Abstract
The unusual basic amino acid, hypusine [Nε-(4-amino-2-hydroxybutyl)-lysine], is a modified lysine with the addition of the 4-aminobutyl moiety from the polyamine spermidine. This naturally occurring amino acid is a product of a unique posttranslational modification that occurs in only one cellular protein, eukaryotic translation initiation factor 5A (eIF5A, eIF-5A). Hypusine is synthesized exclusively in this protein by two sequential enzymatic steps involving deoxyhypusine synthase (DHS) and deoxyhypusine hydroxylase (DOHH). The deoxyhypusine/hypusine synthetic pathway has evolved in archaea and eukaryotes, and eIF5A, DHS and DOHH are highly conserved suggesting a vital cellular function of eIF5A. Gene disruption and mutation studies in yeast and higher eukaryotes have provided valuable information on the essential nature of eIF5A and the deoxyhypusine/hypusine modification in cell growth and in protein synthesis. In view of the extraordinary specificity and functional significance of hypusine-containing eIF5A in mammalian cell proliferation, eIF5A and the hypusine biosynthetic enzymes are novel potential targets for intervention in aberrant cell proliferation.
Keywords: Hypusine, eIF5A, Posttranslational modification, Polyamine, Deoxyhypusine synthase, Deoxyhypusine hydroxylase, Gene inactivation
Introduction
Polyamines, i.e., putrescine, spermidine, and spermine, are ubiquitous in living cells and are essential for cell growth. These polyamines exist as protonated polycations at physiological pH in cells and interact with nucleic acids, DNA and RNA, acidic proteins, and phospholipids. Polyamines regulate a vast array of cellular activities at the level of replication, transcription, translation, posttranslational modification, protein activation, membrane stability, and ion channeling. The role of polyamines in translational regulation may be most important, in that a majority of polyamines are bound to RNA (Igarashi and Kashiwagi 2009) and in that the polyamine spermidine is required for activation of the eukaryotic initiation factor 5A (eIF5A). Thus, polyamines regulate cell growth, proliferation, differentiation, transformation, and apoptosis and their cellular levels are tightly regulated at the level of biosynthesis, metabolism, and transport. The structural requirement for polyamines for the polycationic functions is not strict and a variety of polyamine analogs or derivatives can substitute for the natural polyamines in cells. In eukaryotic organisms, the polyamine spermidine has an independent and specific function as the source of the 4-aminobutyl portion of hypusine [Nε-(4-amino-2-hydroxybutyl)-lysine] in the essential cellular protein eIF5A. Since the structural requirement for spermidine in the synthesis of hypusine is quite strict, this function can only be fulfilled by spermidine or by few close structural analogs. Thus, hypusine synthesis defines an absolute requirement for the polyamine spermidine in eukaryotes (Byers et al. 1994; Chattopadhyay et al. 2003, 2008).
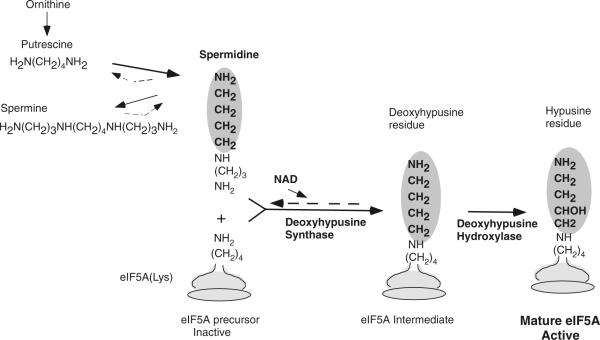
Hypusine was discovered by Shiba et al. (1971) from bovine brain extract and is a natural amino acid occurring in a single cellular protein, eIF5A (see reviews, Chen and Liu 1997; Park 2006; Wolff et al. 2007; Zanelli and Valentini 2007). Hypusine is formed by a unique posttranslational modification involving two enzymatic steps (Fig. 1). In the first step, deoxyhypusine synthase (DHS) catalyzes the formation of an intermediate, deoxyhypusine [Nε-(4-amino-butyl)-lysine] residue, by the transfer of the aminobutyl moiety of polyamine spermidine to one specific lysine residue of the eIF5A precursor. This intermediate is subsequently hydroxylated by deoxyhypusine hydroxylase (DOHH; Abbruzzese et al. 1986) to form hypusine-containing, biologically active eIF5A. The two enzymes have been cloned (Joe et al. 1995; Park et al. 2006) and their structures and reaction mechanism have been partially characterized. Neither enzyme modifies free amino acids (lysine or deoxyhypusine) or short peptides modeled on the eIF5A sequence (Wolff et al. 2007). Results from site-directed mutagenesis or truncation of eIF5A indicate that DHS and DOHH require the specific amino acid sequence surrounding the hypusine modification site and the macro-molecular structure of the eIF5A substrate for the modification reaction (Wolff et al. 2007). Inhibitors of DHS and DOHH exert anti-proliferative effects in mammalian cells (Hanauske-Abel et al. 1994; Park et al. 1994), suggesting an importance of the hypusine modification in cell growth. The most convincing evidence for the essential nature of eIF5A and its deoxyhypusine/hypusine modification was obtained from gene mutation, gene disruption or knock down studies in the yeast, S. cerevisiae and higher eukaryotes. Furthermore, recent evidence from yeast eIF5A temperature-sensitive mutant strains (Zuk and Jacobson 1998; Valentini et al. 2002; Zanelli and Valentini 2005; Chatterjee et al. 2006; Schrader et al. 2006; Gregio et al. 2009; Saini et al. 2009) and a Drosophila DOHH mutant (Patel et al. 2009) provides new insights into the mechanism of eIF5A action at the elongation step of translation (Gregio et al. 2009; Patel et al. 2009; Saini et al. 2009). In this review, we will address the evolutionary and functional significance of eIF5A and hypusine modification pathway with an emphasis on genetic studies targeting eIF5A, DHS or DOHH, including gene disruption, mutation, or knock down using SiRNA.
Fig. 1.
Hypusine biosynthesis in eIF5A. The polyamine spermidine, which is synthesized from putrescine, is the source of the aminobutyl moiety of hypusine, as indicated by shading. Hypusine synthesis occurs at one specific lysine residue of the eIF5A precursor protein, eIF5A(Lys), by two enzymatic steps, involving deoxyhypusine synthase (DHS) and deoxyhypusine hydroxylase (DOHH). Modified from Park 2006
Evolution of eIF5A and the hypusine pathway
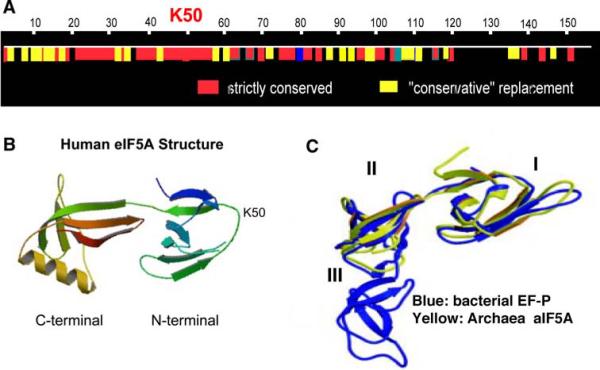
Unlike most other posttranslational modifications that occur in a wide variety of substrate proteins, hypusine synthesis involves only one substrate protein. It is extraordinary that the two enzymes, DHS and DOHH, have been designed just for the modification of eIF5A. eIF5A, DHS, and DOHH are found in all eukaryotes and are highly conserved, suggesting a vital function of eIF5A and the deoxyhypusine/hypusine modification. The amino acid sequence of the exposed hypusine site loop (Lys50 in the human eIF5A is modified to hypusine; Fig. 2a, b) is strictly conserved in eukaryotes (Wolff et al. 2007). Deoxyhypusine and hypusine occur in archaea and eukaryotes, but not in eubacteria. Yet, eubacteria has an elongation factor P (EF-P; Figs. 2c, 3), which is a distant ortholog of eIF5A. EF-P enhances elongation by stimulating the peptidyl transferase activity of the ribosome and is an essential protein in bacteria (Glick and Ganoza 1975). Like eIF5A, EF-P promotes methionyl-puromycin synthesis in vitro. There is a significant amino acid sequence similarity between EF-P, archaeal IF5A (aIF5A), and eIF5A (Hanawa-Suetsugu et al. 2004). Although EF-P does not contain deoxyhypusine/hypusine, EF-P from E. coli undergoes a different posttranslational modification that adds a mass of 144 at K34, the site corresponding to the hypusine modification site of eIF5A (Aoki et al. 2008). Crystal structures have been determined for yeast and human eIF5A (PDB 3er0, 3cpf) four aIF5A proteins (PDB 1eif, 2eif, 1iz6, 1bkb) and two Leishmania proteins (PDB 1x6o, 1xtd), and they exhibit only minor differences (Tong et al. 2009). The eukaryotic proteins have an alpha-helix in the C-terminal domain, not present in aIF5A. These proteins consist of two domains: a basic N-terminal domain containing the hypusine modification site in an exposed loop and an acidic C-terminal domain resembling an oligonucleotide-binding fold (Fig. 2b). The structure of aIF5A is superimposable on that of the first two domains of EF-P (Fig. 2c) (Hanawa-Suetsugu et al. 2004). These structural similarities suggest that there may be a certain functional conservation among the eubacterial, archaeal, and eukaryotic proteins in their role in protein synthesis.
Fig. 2.
Conservation of the amino acid sequence of eIF5A in eukaryotes (a), crystal structure of human eIF5A(Lys) (b) and comparison of crystal structures of aIF5A with EF-P(c). a The numbers on top of the bar indicate the amino acid residue number for human eIF5A. One lysine (Lys50) is converted to hypusine residue and the sequence surrounding this modification site is highly conserved. b crystal structure of truncated human eIF5A(Lys) (aa15–151) protein (PDB:3cpf; Tong et al. 2009), eIF5A consists of two domains, N- and C-terminal domains of β-sheet structure. The hypusine modification site (K50) is located at the tip of an exposed loop in the N-terminal domain. c comparison of bacterial EF-P and aIF5A structure, Thermus thermophilus EF-P consists of three domains (I, II, and III). The aIF5A structure (yellow) is superimposable on the first two domains of EF-P (blue). Modified from (Park 2006)
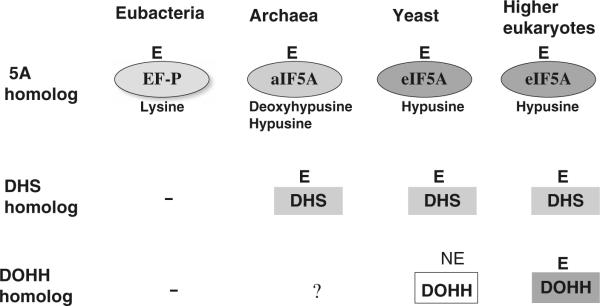
Fig. 3.
Evolution of eIF5A and its hypusine modification pathway. eIF5A orthologs are found in eubacteria and archaea and are essential genes in each organism. The DHS gene exists in archaea, and in all eukaryotes, but not in eubacteria. DOHH gene is found only in eukaryotes. E indicates essential gene, and NE indicates non-essential gene
The requirement for eIF5A and for the hypusine modification enzymes in cell viability and growth shows an interesting progression in the course of evolution (Fig. 3). DHS is not found in most commonly studied bacteria. However, several bacterial species contain DHS cognate genes that might have been transferred from archaea by horizontal gene transfer (Brochier et al. 2004). It is not known whether EF-P undergoes deoxyhypusine modification in those bacteria that contain a DHS cognate (Brochier et al. 2004). Only aIF5A and DHS homolog genes but no DOHH homolog genes have been identified in the genome of archaea kingdom (Wolff et al. 2007). It is not clear how hypusine is synthesized in certain archaea strains reported to contain hypusine (Bartig et al. 1990). In yeast (and also in plants), eIF5A, DHS, and DOHH homolog genes have been identified and eIF5A mainly exists as the hypusine-containing mature form in cells. Interestingly, however, DOHH is not an essential gene in yeast, as a DOHH null S. cerevisiae strain is viable and grows at a rate slightly slower than the wild type strain (Park et al. 2006) suggesting that deoxyhypusine-containing eIF5A is functional in yeast. In contrast, in multicellular (higher) eukaryotes such as Caenorhabditis elegans (C. elegans; Maeda et al. 2001) and Drosophila melanogaster (D. melanogaster), (Spradling et al. 1999; Patel et al. 2009) the DOHH gene is essential. Thus, eIF5A and the two modification enzymes seem to have evolved sequentially to meet the specialized developmental needs of eukaryotic organisms and in an independent manner without a co-evolutionary linkage between them.
Gene disruption studies of eIF5A isoforms
Two or more eIF5A isoforms have been identified in all eukaryotes, including fungi, plants, vertebrates, and mammals (Jenkins et al. 2001). In the yeast, S. cerevisiae, the two eIF5A genes, TIF51A and TIF51B, are reciprocally regulated by oxygen with TIF51A being mainly expressed under aerobic conditions and TIF51B being expressed only under anaerobic condition (Schnier et al. 1991; Wöhl et al. 1993). The two EIF5A genes seem to be co-expressed in fish, amphibians, and chicken (Jenkins et al. 2001; Clement et al. 2003). In contrast, in most mammals, only one isoform, eIF5A-1, is constitutively expressed in all tissues, whereas the eIF5A-2 protein is not. eIF5A-2 mRNA expression shows tissue specific dependency (Jenkins et al. 2001) suggesting differentiated function of the second isoform. eIF5A-2 protein is normally too low to be detected in mammalian cells and tissues (Clement et al. 2003). However, eIF5A-2 is highly expressed in certain human ovarian cancer tissues and colorectal and ovarian cancer cell lines in which the EIF5A-2 gene is amplified (Guan et al. 2001; Clement et al. 2003). Furthermore, overexpression of eIF5A-2 in certain mammalian cells caused cellular transformation. Based on these findings, eIF5A-2 has been proposed as an oncogene (Guan et al. 2001, 2004).
Disruption of the anaerobic gene TIF51B showed no growth defects under aerobic or anaerobic condition, since Tif51a protein (eIF5A-1) is produced under aerobic and to a much lesser extent under anaerobic conditions. However, gene inactivation of TIF51A alone (Wöhl et al. 1993) or both genes TIF51A and TIF51B (Schnier et al. 1991) rendered S. cerevisiae nonviable under aerobic conditions, indicating the essential nature of eIF5A in yeast survival. Expression of either isoform from plasmid-borne TIF51A or TIF51B under the GAL1 promoter supported yeast growth under aerobic and anaerobic conditions, indicating the functional identity of the two isoforms in yeast (Schwelberger et al. 1993; Magdolen et al. 1994). Furthermore, heterologous eIF5A proteins from other eukaryotic species, slime mold, or alfalfa, and the two human eIF5A isoforms were hypusine-modified by the yeast enzymes and could substitute for the yeast eIF5A in supporting growth (Magdolen et al. 1994; Clement et al. 2003), suggesting that they are functionally interchangeable in yeast. In contrast, the archaeal homolog, aIF5A, was neither modified in yeast nor did it support growth of the yeast tif51a null strain (Magdolen et al. 1994).
Whereas the eIF5A isoforms are indistinguishable in their growth supporting activity of yeast, they seem to have differentiated functions in higher multi-cellular eukaryotic organisms. C. elegans contains two eIF5A isoforms, IFF-2 (eIF5A-1) and IFF-1 (eIF5A-2; Hanazawa et al. 2004). IFF-2 (eIF5A-1) is expressed in all tissues throughout development and iff-2 mutation causes somatic defects, including slow larval growth, and some structural abnormalities. Iff-2 mutants either were arrested during the larval stages or grew very slowly to become sterile adults. In contrast to IFF-2, another isoform, IFF-1 (eIF5A-2) is mainly expressed in the gonad. The loss of IFF-1 activity resulted in impaired mitotic proliferation of germ cells, inhibition of gametogenesis, and sterility. Furthermore, localization of the P-granules (large complex of RNA-binding proteins and RNA) was disrupted in the iff-1 mutant. Thus, the two C. elegans eIF5A homologs appear to have distinct functions and tissue localizations. IFF-2 is required in soma for somatic tissue growth and organization. On the other hand, IFF-1 is required in the germline for germ cell proliferation, gametogenesis, and for proper localization of P-granules.
The role of mammalian eIF5A isoforms is also being investigated in mice (K. Nishimura and M. H. Park, unpublished results). Mouse contains two genes, Eif5a1 and Eif5a2, encoding eIF5A-1 and eiF5A-2, respectively. We performed gene targeting of mouse Eif5a1 in mice by using the Eif5a1 gene-trapped ES cell line RRE174 (BayGenomics) and heterozygous mice (Eif5a1+/−) were generated. The Eif5a1+/− mice appeared to be normal and did not show any growth defects or phenotypes. However, when the heterozygous male and female mice were crossed, no pups with the genotype of Eif5a1−/− homozygous deletion were born indicating that eIF5A-1 is essential for embryonic development and that eIF5A-2 does not substitute for eIF5A-1 in the mouse.
Gene inactivation studies of deoxyhypusine synthase (DHS)
The importance of the hypusine/deoxyhypusine modification in yeast viability was initially indicated by the lack of growth supporting activity of the eIF5A-1 mutant (K51R) in an eIF5A null S. cerevisiae strain (Schnier et al. 1991). This mutant eIF5A with substitution of Arg for Lys51 (the specific lysine that is modified to hypusine in the yeast eIF5A) cannot be modified to the hypusine form. eIF5A homolog and DHS homolog genes are found in archaea genomes and deoxyhypusine/hypusine amino acids were isolated from these organisms. Although gene inactivation of aIF5A or DHS has not been performed in archaea, growth was arrested in G1 upon treatment with GC7, a potent inhibitor of DHS (Jansson et al. 2000), indicating that DHS is also essential in archaea (Fig. 3).
After cloning of deoxyhypusine synthase genes in yeast, Neurospora, and mammals (Joe et al. 1995; Tao and Chen 1995; Bevec et al. 1996; Sasaki et al. 1996) the role of deoxyhypusine synthase was directly assessed by gene disruption in S. cerevisiae. There is only a single DHS gene in yeast and most eukaryotes. A haploid S. cerevisiae strain with disruption of the DHS gene was not viable, and could be rescued only by a plasmid encoding the wild type DHS, but not by that encoding inactive DHS enzyme (with Arg substitution at the critical active site lysine K350 of the yeast enzyme; Sasaki et al. 1996; Park et al. 1998). When examined under the microscope, a non-viable DHS-disrupted ascus from a tetrad dissection grew several generations to a colony of 100–200 enlarged cells before growth arrest, presumably due to depletion of hypusine-containing eIF5A. Similar growth arrest of enlarged cells occurred upon depletion of eIF5A in yeast (Kang and Hershey 1994) suggesting a cell cycle arrest at G1 stage upon depletion of eIF5A(Hpu).
The essential requirement for DHS is also indicated from the DHS knock-out studies in mice. There is a single deoxyhypusine synthase gene (Dhps) in the mouse genome. A Dhps gene-trapped ES cell line, RRM039 (BayGenomics), was used to generate heterozygous mice (Dhps+/−). The Dhps+/− mice appeared normal and did not show any growth defects or phenotypes. When, the heterozygous male and female mice were crossed, no pups were born with the genotype of Dhps−/− homozygous deletion (K. Nishimura and M. H. Park, unpublished results). Taken together, these findings suggest that deoxyhypusine modification of eIF5A is vital for the growth and survival of eukaryotes, from yeast to mammals.
Gene inactivation studies of deoxyhypusine hydroxylase (DOHH)
Although deoxyhypusine hydroxylase activity could be detected in mammalian cells and tissues, the identity of this enzyme remained obscure for many years. The DOHH gene was initially cloned by Thompson et al. (2003) from yeast two-hybrid screening in search of eIF5A binding proteins. Two genes were selected: one encoding DHS and the second LIA-1 (ligand of eIF5A) with unknown function. When we cloned the DOHH gene by activity screening of the S. cerevisiae arrayed GST-ORF expression library (Park et al. 2006), it turned out that the DOHH gene was identical to that of Lia-1. DOHH exists as a product of a single gene in all eukaryotes and has a unique superhelical structure termed “HEAT-repeat”. This enzyme has a nonheme diiron active center that activates O2 (Vu et al. 2009). Since a number of metal-chelating inhibitors of DOHH caused growth inhibition and G1 cell cycle arrest in mammalian cells, DOHH has been assumed to be essential for cell growth (Hanauske-Abel et al. 1994). Interestingly, however, the DOHH gene is apparently not essential in the yeast S. cerevisiae, even though endogenous yeast eIF5A mostly exists as the fully modified hypusine form. DOHH seems to be functionally more significant in the fission yeast, Sacchromyces pombe, since a mutation (E66K) in its DOHH homolog gene, Mmd1, caused a temperature-sensitive growth phenotype and altered mitochondrial morphology and distribution (Weir and Yaffe 2004). Based on this observation, a role for DOHH, or eIF5A, in micro-tubule assembly and mitochondrial function was implicated.
In contrast to yeast, inactivation of DOHH is recessively lethal in multicellular eukaryotes such as C. elegans and D. melanogaster (Spradling et al. 1999; Maeda et al. 2001; Patel et al. 2009). The phenotypes resulting from depletion of DOHH and hypusine-modified eIF5A was extensively investigated in D. melanogaster (Patel et al. 2009). During P-element screening that altered bristle number in fruit flies, two nero mutations (nero1 and nero2) that involve deletion of 189 and 123 base pairs in the ORF of the DOHH homolog gene (CG2245) were isolated, and the mutation was complemented by a CG2245 genomic clone. Flies carrying homozygous or in heteroallelic combinations of nero1 and nero2 died as second instar (L2) larvae, indicating an essential requirement of DOHH and eIF5A hydroxylation for D. melanogaster development. Nero mutants displayed small size of cells and tissues, and also decreased level of BrdU incorporation and delay in cell cycle progression, suggesting a role for DOHH in cell growth and proliferation. Furthermore, nero (DOHH) mutant fat bodies displayed autophagic structures, even when the animals were fed, suggesting that nero mutant larvae undergo a constitutive starvation response. eIF5A was highly upregulated in nero mutants probably as a compensatory mechanism to overcome nero (DOHH) mutation by the mutant larvae. Knockdown of eIF5A using RNAi also resulted in similar phenotypes as nero (DOHH) mutations, indicating that the nero phenotype is due to the defect in eIF5A hydroxylation and that nero functions in eIF5A-mediated translational control. Furthermore, RNAi knockdown of either Nero or eIF5A resulted in an increase in polysomes in Drosophila S2 cells (Patel et al. 2009), suggesting a function of eIF5A at the elongation step of protein synthesis.
Functional studies of eIF5A using yeast mutants
Implication of eIF5A with mRNA decay and cell cycle: a secondary role?
Since eIF5A is a highly conserved protein among archaea and eukaryotes, studies in diverse systems have contributed to the understanding of the functional relevance of this protein in both unicellular and more complex organisms (Zanelli and Valentini 2007). Of great importance are those investigations that employed genetic approaches in the budding yeast S. cerevisiae.
As described above, knockout of eIF5A genes (TIF51A and TIF51B) in a S. cerevisiae haploid renders the strain nonviable. Thus, using a double knockout strain, Kang and Hershey (1994) generated an unstable ubiquitin-Arg-eIF5A fusion construct (UBR5A), which permitted analysis of cells during rapid depletion of eIF5A. The eIF5A-depleted cells were enlarged and unbudded, characteristic of a G1 arrest phenotype (Kang and Hershey 1994). This finding is consistent with the arrest of mammalian cells at G1 upon blocking eIF5A maturation by inhibitors of DOHH (Hanauske-Abel et al. 1994). These eIF5A-depleted cells showed a relatively moderate defect in translation initiation, when assayed by both protein synthesis rate (reduction by 30%) and polysome profile analyses (Kang and Hershey 1994).
In a screen for mRNA decay factors, another eIF5A mutant (ts1159) was identified (Zuk and Jacobson 1998). The ts1159 mutant strain showed temperature sensitivity due to the unstable eIF5AS149P protein and accumulated different reporter mRNAs at the restrictive temperature. Curiously, only a modest decrease in protein synthesis rate (by 30%) was observed with no significant alterations in polysome profile in the ts1159 mutant at restrictive temperature, similar to that observed for a mutant strain expressing unstable eIF5A fusion protein (UBR5A; Kang and Hershey 1994). Based on these and additional experiments performed therein, a direct role of eIF5A in mRNA degradation was proposed (Zuk and Jacobson 1998). Subsequently, three other temperature-sensitive eIF5A mutants (tif51A-1, tif51A-2, and tif51A-3 with P83S, P83L, and C39Y/G118D substitutions, respectively) were characterized and were shown to harbor unstable forms of eIF5A at the restrictive temperature (Valentini et al. 2002). These mutants also displayed mRNA decay defects, albeit less severe than that for the ts1159 allele. However, there was no causative correlation between mRNA accumulation and cell growth arrest, disputing a direct role for eIF5A in the degradation of mRNAs (Valentini et al. 2002). Another temperature-sensitive mutant, expressing a human eIF5A allele (V81G) in the eIF5A null yeast strain, displayed very severe growth phenotypes, induction of apoptotic death, accumulation of NMD-targeted mRNAs and shortened telomeres, leading to a suggestion of a direct role of eIF5A in mRNA degradation (Schrader et al. 2006). However, as presented in the next section, eIF5A mutants show defects in the elongation step of translation (Gregio et al. 2009; Saini et al. 2009). The blockage in translation elongation would cause accumulation of mRNAs in polysomes and thereby result in their stabilization (Peltz et al. 1992). Thus, the mRNA decay phenotype in the eIF5A mutants is likely to be a secondary effect of inhibition of translation elongation.
Regarding other aspects of eIF5A function, the tif51A-1, tif51A-2, and tif51A-3 mutant phenotypes were shown to be suppressed by the osmostabilizer sorbitol. High-copy suppressor screening using the tif51A-1 mutant identified several components of the Pkc1 pathway and factors important for proper actin organization during G1/S transition (Valentini et al. 2002; Zanelli and Valentini 2005). In line with this finding, a striking change in actin dynamics was observed in a temperature-sensitive eIF5A mutant strain (with L102A substitution) at a restrictive temperature (Chatterjee et al. 2006). Interestingly, yeast cells deprived of spermidine, and thereby of hypusinated mature eIF5A, also exhibited defects of actin polarity (Balasundaram et al. 1991). Moreover, mutations in eIF5A and Lia1 (yeast DOHH) appear to interact genetically with components of the secretory pathway (Frigieri et al. 2007, 2008), essential for yeast polarized growth during cell cycle progression. While these studies do not demonstrate the exact mechanism by which eIF5A influences actin polarity during cell cycle progression, they provide further insights into the role of eIF5A in cell proliferation (Park et al. 1997).
Evidence for a direct role of eIF5A in translation elongation
Although eIF5A was initially identified as a translation initiation factor (Kemper et al. 1976), later studies conducted in yeast suggest that eIF5A does not have an essential role in the translation initiation step (Kang and Hershey 1994; Zuk and Jacobson 1998). In recent studies employing yeast two-hybrid screening or co-purification with tagged eIF5A, eIF5A binding proteins have been identified, including several components of the translational machinery such as ribosomal proteins (large and small subunits) and elongation factors. The specificity of the physical interaction of eIF5A with the translational machinery was confirmed by co-sedimentation and copurification under different conditions and it was demonstrated that eIF5A binds only to translating ribosomes (Zanelli et al. 2006) and in a hypusine-dependent manner (Jao and Chen 2006; Zanelli et al. 2006).
Moreover, eIF5A dysfunction caused sensitivity to protein synthesis inhibitors and a significant decrease in protein synthesis (Zanelli et al. 2006; Cano et al. 2008; Dias et al. 2008; Gregio et al. 2009; Saini et al. 2009). In recent studies, a rapid decrease in protein synthesis was observed upon depletion of functional eIF5A, unlike the moderate inhibition previously reported (Kang and Hershey 1994; Zuk and Jacobson 1998). The inhibition of protein synthesis preceded growth inhibition at the restrictive condition, strongly suggesting that the growth arrest was attributable to a prior defect in translation (Cano et al. 2008; Dias et al. 2008). The dependence of protein synthesis on eIF5A was also demonstrated in in vitro assays using yeast eIF5A mutant cell extracts, in which the addition of hypusinated wild type eIF5A, but not the unhypusinated K51R mutant, stimulated translation of luciferase reporter mRNA (Saini et al. 2009).
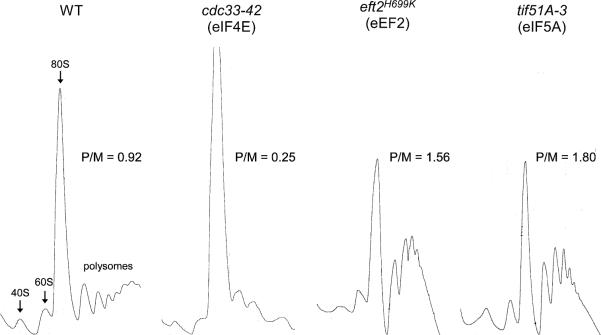
Since the polysome profiles of eIF5A mutant strains from earlier studies (Kang and Hershey 1994; Zuk and Jacobson 1998) were not consistent with a role of eIF5A at the translation initiation step, further experiments were performed to address this question. Careful analyses of polysome profiles of different temperature-sensitive eIF5A mutants revealed an increase in the ratio of polysomes/monosome at the restrictive temperature, a pattern expected in case of a block in translation elongation (Zanelli et al. 2006; Gregio et al. 2009; Saini et al. 2009) (Fig. 4). Indeed, polysome profiles of eIF5A mutants were very similar to that of wild type cells blocked at elongation by treatment with sordarin, an inhibitor of elongation factor 2 (eEF2) (Saini et al. 2009) and that of the eEF2 mutant eft2H699K (Gregio et al. 2009) (Fig. 4). Moreover, a strain harboring both the eIF5A mutant and a translation initiation (eIF4E) mutant showed an intermediate polysome profile pheno-type, further supporting a role for eIF5A at the elongation step of translation (Gregio et al. 2009). Independent evidence for a role of eIF5A in translation elongation was obtained from measurement of the ribosome transit time (the time in which a growing nascent polypeptide chain remains attached to the translating ribosome, i.e., the time of elongation plus the time of termination). The ribosome transit was significantly delayed for different eIF5A temperature-sensitive mutants at the restrictive temperature (Gregio et al. 2009; Saini et al. 2009), supporting the notion that eIF5A is involved in the elongation step of translation.
Fig. 4.
Polysome profiles of S. cerevisiae strains harboring wild type, eIF4E, eEF2, and eIF5A mutants. Ratios between polysomes and 80S monosome peaks (P/80S) were obtained by comparing the respective areas under the graphs. Modified from Gregio et al. 2009
In addition, lack of formation of P-bodies (mRNA-protein aggregates where mRNA degradation takes place) was observed upon impairment of eIF5A function, similar to that observed for cells treated with cycloheximide, an inhibitor of translation elongation. This effect on P-bodies is also consistent with a role for eIF5A at the translation elongation step (Parker and Sheth 2007; Gregio et al. 2009).
Finally, an eIF5A mutant tif51A(D63V) was defective in +1 ribosomal-programmed frame-shifting, similarly to wild type cells treated with the eEF2 inhibitor sordarin. This result and the fact that the eIF5A mutant tif51A(D63V) is sensitive to sordarin led to the suggestion that eIF5A functionally interacts with eEF2 in the ribosome elongation cycle (Saini et al. 2009). In agreement with this idea, the eIF5A mutant tif51A-3 displayed synthetic lethality with the eEF2 dominant negative mutant eft2H699K (Gregio et al. 2009).
Future prospects for eIF5A research
Several lines of evidence described above provide strong support for a role for eIF5A at the elongation step of translation. Although preliminary experimental data suggest genetic, functional, and physical interaction between eIF5A and eEF2, there is little information on the mode and mechanism of eIF5A action on the ribosome. It is not known at which site of the eukaryotic ribosome, the aminoacyl-tRNA (A) site, peptidyl-tRNA (P) site, or exiting tRNA (E) site, eIF5A is binding and what role the hypusine residue and the strictly conserved neighboring amino acids play in ribosome binding. The relevance of an eIF5A-eEF2 interaction in eukaryotic translational control needs to be further investigated at the molecular and cellular level.
The mechanism of action and structure of EF-P (a bacterial ortholog of eIF5A) may shed light on the eIF5A mode of action. The results from a chemical protection foot-printing study by Aoki et al. (2008) using E. coli EF-P and ribosomes suggest that this factor binds to the A-site of the 70S ribosome and thereby may prevent the initiator tRNA from binding at the A-site. On the other hand, a recent crystal structure of Thermus thermophilus EF-P bound to the its 70S ribosome along with the initiator transfer RNA, N-formyl-methionyl-tRNAi (fMet-tRNAi), and a short mRNA (Blaha et al. 2009) shows EF-P binding between the P site and E site of the 70S ribosome. These crystal structure data led to the conclusion that EF-P facilitates the proper positioning of the initiator tRNA to enhance the formation of only the first peptide bond (Blaha et al. 2009). The authors also proposed a model in which eIF5A is positioned similarly on the ribosome with the hypusine side chain extending to the peptidyl transferase center and a similar role for eIF5A in the first peptide bond formation.
However, a functional conservation of EF-P and eIF5A cannot be assumed based solely on the similarities in amino acid sequence and crystal structures. The recent experimental data described above collectively suggest a role for eIF5A as a factor participating in several rounds of peptide bond formation during translation elongation, rather than just at the first peptide bond formation. It is likely that the evolutionary divergence between EF-P and eIF5A protein sequences/structures, including the hypusine modification, may have contributed to a dissimilar function (or mode of action) for eIF5A in the translation process in archaea and eukaryotes. Future studies on the structure of eIF5A bound to eukaryotic ribosomes and the detailed mechanism of eIF5A in translation elongation will resolve this question.
Acknowledgments
The research was supported in part by the Intramural Research Program of National Institute of Dental and Craniofacial Research (NIDCR), NIH, FAPESP (Fundação de Amparo à Pesquisa do Estado de São Paulo) and KANAE Foundation for the Promotion of Medical Science. We thank Dr. Edith C. Wolff (NIDCR, NIH) for critical reading of the manuscript and helpful suggestions.
Glossary
Abbreviations
- aIF5A
Archaeal initiation factor 5A
- EF-P
Elongation factor P
- eIF5A
Eukaryotic translation initiation factor 5A
- eIF5A-1
Primary isoform of eIF5A
- eIF5A-2
Secondary isoform of eIF5A
- eIF5A(Lys)
eIF5A precursor
- eIF5A(Dhp)
eIF5A intermediate containing deoxyhypusine
- eIF5A(Hpu)
eIF5A active form containing hypusine
- UBR5A
Ubiquitin arginine-fusion yeast eukaryotic initiation factor 5A
- DHS
Deoxyhypusine synthase
- DOHH
Deoxyhypusine hydroxylase
- GC7
N1-guanyl-1,7-diaminoheptane
- A site
Aminoacyl-tRNA site
- P site
Peptidyl-tRNA site
- E site
Exiting tRNA site
References
- Abbruzzese A, Park MH, Folk JE. Deoxyhypusine hydroxylase from rat testis: partial purification and characterization. J Biol Chem. 1986;261:3085–3089. [PubMed] [Google Scholar]
- Aoki H, Xu J, Emili A, Chosay JG, Golshani A, Ganoza MC. Interactions of elongation factor EF-P with the Escherichia coli ribosome. FEBS J. 2008;275:671–681. doi: 10.1111/j.1742-4658.2007.06228.x. [DOI] [PubMed] [Google Scholar]
- Balasundaram D, Tabor CW, Tabor H. Spermidine or spermine is essential for the aerobic growth of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1991;88:5872–5876. doi: 10.1073/pnas.88.13.5872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartig D, Schümann H, Klink F. The unique posttranslational modification leading to deoxyhypusine or hypusine is a general feature of the Archebacterial kingdom. System Appl Microbiol. 1990;13:112–116. [Google Scholar]
- Bevec D, Kappel B, Jaksche H, Csonga R, Hauber J, Klier H, Steinkasserer A. Molecular characterization of a cDNA encoding functional human deoxyhypusine synthase and chromosomal mapping of the corresponding gene locus. FEBS Lett. 1996;378:195–198. doi: 10.1016/0014-5793(95)01456-x. [DOI] [PubMed] [Google Scholar]
- Blaha G, Stanley RE, Steitz TA. Formation of the first peptide bond: the structure of EF-P bound to the 70s ribosome. Science. 2009;325:966–970. doi: 10.1126/science.1175800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brochier C, Lopez-Garcia P, Moreira D. Horizontal gene transfer and archaeal origin of deoxyhypusine synthase homologous genes in bacteria. Gene. 2004;330:169–176. doi: 10.1016/j.gene.2004.01.018. [DOI] [PubMed] [Google Scholar]
- Byers TL, Lakanen JR, Coward JK, Pegg AE. The role of hypusine depletion in cytostasis induced by S-adenosyl-L-methionine decarboxylase inhibition: new evidence provided by L-methylspermidine and 1,12-dimethylspermine. Biochem J. 1994;303:363–368. doi: 10.1042/bj3030363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cano VS, Jeon GA, Johansson HE, Henderson CA, Park JH, Valentini SR, Hershey JW, Park MH. Mutational analyses of human eIF5A-1—identification of amino acid residues critical for eIF5A activity and hypusine modification. FEBS J. 2008;275:44–58. doi: 10.1111/j.1742-4658.2007.06172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee I, Gross SR, Kinzy TG, Chen KY. Rapid depletion of mutant eukaryotic initiation factor 5A at restrictive temperature reveals connections to actin cytoskeleton and cell cycle progression. Mol Genet Genomics. 2006;275:264–276. doi: 10.1007/s00438-005-0086-4. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay MK, Tabor CW, Tabor H. Spermidine but not spermine is essential for hypusine biosynthesis and growth in Saccharomyces cerevisiae: spermine is converted to spermidine in vivo by the fms1-amine oxidase. Proc Natl Acad Sci USA. 2003;100:13869–13874. doi: 10.1073/pnas.1835918100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay MK, Park MH, Tabor H. Hypusine modification for growth is the major function of spermidine in Saccharomyces cerevisiae polyamine auxotrophs grown in limiting spermidine. Proc Natl Acad Sci USA. 2008;105:6554–6559. doi: 10.1073/pnas.0710970105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen KY, Liu AY. Biochemistry and function of hypusine formation on eukaryotic initiation factor 5A. Biol Signals. 1997;6:105–109. doi: 10.1159/000109115. [DOI] [PubMed] [Google Scholar]
- Clement PC, Henderson CA, Jenkins ZA, Smit-McBride Z, Wolff EC, Hershey JWB, Park MH, Johansson HE. Identification and characterization of eukaryotic initiation factor 5A-2. Eur J Biochem. 2003;147:4254–4263. doi: 10.1046/j.1432-1033.2003.03806.x. [DOI] [PubMed] [Google Scholar]
- Dias CA, Cano VS, Rangel SM, Apponi LH, Frigieri MC, Muniz JR, Garcia W, Park MH, Garratt RC, Zanelli CF, Valentini SR. Structural modeling and mutational analysis of yeast eukaryotic translation initiation factor 5A reveal new critical residues and reinforce its involvement in protein synthesis. FEBS J. 2008;275:1874–1888. doi: 10.1111/j.1742-4658.2008.06345.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frigieri MC, Thompson GM, Pandolfi JR, Zanelli CF, Valentini SR. Use of a synthetic lethal screen to identify genes related to TIF51a in Saccharomyces cerevisiae. Genet Mol Res. 2007;6:152–165. [PubMed] [Google Scholar]
- Frigieri MC, Joao Luiz MV, Apponi LH, Zanelli CF, Valentini SR. Synthetic lethality between eIF5A and Ypt1 reveals a connection between translation and the secretory pathway in yeast. Mol Genet Genomics. 2008;280:211–221. doi: 10.1007/s00438-008-0357-y. [DOI] [PubMed] [Google Scholar]
- Glick BR, Ganoza MC. Identification of a soluble protein that stimulates peptide bond synthesis. Proc Natl Acad Sci USA. 1975;72:4257–4260. doi: 10.1073/pnas.72.11.4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregio AP, Cano VP, Avaca JS, Valentini SR, Zanelli CF. eIF5A has a function in the elongation step of translation in yeast. Biochem Biophys Res Commun. 2009;380:785–790. doi: 10.1016/j.bbrc.2009.01.148. [DOI] [PubMed] [Google Scholar]
- Guan XY, Sham JS, Tang TC, Fang Y, Huo KK, Yang JM. Isolation of a novel candidate oncogene within a frequently amplified region at 3q26 in ovarian cancer. Cancer Res. 2001;61:3806–3809. [PubMed] [Google Scholar]
- Guan XY, Fung JM, Ma NF, Lau SH, Tai LS, Xie D, Zhang Y, Hu L, Wu QL, Fang Y, Sham JS. Oncogenic role of eIF-5A2 in the development of ovarian cancer. Cancer Res. 2004;64:4197–4200. doi: 10.1158/0008-5472.CAN-03-3747. [DOI] [PubMed] [Google Scholar]
- Hanauske-Abel HM, Park MH, Hanauske AR, Popowicz AM, Lalande M, Folk JE. Inhibition of the G1-S transition of the cell cycle by inhibitors of deoxyhypusine hydroxylation. Biochim Biophys Acta. 1994;1221:115–124. doi: 10.1016/0167-4889(94)90003-5. [DOI] [PubMed] [Google Scholar]
- Hanawa-Suetsugu K, Sekine S, Sakai H, Hori-Takemoto C, Terada T, Unzai S, Tame JR, Kuramitsu S, Shirouzu M, Yokoyama S. Crystal structure of elongation factor P from Thermus thermophilus hb8. Proc Natl Acad Sci USA. 2004;101:9595–9600. doi: 10.1073/pnas.0308667101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanazawa M, Kawasaki I, Kunitomo H, Gengyo-Ando K, Bennett KL, Mitani S, Iino Y. The Caenorhabditis elegans eukaryotic initiation factor 5A homologue, IFF-1, is required for germ cell proliferation, gametogenesis and localization of the P-granule component PGL-1. Mech Dev. 2004;121:213–224. doi: 10.1016/j.mod.2004.02.001. [DOI] [PubMed] [Google Scholar]
- Igarashi K, Kashiwagi K. Modulation of cellular function by polyamines. Int J Biochem Cell Biol. 2009 doi: 10.1016/j.biocel.2009.07.009. in press. [DOI] [PubMed] [Google Scholar]
- Jansson BP, Malandrin L, Johansson HE. Cell cycle arrest in archaea by the hypusination inhibitor N(1)-guanyl-1,7-diaminoheptane. J Bacteriol. 2000;182:1158–1161. doi: 10.1128/jb.182.4.1158-1161.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jao DL, Chen KY. Tandem affinity purification revealed the hypusine-dependent binding of eukaryotic initiation factor 5A to the translating 80s ribosomal complex. J Cell Biochem. 2006;97:583–598. doi: 10.1002/jcb.20658. [DOI] [PubMed] [Google Scholar]
- Jenkins ZA, Haag PG, Johansson HE. Human eIF5A2 on chromosome 3q25-q27 is a phylogenetically conserved vertebrate variant of eukaryotic translation initiation factor 5A with tissue-specific expression. Genomics. 2001;71:101–109. doi: 10.1006/geno.2000.6418. [DOI] [PubMed] [Google Scholar]
- Joe YA, Wolff EC, Park MH. Cloning and expression of human deoxyhypusine synthase cDNA. Structure–function studies with the recombinant enzyme and mutant proteins. J Biol Chem. 1995;270:22386–22392. doi: 10.1074/jbc.270.38.22386. [DOI] [PubMed] [Google Scholar]
- Kang HA, Hershey JW. Effect of initiation factor 5A depletion on protein synthesis and of Saccharomyces cerevisiae. J Biol Chem. 1994;269:3934–3940. [PubMed] [Google Scholar]
- Kemper WM, Berry KW, Merrick WC. Purification and properties of rabbit reticulocyte protein synthesis initiation factors M2Balpha and M2Bbeta. J Biol Chem. 1976;251:5551–5557. [PubMed] [Google Scholar]
- Maeda I, Kohara Y, Yamamoto M, Sugimoto A. Large-scale analysis of gene function in Caenorhabditis elegans by high-throughput RNAi. Curr Biol. 2001;11:171–176. doi: 10.1016/s0960-9822(01)00052-5. [DOI] [PubMed] [Google Scholar]
- Magdolen V, Klier H, Wohl T, Klink F, Hirt H, Hauber J, Lottspeich F. The function of the hypusine-containing proteins of yeast and other eukaryotes is well conserved. Mol Gen Genet. 1994;244:646–652. doi: 10.1007/BF00282755. [DOI] [PubMed] [Google Scholar]
- Park MH. The post-translational synthesis of a polyamine-derived amino acid, hypusine, in the eukaryotic translation initiation factor 5A (eIF5A) J Biochem (Tokyo) 2006;139:161–169. doi: 10.1093/jb/mvj034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park MH, Wolff EC, Lee YB, Folk JE. Antiproliferative effects of inhibitors of deoxyhypusine synthase. Inhibition of growth of Chinese hamster ovary cells by guanyl diamines. J Biol Chem. 1994;269:27827–27832. [PubMed] [Google Scholar]
- Park MH, Lee YB, Joe YA. Hypusine is essential for eukaryotic cell proliferation. Biol Signals. 1997;6:115–123. doi: 10.1159/000109117. [DOI] [PubMed] [Google Scholar]
- Park MH, Joe YA, Kang KR. Deoxyhypusine synthase activity is essential for cell viability in the yeast Saccharomyces cerevisiae. J Biol Chem. 1998;273:1677–1683. doi: 10.1074/jbc.273.3.1677. [DOI] [PubMed] [Google Scholar]
- Park J-H, Aravind L, Wolff EC, Kaevel J, Kim YS, Park MH. Molecular cloning, expression, and structural prediction of deoxyhypusine hydroxylase: a HEAT-repeat-containing metalloenzyme. Proc Natl Acad Sci USA. 2006;103:51–56. doi: 10.1073/pnas.0509348102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R, Sheth U. P bodies and the control of mRNA translation and degradation. Mol Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Patel PH, Costa-Mattiolo M, Schulze KL, Bellen HJ. The Drosophila deoxyhypusine hydroxylase homologue nero and its target eIF5A are required for cell growth and the regulation of autophagy. J Cell Biol. 2009;185:1181–1194. doi: 10.1083/jcb.200904161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltz SW, Donahue JL, Jacobson A. A mutation in the tRNA nucleotidyltransferase gene promotes stabilization of mRNAs in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:5778–5784. doi: 10.1128/mcb.12.12.5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini P, Eyler DE, Green R, Dever TE. Hypusine-containing protein eIF5A promotes translation elongation. Nature. 2009;459:118–121. doi: 10.1038/nature08034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki K, Abid MR, Miyazaki M. Deoxyhypusine synthase gene is essential for cell viability in the yeast Saccharomyces cerevisiae. FEBS Lett. 1996;384:151–154. doi: 10.1016/0014-5793(96)00310-9. [DOI] [PubMed] [Google Scholar]
- Schnier J, Schwelberger HG, Smit-McBride Z, Kang HA, Hershey JW. Translation initiation factor 5A and its hypusine modification are essential for viability in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:3105–3114. doi: 10.1128/mcb.11.6.3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader R, Young C, Kozian D, Hoffmann R, Lottspeich F. Temperature-sensitive eIF5A mutant accumulates transcripts targeted to the nonsense-mediated decay pathway. J Biol Chem. 2006;281:35336–35346. doi: 10.1074/jbc.M601460200. [DOI] [PubMed] [Google Scholar]
- Schwelberger HG, Kang HA, Hershey JW. Translation initiation factor eIF-5A expressed from either of two yeast genes or from human cDNA. Functional identity under aerobic and anaerobic conditions. J Biol Chem. 1993;268:14018–14025. [PubMed] [Google Scholar]
- Shiba T, Mizote H, Kaneko T, Nakajima T, Kakimoto Y. Hypusine, a new amino acid occurring in bovine brain. Isolation and structural determination. Biochim Biophys Acta. 1971;244:523–531. doi: 10.1016/0304-4165(71)90069-9. [DOI] [PubMed] [Google Scholar]
- Spradling AC, Stern D, Beaton A, Rhem EJ, Laverty T, Mozden N, Misra S, Rubin GM. The Berkeley Drosophila genome project gene disruption project: single P-element insertions mutating 25% of vital Drosophila genes. Genetics. 1999;153:135–177. doi: 10.1093/genetics/153.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, Chen KY. Molecular cloning and functional expression of Neurospora deoxyhypusine synthase cDNA and identification of yeast deoxyhypusine synthase cDNA. J Biol Chem. 1995;270:23984–23987. doi: 10.1074/jbc.270.41.23984. [DOI] [PubMed] [Google Scholar]
- Thompson GM, Cano VS, Valentini SR. Mapping eIF5A binding sites for Dys1 and Lia1: in vivo evidence for regulation of eIF5A hypusination. FEBS Lett. 2003;555:464–468. doi: 10.1016/s0014-5793(03)01305-x. [DOI] [PubMed] [Google Scholar]
- Tong Y, Park I, Hong BS, Nedyalkova L, Tempel W, Park HW. Crystal structure of human eIF5A1: insight into functional similarity of human eIF5A1 and eIF5A2. Proteins. 2009;75:1040–1045. doi: 10.1002/prot.22378. [DOI] [PubMed] [Google Scholar]
- Valentini SR, Casolari JM, Oliveira CC, Silver PA, McBride AE. Genetic interactions of yeast eukaryotic translation initiation factor 5A(eIF5A) reveal connections to poly(A)-binding protein and protein kinase C signaling. Genetics. 2002;160:393–405. doi: 10.1093/genetics/160.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu VV, Emerson JP, Martinho M, Kim YS, Munck E, Park MH, Que L., Jr Human deoxyhypusine hydroxylase, an enzyme involved in regulating cell growth, activates O2 with a nonheme diiron center. Proc Natl Acad Sci USA. 2009;106:14814–14819. doi: 10.1073/pnas.0904553106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir BA, Yaffe MP. Mmd1p, a novel, conserved protein essential for normal mitochondrial morphology and distribution in the fission yeast Schizosaccharomyces pombe. Mol Biol Cell. 2004;15:1656–1665. doi: 10.1091/mbc.E03-06-0371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wöhl T, Klier H, Ammer H. The HYP2 gene of Saccharomyces cerevisiae is essential for aerobic growth: characterization of different isoforms of the hypusine-containing protein Hyp2p and analysis of gene disruption mutants. Mol Gen Genet. 1993;241:305–311. doi: 10.1007/BF00284682. [DOI] [PubMed] [Google Scholar]
- Wolff EC, Kang KR, Kim YS, Park MH. Posttranslational synthesis of hypusine: evolutionary progression and specificity of the hypusine modification. Amino Acids. 2007;33:341–350. doi: 10.1007/s00726-007-0525-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanelli CF, Valentini SR. Pkc1 acts through Zds1 and Gic1 to suppress growth and cell polarity defects of a yeast eIF5A mutant. Genetics. 2005;171:1571–1581. doi: 10.1534/genetics.105.048082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanelli CF, Valentini SR. Is there a role for eIF5A in translation? Amino Acids. 2007;33:351–358. doi: 10.1007/s00726-007-0533-0. [DOI] [PubMed] [Google Scholar]
- Zanelli CF, Maragno AL, Gregio AP, Komili S, Pandolfi JR, Mestriner CA, Lustri WR, Valentini SR. eIF5A binds to translational machinery components and affects translation in yeast. Biochem Biophys Res Commun. 2006;348:1358–1366. doi: 10.1016/j.bbrc.2006.07.195. [DOI] [PubMed] [Google Scholar]
- Zuk D, Jacobson A. A single amino acid substitution in yeast eIF-5A results in mRNA stabilization. EMBO J. 1998;17:2914–2925. doi: 10.1093/emboj/17.10.2914. [DOI] [PMC free article] [PubMed] [Google Scholar]