvent).49
In our previous studies of peptides derived from the sequence of the C-terminal β-hairpin of protein G, we also observed low temperature coefficients for amide protons of residues Thr54 and Thr56.22,33 Additionally, for some of these peptides (16 residues and 12 residues long), we also observed low coefficients for the amide protons of Asp51.22,33 All of these amino acid residues (Asp51, Thr54, and Thr56) are located in the turn region in the middle of the sequence of the C-terminal β-hairpin. On the other hand, for the 20-residue peptide studied in our previous work35, we showed that none of the amide protons possessed small temperature coefficients. The results of NMR measurements of peptides of different length show that amide protons with low temperature coefficients are always observed in the turn region22,33-34; however, in some cases, the presence of the turn structure (detected by ROE connectivities) was not associated with low-temperature-coefficient amide protons35. Such an observation could lead to the conclusion that possible hydrogen bonds in the turn region are rather induced by the general shape of the polypeptide chain in the turn region, but are not the interaction that creates the turn of the polypeptide chain22,22-35.
Our previous studies showed that none of the amide protons of the isolated α-helical fragment possessed small temperature coefficients39 (lower than ∣4.5∣ ppb/K).49 As shown for the 34-residue peptide investigated in this work, in Table V of the supplemental material, the amide protons within the α-helical region display high values of the temperature coefficients (as was observed for the isolated α-helical peptide), which indicates lack of well organized structure in this region of the sequence.
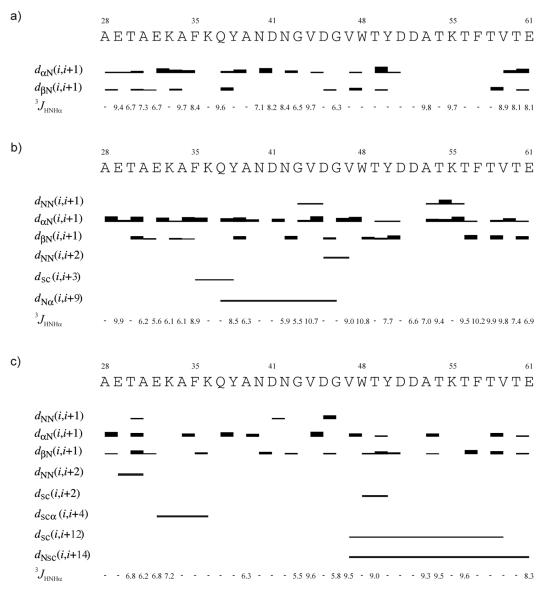
In Figure 4, the ROE effects corresponding to interproton contacts and the values of the 3JNHHα coupling constants obtained in NMR measurements carried out at different temperatures for IG(28-61) are presented.
Figure 4.
ROE effects corresponding to the interproton contacts and the 3JNHHα coupling constants of IG(28-61) measured in H2O at (a) 283 K, (b) 305 K and (c) 313 K. The thickness of the bars reflects the strength of the ROE correlation as strong, medium or weak.
At each temperature (T = 283, 305, and 313 K) the Hα(i) – HN(i+1) and Hβ(i) – HN(i+1) ROE connectivities are present for most residues, and they are usually strong or very strong. At T = 283 K only the Hα(i) – HN(i+1) and Hβ(i) – HN(i+1) ROE connectivities are observed (Figure 4a), whereas at higher temperatures (305 and 313 K) sequential HN(i) – HN(i+1) and long-range connectivities start being observed (Figure 4b, c).
At T = 305 K, sequential HN(i) – HN(i+1) ROEs are present in two regions of the sequence: in the turn region of the C-terminal β-hairpin part and in the part of the sequence which plays a role as a linker (Asn42 – Asp45) between the α-helix and β-hairpin fragments (Figure 4b). The HN(i) – HN(i+1) connectivities are very characteristic of bent conformations (helix/turn);50 thus, it can be seen that these two parts (the turn and linker regions) of the sequence of the IG(28-61) peptide have some propensity to bend, as observed in the native protein.40 At T = 305 K, the following ∣i-j∣ > 1 connectivities are observed: HN(i) – HN(i+2), Hsc(i) - Hsc(i+3), and HN(i) – Hα(i+9) between Asp45 – Val47, Phe35 – Tyr38, and Gln37 – Gly46, respectively (Figure 4b, Table I). The HN(i) – HN(i+2) (Asp45 – Val47) connectivity connects the linker part of the IG(28-61) sequence with the N-terminus of the hairpin sequence (Figure 4b), which additionally supports the appearance of a bent conformation in this part of the sequence. The Hsc(i) - Hsc(i+3) interaction (Phe35 – Tyr38) encompasses the native interactions within the part of the sequence which forms the α-helix in the native structure of the protein.40 Another ∣i-j∣ > 1 interaction occurs between HN(i) – Hα(i+9), residues Gln37 – Gly46, and it is the only interaction which brings the α-helix part of the sequence close in space to the linker sequence. All these data suggest that the peptide part corresponding to the α-helix as well as the linker part become more ordered, similar to the structure observed in the whole protein, but the region corresponding to the native β-hairpin seems to be very flexible, although its turn region is very well defined.
Table I.
Atoms of residues separated by at least 2 residues in sequence (∣i-j∣ > 1) between which ROE peaks were found at 283, 305 and 313 K at pH = 6.02.
| ROE peaks between residues ∣i – j∣ > 1 | ||
|---|---|---|
| 283 K | 305 K | 313 K |
| δF35 – β1Y38 | NE29 – NA31 | |
| NQ37 – α1G46 | γE32 – αK36 | |
| ND45 – NV47 | NV47 – β1E61 | |
| NV47 – β2E61 | ||
| βV47 – γ1V59 | ||
| η2W48 – β2Y50 | ||
N – amide protons
When the temperature increases to T = 313 K, the number of ∣i-j∣ > 1 connectivities increases (Table I). Most of these ∣i-j∣ > 1 connectivities stabilize the C-terminal β-hairpin fragment: Hsc(i) - Hsc(i+2), Hsc(i) - Hsc(i+12), and HN(i) - Hsc(i+14) between Trp48 – Tyr50, Val47 – Val59, and Val47 – Glu61, respectively (Figure 4c, Table I). In the helix part, only one long-range ROE, between Glu32 and Lys36, is observed. Additionally, in the N-terminal part of the helix sequence, sequential HN(i) – HN(i+1) and HN(i) – HN(i+2) ROEs between Thr30 – Ala31, and Glu29 – Ala31, respectively, are observed (Figure 4c). Some of the weak and medium HN(i) – HN(i+1) connectivities are observed in the linker region which suggests formation of a bent structure. (see Figure 4c). Surprisingly, no HN(i) – HN(i+1) ROE is observed in the β-turn region, which suggests that the turn of the polypeptide chain at this temperature is not as sharp as at T = 303K. In our study of the 20-residue peptide, we observed a similar situation of long-range interactions between hydrophobic residues located far from the turn region and an absence of HN(i) – HN(i+1) interactions within the turn region35. The hydrophobic interaction (between Val47 and Val59) observed in the IG(28-61) fragment at 313 K is a non-native one, as was often observed for shorter β-hairpin fragments studied in our previous work [between the “1st pair” (Tyr50 – Phe57)22,33,34 and the “2nd pair” (Trp48 – Val59) of hydrophobic residues].22,35 This means that hydrophobic interactions, as well as any long-range interactions, which stabilize the C-terminal β-hairpin structure, are very dynamic and their pattern is very sensitive to the length of the peptide under investigation. However, in our study, we found that, for β-hairpin forming peptides, the most stable hydrophobic interaction is located farther from the turn region as the peptide length increases33-35. Shifting the most stable hydrophobic interaction further form the turn region is also accompanied by a decrease or disappearance of the HN(i) – HN(i+1) ROE within the turn sequence35. Thus, based on the results presented in this and the previous work,22,33-35 it can be suggested that the turn in the C-terminal part of the sequence is formed first and, once it becomes stable, small nucleation centers, farther from the turn region, are formed step-wise in the next folding stages (such a mechanism is proposed in reference 35).
MD simulations
The structural data summarized in Figure 4 were used to carry out MD simulations of IG(28-61) with time-averaged restraints, in order to determine the structures of this peptide. The structures are discussed below.
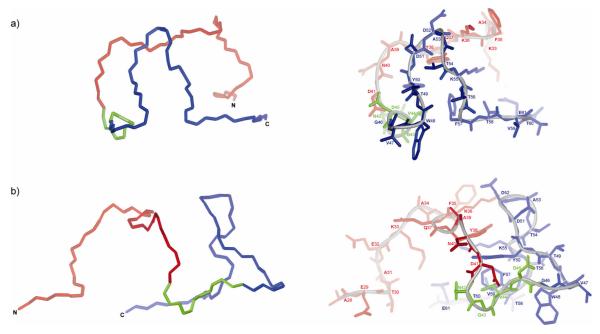
In Figures 5, 6, and 7, representative conformations of the most populated families of conformations of IG(28-61) generated from MD simulations using NMR data recorded at T = 283, 305, and 313 K, respectively, are presented.
Figure 5.
The lowest-energy conformation which is the representative conformation from the most populated family of conformations of IG(28-61) obtained by using time-averaged MD methodology with restraints from NMR measurements at 283 K. (a) The front view of the structure, (b) the side view of the structure.
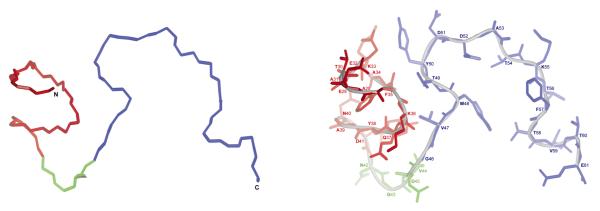
Figure 6.
The lowest-energy conformation which is the representative conformation from the most populated family of conformations of IG(28-61) obtained by using time-averaged MD methodology with restraints from NMR measurements at 305 K.
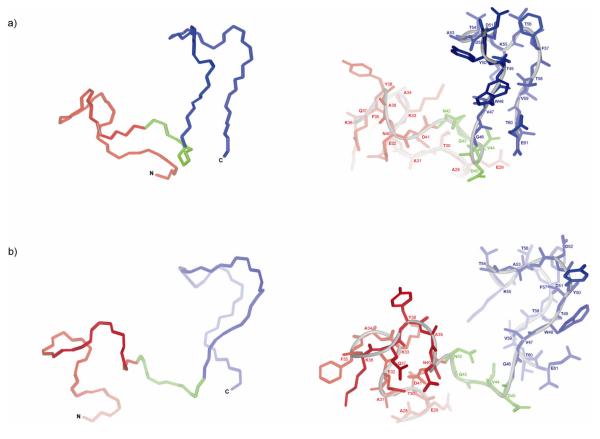
Figure 7.
The lowest-energy conformation which is the representative conformation from the most populated family of conformations of IG(28-61) obtained by using time-averaged MD methodology with restraints from NMR measurements at 313 K. (a) The front view of the structure, (b) the side view of the structure.
As shown in Figure 5, the conformation of the IG(28-61) fragment generated by MD simulations with restraints obtained from the NMR data recorded at T = 283 K exhibits a β-hairpin-like structure (marked in blue in Figure 5) in the C-terminal part of the sequence and an extended structure with some bend in the middle in the N-terminal part corresponding to the native α-helix. The spatial orientation of the α-helical and β-hairpin parts is completely random (no long-range interactions between the two parts of peptide were observed in NMR experiments at 283 K) (see Figure 4a). Because we gather a very limited number of restraints from NMR spectra at T = 283 K, and all of them are only short-range, the structure presented in Figure 5 should be considered with caution.
Using the NMR data recorded at T = 305 K (Figure 4b), the structure of IG(28-61) shown in Figure 6 was generated. In this structure, a well-defined shape of the linker part between the Asn41 – Asp45 residues (marked in green in Figure 6) can be observed. The sequence of the linker bends in a similar manner as in the native protein,40 and the main reason for the formation and stability of this structure is the appearance of strong HN(i) – HN(i+1) and HN(i) – HN(i+2) connectivities (Figure 4b). As well as the linker part, the turn region in the C-terminal β-hairpin is pretty well defined, which is once again connected with the appearance of the HN(i) – HN(i+1) connectivities in this region (Figure 4b). However, the NMR data recorded at T = 305 K do not indicate any long-range interactions inside the β-hairpin sequence (marked in blue in Figure 6). Thus, a bend in the turn region sequence is necessary for the hairpin shape to appear, as observed in Figure 6. The α-helical part (marked in red in Figure 6) is ordered and exhibits the α-helix-like structure. This is caused by the long-range connectivities observed within this part of the sequence as well as by the low value of the coupling constants (below 6.2 Hz) for residues 31 - 34 (see Figure 4b).
In Figure 7, a representative structure of IG(28-61), obtained by using the geometrical restraints based on the NMR spectra recorded at T = 313 K (Figure 4c) is presented. In the structure presented in Figure 7, we can see well-developed β-hairpin-like structure in the C-terminal part (marked in blue in Figure 7), which well reproduces the long-range interactions between two pairs of amino acid residues, namely Val47 – Val59 and Val47 – Glu61 (see Table I and Figure 4c) observed in the NMR spectra. The turn region in the β-hairpin part is very flexible and does not create a regular β-turn which is associated with absence of HN(i) – HN(i+1) ROE connectivities in this region of the sequence, as shown. The N-terminal part of the peptide (marked in red in Figure 7) is more disordered than the C-terminal part. However, one long-range interaction observed in the NMR spectra [Hsc(i) – Hα(i+4) between Glu32 and Lys36 (see Figure 4c)] causes the formation of a loop-like structure (one α-helix-like turn) in the middle of the sequence corresponding to the α-helical fragment in the native protein (marked in red in Figure 7). The linker sequence (marked in green in Figure 7) displays some sort of bent conformation, mainly because of appearance of some HN(i) – HN(i+1) connectivities in this region (see Figure 4c). Similarly, as in the structures presented in Figures 5 and 6, the overall structure is quite compact; however, we should stress that no long-range interactions were observed at T = 313 K between the α-helical and β-hairpin parts, even if the parts related to the secondary structure elements of the IG(28-61) sequence are conformationally well restrained as found from ROESY spectra.
DISCUSSION
Our previous studies showed that, for peptides with length 6, 8, 12, 14, and 16 residues, respectively (all of them based on the sequence of the C-terminal β-hairpin of the B3 domain of protein G), the six-residue fragment Asp51 - Thr56 (corresponding to the turn sequence in the structure of the native protein) is conformationally very rigid and has a bent shape very similar to that observed in the structure of the native protein.22,33,34 The conformational feature of this six-residue fragment remains unchanged in the range of temperatures T = 283 - 313 K for peptides of various length (from 6 to 16 residues). Because of its unusual conformational properties, this six-residue peptide could be a nucleation center for the folding of the whole B3 domain protein G.22,33,34 However, when we studied a longer 20-residue peptide,35 based on the sequence of the C-terminal β-hairpin of the B3 domain of protein G, we found that the unusual conformational rigidity of the turn region is observed only at low temperature (T = 283 K). For the 34-residue peptide studied in this work, we did not observe a rigid conformation of the turn region at any temperature (T = 283, 305 and 313 K) used in our study (see Figure 4a-c). We found that, when the peptide length increases, the most stable long-range hydrophobic interactions are observed further away from the turn position. For the 8-,34 12-33 and 14-residue33 peptides, we observed long-range hydrophobic interactions between residues Tyr50 – Phe57 (the so-called “1st pair” of hydrophobic residues). For the 16-residue peptide, we observed strong Tyr50 – Phe57 and weak Trp48 - Val59 interactions (the so-called “2nd pair” of hydrophobic residues).22 With further increase of the peptide length to 20 residues, we observed a very weak Tyr50 – Phe57 interaction (this interaction is observed only at T= 283 K) and much stronger Trp48-Val59 interactions.35 Finally, for the 34-residue peptide (this work), we observed only one long-range hydrophobic interaction between residues Val47 and Val59 (see Figure 4c) (observed only at 313 K).
The results from this study confirm our conclusion from the previous paper35. The most plausible mechanism could apply to the formation of hairpin structure of the C-terminal β-hairpin of the B3 domain of protein G: initially the turn structure is formed, and it facilitates the formation of subsequent hydrophobic interactions farther and farther from the turn; such a mechanism was already proposed earlier.4,32,51 However, when more hydrophobic interactions are formed farther away from the turn position [Trp48 - Val5935 or Val47 – Val59 (this work)], those hydrophobic interactions which are close to the turn region (Tyr50 – Phe57), start to break down, disrupting the tight conformation of the turn region (Figure 4a-c).35 We could not find a good explanation as to why some long-range interactions (Trp48 - Val59 or Val47 – Val59) are more stable than the local ones which occur in the turn region, or than short-range/medium (Tyr50 – Phe57) interactions. This issue will be the subject of our further studies. Nevertheless, our results show how a series of local/short-range interactions, formation/breaking events, create long-range interactions. Such a mechanism for establishing long-range interactions can be a general feature of protein folding but further studies are required to verify this hypothesis.
Before starting our study, we knew that peptides corresponding to the C-terminal β-hairpin have a tendency to form a stable tertiary structure similar in shape to the β-hairpin. We expected that, by studying longer peptides, we would eventually observe further stabilization of the β-hairpin structure (in the C-terminal part of the peptide), formation of a structure similar to that of the α-helix (in the N-terminal part of the peptide), or cooperative development of stable secondary structures in both parts of the investigated peptide; however, the results of our study do not show any of these features. On the other hand, we observed that the β-hairpin-like structure, which we expected to form in the C-terminal part of the sequence, does form, but it is more irregular (see Figures 5-7) than those structures observed for shorter peptides studied in our earlier work.22,33-35 The N-terminal part of the investigated peptide corresponding to the α-helix shows some helix-like conformation but only at T = 305 K (see Figure 6). At T = 313 K, we observe formation of one helical-like turn in the K33 - Q37 region (see Figure 7), and at T = 283 K the N-terminal part of the IG(28-61) peptide remains unstructured (see Figure 5). Comparison of the results obtained in this study and in our earlier study using a peptide corresponding to the isolated α-helix sequence39 shows that the corresponding sequences are unstructured at low temperature (T = 283 K) in both cases. When the temperature increases, the overall helicity of the corresponding sequences increases, but, for the 34-residue peptide studied in this work, the maximal helicity is observed at T = 305 K, and in the case of the 16-residue helical peptide at 313 K (see Table II of ref 39). There are no large differences between the conformational behavior of a sequence corresponding to the α-helix when this sequence is studied in isolation39 or when it is part of a longer peptide (this work).
In our study, we did not find any long-range interactions between the parts of the sequences corresponding to the β-hairpin and α-helix. The absence of such long-range interactions stands in contrast to the hypothesis for the mechanism of folding proposed for the B domains of protein G by Kmiecik and Kolinski.37 As mentioned in the Introduction, Kmiecik and Kolinski proposed that, in the immunoglobulin binding protein G family, the folding pathway can be described by three consecutive events: (i) formation of the C-terminal β-hairpin; (ii) formation of the middle α-helix structure packed to the C-terminal β-hairpin; (iii) formation of the N-terminal β-hairpin packed to the C-terminal β-hairpin and the middle α-helix (see Figure 1). This mechanism is partially supported by deuterium exchange experiments performed by Kuszewski and coworkers.36 Our study indicates that the mechanism of folding is different from that proposed by Kmiecik and Kolinski. Our data support only the initial stage of their mechanism that the C-terminal β-hairpin-like structure forms first. However, our results show that the middle α-helix does not form or pack to the C-terminal β-hairpin in the second stage of folding. It should be noted that, in all studies mentioned above36,37, the research was conducted on sequences that differ in a few positions rather than those used in this paper. We already showed22,23 that slight differences in the sequences do not change the conformational properties of the peptides dramatically, based on the secondary structure elements of the investigated protein 22,33. However, from the results of our study, we cannot decide how mutations will affect the folding mechanism of the whole protein. In the future, we plan to study more sequences to obtain a clear picture of the impact of naturally-observed mutations on the structure and folding mechanism of the B domain of protein G.
In light of our results, it is possible that the mechanism of folding may consist of the following three stages: (i) formation of the C-terminal β-hairpin-like structure; (ii) formation of an anti-parallel β-structure; (iii) formation of the middle α-helix induced by β-structure. This mechanism is in agreement with our results, but it will be very difficult to explain how many long-range interactions in the anti-parallel β-structure are formed, whereas the Kmiecik-Kolinski mechanism is based on structure formation in sequential order in the direction from the C- to the N-terminus of the sequence, which makes all long-range contacts easier to achieve. We can also propose another folding pathway: (i) formation of the C-terminal β-hairpin-like structure; (ii) cooperative formation of the structure in the N-terminal part of the protein sequence between the N-terminal β-hairpin and the middle α-helix, and (iii) formation of the final structure by assembly of the elements formed in steps (i) and (ii). Such a mechanism was already suggested by Weikl and Dill.52 To eventually prove or discard such a mechanism, additional conformational studies should be performed. In the future, we plan to perform conformational studies of peptides corresponding to the N-terminal β-hairpin as well as peptides corresponding to the N-terminal β-hairpin plus the middle α-helix. Results from such studies can provide some hints about the processes related to the folding mechanism which occur in the N-terminal part of the protein. Earlier studies suggested that the N-terminal β-hairpin does not form any regular structure in solution;38 however, these studies were performed at T = 278 K. On the other hand, the results of our study clearly indicate that the temperature influences structure formation and, as shown in this work, the long-range interactions which stabilized β-hairpin formation, could be observed at higher temperatures (see Figure 4c).
MATERIALS AND METHODS
Peptide synthesis
The peptide H-AETAEKAFKQYANDNGVDGVWTYDDATKTFTVTE-NH2 [IG(28-61); 34 amino acid residues] was synthesized by standard solid-phase Fmoc-amino acid chemistry with a Milipore synthesizer. The resin Tentagel R RAM (1g, capacity 0.19 mmol/g) was treated with piperidine (20%) in DMF, and all amino acids were coupled using DIPCI/HOBt methodology. The coupling reaction time was 2 h. Piperidine (20%) in DMF was used to remove the Fmoc group at all steps. After deprotection of the last Fmoc N-terminal group, the resin was washed with methanol and dried in vacuo. In the final step, the resin was treated with a TFA/water/phenol/triisopropylsilane (8.8/0.5/0.5/0.2) mixture (10 ml per gram of resin) at room temperature for 2 h to remove the peptide from the resin.
The resin was separated from the mother liquid; the excess of solvent was then evaporated to a volume of 2 ml, and the residue was precipitated with diethyl ether. The crude peptide was dissolved in 18.7% CH3CN in TEA/H3PO4 and purified by reverse-phase HPLC using a SupelcosilTM SPLC-ABZ C18 semi-preparative column (10 × 250 mm, 5 μm) with 4 ml/min elution and a 120 min isocratic mixture of 18.7% CH3CN in TEA/H3PO4 to adjust the pH to approximately 7.0. To identify the fractions containing the pure peptide, HPLC was run first with a small amount of the crude peptide and the absorbance at 222 nm was measured for each fraction. A plot of absorbance vs. retention time was constructed, and the interval of the retention time to separate the pure peptide was estimated as that corresponding to the large peak in the plot. Subsequently, a semi-preparative HPLC run was carried out and the fractions containing the pure peptide were collected and lyophilized. The purity of the peptide was confirmed by analytical HPLC and MALDI-TOF analysis (M = 3787 g/mol, calculated 3786.04 g/mol).
Circular dichroism (CD) spectroscopy
CD spectra were recorded on a Jasco J-815 spectropolarimeter with a 100 nm/min scan speed, and data were collected from 260 to 190 nm with a 1 mm path-length quartz cell. The samples were dissolved (a) in water (pH = 6.02) and the CD spectra were measured at 16 different temperatures, i.e., at 5 deg intervals between 278 and 353 K, and (b) in water solution of CF3CH2OH [H2O/CF3CH2OH ratio was 9:1, 1:1, and 1:9 by vol.] and the CD spectra were measured at 305 K. The final concentration of IG(28-61) was 0.02g/100 ml. The secondary structure content was calculated from CD spectra using the CONTINLL method.48
Differential Scanning Calorimetry (DSC)
Calorimetric measurements were carried out with a VP-DSC microcalorimeter (MicroCal) at a scanning rate of 1.5 degree/minute. Scans were obtained at a peptide concentration of 0.053 mM. The cell volume was 0.5 ml. All scans were run at pH = 6.02 in pure water in the range of temperatures from 5 °C to 80 °C. The reversibility of the transition was checked by cooling and reheating the same sample. No histeresis of heat capacity was found in the repeated heating and cooling cycles and, moreover, no largely negative values of heat capacity were observed. This demonstrates that no irreversible processes such as, e.g., aggregation or hydrolysis, occurred during the thermal transition. The data presented are mean values from three independent measurements. Results from the DSC measurements were analyzed with the Origin 7.0 software from MicroCal using the software routines provided with the instrument.53
1H-NMR spectroscopy
The NMR spectra of IG(28-61) were measured on VARIAN 500 MHz and 600 MHz spectrometers. The following spectra were recorded: 1D 1H-NMR (at 283, 289, 297, 305, 313 and 321 K) and 2D 1H-NMR: DQF-COSY,54 TOCSY55 (80 ms), ROESY56 (250 ms) at 283, 305 and 313 K. The sample was dissolved in H2O/2H2O (9:1 by vol.) [pH = 6.02], and the concentration of the sample was 3.3 mM. The spectra were processed using VARIAN 4.3 software (Varian Instruments, PaloAlto, CA, USA) and analyzed with the XEASY program.57 The spectra were calibrated against the DSS (sodium 4,4-dimethyl-4-silapentane-1-sulfonate) signal.58 Proton signals were assigned based on the TOCSY spectra. The sequential analysis of the peptide was confirmed by the ROESY spectra.56 The chemical shifts are reported in Tables I – III in supplemental materials. The coupling constants between NH and Hα protons (3JHNHα) of IG(28-61) were obtained from two-dimensional DQF-COSY and one-dimensional 1H spectra. The intensities of ROE signals were estimated from the ROESY spectra.56 In Figure 1 of supplemental material, the TOCSY spectra, with peak assignments of IG(28-61), are shown.
Three-dimensional structure calculations
The ROE inter-proton cross-peaks of IG(28-61) were derived from 2D 1H-NMR ROESY spectra, and vicinal coupling constants 3JHNHα were obtained from 2D 1H-NMR DQF-COSY and temperature-dependent 1D 1H-NMR spectra. In the first step, the ROESY peak volumes were converted to upper distance bounds by using the CALIBA59 program of the DYANA package.60 In the next step, torsion angles, were generated using the HABAS algorithm of the DYANA package,61 based on the Bystrov-Karplus62 equation. The upper distance limits and torsional angles were used as restraints in molecular dynamics calculations.
Molecular dynamics simulations with the time-averaged methodology (TAV)63-65 were carried out with the AMBER force field66 using the AMBER 8.0 package.65 The interproton distances were restrained with the force constant k = 20 kcal/(mol × Å2), and the dihedral angles with k = 2 kcal/(mol × deg2), respectively. The dihedral angles ω were restrained with a center at 180° and k = 10 kcal/(mol × deg2). The improper dihedral angles centered at the Cα atoms (defining the chirality of amino acid residues) were restrained with k = 50 kcal/(mol × deg2). Three sets of separate simulations were run for IG(28-61), using the restraints from the NMR data collected at 283, 305 and 313 K, respectively. All simulations were carried out in a TIP3P67 periodic water box at constant volume, with the particle-mesh Ewald procedure for long-range electrostatic interactions.68,69 MD simulations with time-averaged restraints at these three different temperatures, were carried out with a time step of 2 fs,70 and the total duration of the run was 5 ns. Coordinates were saved every 2000 steps of MD simulations.
For every NMR restraint set, four independent TAV MD simulations were run at the following temperatures: N, 400 K, 500 K, and 600 K (where N is the temperature of the NMR experiment (i.e., runs at 283 K, 305 K, and 313 K), respectively, for three independent sets of calculations). The purpose of running simulations at many temperatures including elevated temperatures was to enhance sampling. From every trajectory, 300 final conformations were collected for the analysis. The structures from four trajectories, obtained from simulations performed using the same NMR restraint set, were combined together. After TAV MD simulations, we obtained three sets of 1200 conformation each (four runs, with 300 conformations from every run) corresponding to three NMR restraint sets recorded at different temperatures for IG(28-61). All three sets of conformations were clustered separately, with the use of the MOLMOL program.71 An RMS deviation cut-off of 5.0 Å was used in the clustering procedure. The clustering procedure provided five families of conformations for the IG(28-61) peptide at each temperature (283, 305, and 313 K). The one structure which represent the most populated family at each temperature for IG(28-61) was selected for presentation. The RMS deviation was calculated based on the Cα atoms.
Supplementary Material
ACKOWLEDGEMENTS
This research was supported by the Polish Ministry of Science and Higher Education grant 1696/H03/2007/32 and the U.S. National Institutes of Health (GM-14312).
REFERENCES
- 1.Anfinsen CB. Science. 1973;181:223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- 2.Karplus M, Weaver DL. Nature. 1976;260:404–406. doi: 10.1038/260404a0. [DOI] [PubMed] [Google Scholar]
- 3.Tanaka S, Scheraga HA. Macromolecules. 1977;10:291–304. doi: 10.1021/ma60056a015. [DOI] [PubMed] [Google Scholar]
- 4.Matheson RR, Jr, Scheraga HA. Macromolecules. 1978;11:819–829. [Google Scholar]
- 5.Bryngelson JD, Wolynes PG. Proc Natl Acad Sci USA. 1987;84:7524–7528. doi: 10.1073/pnas.84.21.7524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karplus M, Weaver DL. Protein Sci. 1994;3:650–668. doi: 10.1002/pro.5560030413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karplus M, Šali A. Curr Opin Struct Biol. 1995;5:58–73. doi: 10.1016/0959-440x(95)80010-x. [DOI] [PubMed] [Google Scholar]
- 8.Dill KA, Bromberg S, Yue K, Fiebig KM, Yee DP, Thomas PD, Chan HS. Protein Sci. 1995;4:561–602. doi: 10.1002/pro.5560040401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levitt M, Warshel A. Nature. 1975;253:693–698. doi: 10.1038/253694a0. [DOI] [PubMed] [Google Scholar]
- 10.Taketomi H, Kano F, Gô N. Biopolymers. 1988;27:527–560. doi: 10.1002/bip.360270402. [DOI] [PubMed] [Google Scholar]
- 11.Skolnick J, Kolinski A. Science. 1990;250:1121–1125. doi: 10.1126/science.250.4984.1121. [DOI] [PubMed] [Google Scholar]
- 12.Godzik A, Skolnick J, Kolinski A. Proc Natl Acad Sci USA. 1992;89:2629–2633. doi: 10.1073/pnas.89.7.2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Daggett V, Levitt M. J Mol Biol. 1993;232:600–619. doi: 10.1006/jmbi.1993.1414. [DOI] [PubMed] [Google Scholar]
- 14.Šali A, Shakhnovich E, Karplus M. Nature. 1994;369:248–251. doi: 10.1038/369248a0. [DOI] [PubMed] [Google Scholar]
- 15.Kolinski A, Skolnick J. Proteins Struct Funct Genet. 1994;18:338–352. doi: 10.1002/prot.340180405. [DOI] [PubMed] [Google Scholar]
- 16.Baldwin RL. Ann Rev Biochem. 1975;44:453–475. doi: 10.1146/annurev.bi.44.070175.002321. [DOI] [PubMed] [Google Scholar]
- 17.Kim PS, Baldwin RL. Ann Rev Biochem. 1982;51:459–489. doi: 10.1146/annurev.bi.51.070182.002331. [DOI] [PubMed] [Google Scholar]
- 18.Kim PS, Baldwin RL. Ann Rev Biochem. 1990;59:631–660. doi: 10.1146/annurev.bi.59.070190.003215. [DOI] [PubMed] [Google Scholar]
- 19.Scheraga HA, Wedemeyer WJ, Welker E. Meth Enzym. 2001;341:189–221. doi: 10.1016/s0076-6879(01)41153-0. [DOI] [PubMed] [Google Scholar]
- 20.Blanco FJ, Rivas G, Serrano L. Nat Struct Biol. 1994;1:584–590. doi: 10.1038/nsb0994-584. [DOI] [PubMed] [Google Scholar]
- 21.Searle MS, Williams DH, Packman LC. Nat Struct Biol. 1995;2:999–1006. doi: 10.1038/nsb1195-999. [DOI] [PubMed] [Google Scholar]
- 22.Skwierawska A, Ołdziej S, Liwo A, Scheraga HA. Biopolymers. 2009;91:37–51. doi: 10.1002/bip.21080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dill KA. Biochemistry. 1990;29:7133–7155. doi: 10.1021/bi00483a001. [DOI] [PubMed] [Google Scholar]
- 24.Brown JE, Klee WA. Biochemistry. 1971;10:470–476. doi: 10.1021/bi00779a019. [DOI] [PubMed] [Google Scholar]
- 25.Silverman DN, Kotelchuck D, Taylor GT, Scheraga HA. Arch Biochem Biophys. 1972;150:757–766. doi: 10.1016/0003-9861(72)90095-1. [DOI] [PubMed] [Google Scholar]
- 26.Jiménez MA, Herranz J, Nieto JL, Rico M, Santoro J. FEBS Lett. 1987;221:320–324. doi: 10.1016/0014-5793(87)80948-1. [DOI] [PubMed] [Google Scholar]
- 27.Jiménez MA, Rico M, Herranz J, Santoro J, Nieto JL. Eur J Biochem. 1988;175:101–109. doi: 10.1111/j.1432-1033.1988.tb14171.x. [DOI] [PubMed] [Google Scholar]
- 28.Dyson HJ, Merutka G, Waltho JP, Lerner RA, Wright PE. J Mol Biol. 1992;226:795–817. doi: 10.1016/0022-2836(92)90633-u. [DOI] [PubMed] [Google Scholar]
- 29.Kuroda Y. Biochemistry. 1993;32:1219–1224. doi: 10.1021/bi00056a004. [DOI] [PubMed] [Google Scholar]
- 30.Muñoz V, Serrano L, Jiménez MA, Rico M. J Mol Biol. 1995;4:648–669. doi: 10.1016/s0022-2836(05)80145-7. [DOI] [PubMed] [Google Scholar]
- 31.Hill R, Degrado W. J Am Chem Soc. 1998;120:1138–1145. [Google Scholar]
- 32.Muñoz V, Thompson PA, Hofrichter J, Eaton WA. Nature. 1997;390:196–199. doi: 10.1038/36626. [DOI] [PubMed] [Google Scholar]
- 33.Skwierawska A, Makowska J, Ołdziej S, Liwo A, Scheraga HA. Proteins Struct Funct Bioinform. 2009;75:931–953. doi: 10.1002/prot.22304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Skwierawska A, Żmudzińska W, Ołdziej S, Liwo A, Scheraga HA. Proteins Struct Funct Bioinform. 2009;76:637–654. doi: 10.1002/prot.22377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lewandowska A, Ołdziej S, Liwo A, Scheraga HA. Proteins Struct Funct Bioinform. 2009 in press. [Google Scholar]
- 36.Kuszewski J, Clore GM, Gronenborn AM. Protein Sci. 1994;3:1945–1952. doi: 10.1002/pro.5560031106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kmiecik S, Kolinski A. Biophys J. 2008;94:726–736. doi: 10.1529/biophysj.107.116095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blanco FJ, Serrano L. Eur J Biochem. 1995;230:634–649. doi: 10.1111/j.1432-1033.1995.tb20605.x. [DOI] [PubMed] [Google Scholar]
- 39.Skwierawska A, Rodziewicz-Motowidło S, Ołdziej S, Liwo A, Scheraga HA. Biopolymers. 2008;89:1032–1044. doi: 10.1002/bip.21056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Derrick JP, Wigley DB. J Mol Biol. 1994;243:906–918. doi: 10.1006/jmbi.1994.1691. [DOI] [PubMed] [Google Scholar]
- 41.Dill KA. Biochemistry. 1990;29:7133–7155. doi: 10.1021/bi00483a001. [DOI] [PubMed] [Google Scholar]
- 42.Dzwolak W, Ravindra R, Lendermann J, Winter R. Biochemistry. 2003;42:11347–11355. doi: 10.1021/bi034879h. [DOI] [PubMed] [Google Scholar]
- 43.Honda S, Kobayashi N, Munekata E, Uedaira H. Biochemistry. 1999;38:1203–1213. doi: 10.1021/bi982271g. [DOI] [PubMed] [Google Scholar]
- 44.Huyghues-Despointes BM, Qu X, Tsai J, Scholtz JM. Proteins Struct Funct Bioinform. 2006;63:1005–1017. doi: 10.1002/prot.20916. [DOI] [PubMed] [Google Scholar]
- 45.Wei Y, Huyghues-Despointes BM, Tsai J, Scholtz M. Proteins Struct Funct Bioinform. 2007;69:258–296. [Google Scholar]
- 46.Fasman GD. Circular dichroism and the conformational analysis of biomolecules. Plenum Press; New York: 1996. p. 738. [Google Scholar]
- 47.Greenfield NJ. Anal Biochem. 1996;235:1–10. doi: 10.1006/abio.1996.0084. [DOI] [PubMed] [Google Scholar]
- 48.Provencher SW, Glockner J. 1981. pp. 33–37. [DOI] [PubMed]
- 49.Baxter NJ, Williamson MP. J Biomol NMR. 1997;9:359–369. doi: 10.1023/a:1018334207887. [DOI] [PubMed] [Google Scholar]
- 50.Wüthrich K. NMR of Proteins and Nucleic Acids. John Wiley Press; New York: 1986. [Google Scholar]
- 51.Muñoz V, Ghirlando R, Blanco JF, Jas GS, Hofrichter J, Eaton WA. Biochemistry. 2006;45:7023–7035. doi: 10.1021/bi052556a. [DOI] [PubMed] [Google Scholar]
- 52.Weikl TR, Dill KA. J Mol Biol. 2003;329:585–598. doi: 10.1016/s0022-2836(03)00436-4. [DOI] [PubMed] [Google Scholar]
- 53.Plotnikov V, Rochalski A, Brandts M, Brandts JF, Williston S, Frasca V, Lin LN. Assay Drug Dev Technol. 2002;1:83–90. doi: 10.1089/154065802761001338. [DOI] [PubMed] [Google Scholar]
- 54.Piantini U, Sørensen OW, Ernst RR. J Am Chem Soc. 1982;104:6800–6801. [Google Scholar]
- 55.Bax A, Freeman R. J Magn Reson. 1985b;65:355–360. [Google Scholar]
- 56.Bax A, Davis DG. J Magn Reson. 1985a;63:207–213. [Google Scholar]
- 57.Bartels C, Xia T, Billeter M, Güntert P, Wüthrich K. J Biomol NMR. 1995;6:1–10. doi: 10.1007/BF00417486. [DOI] [PubMed] [Google Scholar]
- 58.Tiers GVD, Coon RI. J Org Chem. 1961;26:2097–2098. [Google Scholar]
- 59.Güntert P, Braun W, Wüthrich K. J Mol Biol. 1991;217:517–530. doi: 10.1016/0022-2836(91)90754-t. [DOI] [PubMed] [Google Scholar]
- 60.Güntert P, Mumenthaler C, Wüthrich K. J Mol Biol. 1997;273:283–298. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- 61.Güntert P, Wüthrich K. J Biomol NMR. 1991;1:447–456. doi: 10.1007/BF02192866. [DOI] [PubMed] [Google Scholar]
- 62.Bystrov VF. Progr NMR Spectrosc. 1976;10:41–81. [Google Scholar]
- 63.Torda AE, Scheek RM, van Gunsteren WF. Chem Phys Lett. 1989;157:289–294. [Google Scholar]
- 64.Pearlman DA, Kollman PA. J Mol Biol. 1991;220:457–479. doi: 10.1016/0022-2836(91)90024-z. [DOI] [PubMed] [Google Scholar]
- 65.Case DA, Darden TA, Cheatham TE, III, Simmerling CL, Wang J, Duke RE, Luo R, Merz KM, Wang B, Pearlman DA, Crowley M, Brozell S, Tsui V, Gohlke H, Mongan J, Hornak V, Cui G, Beroza P, Schafmeister C, Caldwell JW, Ross WS, Kollman PA. AMBER8. University of California; San Francisco, CA: 2004. [Google Scholar]
- 66.Weiner SJ, Kollman PA, Nguyen DT, Case DA. J Comput Chem. 1987;7:230–252. doi: 10.1002/jcc.540070216. [DOI] [PubMed] [Google Scholar]
- 67.Mahoney MW, Jorgensen WL. J Chem Phys. 2000;112:8910–8922. [Google Scholar]
- 68.Ewald PP. Ann Phys. 1921;64:253–287. [Google Scholar]
- 69.Darden T, York D, Pedersen L. J Chem Phys. 1993;98:10089–10092. [Google Scholar]
- 70.Ryckaert JP, Ciccotti G, Berendsen HJC. J Comput Phys. 1977;23:327–341. [Google Scholar]
- 71.Koradi R, Billeter M, Wüthrich K. J Mol Graphics. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.