Summary
Microtubules are organized by the centrosome, a dynamic organelle that exhibits changes in both size and number during the cell cycle. Here we show that SZY-20, a putative RNA-binding protein, plays a critical role in limiting centrosome size in C. elegans. SZY-20 localizes in part to centrosomes and in its absence centrosomes possess increased levels of centriolar and pericentriolar components including γ-tubulin and the centriole duplication factors ZYG-1 and SPD-2. These enlarged centrosomes possess normal centrioles, nucleate more microtubules, and fail to properly direct a number of microtubule-dependent processes. Depletion of ZYG-1 restores normal centrosome size and function to szy-20 mutants whereas loss of szy-20 suppresses the centrosome duplication defects in both zyg-1 and spd-2 mutants. Our results describe a pathway that determines centrosome size and implicate centriole duplication factors in this process.
Keywords: centrosome, pericentriolar material, ZYG-1, SZY-20, C. elegans
Introduction
The centrosome serves as the primary microtubule-organizing center (MTOC) in animal cells. In this capacity, the centrosome participates in many processes including assembly and positioning of the mitotic spindle, chromosome segregation, and cytokinesis. The centrosome consists of a pair of centrioles surrounded by a mass of pericentriolar material (PCM) (Luders and Stearns, 2007). Centrioles are barrel-shaped structures, composed of a nine-fold radial symmetric array of microtubules and are structurally similar to the basal bodies that organize the microtubules of cilia and flagella. By contrast, the structure of the PCM is not well defined, but is enriched in coiled-coil proteins that are thought to form a framework that supports microtubule nucleation and anchoring (Azimzadeh and Bornens, 2007).
To ensure spindle bipolarity and the proper transmission of centrioles, the centrosome must be duplicated precisely once per cell cycle. The central event in centrosome duplication is the assembly of a single daughter centriole next to each preexisting (mother) centriole. Recent work indicates that the molecular pathway of centriole assembly is similar in humans and worms, and relies upon a conserved set of core duplication factors (Delattre et al., 2006; Kleylein-Sohn et al., 2007; Pelletier et al., 2006). In C. elegans, the kinase ZYG-1 (O'Connell et al., 2001), a relative of human Plk4 and Drosophila Sak (Bettencourt-Dias et al., 2005; Habedanck et al., 2005) localizes early to the site of centriole assembly (Delattre et al., 2006; Kleylein-Sohn et al., 2007; Pelletier et al., 2006) and is required to recruit a series of coiled-coil proteins including SAS-4 (Kirkham et al., 2003; Leidel and Gonczy, 2003), SAS-5 (Delattre et al., 2004), and SAS-6 (Dammermann et al., 2004; Leidel et al., 2005). Likewise, during centriole assembly in human cells, Plk4 recruits orthologs of SAS-4 and SAS-6 (Kleylein-Sohn et al., 2007).
Centrosome duplication in C. elegans also requires the activity of another conserved factor called SPD-2 (Kemp et al., 2004; Pelletier et al., 2004). SPD-2 is a component of both PCM and centrioles, and has a conserved role in PCM assembly (Dix and Raff, 2007; Giansanti et al., 2008; Gomez-Ferreria et al., 2007; Kemp et al., 2004; Pelletier et al., 2004; Zhu et al., 2008). During centrosome duplication in C. elegans, SPD-2 is required to localize ZYG-1 to sites of centriole assembly (Delattre et al., 2006; Pelletier et al., 2006). Yet SPD-2 does not appear to be required for centrosome duplication in flies (Dix and Raff, 2007; Giansanti et al., 2008) and in humans this role remains controversial (Gomez-Ferreria et al., 2007; Zhu et al., 2008). Thus, the role of SPD-2 in centrosome duplication does not appear to be absolutely conserved, suggesting that in some species ZYG-1 homologs can localize independently of SPD-2.
Recent studies indicate that the PCM plays a role in daughter centriole assembly. In worms, depletion of PCM components such as γ-tubulin and the coiled-coil protein SPD-5 inhibits daughter centriole formation (Dammermann et al., 2004) while in vertebrates experimental expansion of the PCM promotes the formation of multiple daughter centrioles (Loncarek et al., 2008). These results indicate that the PCM provides an environment favorable for centriole assembly. Conversely centrioles organize PCM. Disruption of centrioles in human cells results in dispersion of the PCM, suggesting that the centriole possesses a PCM-organizing activity (Bobinnec et al., 1998). This is supported by work in C. elegans where partial depletion of centriole duplication factors has been shown to lead to partial assembly of centrioles and a corresponding reduction in PCM, suggesting that a centriole-associated activity determines centrosome size (Delattre et al., 2004; Kirkham et al., 2003; Leidel et al., 2005). What this activity is and how centrosome size is determined remain unclear.
Here we have identified a conserved protein SZY-20 that limits the size of centrosomes in C. elegans embryos. Loss of SZY-20 activity results in an approximately two-fold increase in the amounts of centrosome-associated ZYG-1, SPD-2, SPD-5 and γ-tubulin. This increase in centrosome size is associated with an increase in microtubule nucleating capacity, defects in microtubule-dependent processes, and embryonic lethality. Reducing ZYG-1 activity in szy-20 mutants restores centrosome size, normal microtubule function, and to some extent embryonic viability, indicating that the level of centriole-associated ZYG-1 determines centrosome size. We also find that centrosome size is influenced by SAS-6 activity.
Results
SZY-20 is a negative regulator of centrosome duplication
The szy-20 gene was identified in a screen for genetic suppressors of the temperature-sensitive (ts) lethality of the zyg-1(it25) mutation (Kemp et al., 2007). In zyg-1(it25) embryos raised at the restrictive temperature (24°C), duplication of the sperm-derived centriole pair invariably fails. This initial centriole pair, however, is able to separate and direct formation of a bipolar spindle. Such embryos divide once to form a two-cell embryo but during the second round of mitosis, each blastomere assembles a monopolar spindle (Figure 1A). As a result, 100% of the embryos die (O'Connell et al., 2001). The szy-20(bs52) mutation however suppresses the embryonic lethality of zyg-1(it25) such that a percentage of double mutant embryos are able to complete development and hatch at the restrictive temperature (Table 1A). Importantly, the szy-20(bs52) mutation is unable to suppress a complete loss of zyg-1 activity indicating that suppression might involve upregulation of ZYG-1 (Kemp et al., 2007).
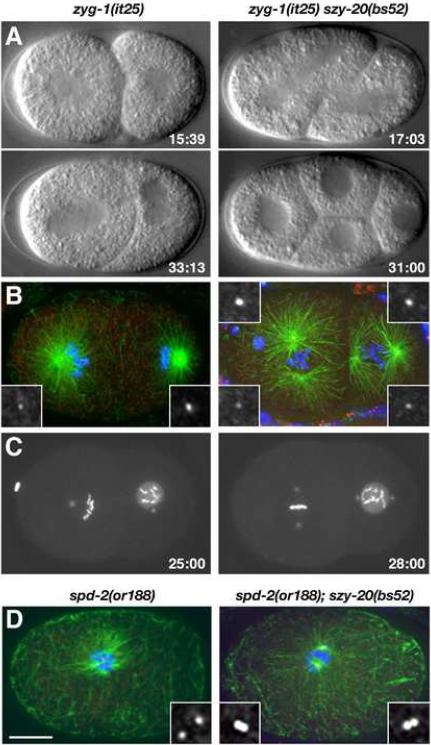
Figure 1.
Loss of szy-20 activity suppresses centrosome duplication defects in zyg-1 and spd-2 mutants. (A–C) Centrosome duplication and bipolar spindle formation are restored in zyg-1(it25) szy-20(bs52) embryos. (A) Selected images from 4D-DIC recordings. Only the double mutant embryo progresses to the four-cell stage. (B) Two-cell stage embryos stained for microtubules (green), centrioles (SAS-4, red) and DNA (blue). (C) Selected images from recordings of embryos expressing GFP-SPD-2 and GFP-histone. Time (minutes) is relative to first metaphase. (D) One-cell embryos stained for microtubules (green), SAS-4 (red) and DNA (blue). The spd-2(or188) embryo has two centrioles and the spd-2(or188) szy-20(bs52) embryo four. Each image is a Z-projection. Insets are magnified 3-fold. Bar, 10 μm.
Table 1.
Genetic Analysis of szy-20
| A. Inhibition of szy-20 suppresses zyg-1 mutations | |||
|---|---|---|---|
| °C | % Embryonic Viability (ave ± s.d.) | Na | |
| zyg-1(it25) | 24 | 0 ± 0 | 352 |
| zyg-1(it25) szy-20(bs52) | 4.7 ± 2.1 | 618 | |
| zyg-1(it25) RNAi neg controlb | 0 ± 0 | 200 | |
| zyg-1(it25) C18E9.3 RNAi | 2.6 ± 2.6 | 2467 | |
| zyg-1(or409) RNAi neg controlb | 0 ± 0 | 466 | |
| zyg-1(or409) C18E9.3 RNAi | 8.2 ± 9.7 | 1952 | |
| zyg-1(it25) RNAi neg controlb | 23.5 | 0.3 ± 0.003 | 752 |
| zyg-1(it25) C18E9.3 RNAi | 27.2 ± 18.7 | 1123 | |
| zyg-1(it25) | 20 | 96.2 ± 2.5 | 909 |
| szy-20(bs52) | 55.0 ± 8.7c | 909 | |
| zyg-1(it25) szy-20(bs52) | 92.1 ± 3.3c | 900 | |
| B. szy-20 is required for embryogenesis | |||
|---|---|---|---|
| °C | % Embryonic Viability (ave ± s.d.) | N | |
| szy-20(bs52) | 20 | 55.0 ± 8.7 | 909 |
| szy-20(tm1997) | 63.2 ± 10.0 | 199 | |
| C18E9.3 RNAi | 92.3 ± 8.4 | 1315 | |
| szy-20(bs52) C18E9.3 RNAi | 4.8 ± 6.6 | 380 | |
| szy-20(bs52) | 24 | 24.6 ± 13.0 | 668 |
| szy-20(tm1997) | 23.1 ± 17.9 | 324 | |
| szy-20(bs52) | 25 | 1.3 ± 0.7 | 152 |
| szy-20(bs52)/+ | 99.6 ± 0.9 | 224 | |
| szy-20(tm1997) | 0.4 ± 0.9 | 261 | |
| szy-20(bs52)/szy-20(tm1997) | 3.9 ± 4.8 | 573 | |
| +/mnDf68d | 55.4 ± 24.2 | 428 | |
| szy-20(bs52)/mnDf68 | 4.5 ± 8.3 | 620 | |
| +/mnDf104e | 66.7 ± 7.6 | 696 | |
| szy-20(bs52)/mnDf104 | 61.1 ± 9.5 | 1657 | |
| N2 | 99.9 ± 0.1 | 202 | |
| RNAi neg controlb | 99.7 ± 0.5 | 505 | |
| C18E9.3 RNAi | 13.6 ± 8.1 | 902 | |
Number of embryos scored.
For negative controls for RNAi soaking experiments, animals were incubated in M9 buffer lacking RNA.
P-value=0.0069 (Student t-test; two-tailed distribution and two-sample equal variance)
The deficiency chromosome mnDf68 carries a large deletion of LG II that encompasses ORF C18E9.3.
The deficiency chromosome mnDf104 carries a large deletion of LG II that does not encompass ORF C18E9.3
To investigate the basis of suppression, we analyzed bipolar spindle formation and centrosome duplication directly in zyg-1(it25) szy-20(bs52) double mutant embryos. By four-dimensional differential interference contrast (4D-DIC) imaging (Figure 1A and Movie S1), we found that zyg-1(it25) szy-20(bs52) embryos are often successful in assembling bipolar spindles at the two-cell stage (13/24 events), unlike zyg-1(it25) embryos (0/28 events). The ability of the double mutant to assemble bipolar spindles appears to result from suppression of the centrosome duplication defect, as spindle poles in zyg-1(it25) szy-20(bs52) embryos invariably stained with the centriole marker SAS-4 (Figure 1B). The ability of the szy-20(bs52) mutation to suppress the centrosome duplication defect was further confirmed by fluorescence imaging of live embryos expressing GFP-SPD-2 to mark centrosomes and GFP-histone to mark DNA (Figure 1C and Movie S2). As szy-20(bs52) is a loss-of-function allele (see below), our results demonstrate that szy-20 negatively regulates centriole assembly.
The szy-20(bs52) mutation restores centrosome duplication in spd-2 mutants
We next asked if the szy-20(bs52) mutation could also suppress mutations in other genes required for centrosome duplication. spd-2 is required for both centrosome duplication and PCM assembly. In spd-2 mutants, the sperm-derived centriole pair fails to duplicate and only weakly organizes microtubules (Kemp et al., 2004; Pelletier et al., 2004). As a result, spd-2 mutants fail to assemble the first mitotic spindle and arrest at the one-cell stage. We generated a double mutant line and found that the szy-20(bs52) mutation only weakly suppresses the embryonic lethality of the hypomorphic spd-2(or188) mutation (data not shown). When we examined the spd-2(or188); szy-20(bs52) double mutant by immunofluorescence microscopy, we found that the microtubule organization and one-cell arrest phenotypes were still present. However, as judged by SAS-4 immunostaining, the double mutant embryos exhibited evidence of centrosome duplication (Figure 1D). While only seven percent of one-cell spd-2(or188) embryos (n=28) possessed more than the two original sperm-derived centrioles, 65% of spd-2(or188); szy-20(bs52) embryos (n=45) possessed three or more centrioles. Thus, szy-20 genetically interacts with zyg-1 and spd-2, two genes whose products function at the earliest known step in centrosome duplication (Delattre et al., 2006; Pelletier et al., 2006).
As a control we assayed the ability of the szy-20(bs52) mutation to suppress an unrelated ts mutant. At 24°C, the mat-3(or180) mutation blocks the function of the anaphase-promoting complex, producing an embryonic lethal phenotype marked by a one-cell arrest (Stein et al., 2007). Analysis of a szy-20(bs52); mat-3(or180) double mutant revealed that szy-20(bs52) suppresses neither the embryonic lethal phenotype nor the one-cell arrest of mat-3(or180) mutants (data not shown), indicating that szy-20(bs52) is not a general suppressor of ts mutations.
Maternally supplied SZY-20 is essential for embryonic development
The szy-20(bs52) mutation confers its own recessive ts embryonic lethal phenotype (Table 1B and Kemp et al., 2007). At 20°C greater than 50% of the progeny of szy-20(bs52) hermaphrodites are viable, but at 25°C nearly all of the progeny fail to hatch. As the embryonic lethality of szy-20(bs52) hermaphrodites can not be rescued by mating to wild-type males (data not shown), maternally-expressed SZY-20 is essential for embryogenesis.
Preliminary analysis of szy-20(bs52) embryos revealed defects in cytokinesis and in attachment of the centrosome to the nucleus (Kemp et al., 2007). To further characterize the consequences of loss of szy-20, we analyzed a large number of szy-20(bs52) embryos stained for microtubules, DNA and centrosomes (Figure 2). At 25°C, szy-20(bs52) embryos exhibit multiple defects in cell division. As observed previously, some embryos contain extra nuclei and centrosomes, a phenotype suggestive of cytokinesis failure (20%, n=135; Figure 2A), while in others, centrosomes lose contact with the nuclear envelope (12%, n=135; Figure 2B). We also observed defects that had not been previously observed. szy-20(bs52) embryos exhibit a 10% decrease in the length of metaphase spindles compared to wild-type embryos (n=12; Figure 4D). We also noticed the presence of more than one sperm pronucleus-centrosome pair in a small fraction (<5%) of embryos, a phenotype suggestive of polyspermy (Figure 2C).
Figure 2.
SZY-20 is required for cell division. szy-20(bs52) embryos stained for microtubules (green), centrosomes (red) and DNA (blue). (A) A one-cell embryo with four centrosomes stained for ZYG-1. (B) A two-cell embryo with detached centrosomes (arrows) stained for SPD-2. (C) A zygote with two sperm pronuclei and centrosomes (boxed) stained for SAS-4. Each image is Z-projection. Insets are magnified 3-fold. Bar, 10 μm.
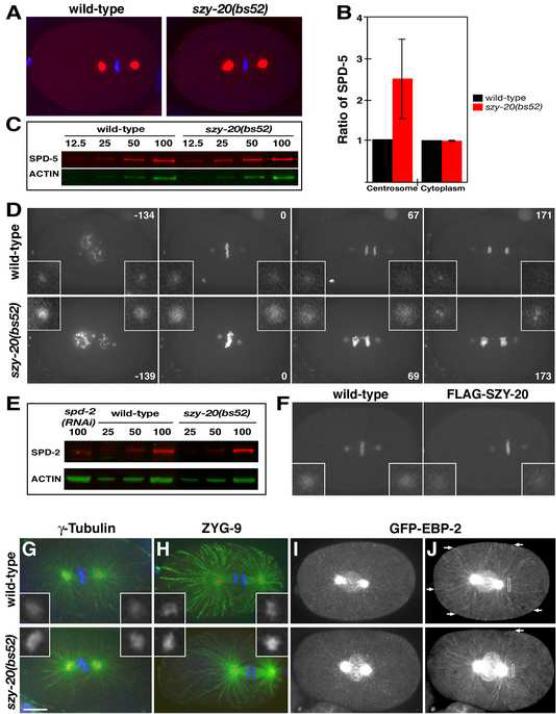
Figure 4.
SZY-20 limits the size and the microtubule-nucleating capacity of the centrosome. (A) Embryos at first metaphase stained for SPD-5 (red) and DNA (blue). (B) Fluorescence intensity measurements of centrosomal and cytoplasmic SPD-5. Vertical bars indicate the standard deviation. (C) Quantitative immunoblot of SPD-5. Relative loading volumes are indicated above lanes. Actin served as a loading control (D) Still images from recordings of embryos expressing GFP-SPD-2 and GFP-histone. Note that in the szy-20(bs52) embryo, more chromosomes are present due to polar body extrusion failure and that the metaphase spindle is shorter. Time (in seconds) is relative to metaphase. (E) Quantitative immunoblot of SPD-2. (F) Overexpression of wild-type FLAG-SZY-20 reduces GFP-SPD-2 fluorescence at centrosomes. (G, H) Embryos stained for microtubules (green), centrosomes (red) and DNA (blue). The levels of (G) γ-tubulin and (H) ZYG-9 are increased at szy-20(bs52) centrosomes compared to the wild type. (I, J) Time-lapse recordings of embryos expressing GFP-EBP-2. (I) A Z-projection of a single time point illustrates the presence of more GFP-EBP-2 at szy-20(bs52) centrosomes. (J) Time projections of a 10-second interval show the tracks of growing microtubules. During this period, more microtubules grow to reach the cortex (arrows) in the wild type than in the mutant. Boxes (6 μm2) indicate area used to measure GFP-EBP-2 fluorescence. Each image is a Z-projection. Insets are magnified 3-fold. Bar, 10 μm.
To further define szy-20 function, we performed live imaging of szy-20(bs52) embryos grown at 25°C (Movie S3 and S4). Time-lapse recordings confirmed a cytokinesis defect in szy-20(bs52) embryos. In these embryos, the furrow initiates normally but fails to ingress completely, resulting in the formation of a multinucleated cell with four centrosomes. szy-20(bs52) embryos also frequently fail to extrude polar bodies and exhibit a slight (1.2–1.4 fold) lengthening of the cell cycle. While these defects are observed at all temperatures, the severity and penetrance of each defect increases at higher temperature. We also analyzed the deletion allele szy-20(tm1997) and found that it confers a ts embryonic lethal phenotype and many of the cytological defects described for szy-20(bs52) (Table 1B and data not shown).
Molecular characterization of szy-20
To further define the role of SZY-20, we sought to clone the corresponding gene located between dpy-10 and unc-4 on chromosome II (Kemp et al., 2007). Using a physical mapping approach, we localized szy-20 to a small interval of 54.2 Kb that encompasses 14 predicted open reading frames (ORFs; Figure S1). Sequencing detected a nonsense (C-to-T) mutation in exon 6 of ORF C18E9.3 that presumably results in truncation of the gene product (Figures 3A and S1).
Figure 3.
szy-20 encodes a novel conserved protein. (A) Schematic of wild-type and mutant forms of SZY-20. Due to a frame shift, the product of the szy-20(tm1997) allele is expected to contain a novel 36-amino acid extension (blue box). (B) Immunoblot of wild-type and szy-20(bs52) embryonic extracts probed for SZY-20 (red), and α-tubulin (green) as a loading control. (C–G) SZY-20 localizes to the centrosome. Embryos stained for microtubules (green), SZY-20 (αS20N, red) and DNA (blue): Wild-type embryos at (C) meiosis, (D) first metaphase, (E) first anaphase, (F) second interphase, and (H) a szy-20(RNAi) embryo at metaphase. (G) A metaphase embryo co-stained for SZY-20 (red) and GFP-SAS-4 (green). Boxes highlight centrosomes. Insets are magnified 3-fold. Bar, 10 μm.
To confirm the identity of szy-20, we used RNAi to inhibit expression of C18E9.3 in both wild-type and zyg-1(it25) animals. In all respects, C18E9.3 RNAi phenocopied the szy-20(bs52) mutation. C18E9.3 RNAi partially suppressed zyg-1(it25) embryonic lethality (Table 1A); this was most obvious at 23.5°C where C18E9.3 RNAi of zyg-1(it25) animals led to a 100-fold increase in the frequency of viable progeny (27.2 % versus 0.27% in controls). C18E9.3 RNAi also suppressed the embryonic lethality of the zyg-1(or409) allele. In wild-type animals, C18E9.3 RNAi produced a ts embryonic lethal phenotype reminiscent of szy-20(bs52): <10% embryonic lethality at 20°C, but ~90% at 25°C (Table 1B). Further, combining the mutation and RNAi had a synergistic effect at 20°C: only ~5% of the progeny of szy-20(bs52) C18E9.3 RNAi mothers survived, compared to 55% for szy-20(bs52) and 92.3 % for C18E9.3 RNAi. Finally, szy-20(bs52) failed to complement szy-20(tm1997), a partial deletion of C18E9.3 (Table 1B). We conclude that szy-20 corresponds to ORF C18E9.3. Based on the ability to phenocopy szy-20(bs52) by RNAi as well as the failure of szy-20(bs52) to complement mnDf68, a chromosomal deficiency that removes the szy-20 locus (Table 1B), we conclude that szy-20(bs52) is a loss-of-function allele.
Wormbase (http://www.wormbase.org/) lists multiple ESTs for C18E9.3. We sequenced four of these cDNA clones and identified two alternatively spliced transcripts (Figure S1). The longer transcript encodes a protein of 558 amino acids (Isoform A; GenBank accession EF656060) and the shorter transcript a protein of 457 amino acids (Isoform B; GenBank accession EF656061). Both messages are trans-spliced to the SL1 leader sequence and appear to utilize the same initiation and termination codons.
szy-20 encodes a conserved protein
A BLAST search (Experimental Procedures) identified SZY-20 orthologs in other species including flies and humans. All animal genomes with significant sequence data contain one ortholog of SZY-20, indicating a potentially conserved role across animals. A multiple alignment of SZY-20 with its homologs showed that they all share three prominent blocks of conservation (Figure 3A and S2). The N-terminal block is a short element with a characteristic hx[DE][SN]W[DE][DE] signature (where h is any hydrophobic residue and x is any residue), which is found exclusively in orthologs of SZY-20 (Figure S2A). The second block in the center of the protein, termed the SUZ domain, is the strongest stretch of conservation and is enriched in charged residues (Figure S2B). The third block, the SUZ-C domain, is at the extreme C-terminus and is defined by a characteristic pattern of two highly conserved glycines and one absolutely conserved proline (Figure S2C). In nematodes, however, this domain has rapidly diverged.
We noticed that the SUZ and SUZ-C domains occur independently in proteins outside of the SZY-20 family (Figure S3 and Supplemental Results). Interestingly, many of these proteins contain known RNA-binding domains suggesting that SZY-20 might function in RNA metabolism. We tested SZY-20 for RNA-binding in vitro. GST-SZY-20 specifically and reproducibly co-precipitates with polyuridine-coupled beads, indicating that SZY-20 can directly bind RNA (Figure S4A). Both the SUZ and SUZ-C domains contribute to this activity as mutation of either domain significantly reduces the ability of the protein to co-precipitate with the beads. Furthermore, RNA-binding is nearly eliminated in a double mutant (GST-SZY-20dm) that carries mutations in both domains (Figure S4B). To investigate the function of these putative RNA-binding domains in vivo, we expressed FLAG-tagged versions of wild-type and SZY-20dm in worms (Figure S4C). When expressed in szy-20(bs52) animals grown at 25°C, the wild-type FLAG-SZY-20 protein provided strong rescue of the embryonic lethality, reducing it to 52% (n=2615) from 96% in controls (n=936). In contrast, the FLAG-SZY-20dm mutant had little effect (84% embryonic lethality, n=1215), consistent with the notion that RNA-binding is involved in SZY-20 function.
SZY-20 localizes to nuclei, cytoplasm and centrosomes
To determine how SZY-20 might function, we examined its subcellular distribution using affinity-purified antibodies. On immunoblots, the αS20N antibody detected a single band in wild-type embryonic extracts. The migration of this band was consistent with the size of isoform A (Figure 3B). This band was absent in szy-20(bs52) embryonic extracts. Instead a faster migrating band, presumably corresponding to the truncated product, was detected.
In immunostained embryos, SZY-20 is found in small foci in the cytoplasm and at centrosomes where it coincides with GFP-SAS-4, a marker of centrioles (Figure 3C–G). In addition, SZY-20 localizes to nucleoli during interphase and to the surface of chromosomes during meiosis and mitosis (Figure S5). RNAi of szy-20 significantly reduces staining of all structures, confirming the specificity of the antibody (Figure 3H). SZY-20 is first detected at centrosomes during meiosis where it often appears as two distinct dots adjacent to the male pronucleus (Figure 3C). While SZY-20 localizes to the centrosome throughout the cell cycle, the extent of its association is regulated in a cell cycle-dependent manner (Figure 3C–F). The level of centrosome-associated SZY-20 peaks at prometaphase and metaphase, slowly declines, and reaches a minimal level during interphase. In contrast, the level of ZYG-1 at centrosomes is lowest at metaphase (Delattre et al., 2006), the time when the level of SZY-20 peaks. ZYG-1 then rapidly increases during anaphase while SZY-20 levels drop. This reciprocal relationship between centrosomal levels of SZY-20 and ZYG-1 suggests that SZY-20 acts locally to regulate ZYG-1.
SZY-20 limits centrosome size
During our cytological characterization, we noticed that centrosomes appear larger in szy-20(bs52) than in wild-type embryos. This difference was apparent in embryos stained for the pericentriolar component SPD-5 (Figure 4A). We measured the fluorescence intensity of stained centrosomes at first metaphase (Figure 4B) and found that szy-20(bs52) embryos (n=31) contain more than twice as much centrosomal SPD-5 as controls (n=10). In contrast, cytoplasmic pools of SPD-5 appear similar in the two strains, suggesting that the overall levels of SPD-5 are unaffected by the szy-20(bs52) mutation. This was confirmed by quantitative immunoblotting (Figure 4C), which showed that wild-type and mutant embryos possessed similar amounts of SPD-5 (mutant/wt = 1.12 ± 0.09, n=4). This result suggests that the szy-20(bs52) mutation might enhance recruitment of SPD-5 to centrosomes.
SPD-2 and SPD-5 function at the top of a hierarchy in the centrosome maturation pathway (Hamill et al., 2002; Kemp et al., 2004; Pelletier et al., 2004). In addition to recruiting downstream components, SPD-2 and SPD-5 localize to PCM in a mutually dependent manner. We postulated that the szy-20(bs52) mutation would affect the level of SPD-2 at centrosomes and analyzed embryos expressing GFP-SPD-2 (Figure 4D). As predicted, the level of centrosome-associated GFP-SPD-2 is significantly increased in the szy-20(bs52) mutant at all stages of the cell cycle. The increase in centrosomal levels of GFP-SPD-2 reflects increases in both density (1.4 ± 0.6 fold) and cross-sectional area (1.4 ± 0.5 fold). However, we detected no significant difference in overall levels of endogenous SPD-2 between wild-type and szy-20(bs52) embryos (mutant/wt = 1.08 ± 0.14, n=5) as shown in Figure 4E. Thus, the elevated levels of SPD-5 and SPD-2 at mutant centrosomes most likely result from enhanced recruitment.
To further confirm the role of SZY-20 in regulating centrosome size, we overexpressed SZY-20 in wild-type embryos (Figure 4F). Consistent with a role as a negative regulator, overexpression of wild-type FLAG-SZY-20 reduces the centrosomal level of GFP-SPD-2 by half (0.48 ± 0.1 fold, n=11) compared to controls (n=14). Interestingly, we did not observe an effect following overexpression of FLAG-SZY-20dm (1.2 ± 0.3 fold, n=12). Thus, the conserved domains that mediate RNA-binding in vitro are important for regulating centrosome size.
SZY-20 limits the microtubule-nucleating capacity of the centrosome
The microtubule-nucleating capacity of the centrosome positively correlates with the amount of PCM. Since centrosomes in szy-20(bs52) embryos contain elevated levels of PCM components, we wondered if such centrosomes nucleate more microtubules. First we analyzed the centrosome level of γ-tubulin, a PCM component directly involved in microtubule nucleation (Figure 4G). Similar to other centrosome proteins, γ-tubulin levels at centrosomes are elevated in the mutant (2.5 ± 1.2 fold, n=14) relative to the wild type (n=13).
We next investigated microtubule nucleation using strains expressing GFP-EBP-2, a marker for microtubule plus ends (Srayko et al., 2005). Compared to controls, szy-20(bs52) centrosomes exhibit a significant increase in GFP-EBP-2 fluorescence (Figure 4I). We then made time-lapse recordings of embryos expressing GFP-EBP-2. In projected images from these recordings, it was evident that more microtubules grow out from centrosomes in szy-20(bs52) than in wild-type embryos (Figure 4J and S6A, Movie S5). As a means of quantifying the difference in the outgrowth of microtubules between the two strains, we measured the intensity of GFP-EBP-2 in an area adjacent to the centrosome. We found that GFP-EBP-2 fluorescence is increased in the mutant (2.0 ± 0.6 fold, n=8) compared to controls (n=8), consistent with the presence of more centrosome-associated microtubules in the mutant. Interestingly, microtubule growth appeared stunted in the mutant. In a ten-second interval, many microtubules nucleated by wild-type centrosomes grow out to meet the cortex, while most microtubules nucleated by szy-20(bs52) centrosomes grow only a short distance (Figure 4J and S6B, Movie S6). In fact, only about half as many microtubules reach the cortex in szy-20(bs52) embryos as in controls. The lack of long microtubules in the mutant is not due to insufficient recruitment of the microtubule-stabilizing protein ZYG-9 (Matthews et al. 1998) as szy-20(bs52) centrosomes (n=16) actually possess more ZYG-9 (1.7 ± 0.7 fold) than controls (n= 24) (Figure 4H). Instead, microtubules grow slower in the mutant (1.1 μm/s) than in the wild type (1.3 μm/s). Our results are reminiscent of those of (Srayko et al., 2005) who found conditions that favor microtubule polymerization slow the growth rate of individual microtubules, presumably by depleting free tubulin subunits.
SZY-20 regulates the level of centrosome-associated ZYG-1
Since szy-20(bs52) suppresses zyg-1 mutations, we wondered if the loss of SZY-20 affects the localization of ZYG-1. In immunostained szy-20(bs52) embryos, we found that the level of ZYG-1 at the centrosome varies during the cell cycle as it does in the wild type, with the lowest level at metaphase and highest level at anaphase. However, at all cell cycle stages, the level of ZYG-1 is higher at szy-20(bs52) centrosomes than at wild-type centrosomes (Figure 5A and S7). This is evident during prometaphase and metaphase where ZYG-1 was detected in 64% of szy-20(bs52) centrosomes (n=117) but in only 23% of wild-type centrosomes (n=86). Quantitative fluorescence microscopy at metaphase revealed that szy-20(bs52) mutants contain more than twice as much centrosome-associated ZYG-1 as wild-type embryos (Figure 5B). However, we found no difference in the cytoplasmic levels of ZYG-1 between the two strains, suggesting that loss of szy-20 activity does not simply result in overexpression of ZYG-1. This was confirmed by quantitative immunoblotting (Figure 5C) which found that wild-type and mutant embryos possess equivalent amounts of ZYG-1 (mutant/wt = 1.03 ± 0.12, n=6). In contrast, we found that depletion of ZYG-1 did not affect SZY-20 localization (data not shown). Thus, SZY-20 regulates ZYG-1 levels at the centrosome but not vice-versa.
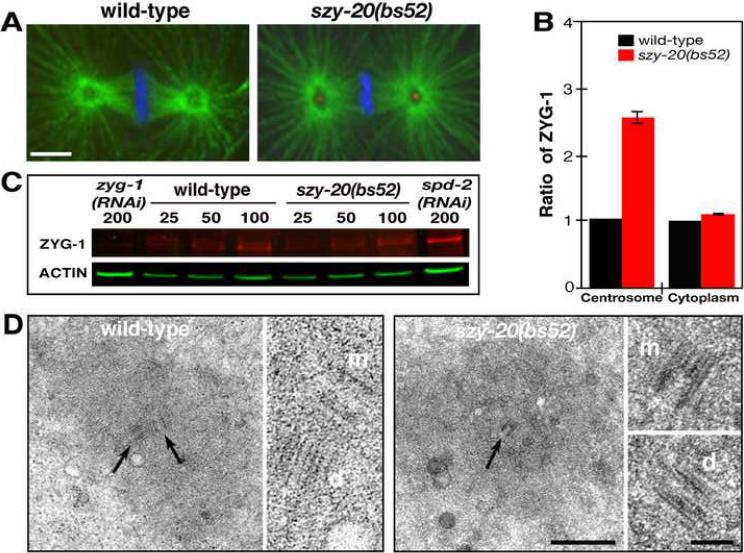
Figure 5.
Loss of SZY-20 activity enhances ZYG-1 localization at centrosomes. (A) a Z-projections of embryos at first metaphase stained for microtubules (green), ZYG-1 (red) and DNA (blue). ZYG-1 is detectable only at szy-20(bs52) centrosomes. Bar, 5 μm. (B) Fluorescence intensity measurements of centrosomal and cytoplasmic ZYG-1. Vertical bars indicate the standard deviation. (C) Quantitative immunoblot of ZYG-1. Relative loading volumes are indicated above lanes. Actin served as a loading control. (D) TEM of centrosomes at anaphase. In the large panels, a wild-type centriole pair (arrows) and the mother centriole (arrow) of a szy-20(bs52) centriole pair are shown. In the small panels, all mother (m) and daughter (d) centrioles are shown at higher magnification. Bars, 500 nm for lower and 100 nm for higher magnification images.
As the enlarged centrosomes in szy-20(bs52) mutants possess elevated levels of the centriole duplication factor ZYG-1, we wanted to determine if they also possess centrioles of altered structure. Thin section TEM revealed that centrioles are of similar size and structure in wild-type (n=2) and szy-20(bs52) centrosomes (n=10) (Figure 5D and S8). Thus, the elevated level of PCM observed in szy-20 mutants appears to occur in the absence of centriole duplication defects.
Centriole duplication factors regulate centrosome size
In the course of our analysis, we uncovered an unusual genetic relationship between zyg-1 and szy-20. Not only does szy-20(bs52) suppress zyg-1(it25) but the converse is also true (Table 1A). At 20°C, nearly 100% of the progeny produced by zyg-1(it25) hermaphrodites are viable while only 55% of szy-20(bs52) embryos are viable. Remarkably, over 90% of the progeny of zyg-1(it25) szy-20(bs52) double mutants are viable. This difference between the szy-20(bs52) and zyg-1(it25) szy-20(bs52) strains is statistically significant (p <0.01), thus at 20°C loss of zyg-1 activity restores proper embryonic development to szy-20(bs52) mutants.
The mutual suppression exhibited by zyg-1 and szy-20 loss-of-function mutations suggests a simple model. A reduction in SZY-20 activity results in increased levels of ZYG-1 at the centrosome, which in turn has deleterious consequences for the embryo. Reducing ZYG-1 activity in the szy-20(bs52) mutant could restore an appropriate level of centrosome-associated ZYG-1 and thus normal cellular processes. To analyze the interactions between zyg-1 and szy-20 at a cytological level, we performed time-lapse microscopy. At 20°C, szy-20(bs52) zygotes exhibit highly penetrant defects in microtubule-dependent processes such as pronuclear rotation (87%, n=23; Figure 6A) and cytokinesis (63%, n=19; Movie S3 and S4). In wild-type embryos, the two apposed pronuclei rotate 90° to position the centrosomes and ultimately the spindle on the A–P axis. While this rotation is normally completed by prometaphase in wild-type embryos, it often does not initiate until metaphase in szy-20(bs52) embryos. We observed robust rescue of both defects in szy-20(bs52) mutants when zyg-1 activity was reduced. In the zyg-1(it25) szy-20(bs52) double mutant, the pronuclear rotation defect is reduced to 12.5% (n=16) and the cytokinesis defect to 17% (n=12).
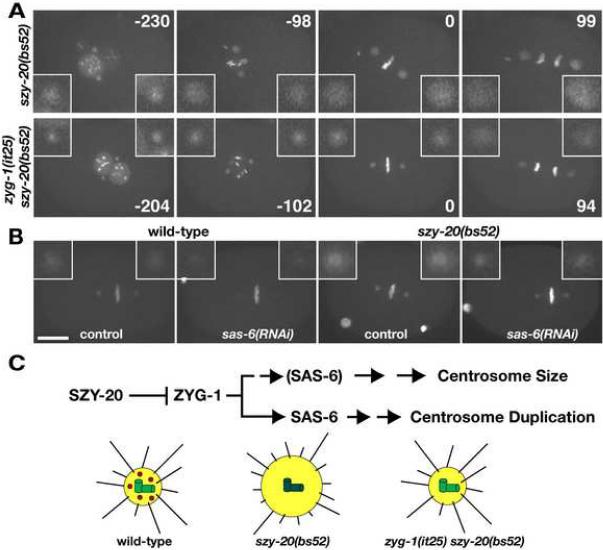
Figure 6.
Centriole duplication factors regulate centrosome size. (A, B) Images from recordings of embryos expressing GFP-SPD-2 and GFP-histone. (A) Reducing zyg-1 activity restores normal centrosome size and function to szy-20(bs52) mutants. Note that the delay in pronuclear rotation observed in szy-20(bs52) is rescued by the zyg-1(it25) mutation. Time (seconds) is relative to first metaphase. (B) sas-6(RNAi) reduces centrosome size in both wild-type and szy-20(bs52) embryos. Shown are Z-projections at each time point. Insets are magnified 3-fold. Bar, 10 μm. (C) A molecular pathway for controlling centrosome size (Top). SZY-20 acts upstream and controls the centrosomal level of ZYG-1, which regulates both centrosome duplication and size. The centriole duplication factor SAS-6 also influences centrosome size. It is shown in parentheses in the `Centrosome Size' Pathway to indicate that its role here has not yet been firmly established. A depiction of the effects of SZY-20 on the centrosome (Bottom). In the wild type, SZY-20 (red) restricts the amount of ZYG-1 (green) at centrioles, thereby establishing the proper level of PCM (yellow) and microtubules (black lines). In szy-20 mutants, ZYG-1 levels at centrioles increase, leading to enhanced recruitment of PCM and the nucleation of more microtubules (on average these are shorter than those nucleated by wild-type centrosomes). Reducing ZYG-1 activity in szy-20 mutants restores normal levels of PCM and microtubule-nucleation.
The pronuclear rotation and cytokinesis defects of szy-20(bs52) embryos could be a consequence of the enlarged centrosomes. That is, by nucleating more microtubules, the enlarged centrosomes deplete free tubulin and impede the formation of long microtubules that are needed for these processes. If true, suppression of both defects by the zyg-1(it25) allele might involve the restoration of centrosome size. To test this, we compared the level of centrosome-associated GFP-SPD-2 in szy-20(bs52) and zyg-1(it25) szy-20(bs52) zygotes grown at 20°C (Figure 6A). Consistent with our prediction, GFP-SPD-2 fluorescence was reduced by nearly half (0.59 ± 0.07) at zyg-1(it25) szy-20(bs52) centrosomes (n=12) compared to szy-20(bs52) centrosomes (n=14). Importantly, these experiments were conducted at 20°C where centrosome duplication is not blocked by the zyg-1(it25) mutation. Thus, the reduction in centrosome size is not a consequence of blocking daughter centriole formation. We further found that in the wild type, depletion of ZYG-1 by RNAi reduces the level of GFP-SPD-2 at the centrosome by more than half (0.4 ± 0.26 fold, n=11) relative to controls (n=10). Therefore, ZYG-1 has two separable functions. It regulates centrosome duplication and controls centrosome size. These results also show that inappropriately high levels of ZYG-1 at the centrosome can perturb centrosome function.
Finally, we investigated if the ability to regulate centrosome size is shared by other centriole duplication factors. We used RNAi to deplete SAS-6 in both wild-type and szy-20(bs52) embryos expressing GFP-SPD-2 (Figure 6B and S9, Movie S7 and S8). Compared to controls (n=20), depletion of SAS-6 in szy-20(bs52) mutants reduced the amount of GFP-SPD-2 at centrosomes by nearly half (54%, n=23), an effect similar in magnitude to that produced by depleting ZYG-1 activity. In wild-type embryos, sas-6(RNAi) (n=14) produced an even greater effect reducing the amount of centrosome-associated GFP-SPD-2 to 40% of controls (n=11). Interestingly, while sas-6(RNAi) invariably blocked centrosome duplication in wild-type embryos, we found it to be much less potent in szy-20(bs52) embryos where centrosomes were often able to duplicate (data not shown). This suggests that the szy-20(bs52) mutation can also partially suppress the duplication defect in SAS-6 deficient embryos. Our results thus uncover a second function for ZYG-1, SAS-6, and possibly other centriole assembly factors: to regulate centrosome size.
Discussion
Many factors that participate in assembly of a functional centrosome have been identified. However, the precise roles played by these factors, the order in which they act, and the mechanisms by which the cell determines an appropriate amount of PCM and microtubule-nucleating capacity at each stage of the cell cycle are not completely understood. SZY-20 plays an unusual role in centrosome morphogenesis. Whereas all factors identified to date play a positive role in centrosome assembly, SZY-20 negatively regulates this process.
SZY-20 links centrosome duplication and PCM recruitment
Our data shows that SZY-20 is involved in both centrosome duplication and PCM recruitment. These results confirm and extend previous work on the complex interplay between centrioles and PCM. Disruption of centrioles causes PCM to disperse (Bobinnec et al., 1998) while disruption of PCM hinders centriole duplication (Dammermann et al., 2004). Recently Loncarek et al. (2008) showed that experimentally increasing the size of PCM in vertebrate somatic cells stimulates the overproduction of centrioles. These centrioles arise as singlets within localized densities of PCM, similar to how centrioles arise de novo (Khodjakov et al. 2002). These studies suggest that PCM provides a micro-environment that facilitates centriole assembly, perhaps by concentrating centriole duplication factors around the mother centriole (Dammermann et al., 2004). The centriole in turn, acts to organize the PCM into a tight focus thereby restricting the number of daughter centrioles that can form during S phase (Loncarek et al. 2008).
A key finding of our work is that loss of SZY-20 results in both expansion of the PCM and elevated levels of centrosome-associated ZYG-1. This result suggests two possible models for how SZY-20 suppresses centrosome duplication defects. First, suppression may result from concentrating ZYG-1 at centrosomes, thereby facilitating the ability of the mutant protein to execute its function. Since SPD-2 is required to recruit ZYG-1 to the site of centriole assembly (Delattre et al., 2006; Pelletier et al., 2006), the enhanced localization of ZYG-1 may also compensate for a partial loss of SPD-2 activity. Second, suppression could be due to the expansion of PCM, which could enhance the ability of the centrosome to concentrate the depleted factors around mother centrioles. Although both models are possible, our finding that ZYG-1 is required for the expansion of PCM in szy-20 mutants argues in favor of the first model. Most likely, loss of SZY-20 results in an elevated level of centrosome-associated ZYG-1, which then enhances recruitment of downstream factors such as SAS-6 and ultimately PCM components. These results highlight the attractive interactions between centrioles and PCM; not only does the PCM attract centriole duplication factors, but as our results show, the reverse is also true.
If centriole amplification can be driven in other systems by the expansion of PCM or Plk4/Sak overexpression, why doesn't the szy-20 mutant overduplicate its centrioles? One obvious answer is that centrosome duplication might be perturbed more easily in some tissues than in others. Consistent with this idea, Peel et al. (2007) found tissue-specific variations in the extent to which centriole amplification could be induced by overexpressing various duplication factors, suggesting that cells differ with respect to how tightly they regulate centriole duplication. It thus seems likely that centrosome duplication in the worm embryo is stringently regulated and that perturbations in ZYG-1 levels and PCM size cannot disrupt the fidelity of this process.
Centriole duplication factors also function to determine centrosome size
An unexpected outcome of the current study is that control of centrosome size involves the activities of ZYG-1 and SAS-6, two factors whose only known roles are in centriole assembly (Dammermann et al., 2004; Leidel et al., 2005; O'Connell et al., 2001). Partial depletion of either factor reduces the size of centrosomes that form during the first cell cycle of the embryo. This small-centrosome phenotype is distinct from those reported earlier when ZYG-1 and other centriole duplication factors are partially depleted (Delattre et al., 2004; Kirkham et al., 2003; Leidel et al., 2005). In these cases, dramatically undersized centrosomes are observed following a round of defective centrosome duplication in which centrioles of altered structure and reduced size are formed (Kirkham et al., 2003). This particular effect on centrosome size is likely an indirect consequence of structurally defective centrioles.
Our results suggest that components of the centriole duplication pathway have two separable functions: to assemble centrioles and to endow centrioles with the ability to recruit PCM (Figure 6C). This is most evident from the results on partial depletion of ZYG-1 in szy-20(bs52) embryos where we could significantly reduce centrosome size without affecting centrosome duplication (Figure 6A). SAS-6 also appears to be involved in this pathway as its depletion reduces centrosome size by more than half (Figures 6B and S9). Although some of this reduction might be due to a block in daughter centriole formation, the magnitude of the effect suggests that ability of the mother centriole to recruit PCM has been impaired. Further, szy-20(bs52) embryos contain centrioles of normal structure and size, indicating that the ZYG-1-dependent increase in centrosome size is unlikely due to perturbations in centriole assembly. Finally, ZYG-1 regulates centrosome size through recruitment of SPD-2 a factor that functions both in centrosome duplication and maturation. This is in contrast to recent studies, showing that during centriole assembly SPD-2 is required to recruit ZYG-1 but not vice versa (Delattre et al., 2006; Pelletier et al., 2006). This discrepancy likely reflects different pathways under study: the earlier studies dealing with SPD-2 recruitment to the centriole during duplication and our study dealing with recruitment of SPD-2 to the PCM during maturation.
SZY-20 possibly functions in RNA metabolism
Cytological and genetic evidence favor the site of SZY-20 action to be the centrosome itself. Although SZY-20 localizes to a number of structures, its temporal pattern of association with the centrosome is suggestive; at the centrosome, SZY-20 levels peak when ZYG-1 levels are lowest, suggesting that SZY-20 regulates ZYG-1 directly. However, we have not detected a direct physical interaction between ZYG-1 and SZY-20 (unpublished data). While it is not yet known how SZY-20 controls centrosome size and duplication, it seems likely that SZY-20 participates in regulating some aspect of RNA metabolism. SZY-20 contains domains that are found in known RNA-binding proteins. For instance, the Drosophila Encore protein which contains a SUZ domain, associates with other RNA-binding proteins and has been shown to be required for the localization and translation of different maternal mRNAs (Hawkins et al., 1997; Van Buskirk and Schupbach, 2002). In support of a role in RNA metabolism, mutation of the SUZ and SUZ-C domains of SZY-20 disrupts both in vitro RNA-binding activity and centrosome size control.
How would a role in RNA metabolism allow SZY-20 to regulate centrosome size? Recently, a number of RNA and RNA-binding proteins have been shown to be associated with centrosomes and microtubules (Alliegro et al., 2006; Blower et al., 2007; Blower et al., 2005; Groisman et al., 2000; Lambert and Nagy, 2002; Lecuyer et al., 2007). Rae1, an RNA export factor, binds directly to microtubules and plays a role in mitotic spindle assembly (Blower et al., 2005). In Xenopus, the protein Maskin, a member of a family of spindle assembly proteins regulates translation of centrosome-associated cyclin B1 message (Groisman et al., 2000; O'Brien et al., 2005). Also, the transcript of a centrosomal component, D-PLP/CP309 in Drosophila has been reported to localize to centrosomes (Lecuyer et al., 2007). Thus, one intriguing possibility is that SZY-20 might function in the localization, stability, or translation of RNA at the centrosome to locally regulate expression of one or more proteins. Future studies, aimed at identifying interacting molecules, will help elucidate the molecular targets and mechanism of action of SZY-20.
A balance of SZY-20 and ZYG-1 activities regulates the microtubule-nucleating capacity of the centrosome
szy-20 mutant embryos improperly execute a number of microtubule-dependent processes. As inhibition of zyg-1 restores both normal centrosome size and microtubule function to szy-20 mutants, there appears to be a causal relationship between the two. It has been shown that depletion of microtubule destabilizing factors depresses the rate of microtubule growth in the C. elegans embryo (Schlaitz et al., 2007; Srayko et al., 2005). Similarly when treated with the microtubule-stabilizing agent taxol, C. elegans embryos form more but shorter astral microtubules and fail to properly align spindles (Hyman and White, 1987). These defects, which are similar to those observed in szy-20 mutants, are thought to be due to depletion of free tubulin. This suggests that the cell must carefully balance the levels of free and polymerized tubulin to maintain normal microtubule-dependent processes. SZY-20 appears to regulate this balance at the level of microtubule nucleation and possibly stability, as centrosomes in szy-20 mutants possess elevated levels of microtubule nucleating (γ-tubulin) and stabilizing (ZYG-9) factors. Our work shows that SZY-20 regulates the level of ZYG-1 at centrosomes and that a delicate balance in the activity of these two factors ensures proper centrosome size and function. When this balance is upset either by inhibiting SZY-20 or ZYG-1 or overexpressing SZY-20, centrosome size is affected in predictable ways.
In summary, our work describes a molecular pathway that is important for regulating centrosome size. This pathway, which depends upon the conserved protein SZY-20, the kinase ZYG-1, and likely other core centriole duplication factors, is functionally distinct from the mechanism of centriole assembly. Our work thus identifies a new function for centriole duplication factors and provides insight into a molecular mechanism for controlling centrosome size.
Experimental Procedures
Strain Maintenance and Genetics
Worms were cultured on MYOB plates seeded with E. coli OP50 (Church et al., 1995). Except for the Hawaiian variant CB4856, all strains were derived from the wild-type Bristol N2 strain. The following mutations were used: LGI: spd-2(or188ts); LGII: zyg-1(it25ts), zyg-1(or409ts), dpy-10(e128), mnDf68, mnDf104, szy-20(bs52ts), szy-20 (tm1997ts), unc-4(e120). The szy-20(tm1997) deletion allele was produced by The National BioResource Project-Japan. The following integrated transgenes were used: bsIs2 [pCK6.1: unc-119(+) pie-1-gfp-spd-2] (Kemp et al., 2004), ojIs2 [pLM6:unc-119(+) pie-1-gfp-tba-1], ruIs32 [pAZ132: unc-119(+) pie-1-gfp-his-58] (Praitis et al., 2001). All strains were grown at 16 or 20°C unless otherwise indicated.
Cloning of szy-20
For single nucleotide polymorphism (SNP) mapping, dpy-10(e128) szy-20(bs52) unc-4(e120) hermaphrodites were crossed with CB4856 males. A total of 124 independent Dpy-nonUnc or Unc-nonDpy recombinants were isolated from the F2 generation. Homozygous recombinant lines were established and scored for the presence of szy-20(bs52) by examining embryonic viability at 25°C. SNPs were analyzed by sequencing or by the method of Wicks et al., (2001). Selected ORFs within a region defined by SNP mapping were amplified from szy-20(bs52) genomic DNA and sequenced.
Molecular Biology
The EST clones yk658b1, yk1428h04, yk1555h10 and yk1636d07 (gifts from Yuji Kohara) were sequenced to confirm the molecular structure of szy-20. The 5' end of the szy-20 transcript was amplified from the cDNA library pHS1 (gift of Harold Smith) using primers directed against the trans-spliced SL1 leader, 5'-GGTTTAATTACCCAAGTTTGAG-3' and C18E9.3, 5'-CTCTCCATAGTGCTCTACCATG-3'.
szy-20(RNAi) was performed by soaking and/or feeding (Wang and Barr, 2005). To produce dsRNA for soaking, we used the primers 5'-ATGAGTAAAGAAAATGTTGTCG-3' and 5'-CTATTGAGGCCGATTCTGCTGC-3' to amplify a DNA template from N2 genomic DNA or yk1428h04. Each primer contained the T7 promoter sequence at its 5' end. Templates were transcribed in vitro using the T7-MEGAscript kit (Ambion). For soaking, L1-L4 larvae were incubated overnight in M9 buffer containing either 0.1–0.4 mg dsRNA/ml or no dsRNA (control). For feeding, larvae were grown on E. coli HT115 carrying pMS1.3, an RNAi feeding vector containing the entire szy-20 genomic sequence. Modifications of the szy-20 coding region were made using the QuikChange® II XL Site-Directed Mutagenesis Kit (Stratagene) according to the manufacturer's instructions. Transgenic lines were produced as described (Praitis et al., 2001).
Protein Sequence and Structure Analysis
Protein complexity analysis was performed using the SEG program (Wootton and Federhen, 1996). The non-redundant (NR) database of protein sequences (National Center for Biotechnology Information, NIH, Bethesda) was searched using the BLASTP program (Altschul et al., 1997). Profile searches were conducted using the PSI-BLAST program with either a single sequence or an alignment used as the query, with a default profile inclusion expectation (E) value threshold of 0.01 (unless specified otherwise) and was iterated until convergence (Altschul et al., 1997). The query sequences were subject to a statistical correction for compositional bias to avoid inflation of significance by low complexity regions (Schaffer et al., 2001).
Cytology
The following antibodies were used at a 1:500–2000 dilution: DM1A (Sigma), α-GFP (Roche), α-SPD-2 (Kemp et al., 2004), α-SPD-5 (Hamill et al., 2002), α-ZYG-1 (O'Connell et al., 2001), α-ZYG-9 (Matthews et al., 1998), and Alexa Fluor 488 and 568 secondary antibodies (Invitrogen). Affinity-purified rabbit polyclonal antibodies were prepared against the following peptides: for SZY-20 (αS20N): Ac-HHKLNQKEKQPAPTYEERQAC-amide; for SAS-4: Ac-MASDENIGADGEQKPSC-amide; for γ-tubulin: Ac-CLSKYDKLRSKRAFIDK-amide.
Indirect immunofluorescence, spinning disk confocal microscopy and 4D-DIC microscopy were performed as described (Kemp et al., 2007). For confocal microscopy, Openlab software (Improvision, Inc) was used to acquire images from a Nikon Eclipse E800 microscope equipped with a Plan Apo 100X 1.4 NA lens, a PerkinElmer UltraVIEW spinning disk unit and a Hamamatsu C9100-12 EM-CCD camera. Image processing was done with Photoshop CS. For 4D-DIC, we used IPLab software to acquire images from a Zeiss Axiovert 200M microscope equipped with a Hamamatsu ORCA-ER camera. Fluorescence intensity measurements were made using ImageJ v1.39e. Electron microscopy was performed as described (Muller-Reichert et al., 2007).
Immunoblotting
Embryos were dissolved in sample buffer, fractionated on a NuPAGE Bis-Tris Gel (Invitrogen) and blotted to nitrocellulose using the iBlot Gel Transfer System (Invitrogen). αS20N, α-SPD-2 (Kemp et al., 2004), α-SPD-5 (Dammermann et al., 2004), α-ZYG-1 (Kemp et al., 2007), DM1A (Sigma) and α-Actin (Novus Biologicals) were used at a 1:500–1500 dilution. IRDye secondary antibodies (LI-COR Biosciences) were used at 1:15,000 dilution. Blots were analyzed using the Odyssey Infrared Imaging System (LI-COR Biosciences).
Supplementary Material
Acknowledgements
We thank So Jung Kim and O'Connell lab members for their support; David Weisblat, Andy Golden, Bob Goldstein, Nick Miliaras and Nina Peel for comments on the manuscript; Harold Smith, Martin Srayko, Alexander Dammerman, Anjon Audhya, Karen Oegema, Ken Kemphues and Yuji Kohara for reagents; In-Geol Choi and Kevin Eliceiri for technical assistance. Some strains were provided by The Caenorhabditis Genetics Center and The National Bioresource Project- Japan. This work was supported by the Intramural Research Program of the National Institutes Health (NIH) and by the National Institute of Diabetes and Digestive and Kidney Diseases.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental Experimental Procedures Additional experimental details on protein structure analysis, immunostaining, protein expression, and RNA-binding assays can be found in Supplemental Materials.
References
- Alliegro MC, Alliegro MA, Palazzo RE. Centrosome-associated RNA in surf clam oocytes. Proc Natl Acad Sci U S A. 2006;103:9034–9038. doi: 10.1073/pnas.0602859103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azimzadeh J, Bornens M. Structure and duplication of the centrosome. J Cell Sci. 2007;120:2139–2142. doi: 10.1242/jcs.005231. [DOI] [PubMed] [Google Scholar]
- Bettencourt-Dias M, Rodrigues-Martins A, Carpenter L, Riparbelli M, Lehmann L, Gatt MK, Carmo N, Balloux F, Callaini G, Glover DM. SAK/PLK4 is required for centriole duplication and flagella development. Curr Biol. 2005;15:2199–2207. doi: 10.1016/j.cub.2005.11.042. [DOI] [PubMed] [Google Scholar]
- Blower MD, Feric E, Weis K, Heald R. Genome-wide analysis demonstrates conserved localization of messenger RNAs to mitotic microtubules. J Cell Biol. 2007;179:1365–1373. doi: 10.1083/jcb.200705163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blower MD, Nachury M, Heald R, Weis K. A Rae1-containing ribonucleoprotein complex is required for mitotic spindle assembly. Cell. 2005;121:223–234. doi: 10.1016/j.cell.2005.02.016. [DOI] [PubMed] [Google Scholar]
- Bobinnec Y, Khodjakov A, Mir LM, Rieder CL, Edde B, Bornens M. Centriole disassembly in vivo and its effect on centrosome structure and function in vertebrate cells. J Cell Biol. 1998;143:1575–1589. doi: 10.1083/jcb.143.6.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church DL, Guan KL, Lambie EJ. Three genes of the MAP kinase cascade, mek-2, mpk-1/sur-1 and let-60 ras, are required for meiotic cell cycle progression in Caenorhabditis elegans. Development. 1995;121:2525–2535. doi: 10.1242/dev.121.8.2525. [DOI] [PubMed] [Google Scholar]
- Dammermann A, Muller-Reichert T, Pelletier L, Habermann B, Desai A, Oegema K. Centriole assembly requires both centriolar and pericentriolar material proteins. Dev Cell. 2004;7:815–829. doi: 10.1016/j.devcel.2004.10.015. [DOI] [PubMed] [Google Scholar]
- Delattre M, Canard C, Gonczy P. Sequential protein recruitment in C. elegans centriole formation. Curr Biol. 2006;16:1844–1849. doi: 10.1016/j.cub.2006.07.059. [DOI] [PubMed] [Google Scholar]
- Delattre M, Leidel S, Wani K, Baumer K, Bamat J, Schnabel H, Feichtinger R, Schnabel R, Gonczy P. Centriolar SAS-5 is required for centrosome duplication in C. elegans. Nat Cell Biol. 2004;6:656–664. doi: 10.1038/ncb1146. [DOI] [PubMed] [Google Scholar]
- Dix CI, Raff JW. Drosophila Spd-2 Recruits PCM to the Sperm Centriole, but Is Dispensable for Centriole Duplication. Curr Biol. 2007 doi: 10.1016/j.cub.2007.08.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giansanti MG, Bucciarelli E, Bonaccorsi S, Gatti M. Drosophila SPD-2 is an essential centriole component required for PCM recruitment and astral-microtubule nucleation. Curr Biol. 2008;18:303–309. doi: 10.1016/j.cub.2008.01.058. [DOI] [PubMed] [Google Scholar]
- Gomez-Ferreria MA, Rath U, Buster DW, Chanda SK, Caldwell JS, Rines DR, Sharp DJ. Human Cep192 is required for mitotic centrosome and spindle assembly. Curr Biol. 2007;17:1960–1966. doi: 10.1016/j.cub.2007.10.019. [DOI] [PubMed] [Google Scholar]
- Groisman I, Huang YS, Mendez R, Cao Q, Theurkauf W, Richter JD. CPEB, maskin, and cyclin B1 mRNA at the mitotic apparatus: implications for local translational control of cell division. Cell. 2000;103:435–447. doi: 10.1016/s0092-8674(00)00135-5. [DOI] [PubMed] [Google Scholar]
- Habedanck R, Stierhof YD, Wilkinson CJ, Nigg EA. The Polo kinase Plk4 functions in centriole duplication. Nat Cell Biol. 2005;7:1140–1146. doi: 10.1038/ncb1320. [DOI] [PubMed] [Google Scholar]
- Hamill DR, Severson AF, Carter JC, Bowerman B. Centrosome maturation and mitotic spindle assembly in C. elegans require SPD-5, a protein with multiple coiled-coil domains. Dev Cell. 2002;3:673–684. doi: 10.1016/s1534-5807(02)00327-1. [DOI] [PubMed] [Google Scholar]
- Hawkins NC, Van Buskirk C, Grossniklaus U, Schupbach T. Post-transcriptional regulation of gurken by encore is required for axis determination in Drosophila. Development. 1997;124:4801–4810. doi: 10.1242/dev.124.23.4801. [DOI] [PubMed] [Google Scholar]
- Hyman AA, White JG. Determination of cell division axes in the early embryogenesis of Caenorhabditis elegans. J Cell Biol. 1987;105(5):2123–2135. doi: 10.1083/jcb.105.5.2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp CA, Kopish KR, Zipperlen P, Ahringer J, O'Connell KF. Centrosome Maturation and Duplication in C. elegans Require the Coiled-Coil Protein SPD-2. Dev Cell. 2004;6:511–523. doi: 10.1016/s1534-5807(04)00066-8. [DOI] [PubMed] [Google Scholar]
- Kemp CA, Song MH, Addepalli MK, Hunter G, O'Connell K. Suppressors of zyg-1 Define Regulators of Centrosome Duplication and Nuclear Association in Caenorhabditis elegans. Genetics. 2007;176:95–113. doi: 10.1534/genetics.107.071803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodjakov A, Rieder CL, Sluder G, Cassels G, Sibon O, Wang CL. De novo formation of centrosomes in vertebrate cells arrested during S phase. J Cell Biol. 2002;158(7):1171–1181. doi: 10.1083/jcb.200205102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkham M, Muller-Reichert T, Oegema K, Grill S, Hyman AA. SAS-4 is a C. elegans centriolar protein that controls centrosome size. Cell. 2003;112:575–587. doi: 10.1016/s0092-8674(03)00117-x. [DOI] [PubMed] [Google Scholar]
- Kleylein-Sohn J, Westendorf J, Le Clech M, Habedanck R, Stierhof YD, Nigg EA. Plk4-induced centriole biogenesis in human cells. Dev Cell. 2007;13:190–202. doi: 10.1016/j.devcel.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Lambert JD, Nagy LM. Asymmetric inheritance of centrosomally localized mRNAs during embryonic cleavages. Nature. 2002;420:682–686. doi: 10.1038/nature01241. [DOI] [PubMed] [Google Scholar]
- Lecuyer E, Yoshida H, Parthasarathy N, Alm C, Babak T, Cerovina T, Hughes TR, Tomancak P, Krause HM. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell. 2007;131:174–187. doi: 10.1016/j.cell.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Leidel S, Delattre M, Cerutti L, Baumer K, Gonczy P. SAS-6 defines a protein family required for centrosome duplication in C. elegans and in human cells. Nat Cell Biol. 2005;7:115–125. doi: 10.1038/ncb1220. [DOI] [PubMed] [Google Scholar]
- Leidel S, Gonczy P. SAS-4 is essential for centrosome duplication in C elegans and is recruited to daughter centrioles once per cell cycle. Dev Cell. 2003;4:431–439. doi: 10.1016/s1534-5807(03)00062-5. [DOI] [PubMed] [Google Scholar]
- Loncarek J, Hergert P, Magidson V, Khodjakov A. Control of daughter centriole formation by the pericentriolar material. Nat Cell Biol. 2008;10:322–328. doi: 10.1038/ncb1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luders J, Stearns T. Microtubule-organizing centres: a re-evaluation. Nat Rev Mol Cell Biol. 2007;8:161–167. doi: 10.1038/nrm2100. [DOI] [PubMed] [Google Scholar]
- Matthews LR, Carter P, Thierry-Mieg D, Kemphues K. ZYG-9, a Caenorhabditis elegans protein required for microtubule organization and function, is a component of meiotic and mitotic spindle poles. J Cell Biol. 1998;141(5):1159–1168. doi: 10.1083/jcb.141.5.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller-Reichert T, Srayko M, Hyman A, O'Toole ET, McDonald K. Correlative light and electron microscopy of early Caenorhabditis elegans embryos in mitosis. Methods Cell Biol. 2007;79:101–119. doi: 10.1016/S0091-679X(06)79004-5. [DOI] [PubMed] [Google Scholar]
- O'Brien LL, Albee AJ, Liu L, Tao W, Dobrzyn P, Lizarraga SB, Wiese C. The Xenopus TACC homologue, maskin, functions in mitotic spindle assembly. Mol Biol Cell. 2005;16:2836–2847. doi: 10.1091/mbc.E04-10-0926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell KF, Caron C, Kopish KR, Hurd DD, Kemphues KJ, Li Y, White JG. The C. elegans zyg-1 gene encodes a regulator of centrosome duplication with distinct maternal and paternal roles in the embryo. Cell. 2001;105:547–558. doi: 10.1016/s0092-8674(01)00338-5. [DOI] [PubMed] [Google Scholar]
- Peel N, Stevens NR, Basto R, Raff JW. Overexpressing centriole-replication proteins in vivo induces centriole overduplication and de novo formation. Curr Biol. 2007;17:834–843. doi: 10.1016/j.cub.2007.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier L, O'Toole E, Schwager A, Hyman AA, Muller-Reichert T. Centriole assembly in Caenorhabditis elegans. Nature. 2006;444:619–623. doi: 10.1038/nature05318. [DOI] [PubMed] [Google Scholar]
- Pelletier L, Ozlu N, Hannak E, Cowan C, Habermann B, Ruer M, Muller-Reichert T, Hyman AA. The Caenorhabditis elegans centrosomal protein SPD-2 is required for both pericentriolar material recruitment and centriole duplication. Curr Biol. 2004;14:863–873. doi: 10.1016/j.cub.2004.04.012. [DOI] [PubMed] [Google Scholar]
- Praitis V, Casey E, Collar D, Austin J. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics. 2001;157:1217–1226. doi: 10.1093/genetics/157.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues-Martins A, Riparbelli M, Callaini G, Glover DM, Bettencourt-Dias M. Revisiting the role of the mother centriole in centriole biogenesis. Science. 2007;316:1046–1050. doi: 10.1126/science.1142950. [DOI] [PubMed] [Google Scholar]
- Schaffer AA, Aravind L, Madden TL, Shavirin S, Spouge JL, Wolf YI, Koonin EV, Altschul SF. Improving the accuracy of PSI-BLAST protein database searches with composition-based statistics and other refinements. Nucleic Acids Res. 2001;29:2994–3005. doi: 10.1093/nar/29.14.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlaitz AL, Srayko M, Dammermann A, Quintin S, Wielsch N, MacLeod I, de Robillard Q, Zinke A, Yates JR, 3rd, Muller-Reichert T, et al. The C. elegans RSA complex localizes protein phosphatase 2A to centrosomes and regulates mitotic spindle assembly. Cell. 2007;128:115–127. doi: 10.1016/j.cell.2006.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srayko M, Kaya A, Stamford J, Hyman AA. Identification and characterization of factors required for microtubule growth and nucleation in the early C. elegans embryo. Dev Cell. 2005;9:223–236. doi: 10.1016/j.devcel.2005.07.003. [DOI] [PubMed] [Google Scholar]
- Stein KK, Davis ES, Hays T, Golden A. Components of the spindle assembly checkpoint regulate the anaphase-promoting complex during meiosis in Caenorhabditis elegans. Genetics. 2007;175:107–123. doi: 10.1534/genetics.106.059105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Buskirk C, Schupbach T. Half pint regulates alternative splice site selection in Drosophila. Dev Cell. 2002;2:343–353. doi: 10.1016/s1534-5807(02)00128-4. [DOI] [PubMed] [Google Scholar]
- Wang J, Barr MM. RNA interference in Caenorhabditis elegans. Methods Enzymol. 2005;392:36–55. doi: 10.1016/S0076-6879(04)92003-4. [DOI] [PubMed] [Google Scholar]
- Wicks SR, Yeh RT, Gish WR, Waterston RH, Plasterk RH. Rapid gene mapping in Caenorhabditis elegans using a high density polymorphism map. Nat Genet. 2001;28:160–164. doi: 10.1038/88878. [DOI] [PubMed] [Google Scholar]
- Wootton JC, Federhen S. Analysis of compositionally biased regions in sequence databases. Methods Enzymol. 1996;266:554–571. doi: 10.1016/s0076-6879(96)66035-2. [DOI] [PubMed] [Google Scholar]
- Zhu F, Lawo S, Bird A, Pinchev D, Ralph A, Richter C, Muller-Reichert T, Kittler R, Hyman AA, Pelletier L. The mammalian SPD-2 ortholog Cep192 regulates centrosome biogenesis. Curr Biol. 2008;18:136–141. doi: 10.1016/j.cub.2007.12.055. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.