Abstract
Regulated expression of the genes for anthrax toxin proteins is essential for the virulence of the pathogenic bacterium Bacillus anthracis. Induction of toxin gene expression depends on several factors, including temperature, bicarbonate levels, and metabolic state of the cell. To identify factors that regulate toxin expression, transposon mutagenesis was performed under non-inducing conditions and mutants were isolated that untimely expressed high levels of toxin. A number of these mutations clustered in the heme biosynthetic and cytochrome c maturation pathways. Genetic analysis revealed that two heme-dependent, small c-type cytochromes, CccA and CccB, located on the extracellular surface of the cytoplasmic membrane, regulate toxin gene expression by affecting the expression of the master virulence regulator AtxA. Deregulated AtxA expression in early exponential phase resulted in increased expression of toxin genes in response to loss of the CccA-CccB signaling pathway. This is the first function identified for these two small c-type cytochromes of Bacillus species. Extension of the transposon screen identified a previously uncharacterized protein, BAS3568, highly conserved across many bacterial and archeal species, as involved in cytochrome c activity and virulence regulation. These findings are significant not only to virulence regulation in B. anthracis, but also to analysis of virulence regulation in many pathogenic bacteria and to the study of cytochrome c activity in Gram-positive bacteria.
Keywords: Bacillus anthracis, cytochrome, AtxA, pagA, resB, heme, CccB, CccA
Introduction
Bacteria are able to sense external signals and integrate these signals into regulatory circuits in order to adapt to and survive changes in environmental conditions. Environmental regulation in pathogenic bacteria is particularly important as they must recognize both metabolic and host-derived signals in order to promote bacterial growth in the host conditions. Bacillus anthracis presents an especially interesting system of environmental regulation as the organism leads a complex life cycle that includes periods of quiescence as spores in the external environment and periods of growth and pathogenesis once the bacteria finds itself in a suitable host. But even in the host, the bacteria must carefully balance growth, development, and virulence in order to propagate and disseminate. The complex regulatory networks controlling B. anthracis virulence and development are slowly being revealed (Perego and Hoch, 2008), but far too little is known of the mechanisms underlying the detection and propagation of host signals.
Both the anthrax toxin and the antiphagocytic capsule are essential to B. anthracis virulence. The tripartite anthrax toxin is encoded by three non-contiguous genes, lef, cya and pagA, carried on the virulence plasmid pXO1 (Okinaka et al., 1999). lef encodes Lethal Factor (LF), a zinc metalloprotease targeting host MAP-kinase signaling (Duesbery et al., 1998), cya encodes dema Factor (EF), an adenylate cyclase that increases cellular cAMP levels (Leppla, 1982), and pagA encodes Protective Antigen (PA), which forms a pore allowing entry of toxin components (Milne et al., 1994). The antiphagocytic poly-D-glutamic acid capsule, which is essential for bacterial dissemination in the host (Drysdale et al., 2005), is encoded by genes in the cap operon carried on virulence plasmid pXO2 (Makino et al., 1989). Transcription of the toxin and capsule genes is regulated by AtxA, encoded by the atxA gene on pXO1 (Dai et al., 1995;Uchida et al., 1997). Activity of AtxA is regulated by multiple regulatory circuits. Transcription of atxA is dependent on a dual promoter and repressed by the transition state regulator AbrB, which, in turn, is regulated by components of the sporulation phosphorelay (Saile and Koehler, 2002;Bongiorni et al., 2008). Post-translationally, the activity of AtxA protein is regulated through phosphorylation and dephosphorylation of two histidine residues (Tsvetanova et al., 2007).
Expression of B. anthracis virulence factors is stimulated by conditions suggestive of the host environment. Optimal toxin expression levels occur at 37°C in media supplemented with bicarbonate, in vitro conditions thought to mimic those of the mammalian host (Sirard et al., 1994;Ristroph and Ivins, 1983). The bicarbonate transporter is a required component of the regulatory pathway that controls the induction of toxin gene expression by elevated bicarbonate levels (Wilson et al., 2008b). However, toxin gene expression is regulated in the absence of bicarbonate by other, unknown factors.
In order to identify other regulatory pathways that regulate toxin expression, a transposon mutagenesis screen was performed that identified mutants overexpressing the protective antigen gene, pagA, under non-toxin inducing conditions. Several toxin-overexpressing mutations clustered in genes of the heme synthesis and cytochrome c maturation pathways. By extending the results of the mutagenesis screen through a combination of biochemical and genetic approaches, a novel regulatory pathway was identified that requires extracellular presentation of two small c-type cytochromes. These two cytochromes are required to repress transcription of the AtxA regulatory protein which, in turn, results in down regulation of toxin genes expression. Additionally, the transposon mutagenesis identified a highly conserved but previously uncharacterized protein that affects both cytochrome c activity and toxin gene expression.
Results
Identification of transposon mutants affecting toxin gene expression
The regulatory circuit required for control of virulence gene expression in B. anthracis is not fully understood as several regulatory inputs known to control toxin and capsule gene expression currently have no mechanistic basis. In order to probe for regulators of toxin gene expression in B. anthracis, the Sterne 3F42 strain (pXO1+ pXO2−) carrying a fusion of the protective antigen gene promoter (pagA) to the lacZ reporter on the replicative vector pTCVlac (Poyart and Trieu-Cuot, 1997) was mutagenized using the transposon delivery vector pAW016 (Wilson et al., 2007). pAW016 carries a mini-Tn10 transposon with a spectinomycin-resistance cassette on a temperature-sensitive replicative plasmid. Spectinomycin-resistant transposon insertion clones were screened for altered pagA expression by scoring for blue color on X-Gal plates in growth conditions non-inductive of toxin production (i.e. 37°C, LB in air without bicarbonate) (Leppla, 1988).
Among the pool of mutants that overexpressed pagA relative to the parental strain were a number of disruptions of genes predicted to be involved directly or indirectly in heme production or cytochrome c activity and one insertion in a gene encoding a hypothetical protein of unknown function (Table 2 and Fig. S1).
Table 2.
mini-Tn10 transposon mutants identified in the cytochrome c/heme pathways
| Strain Number |
Locus | Name/Annotation | Function |
pagA-lacZ Increase a |
atxA- lacZ Increase b |
TMPD staining c |
|---|---|---|---|---|---|---|
| tB3 | BAS4358* | glutamate-1-semialdehyde-2,1-aminomutase (HemL) |
heme synthesis | + | E | +/− |
| tB17 | BAS4358* | glutamate-1-semialdehyde-2,1-aminomutase (HemL) |
heme synthesis | + | E | +/− |
| tB18 | BAS1385 | ResC | cytochrome c maturation |
++ | E | − |
| tB21 | BAS1384* | ResB | cytochrome c maturation |
++ | E | − |
| tB23 | BAS3568 | conserved hypothetical protein | unknown | ++ | E | +/− |
| tB28 | BAS1384* | ResB | cytochrome c maturation |
++ | E | − |
multiple strains with unique transposon insertions into the same gene
: + = pagA-lacZ expression higher than parental strain; ++ = pagA-lacZ expression much higher than parental strain
: E = induced atxA-lacZ expression in exponential phase
: +/− =staining reduced relative to parental strain; − = no staining
Insertions were found in:
BAS4358 (GenBank: AAT56656.1) encoding a protein that shares 80% amino acid identity with the product of the B. subtilis hemL gene (GenBank: AAA22515.1). In B. subtilis, HemL converts glutamate-1-semialdehyde to 5-aminolevulinic acid (5-ALA) required for heme biosynthesis. In the absence of hemL, a small amount of 5-ALA is produced spontaneously from glutamate-1-semialdehyde resulting in reduced heme production (Hansson et al., 1991). As in B. subtilis, BAS4358 is the last gene in the heme biosynthetic cluster. Two different insertions into BAS4358 were isolated and both insertions resulted in increased pagA expression.
BAS1384 (GenBank: AAT53704.1) encoding a protein with 62% amino acid identity with the product of the B. subtilis resB gene (GenBank: AAA67495.1). ResB is an integral membrane protein that is required for cytochrome c assembly and maturation (Sun et al., 1996;Le Brun et al., 2000). ResB is similar to other System II cytochrome c maturation proteins, such as the Ccs1 protein of Chlamydomonas reinhardtii and CcsB of Bordetella pertussis, and likely functions in a complex with ResC to export and process heme for extracellular attachment to cytochrome c (Beckett et al., 2000;Hamel et al., 2003). Two independent insertions into BAS1384 were isolated occurring at two unique locations, and both insertions resulted in increased pagA expression. As in B. subtilis, BAS1384 is located between the genes resA and resC.
BAS1385 (GenBank: AAT53705.1) encoding a protein with 71% amino acid identity to the product of the B. subtilis resC gene (GenBank: CAB14245.1). ResC, like ResB, is an integral membrane protein similar to other System II cytochrome c maturation proteins (Le Brun et al., 2000). As in B. subtilis, BAS1385 is located downstream of the genes annotated as resA and resB.
BAS3568 (GenBank: AAT55872.1) encoding a predicted membrane protein with four transmembrane helices containing the PFam DUF420 domain. This gene is well conserved among many Gram-positive bacteria, and its closest ortholog in B. subtilis is the uncharacterized yozB gene (amino acid identity 52.4%, similarity 69.8%).
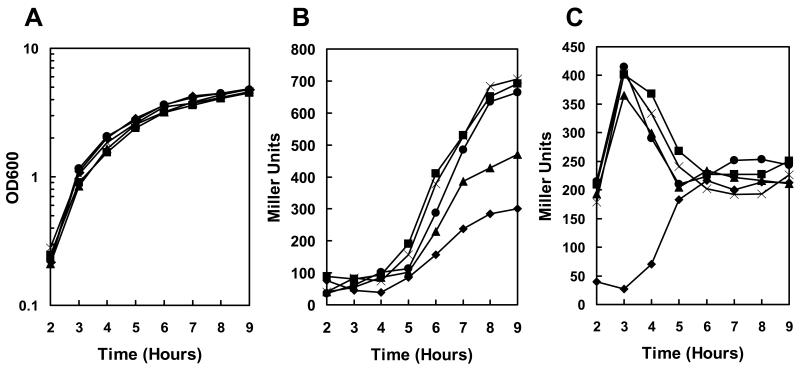
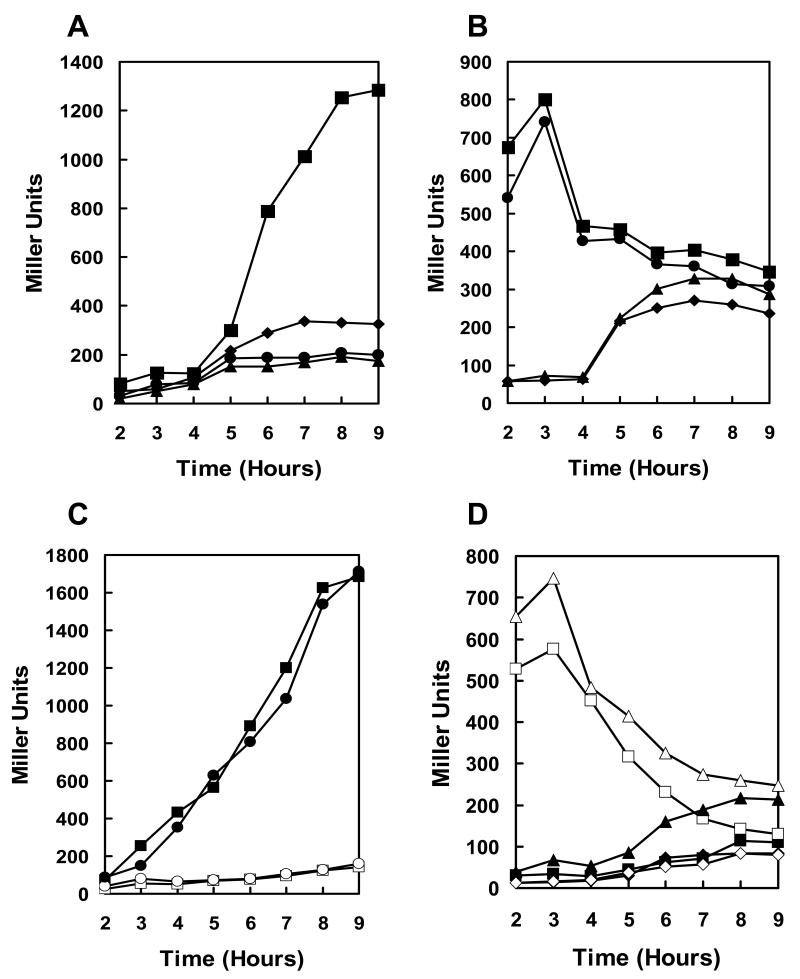
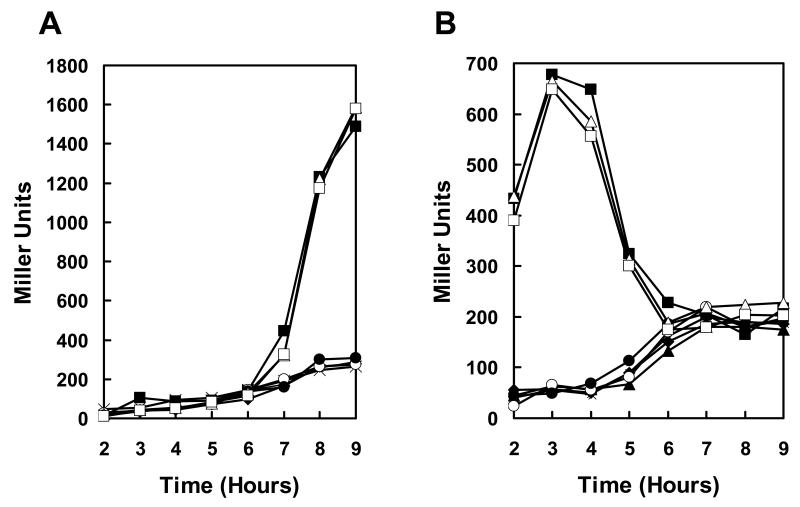
Quantitation of expression in these transposon mutant strains by means of β-galactosidase assays showed that the rate of induction of pagA was 4 fold higher in the resB, resC, and BAS3568 mutants and 2.5 fold higher in the hemL mutant compared to the parental strain while the growth rate was unaffected (Fig. 1A and B).
Figure 1.
Transcription analysis of pagA and atxA expression in transposon mutant strains. Strains carrying a pagA-lacZ or atxA-lacZ fusion on the replicative vector pTCV-lac were grown in LB broth supplemented with kanamycin at 37°C. β-galactosidase assays were carried out on samples taken at hourly intervals as indicated. A. Cell growth of pagA-lacZ reporter strains (cell growth of atxA-lacZ reporter strains was similar) B. β-galactosidase activity of pagA-lacZ reporter strains. C. β-galactosidase activity of atxA-lacZ reporter strains. Symbols in all three panels: -◆- 34F2; -▪- 34F2tB18 (resC); -X- 34F2tB21 (resB); -●- 34F2tB23 (BAS3568); -▲- 34F2tB24 (hemL).
The clustering of transposon mutations deregulating pagA expression in genes involved in heme synthesis and cytochrome c maturation strongly suggested a role for respiration in environmental sensing and toxin production.
Heme-defective mutants overexpress atxA during exponential growth
Experiments were undertaken to distinguish whether the transposon mutants affected pagA expression directly or were influencing the global virulence regulator AtxA required for toxin gene expression in B. anthracis (Koehler et al., 1994;Uchida et al., 1993). To investigate the effect of each transposon mutation on atxA transcription, the mutant strains were cured of the pTCVlac-pagA plasmid and re-transformed with the pTCVlac-atxA reporter plasmid, resulting in strains expressing β-galactosidase from the atxA promoter. Results of a time course experiment monitoring atxA expression in the transposon mutants and parental strain grown in LB broth containing kanamycin are shown in Figure 1C. The parental strain expressed atxA at low level in early exponential phase and an increase in expression occurred at mid exponential phase (Dai et al., 1995). The resB (34F2tB21), resC (34F2tB18), hemL (34F2tB3) and BAS3568 (34F2tB23) transposon mutants expressed atxA at very high levels during early exponential growth but expression dropped to near parental levels once the cells reached stationary phase. To the best of our knowledge, this pattern of exponential phase deregulation of atxA transcription followed by a return to normal expression levels in stationary phase has not previously been observed and is unique to these mutants.
Characterization of markerless, in-frame deletion mutants
The genetic characterization of transposon-generated mutants could be affected by stability or polarity issues that may interfere with the interpretation of the results. Thus, strains containing markerless deletions of transposon-identified genes were constructed to confirm the pagA- and atxA-overexpression phenotypes identified with the transposon mutants. All deletion strains were produced using a modification of the method of Janes and Stibitz (Janes and Stibitz, 2006). This method results in a markerless, in-frame deletion of the gene of interest, thereby reducing the possibility of polar effects on the expression of downstream genes that can be associated with transposon insertions.
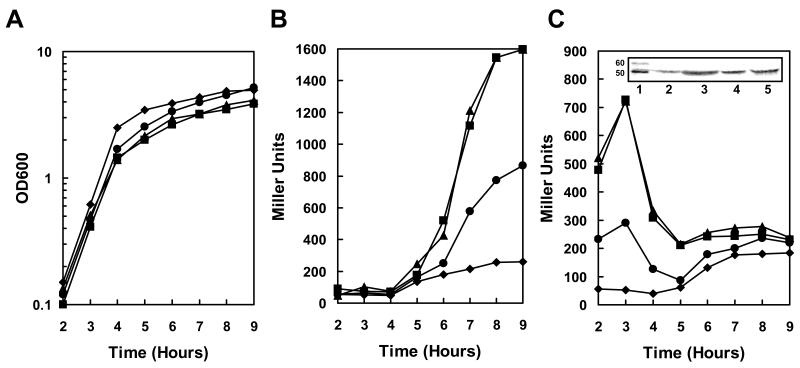
A markerless deletion of B. anthracis BAS1384, resulting in strain 34F2△resB, was transformed with the pTCVlac-pagA or pTCVlac-atxA plasmids to test the effect of resB deletion on pagA and atxA expression. Unlike the transposon mutant 34F2 tB21(Figure 1A), the markerless deletion mutant 34F2△resB showed a modest growth defect relative to the parental strain (Figure 2A). 34F2△resB also expressed pagA at a 4 fold higher rate than the parental strain after induction at the transition phase (Figure 2B) and approximately 10-fold more atxA than the parental strain during early exponential phase (Figure 2C). Consistently, the AtxA protein was overexpressed in the 34F2△resB strain during exponential phase as shown by Western Blot analysis (Fig. 2C and Fig. S2). These data confirmed the contribution of resB to the atxA and pagA overexpression phenotypes observed in the transposon mutants.
Figure 2.
Transcription analysis of pagA and atxA expression in resB, resBC and hemL mutant strains. Strains carrying a pagA-lacZ or atxA-lacZ fusion on the replicative vector pTCV-lac were grown in LB broth supplemented with kanamycin at 37°C. β-galactosidase assays were carried out on samples taken at hourly intervals as indicated. A. Cell growth of pagA-lacZ reporter strains (cell growth of atxA-lacZ reporter strains were similar) B. β-galactosidase activity of pagA-lacZ reporter strains. C. β-galactosidase activity of atxA-lacZ reporter strains. Symbols in all three panels: -◆- 34F2; -▪- 34F2△resB; -▲- 34F2△ resBC; -●- 34F2△hemL. The inset in panel C represents the Western blot analysis of AtxA on B. anthracis cell lysates collected after 3 and 8 hr of growth in LB Broth at 37°C. The amount of sample loaded on a 10% SDS-PAGE was normalized relative to cell growth. Lane 1: Magic Mark XP (Invitrogen); Lane 2: 34F2 after 3 hr of growth, Lane 3: 34F2△resB after 3 hr of growth; Lane 4: 34F2 after 8 hr of growth; Lane 5: 34F2△resB after 8 hr of growth. A full size of this Western blot is shown in Fig. S1.
Transposon insertions obtained in both resB and resC resulted in similar levels of pagA and atxA overexpression. By analogy to the similar cytochrome c maturation proteins in B. subtilis, expression of both ResB and ResC in B. anthracis is most likely required to form an active heme delivery complex and ResC expressed in the absence of ResB should be inactive (Hamel et al., 2003). To test for the effect of resC deletion in B. anthracis, a markerless resBC deletion strain was constructed that completely deleted the coding regions of both BAS1384 and BAS1385. 34F2△resBC grew similarly to the 34F2△resB strain, both growing to a final density less than the parental strain (Figure 2A). The expression of pagA (Fig. 2B) and atxA (Fig. 2C) was identical to the 34F2△resB single deletion strain, demonstrating that deletion of resBC had no additional effect over deletion of resB alone.
A strain containing a deletion of resA, a gene in the resABC operon encoding a thiol-disulfide oxidoreductase involved in cytochrome c maturation (Erlendsson et al., 2003), was also constructed (34F2△resA). When compared to 34F2△resB, this strain showed a similar pattern of atxA- and pagA-overexpression (data not shown), further indicating that disruption of cytochrome c maturation contributes to the observed virulence expression phenotypes.
The markerless deletion of hemL (BAS4358) resulted in increased pagA expression during stationary phase (Figure 2B) and increased atxA expression during early exponential growth (Figure 2C), similar to the transposon insertion mutants isolated in hemL. Though the pattern of overexpression in 34F2△hemL was similar to 34F2△resB, the level of β-galactosidase expression was lower in the 34F2△hemL, consistent with a reduction, but not elimination, of heme production (Hansson et al., 1991).
The analysis of markerless, in-frame deleted strains confirmed the original phenotypes of the resB, resC, and hemL transposon-generated mutants and indicated they were indeed the result of loss-of-function mutations.
Activity of cytochrome c oxidase in the transposon mutants affected in toxin gene expression
The B. anthracis orthologs of B. subtilis heme synthesis and cytochrome c maturation proteins identified in our transposon mutagenesis screen had not been studied previously, so it was essential to demonstrate that these gene products contribute to cytochrome c activity in this organism. A simple test for cytochrome c oxidase activity involves N,N,N’,N’-tetramethyl-p-phenylenediamine (TMPD) staining of B. anthracis colonies grown on solid media plates. TMPD is an artificial electron donor that interacts specifically with cytochrome c oxidase (cytochrome caa3) and is oxidized to a blue colored product that stains colonies in the presence of active cytochrome c oxidase in the membrane (Le Brun et al., 2000).
B. anthracis parental and mutant strains were grown on NSMP and visually scored for TMPD oxidase staining (Table 2 and Fig. 3). A mutant in the CtaC subunit of cytochrome caa3 was also constructed as a negative control (see below). As expected for a subunit of cytochrome c oxidase, disruption of ctaC (BAS3856) resulted in no TMPD oxidase staining, demonstrating a lack of cytochrome c oxidase activity. The resB (BAS1384) and resC (BAS1385) mutants also did not produce active cytochrome c oxidase, consistent with observations of orthologous gene disruptions in B. subtilis (Le Brun et al., 2000) (Table 2). The hemL (BAS4358) mutant showed a reduction in TMPD oxidase staining indicative of reduced, rather than abolished, cytochrome c oxidase activity, consistent with B. subtilis observations that heme production occurs at reduced levels in the absence of hemL (Hansson et al., 1991) (Table 2 and Fig. 3). As heme is required for cytochrome c oxidase activity, a reduction in cellular levels of the heme precursor 5-aminolevulinic acid (5-ALA) (Hansson et al., 1991) in the absence of hemL would result in reduced TMPD oxidase staining. Consequently, complementation of the oxidase defect and pagA overexpression phenotype in the hemL mutant was obtained by the addition of 5-ALA to the medium, while no complementation was observed for the 34F2△ctaC mutant (Fig. 3 and Fig. S1). These results confirmed that the hemL, resB, and resC genes of B. anthracis are required for cytochrome c maturation and their deletion results in toxin gene overexpression.
Figure 3.
Analysis of cytochrome c oxidase activity in B. anthracis parental and hemL mutant strains using TMPD oxidase staining. Strains were isolated on NSMP-agar plates in the absence (−) or presence (+) of 5 mg/ml 5-ALA. Upon exposure to TMPD, the staining for oxidase activity of strain 34F2 (full activity) or 34F2△ctaC (lack of activity) was not affected by the addition of 5-ALA. The partial staining of strain 34F2△hemL in the absence of 5-ALA was increased to the level of the parental strain when 5-ALA was present in the medium.
Increased pagA expression is AtxA-dependent
Inactivation of hemL, resB, or resC resulted in increased transcription of both pagA and atxA (Figs. 1 and 2). Because AtxA is required to activate transcription of pagA (Koehler et al., 1994;Uchida et al., 1993), increased atxA transcription in these mutant strains may directly lead to increased pagA transcription. However, it was reported that when AtxA production was increased by expressing the gene from a multi-copy plasmid, pagA expression actually decreased (Dai and Koehler, 1997), indicating that, under some conditions, increased AtxA expression did not lead directly to increased pagA expression.
To evaluate the role of AtxA in pagA overexpression in these mutants, a markerless deletion of resB was made in a strain containing an insertional inactivation of the atxA gene (Tsvetanova et al., 2007). This strain was transformed with the pagA-lacZ and atxA-lacZ reporter plasmids, and pagA and atxA expression was compared to the parental and single deletion strains. Deletion of atxA alone resulted in decreased pagA expression relative to the parental strain (Figure 4A) but did not significantly effect atxA expression (Figure 4B). The resB-atxA double deletion strain expressed pagA at reduced levels similar to the atxA deletion alone (Figure 4A), suggesting that AtxA is required for pagA overexpression in the resB mutant. Moreover, the resB-atxA double deletion strain expressed atxA at increased levels during exponential growth similar to the resB deletion alone (Figure 4B), indicating that AtxA is not required for its own overexpression in the resB mutant.
Figure 4.
Transcription analysis of pagA and atxA expression in resB and atxA-resB mutant strains. β-galactosidase assays were carried out on samples taken at hourly intervals as indicated. A. and B. Strains carrying a pagA-lacZ or atxA-lacZ fusion on the replicative vector pTCV-lac were grown in LB broth supplemented with kanamycin at 37°C. A. pagA-lacZ reporter strains. B. atxA-lacZ reporter strains. Symbols in both panels: -◆- 34F2; -▪- 34F2△resB; -▲- 34F2△atxA; -●- 34F2△atxA△resB. C. The 34F2△atxA strain carrying an isotopically integrated pagA-lacZ reporter was transformed with plasmids pTCV-lac-spac or the pTCV-spac-AtxA and β-galactosidase activity was determined. Symbols: -▪- 34F2△atxA::pagA-lacZ/pTCV-spac-AtxA; -□- 34F2△atxA::pagA-lacZ/pTCV-spac; -●- 34F2△atxA△resB::pagA-lacZ/pTCV-spac-AtxA; -□- 34F2△atxA△resB::pagA-lacZ/pTCV-spac. D. The resB deletion affects only the P1 promoter of atxA. The 34F2 and 34F2△resB strains were transformed with different atxA-lacZ fusion constructs in pTCV-lac (Bongiorni et al., 2008) and β-galactosidase activity was measured. Symbols: -▪- 34F2/pAtxA10; -□- 34F2△resB/pAtxA10; -▲- 34F2/pAtxA12; -△- 34F2△resB/pAtxA12; -◆- 34F2/pAtxA20; -◇- 34F2△resB/pAtxA20.
resB mutant effect on AtxA
It was important to determine whether the resB-dependent induction of pagA expression was dependent on atxA induced expression or whether it depended on an effect on AtxA activity. To test these possibilities, the atxA coding gene was placed under control of a non-inducible (constitutively expressed) spac promoter in a modified pTCV-lac plasmid (see Experimental Procedures). This construct was then transformed in a strain carrying a pagA-lacZ reporter fusion integrated isotopically in the pX01 plasmid of strains 34F2 △atxA and 34F2△atxA△resB (see Experimental Procedures). Transcription analysis by β-galactosidase assays (Fig. 4C) indicated that expression of AtxA from the spac promoter complemented the atxA deletion on pX01, but did not result in pagA overexpression in the absence of resB compared to the parental strain. These results indicated that the resB-dependent induction of pagA appears solely due to the induction of atxA transcription and not to a modification of its activity.
Because atxA is transcribed from two independent promoters, we determined which promoter(s) was affected in the resB mutant. Transcriptional analysis was carried out using atxA-lacZ reporter constructs carrying only the proximal P1 promoter (pAtxA10), both the proximal and distal P1 and P2 promoters (pAtxA12) or only the distal P2 promoter (pAtxA20) (Bongiorni et al., 2008). The results shown in Fig. 4C indicated that induction of atxA transcription in early exponential phase in the resB mutant occurred only at the proximal P1 promoter.
These results showed that the induction of pagA expression resulting from a deletion of resB was solely the consequence of deregulated atxA expression occurring at the P1 promoter and did not involve regulation of AtxA activity.
Inactivation of two B. anthracis small c-type cytochromes is required for atxA-overexpression
In B. subtilis, 5 membrane-bound proteins with covalently bound heme have been identified: the 39 kDa subunit II of cytochrome caa3 (CtaC), the 28 kDa cytochrome c of the bc complex (QcrC), the 25 kDa cytochrome b subunit of the cytochrome bc complex (QcrB), the 13 kDa cytochrome c550 (CccA), and the 10 kDa cytochrome c551 (CccB) (von Wachenfeldt and Hederstedt, 1990;Bengtsson et al., 1999). ResBC is required for synthesis of all of these cytochromes except the 25 kDa cytochrome b subunit of the cytochrome bc complex (QcrB) (von Wachenfeldt and Hederstedt, 2002). By BlastP analysis we have identified B. anthracis orthologs of all 5 B. subtilis membrane-bound proteins with covalently bound heme. BAS3856 (AAT56157) shares 53% amino acid identity with CtaC (NP_389372), BAS1433 (AAT53753) shares 94% amino acid identity with QcrB (NP_390136), BAS1434 (AAT53754) shares 71% amino acid identity with QcrC (NP_390135), BAS4193 (AAT56492) shares 46% amino acid identity with CccA (NP_390398), and BAS5035 (AAT57324) shares 41% amino acid identity with CccB (NP_390135).
To investigate the roles of these proteins in virulence gene regulation, B. anthracis 34F2 derivative strains were generated containing markerless deletions of each gene (henceforth, each B. anthracis deletion strain will be annotated after the orthologous gene in B. subtilis). Heme attachment to QcrB does not appear to be ResBC-dependent, but given the paucity of information of QcrB and QcrC function and their operon organization on the chromosome, both coding genes were deleted in a single strain (34F2△qcrBC). Single deletion strains showed no significant growth defect and only the 34F2△ctaC strain was deficient in TMPD-oxidase staining (Fig 3 and data not shown). When transformed with the lacZ reporter plasmids, none of the single deletion strains overexpressed pagA or atxA relative to the parental strain (Figure 5).
Figure 5.
Transcription analysis of pagA and atxA expression in cytochrome c deletion strains. Strains carrying a pagA-lacZ or atxA-lacZ fusion on the replicative vector pTCV-lac were grown in LB broth supplemented with kanamycin at 37°C. β-galactosidase assays were carried out on samples taken at hourly intervals as indicated. A. pagA-lacZ reporter strains. B. atxA-lacZ reporter strains. Symbols in both panels: -◆- 34F2; -▪- 34F2△resB; -▲- 34F2△ctaC; -X- 34F2△qcrBC; -○- 34F2△cccA; -●- 34F2△cccB; -△- 34F2△cccA-B; -□- 34F2△5cyt
Exploiting the advantages of the markerless deletion method, a single strain containing deletions of all 5 cytochrome genes (34F2△5cyt) was generated. This strain, unlike the single deletion strains, overexpressed atxA and pagA in a pattern identical to the 34F2△resB strain, suggesting that loss of multiple cytochromes is required for deregulation of atxA and pagA expression (Fig. 5).
The small c-type cytochromes CccA and CccB share 58% amino acid similarity overall, with the extracellular heme-attachment domains being the most conserved region, (Fig. S3), hinting that these two proteins may functionally overlap. A double deletion strain, cccA cccB (34F2△cccA-B) fully mimicked the pagA- and atxA-overexpression phenotype of both the 34F2△resB and 34F2△5cyt strains. While loss of either cccA or cccB did not affect toxin gene expression, the deletion of both genes simultaneously was necessary to deregulate pagA and atxA, suggesting that CccA and CccB can functionally complement one another.
A previously uncharacterized transmembrane protein contributes to regulation of atxA and pagA expression
One additional strain, 34F2tB23, was identified which overexpressed pagA in stationary phase and overexpressed atxA in exponential phase (Table 2 and Figure 1). This strain was partially deficient in TMPD-oxidase staining, suggesting a deficiency in cytochrome c oxidase function (data not shown). The transposon insertion in strain 34F2tB23 disrupted the BAS3568 gene. A markerless deletion strain (34F2△BAS3568) retained the atxA- and pagA-overexpression phenotype and reduction of TMPD-oxidase staining of the transposon mutant strain (Fig. 6 and data not shown), demonstrating that disruption of BAS3568 is responsible for the observed phenotypes. As with a resB-atxA double mutant strain, a BAS3568-atxA double disruption strain expressed pagA at levels identical to the atxA disruption strain (data not shown), which indicated that pagA overexpression in the BAS3568 strain is also atxA-dependent. Deletion of both BAS3568 and resB resulted in overexpression of pagA and atxA at levels similar to either of the single deletion strains alone (Fig. 6). The lack of an additive effect of resB and BAS3568 deletions suggests that both protein products function in the same pathway. Thus, the product of the BAS3568 gene is a newly identified component required for full cytochrome c activity whose function remains to be determined.
Figure 6.
Transcription of pagA and atxA in BAS3568 and resB mutant strains. Strains carrying a pagA-lacZ or atxA-lacZ fusion on the replicative vector pTCV-lac were grown in LB broth supplemented with kanamycin at 37°C. β-galactosidase assays were carried out on samples taken at hourly intervals as indicated. A. pagA-lacZ reporter strains. B. atxA-lacZ reporter strains. Symbols in both panels: -▲- 34F2; -▪- 34F2△resB; -▲- 34F2△BAS3568; -●- 34F2△resB△BAS3568
Bicarbonate induction of toxin expression eliminates the cytochrome-deficient phenotype
The initial transposon mutagenesis screen and subsequent analyses were performed on strains grown in the absence of added bicarbonate or CO2, conditions which do not induce high level of toxin gene expression.
To investigate the role of this CccA-CccB newly identified heme-dependent regulatory pathway during bicarbonate-induced toxin induction, pagA and atxA expression levels were monitored in 34F2 and 34F2△resB strains grown in R-Media (Ristroph and Ivins, 1983) both with and without added bicarbonate. As shown in Figure 7, when grown in R-Media without added bicarbonate at 37° under 5% atmospheric CO2, the 34F2△resB strain overexpressed pagA relative to the parental strain. The timing and extent of pagA overexpression was different from what was observed in strains grown in LB broth under air as the overexpression began much earlier and the extent of overexpression was reduced (approximately 2-fold in R-Media without NaHCO3 versus approximately 4-fold in LB-broth). In the parental strain, expression of pagA was increased and expression was induced earlier in R-Media without bicarbonate, compared to growth in LB. When these strains were grown in R-Media with added 0.8% NaHCO3, pagA expression was strongly induced, as expected, increasing almost 15-fold over the parental strain grown in the absence of added NaHCO3. With bicarbonate induction, however, there was no difference in expression of pagA between the parental and resB mutant strains.
Figure 7.
Transcription analysis of pagA and atxA expression under toxin-inducing growth conditions. Strains carrying a pagA-lacZ or atxA-lacZ fusion on the replicative vector pTCV-lac were grown in R-Media with or without added NaHCO3 supplemented with kanamycin at 37°C under 5% atmospheric CO2. β-galactosidase assays were carried out on samples taken at hourly intervals as indicated. A. pagA-lacZ reporter strains. B. atxA-lacZ reporter strains. Symbols in both panels: -▪- 34F2 without added NaHCO3; -□- 34F2 with 0.8% NaHCO3; -○- 34F2△resB without added NaHCO3; -●- 34F2△resB with 0.8% NaHCO3.
The expression of atxA in 34F2△resB was elevated early in R-Media without bicarbonate, similar to the expression pattern seen in LB broth in air. However, early overexpression of atxA in the 34F2△resB strain was lost when cells were grown in the presence of bicarbonate. Expression of atxA is not induced by bicarbonate (Dai and Koehler, 1997;Bongiorni et al., 2008), but these expression data clearly demonstrated that addition of bicarbonate to growth media eliminates the atxA overexpression phenotype of a resB mutant.
Discussion
B. anthracis regulates virulence gene expression by recognizing signals from the host environment. The mechanistic basis of in vivo and ex vivo toxin induction by elevated temperature (Dai and Koehler, 1997) and bicarbonate (Wilson et al., 2008b;Bartkus and Leppla, 1989) have become clearer in recent years, though much remains unknown about the connections between signals and regulatory pathways. While induction of virulence gene expression is vital to pathogenesis, repression of virulence gene expression may also be important for survival and replication of the organism in other conditions. In this work, we investigated a novel regulatory pathway that represses virulence gene expression under ex vivo non-host conditions.
This study has revealed a specific function in virulence regulation for two small highly conserved c-type cytochromes, CccA and CccB, in the Bacillus genus. Deletion of the genes for CccA and CccB of B. anthracis resulted in a transient deregulation of expression of the gene for the AtxA virulence regulator in the early exponential phase of growth. In turn, this deregulation gave rise to an increased rate of toxin gene expression (pagA). Increased expression was not limited to toxin genes but extended to other AtxA-regulated genes such as pX02-61 (Bourgogne et al., 2003;White et al., 2006) and was also detected when cells were grown at 25°C (Dai and Koehler, 1997) (data not shown). Moreover, the study identified a new putative component of the cytochrome c maturation pathway in the product of the BAS3568 gene which is well conserved among Gram-positive bacteria but whose function has remained unknown until now (Fig. 8).
Figure 8.
Schematic representation of the small c-type cytochrome pathway regulating virulence gene expression in B. anthracis. Intracellularly synthesized heme, through a pathway that requires the Hem proteins including HemL, is transported across the membrane by the ResBC proteins (Ahuja et al., 2007) and is covalently attached to CccA and CccB with the involvement of ResA (Le Brun et al., 2000). Either CccA or CccB can then act to indirectly repress atxA transcription at the P1 promoter and their function is redundant. The BAS3568 protein, which is required for full cytochrome oxidase activity, is also involved in repression of atxA transcription in early exponential phase by an unknown mechanism (indicated by the broken arrow) likely to act on the cytochrome c maturation pathway. The BAS3568 protein does not share amino acid similarity with the Res proteins. The sidedness of the membrane is indicated by “in” and “out”.
Little is known about cytochromes and their maturation in B. anthracis but cytochrome c maturation systems (ccm) have been studied in other organisms. Production of cytochrome c has recently been shown to contribute to a variety of other processes in specific bacteria, for example iron acquisition in Rhizobium leguminosarum (Yeoman et al., 1997) and virulence in Legionella pneumophila (Naylor and Cianciotto, 2004). Ccm systems in Gram-positive bacteria are distinct from those found in Gram-negative bacteria and more closely resemble the ccm systems in chloroplasts and cyanobacteria (Kranz et al., 1998). In B. subtilis, apo-cytochrome c is produced inside the cell and exported to the extracellular face of the cytoplasmic membrane (Fig. S4) (Crow et al., 2005). Once at the extracellular surface, the ResA, CcdA, and BdbD proteins cooperate to maintain the two cysteine residues in the CXXCH heme-attachment motif in the reduced state (Schiött et al., 1997;Erlendsson and Hederstedt, 2002). ResB and ResC likely cooperate to transport heme across the cytoplasmic membrane and attach the heme via two thioester bonds to reduced apo-cytochrome c to generate the mature cytochrome c (Le Brun et al., 2000).
CccA and CccB are similar in the amino acid sequence of their cytochrome c domains (Fig. S3) but differ in their membrane anchoring systems: CccA has a predicted α-helical transmembrane polypeptide membrane anchor while CccB is membrane attached by a diacylglycerol membrane anchor (Bengtsson et al., 1999). The reason for conservation of two highly similar small c-type cytochromes with no known function in Bacillus species when such arrangements are uncommon in other bacteria is still a mystery. In B. anthracis, the two cytochromes are clearly involved in a signaling pathway to virulence gene expression but their precise function is still unknown (Fig. 8). As is typical for c-type cytochromes, the function of CccA and CccB is most likely redox-related and associated with electron transfer (von Wachenfeldt and Hederstedt, 2002), but they are not required for aerobic respiration as growth is not affected. Deletion of both genes is required for atxA overexpression, suggesting one cytochrome can compensate for the loss of the other for this pathway. None of the remaining c-type cytochromes (CtaC and QcrBC) are involved, on an individual basis, in the regulation of atxA expression indicating a specific participation of CccA and CccB in virulence gene expression, at least, in B. anthracis.
The activity of CccA and CccB on virulence gene expression requires heme and its covalent attachment to these two cytochromes as indicated by the requirement for HemL and ResABC to regulated atxA expression in the early exponential phase of growth. A deletion of the hemL gene only partially affected cytochrome c oxidase activity but deregulated atxA expression suggesting that in conditions of low heme availability (as in the case of an hemL mutation) the CccA and CccB cytochromes may not have the highest affinity for the enzymes, presumably ResBC, carrying out the heme binding reaction to the apo-cytochromes. Complementation of the hemL deficiency by the 5-ALA intermediate in heme biosynthesis fully complemented the mutation, confirming the requirement of heme for the cytochrome activity on atxA transcription. The transient deregulation of atxA expression in the resBC mutants also confirmed that not only heme but also its presentation and covalent attachment to the cytochromes are necessary to regulate virulence gene transcription.
Our results also revealed that the protein encoded by BAS3568 is involved in cytochrome c maturation and, consequently, virulence regulation. BAS3568 is required for full cytochrome c oxidase function as its deletion resulted in partial loss of TMPD-oxidase activity. The BAS3568 product likely functions more generally in cytochrome c maturation as both cytochrome c oxidase and virulence expression were disrupted when the gene was deleted, indicative of loss of caa3, c550 and c551 activity. As atxA- and pagA-overexpression levels were lower than in the resB mutant and a very low level of TMPD-oxidase staining was maintained, loss of BAS3568 strongly reduced but did not completely eliminate cytochrome c maturation. The role of the BAS3568 ortholog yozB in B. subtilis is unknown but our results may reveal much more about the function of this previously uncharacterized protein.
In B. subtilis, the ResDE two-component signal transduction system regulates a number of genes involved in aerobic and anaerobic respiration, including the resABC operon (Sun et al., 1996). Deletion of resDE, in our hands, had no effect on either pagA or atxA transcription, demonstrating that this two-component regulatory system plays no role in the phenotype observed for resB deletion or in regulation of toxin expression (Wilson et al., 2008a;Vetter and Schlievert, 2007). Either the ResDE regulatory pathway in B. anthracis differs from B. subtilis or resABC expression in the absence of the two-component system is sufficient for maturation of the CccA and/or CccB in the growth conditions tested.
The question arising from this work is the mechanism by which elevated early exponential phase transcription of atxA leads to elevated stationary phase transcription of pagA. When atxA was expressed in its normal context from a multi-copy plasmid, overexpression of AtxA led to decreased pagA expression (Koehler et al., 1994;Dai and Koehler, 1997). In contrast, overexpression of atxA early in exponential phase induced by cytochrome c deficiency led to increased pagA expression but not changes in the timing of pagA expression. Also, increased atxA expression in an abrB mutant resulted in increased pagA expression throughout the growth cycle although the transcription patterns are significantly different from the ones observed in the cytochrome pathway deletion mutants (Fig. S5) (Saile and Koehler, 2002). These observations suggest that the timing or context of AtxA expression, not necessarily the total amount of AtxA produced, influences the regulatory activity of AtxA. AtxA produced at different stages of growth may become differentially modified in a way that influences activity, perhaps via phosphorylation/dephosphorylation of the histidine residues we have previously investigated (Tsvetanova et al., 2007). Notably, a resB mutation results in overexpression of pagA even in the presence of the AtxA H199D and H379A mutant proteins confirming that the cytochrome c deficiency only affects atxA gene expression (data not shown).
The response to bicarbonate is clearly essential to B. anthracis virulence regulation and the present results indicate that bicarbonate signals both activation and repression of toxin expression. The bicarbonate activation pathway, which relies on bicarbonate import through an ABC transporter (Wilson et al., 2008b), eliminated the atxA and pagA overexpression phenotype of the resB mutant, suggesting that the bicarbonate activation pathway completely overrides the regulatory pathway exerted by the CccA-B cytochromes. The transient nature of the overexpression phenotype resulting from the absence of the CccA and CccB cytochromes potentially suggests the involvement of an inducer of transcription which the bicarbonate response would neutralize. Alternatively, increased AtxA expression may induce a gene that feeds back to repress AtxA. However, the involvement of multiple negative regulators cannot be ruled out at this time. Further, activation requires the intracellular presence of bicarbonate (Wilson et al., 2008b) while these data suggest the involvement of extracellular signal(s) propagated by the two cytochromes, indicating B. anthracis can regulate the response to bicarbonate from both intra- and extra-cellular signals.
The pathway of virulence gene expression regulation identified by this study adds another level of control to the already complex AtxA regulatory circuit. AtxA plays an important role in the induction of pagA expression, but transcription of atxA is not induced by bicarbonate (Dai and Koehler, 1997;Bongiorni et al., 2008;Wilson et al., 2008b). Conversely, repression of pagA in the cytochrome c-dependent pathway and non-host conditions does operate through regulation of atxA transcription.
Our findings highlight the role of integrated metabolic systems in directing the expression of virulence genes. Rather than commit to a dedicated and metabolically expensive virulence regulatory system, many bacteria use or enhance existing metabolic pathways that already sense changes in the environment to regulate expression of genes required for growth in the host environment. There are already several examples of metabolic integration of virulence regulation in B. anthracis, beyond the new system presented here, such as the connection of toxin expression to the sporulation phosphorelay through AbrB (Saile and Koehler, 2002), regulation of toxin in response to bicarbonate import (Wilson et al., 2008b), and the influence of the histidine phosphorylation on AtxA activity (Tsvetanova et al., 2007). The mechanism connecting cytochrome c activity to virulence regulation is an on-going topic of investigation, but these studies could prove valuable in the study of other Gram-positive pathogenic bacteria that share conserved regulatory circuits.
Experimental Procedures
Bacterial strains, plasmids, and growth conditions
The B. anthracis strains used in this study are listed in Table 1. B. anthracis Sterne 34F2 (pXO1+ pXO2−) and its derivatives were routinely grown in LB broth supplemented with the appropriate antibiotics at the following concentrations: spectinomycin (100 μg/ml), chloramphenicol (7.5 μg/ml), tetracycline (5 μg/ml), or kanamycin (7.5 μg/ml). 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-Gal) (40 μg/ml) was added to LB agar to monitor β-galactosidase activity as necessary. As indicated, 5 μg/ml 5-aminolevulinic acid (5-ALA) (Sigma) was added to growth media. To induce high-level toxin expression, LB-agar plates or LB liquid media containing 0.8% sodium bicarbonate and 100 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) pH 8.0 were incubated in a 5% CO2 atmosphere. As indicated, B. anthracis was grown in R-Media with or without 0.8% NaHCO3 (Ristroph and Ivins, 1983) under 5% CO2. Competent cells of B. anthracis were prepared following the method of Koehler et al (Koehler et al., 1994).
Table 1.
B. anthracis strains used in this study
| Strain | Relevant Characteristics | Source |
|---|---|---|
| 34F2 | pXO1+ pXO2− | Laboratory stock |
| 34F2 pagA-lacZ | pagA-lacZ on pTCV-lac, KanR | (Tsvetanova et al, 2007) |
| 34F2 atxA-lacZ | pATXA12 on pTCV-lac, KanR | (Bongiorni et al, 2008) |
| 34F2 tB3 | BAS4358::mini-Tn10, SpcR | pAW016→34F2 |
| 34F2 tB17 | BAS4358::mini-Tn10, SpcR | pAW016→34F2 |
| 34F2 tB18 | BAS1385::mini-Tn10, SpcR | pAW016→34F2 |
| 34F2 tB21 | BAS1384::mini-Tn10, SpcR | pAW016→34F2 |
| 34F2 tB23 | BAS3568::mini-Tn10, SpcR | pAW016→34F2 |
| 34F2 tB28 | BAS1384::mini-Tn10, SpcR | pAW016→34F2 |
| 34F2 △atxA | atxA::spc | (Tsvetanova et al, 2007) |
| 34F2 △resB | GGATCC in place of BAS1384 | pAW050→34F2 |
| 34F2 △resBC | GGATCC in place of BAS1384-5 | pAW073→34F2 |
| 34F2 △hemL | GTCGAC in place of BAS4358 | pAW057→34F2 |
| 34F2 △BAS3568 | GTCGAC in place of BAS3568 | pAW066→34F2 |
| 34F2 △abrB | ACGCGT in place of BAS0036 | pORISceI-abrB→34F2 |
| 34F2 △atxA△resB | atxA::spc, GGATCC in place of BAS1384 | pAW050→34F2△atxA |
| 34F2 △atxA△BAS3568 | atxA::spc, GGATCC in place of BAS3568 | pAW066→34F2△atxA |
| 34F2 △resB△BAS3568 | GGATCC in place of BAS3568 and BAS1384 | pAW066→34F2△resB |
| 34F2 △ctaC | GTCGAC in place of BAS3856 | pAW121→34F2 |
| 34F2 △qcrBC | GTCGAC in place of BAS1433-4 | pAW125→34F2 |
| 34F2 △cccA | GTCGAC in place of BAS4193 | pAW131→34F2 |
| 34F2 △cccB | GTCGAC in place of BAS5035 | pAW135→34F2 |
| 34F2 △resA | GTCGAC in place of BAS1383 | pAW148→34F2 |
| 34F2 △3cyt | GTCGAC in place of BAS3856 and BAS1433-4 | pAW125→34F2△ctaC |
| 34F2 △4cyt | GTCGAC in place of BAS3856, BAS1433-4, and BAS4193 |
pAW131→34F2△3cyt |
| 34F2 △5cyt | GTCGAC in place of BAS3856, BAS1433-4, BAS4193, and BAS5035 |
pAW131→34F2△4cyt |
| 34F2 △cccA-B | GTCGAC in place of BAS4193 and BAS5035 | pAW135→34F2△cccA |
E. coli K-12 TG1, SCS110 and DH5α competent cells were used for the propagation and isolation of all plasmid constructs. E. coli transformation was performed by electroporation using the Bio-Rad-Gene Pulser according to the supplier. Transformants were selected on LB broth supplemented with ampicillin (100 μg/ml), spectinomycin (100 μg/ml), chloramphenicol (10 μg/ml), or kanamycin (30 μg/ml).
Plasmid pTCVlac-pagA is a derivative of the pagA-lacZ reporter plasmid described in Tsvetanova et al (Tsvetanova et al., 2007) obtained by cloning the 580 bp EcoRI-BamHI fragment from pJM115 into pTCVlac (Poyart and Trieu-Cuot, 1997). Plasmid pTCVSpac-AtxA is a derivative of pTCV-lac (Poyart and Trieu-Cuot, 1997) carrying the spac promoter from pMutin2 (Vagner et al., 1998), the atxA coding region and a deletion of the lacZ gene. The lack of the lacI gene results in constitutive expression of the spac promoter once this plasmid is introduced in B. anthracis. The details of its construction will be described elsewhere. The construction of plasmids pAtxA10, pAtxA12, and pAtxA20 was described in Bongiorni et al (Bongiorni et al., 2008). Plasmid pORI-pagA-lacZ is a derivative of pORI-Cm (Brunsing et al., 2005) carrying the pagA-lacZ fragment from pTCV-lac-pagA. The details of its construction will be described elsewhere.
Transposon mutagenesis
The temperature sensitive mini-Tn10 transposon delivery plasmid pAW016 (Wilson et al., 2007) was electroporated into the B. anthracis 34F2 strain containing the replicative vector pTCVlac-pagA (Table S1) and plated on LB with kanamycin, spectinomycin, and chloramphenicol. Resulting transformants were screened for lack of premature transposition and for plasmid loss at non-permissive temperature as previously described (Wilson et al., 2007). Plasmid-containing strains were used to inoculate LB broth containing kanamycin and spectinomycin, and cultures were incubated overnight at the permissive temperature of 28°C. The following morning, the overnight cultures were diluted 1:1000 in fresh LB broth with kanamycin and spectinomycin and incubated 6 hours at the non-permissive temperature of 37°C. Cultures were then serially diluted, the dilutions plated on LB-agar containing kanamycin, spectinomycin, and X-Gal, and the plates incubated overnight at 37°C. The following day, the spectinomycin- and kanamycin-resistant colonies were screened for alterations in pagA-lacZ expression. Mutants that showed a significant alteration in color were re-streaked on LB-agar with kanamycin, spectinomycin, and X-Gal to confirm phenotype, and on LB-agar with chloramphenicol to confirm loss of the transposon delivery plasmid. Mutants that retained their pagA-lacZ expression phenotype and were unable to grow on chloramphenicol were retained for further analysis.
Transposon analysis and sequencing
Spectinomycin-resistant and chloramphenicol-sensitive mutant strains were grown overnight in BHI containing 0.5% glycerol and spectinomycin at 28°C. Genomic DNA was extracted from the overnight cultures using UltraClean Microbial DNA Isolation Kit (MoBio, Carlsbad, CA). The site of insertion was identified by restriction digestion of genomic DNA using a panel of restriction enzymes, including SalI, NsiI, EcoRI, and SacI. Digested genomic DNA was then re-ligated and used to transform E. coli. The presence of the pUC origin of replication allowed re-ligated DNA containing the transposed sequence to replicate in E. coli as a spectinomycin-resistant plasmid. Following isolation of plasmid from E. coli, sequencing of transposon-flanking DNA was performed using the transposon-specific primer TSP3E (Table S2). Genomic DNA was screened for retention of virulence plasmid pXO1 by PCR amplification using the atxA-specific primer set AtxA5′promEco and AtxA3′Bam (Table S2).
β-Galactosidase assays
B. anthracis strains harboring the pagA-lacZ (Tsvetanova et al., 2007) or atxA-lacZ (pAtxA12) (Bongiorni et al., 2008) fusions on the replicative vector pTCV-lac (Poyart and Trieu-Cuot, 1997) were grown at 37°C in LB or R medium supplemented with the appropriate antibiotics. β-galactosidase activity was assayed as described previously and specific activity was expressed in Miller units (Miller, 1972;Wilson et al., 2008a).
TMPD Oxidase Staining
N,N,N’,N’-tetramethyl-p-phenylenediamine (TMPD) is an artificial electron donor that can be oxidized by cytochrome caa3, resulting in a colorimetric change indicative of the presence of functional cytochrome c oxidase. TMPD oxidase staining was performed with B. anthracis colonies grown on NSMP agar plates (nutrient sporulation medium phosphate) (Fortnagel and Freese, 1968) as previously described (Le Brun et al., 2000).
Markerless Gene Deletion
Gene deletions in B. anthracis were generated through a modification of the technique of Janes and Stibitz (Janes and Stibitz, 2006). Regions upstream and downstream of the gene to be deleted were cloned in the temperature sensitive plasmid pORI-I-SceI (Bongiorni et al., 2007) using the PCR primers listed in Table S2. The resulting plasmids (listed in Table S1) were electroporated into B. anthracis 34F2 and grown at the permissive temperature of 28°C in the presence of chloramphenicol. Bacteria were then shifted to the non-permissive temperature of 37°C in the presence of chloramphenicol to achieve targeted plasmid integration by homologous recombination. Following plasmid integration, the protocol of Janes and Stibitz (Janes and Stibitz, 2006) was followed to generate the markerless deletion. Diagnostic PCR was carried out to ensure that the entire coding sequence had been correctly deleted and that plasmid pXO1 was retained.
SDS-PAGE and Western blotting
B. anthracis strains were grown in LB Broth at 37°, and cell pellets were isolated at time points indicated by microcentrifugation of cell suspensions. Cell pellets were lysed following resuspension in buffer (10 mM Tris-HCl [pH 8.0], 10 mM MgCl2, 300 mM NaCl, and 10 mM 2-mercaptoethanol) by sonication for 20 seconds for 3 cycles and then centrifuged. SDS sample buffer was added to each supernatant, and samples were boiled for 5 minutes and loaded on 10% SDS-PAGE gels. The amount loaded was normalized relative to cell growth. The gels were run at 30mA for approximately 2 hr. The proteins in the gel were transferred to a PVDF membrane (BioRad) in transfer buffer (25mM Tris base, 192mM glycine, 20% methanol) at 20V overnight. The membranes were incubated for 30 minutes at room temperature in blocking buffer (5% dried milk in TBST (20 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.1% Tween 20)) followed by addition of a rabbit polyclonal α-AtxA antibody diluted 1:5,000. The blots were washed 5 times in TBST and then incubated for 1hr at RT with horseradish peroxidase-conjugated goat anti-rabbit antibody (BioRad) diluted 1:10,000 in blocking buffer. Following washing of the membrane, binding of the antibodies was probed using the ECL Plus kit (GE), and the protein bands were visualized by PhosphorImager analysis (Molecular Dynamics).
Supplementary Material
Acknowledgements
This work was supported in part by grant AI055860 from the National Institute of Allergy and Infectious Diseases, and GM19416 from the Institute of General Medical Sciences, National Institutes of Health.
Oligonucleotide synthesis and sequencing reactions were supported in part by the Stein Beneficial Trust.
We are indebted to Lars Hederstedt (Lund University, Sweden) for helpful discussion.
We thank Cristina Bongiorni for the construction of plasmid pTCV-Spac-AtxA and Christina Chiang for the construction of plasmid pORI-pag-lacZ.
This is manuscript #19905 from The Scripps Research Institute.
References
- Ahuja U, Kjelgaard P, Schulz B, Thony-Meyer L, Hederstedt L. ResB and ResC of Bacillus subtilis are heme-binding proteins with transport or chaperone function in cytochrome c maturation. 4th Conference on Functional Genomics of Gram-Positive Microorganisms; Tirrenia (Pisa), Italy. 2007. Abstract T62. [Google Scholar]
- Bartkus JM, Leppla SH. Transcriptional regulation of the protective antigen gene of Bacillus anthracis. Infect Immun. 1989;57:2295–2300. doi: 10.1128/iai.57.8.2295-2300.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckett CS, Loughman JA, Karberg KA, Donato GM, Goldman WE, Kranz RG. Four genes are required for the system II cytochrome c biogenesis pathway in Bordetella pertussis, a unique bacterial model. Mol Microbiol. 2000;38:465–481. doi: 10.1046/j.1365-2958.2000.02174.x. [DOI] [PubMed] [Google Scholar]
- Bengtsson J, Rivolta C, Hederstedt L, Karamata D. Bacillus subtilis contains two small c-type cytochromes with homologous heme domains but different types of membrane anchors. J Biol Chem. 1999;274:26179–26184. doi: 10.1074/jbc.274.37.26179. [DOI] [PubMed] [Google Scholar]
- Bongiorni C, Fukushima T, Wilson AC, Chiang C, Mansilla MC, Hoch JA, Perego M. Dual promoters control the expression of the Bacillus anthracis virulence factor AtxA. J Bacteriol. 2008;190:6483–6492. doi: 10.1128/JB.00766-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bongiorni C, Stoessel R, Perego M. Negative regulation of Bacillus anthracis sporulation by the Spo0E family of phosphatases. J Bacteriol. 2007;189:2637–2645. doi: 10.1128/JB.01798-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgogne A, Drysdale M, Hilsenbeck SG, Peterson SN, Koehler TM. Global effects of virulence gene regulators in a Bacillus anthracis strain with both virulence plasmids. Infect Immun. 2003;71:2736–2743. doi: 10.1128/IAI.71.5.2736-2743.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunsing RL, La Clair C, Tang S, Chiang C, Hancock LE, Perego M, Hoch JA. Characterization of sporulation histidine kinases of Bacillus anthracis. J Bacteriol. 2005;187:6972–81. doi: 10.1128/JB.187.20.6972-6981.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow A, Le Brun NE, Oubrie A. The role of ResA in type II cytochrome c maturation. Biochem Soc Trans. 2005;33:149–151. doi: 10.1042/BST0330149. [DOI] [PubMed] [Google Scholar]
- Dai Z, Koehler TM. Regulation of anthrax toxin activator gene (atxA) expression in Bacillus anthracis: temperature, not CO2/bicarbonate, affects AtxA synthesis. Infect Immun. 1997;65:2576–2582. doi: 10.1128/iai.65.7.2576-2582.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Z, Sirard JC, Mock M, Koehler TM. The atxA gene product activates transcription of the anthrax toxin genes and is essential for virulence. Mol Microbiol. 1995;16:1171–1181. doi: 10.1111/j.1365-2958.1995.tb02340.x. [DOI] [PubMed] [Google Scholar]
- Drysdale M, Heninger S, Hutt J, Chen Y, Lyons CR, Koehler TM. Capsule synthesis by Bacillus anthracis is required for dissemination in murine inhalation anthrax. EMBO J. 2005;24:221–227. doi: 10.1038/sj.emboj.7600495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duesbery NS, Webb CP, Leppla SH, Gordon VM, Klimpel KR, Copeland TD, et al. Proteolytic inactivation of MAP-kinase-kinase by anthrax lethal factor. Science. 1998;280:734–737. doi: 10.1126/science.280.5364.734. [DOI] [PubMed] [Google Scholar]
- Erlendsson LS, Acheson RM, Hederstedt L, Le Brun NE. Bacillus subtilis ResA is a thiol-disulfide oxidoreductase involved in cytochrome c synthesis. J Biol Chem. 2003;278:17852–17858. doi: 10.1074/jbc.M300103200. [DOI] [PubMed] [Google Scholar]
- Erlendsson LS, Hederstedt L. Mutations in the thiol-disulfide oxidoreductases BdbC and BdbD can suppress cytochrome c deficiency of CcdA-defective Bacillus subtilis cells. J Bacteriol. 2002;184:1423–1429. doi: 10.1128/JB.184.5.1423-1429.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortnagel P, Freese E. Analysis of sporulation mutants. II. Mutants blocked in the citric acid cycle. J Bacteriol. 1968;95:1431–1438. doi: 10.1128/jb.95.4.1431-1438.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamel PP, Dreyfuss BW, Xie Z, Gabilly ST, Merchant S. Essential histidine and tryptophan residues in CcsA, a system II polytopic cytochrome c biogenesis protein. J Biol Chem. 2003;278:2593–2603. doi: 10.1074/jbc.M208651200. [DOI] [PubMed] [Google Scholar]
- Hansson M, Rutberg L, Schröder I, Hederstedt L. The Bacillus subtilis hemAXCDBL gene cluster, which encodes enzymes of the biosynthetic pathway from glutamate to uroporphyrinogen III. J Bacteriol. 1991;173:2590–2599. doi: 10.1128/jb.173.8.2590-2599.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janes BK, Stibitz S. Routine markerless gene replacement in Bacillus anthracis. Infect Immun. 2006;74:1949–1953. doi: 10.1128/IAI.74.3.1949-1953.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler TM, Dai Z, Kaufman-Yarbray M. Regulation of the Bacillus anthracis protective antigen gene: CO2 and a trans-acting element activate transcription from one of two promoters. J Bacteriol. 1994;176:586–595. doi: 10.1128/jb.176.3.586-595.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranz R, Lill R, Goldman B, Bonnard G, Merchant S. Molecular mechanisms of cytochrome c biogenesis: three distinct systems. Mol Microbiol. 1998;29:383–396. doi: 10.1046/j.1365-2958.1998.00869.x. [DOI] [PubMed] [Google Scholar]
- Le Brun NE, Bengtsson J, Hederstedt L. Genes required for cytochrome c synthesis in Bacillus subtilis. Mol Microbiol. 2000;36:638–650. doi: 10.1046/j.1365-2958.2000.01883.x. [DOI] [PubMed] [Google Scholar]
- Leppla SH. Anthrax toxin edema factor: a bacterial adenylate cyclase that increases cyclic AMP concentrations of eukaryotic cells. Proc Natl Acad Sci U S A. 1982;79:3162–3166. doi: 10.1073/pnas.79.10.3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leppla SH. Production and purification of anthrax toxin. Methods Enzymol. 1988;165:103–116. doi: 10.1016/s0076-6879(88)65019-1. [DOI] [PubMed] [Google Scholar]
- Makino S, Uchida I, Terakado N, Sasakawa C, Yoshikawa M. Molecular characterization and protein analysis of the cap region, which is essential for encapsulation in Bacillus anthracis. J Bacteriol. 1989;171:722–730. doi: 10.1128/jb.171.2.722-730.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1972. pp. 352–355. [Google Scholar]
- Milne JC, Furlong D, Hanna PC, Wall JS, Collier RJ. Anthrax protective antigen forms oligomers during intoxication of mammalian cells. J Biol Chem. 1994;269:20607–20612. [PubMed] [Google Scholar]
- Naylor J, Cianciotto NP. Cytochrome c maturation proteins are critical for in vivo growth of Legionella pneumophila. FEMS Microbiol Lett. 2004;241:249–256. doi: 10.1016/j.femsle.2004.10.028. [DOI] [PubMed] [Google Scholar]
- Okinaka RT, Cloud K, Hampton O, Hoffmaster AR, Hill KK, Keim P, et al. Sequence and organization of pXO1, the large Bacillus anthracis plasmid harboring the anthrax toxin genes. J Bacteriol. 1999;181:6509–6515. doi: 10.1128/jb.181.20.6509-6515.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perego M, Hoch JA. Commingling regulatory systems following acquisition of virulence plasmids by Bacillus anthracis. Trends Microbiol. 2008;16:215–221. doi: 10.1016/j.tim.2008.01.010. [DOI] [PubMed] [Google Scholar]
- Poyart C, Trieu-Cuot P. A broad-host-range mobilizable shuttle vector for the construction of transcriptional fusions to β-galactosidase in Gram-positive bacteria. FEMS Microbiol Lett. 1997;156:193–198. doi: 10.1111/j.1574-6968.1997.tb12726.x. [DOI] [PubMed] [Google Scholar]
- Ristroph JD, Ivins BE. Elaboration of Bacillus anthracis antigens in a new, defined culture medium. Infect Immun. 1983;39:483–486. doi: 10.1128/iai.39.1.483-486.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saile E, Koehler TM. Control of anthrax toxin gene expression by the transition state regulator abrB. J Bacteriol. 2002;184:370–380. doi: 10.1128/JB.184.2.370-380.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiött T, Throne-Holst M, Hederstedt L. Bacillus subtilis CcdA-Defective Mutants Are Blocked in a Late Step of Cytochrome c Biogenesis. J Bacteriol. 1997;179:4523–4529. doi: 10.1128/jb.179.14.4523-4529.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirard JC, Mock M, Fouet A. The three Bacillus anthracis toxin genes are coordinately regulated by bicarbonate and temperature. J Bacteriol. 1994;176:5188–5192. doi: 10.1128/jb.176.16.5188-5192.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun G, Sharkova E, Chesnut R, Birkey S, Duggan MF, Sorokin A, et al. Regulators of aerobic and anaerobic respiration in Bacillus subtilis. J Bacteriol. 1996;178:1374–1385. doi: 10.1128/jb.178.5.1374-1385.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsvetanova B, Wilson AC, Bongiorni C, Chiang C, Hoch JA, Perego M. Opposing effects of histidine phosphorylation regulate the AtxA virulence transcription factor in Bacillus anthracis. Mol Microbiol. 2007;63:644–655. doi: 10.1111/j.1365-2958.2006.05543.x. [DOI] [PubMed] [Google Scholar]
- Uchida I, Hornung JM, Thorne CB, Klimpel KR, Leppla SH. Cloning and characterization of a gene whose product is a trans-activator of anthrax toxin synthesis. J Bacteriol. 1993;175:5329–5338. doi: 10.1128/jb.175.17.5329-5338.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida I, Makino S, Sekizaki T, Terakado N. Cross-talk to the genes for Bacillus anthracis capsule synthesis by atxA, the gene encoding the trans-activator of anthrax toxin synthesis. Mol Microbiol. 1997;23:1229–1240. doi: 10.1046/j.1365-2958.1997.3041667.x. [DOI] [PubMed] [Google Scholar]
- Vagner V, Dervyn E, Ehrlich SD. A vector for systematic gene inactivation in Bacillus subtilis. Microbiol. 1998;144:3097–3104. doi: 10.1099/00221287-144-11-3097. [DOI] [PubMed] [Google Scholar]
- Vetter SM, Schlievert PM. The two-component system Bacillus respiratory response A and B (BrrA-BrrB) is a virulence factor regulator in Bacillus anthracis. Biochemistry. 2007;46:7343–7352. doi: 10.1021/bi700184s. [DOI] [PubMed] [Google Scholar]
- von Wachenfeldt C, Hederstedt L. Bacillus subtilis 13-kilodalton cytochrome c-550 encoded by cccA consists of a membrane-anchor and a heme domain. J Biol Chem. 1990;265:13939–13948. [PubMed] [Google Scholar]
- von Wachenfeldt C, Hederstedt L. Respiratory cytochromes, other heme, proteins, and heme biosynthesis. In: Sonenshein AL, Hoch JA, Losick R, editors. Bacillus subtilis and its closest relatives: from genes to cells. ASM Press; Washington , D.C.: 2002. pp. 163–179. [Google Scholar]
- White AK, Hoch JA, Grynberg M, Godzik A, Perego M. Sensor domains encoded in Bacillus anthracis virulence plasmids prevent sporulation by hijacking a sporulation sensor histidine kinase. J Bacteriol. 2006;188:6354–6360. doi: 10.1128/JB.00656-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson AC, Hoch JA, Perego M. Virulence gene expression is independent of ResDE-regulated respiration control in Bacillus anthracis. J Bacteriol. 2008a;190:5522–5525. doi: 10.1128/JB.00312-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson AC, Perego M, Hoch JA. New transposon delivery plasmids for insertional mutagenesis in Bacillus anthracis. J Microbiol Methods. 2007;71:332–335. doi: 10.1016/j.mimet.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson AC, Soyer M, Hoch JA, Perego M. The bicarbonate transporter essential for Bacillus anthracis lethality. PLoS Biol. 2008b;4:e1000210. doi: 10.1371/journal.ppat.1000210. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Yeoman KH, Delgado MJ, Wexler M, Downie JA, Johnston AW. High affinity iron acquisition in Rhizobium leguminosarum requires the cycHJKL operon and the feuPQ gene products, which belong to the family of two-component transcriptional regulators. Microbiol. 1997;143:127–134. doi: 10.1099/00221287-143-1-127. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.