Abstract
Thymidylate synthase (TS) inhibitors, such as 5-fluorouracil (5-FU) and 5-fluorodeoxyuridine (5-FUdR), are amongst the most frequently used chemotherapeutic drugs available, although their efficacy is often limited by myelotoxicity. An emerging strategy for overcoming bone marrow toxicity involves ex vivo genetic transfer of drug resistance to autologous hematopoietic progenitor cells, followed by reimplantation of the transfected cells before chemotherapy. Here we establish that expression of mutant TS genes, selected from millions of engineered variants, renders human hematopoietic cells resistant to 5-FUdR, and identify the most efficacious variant for gene therapeutic rescue of drug-induced myelosuppression.
Introduction
Protection of bone marrow from the toxicity of chemotherapeutic drugs may be one of the earliest achievable goals of cancer gene therapy, posing fewer obstacles than, for example, tumor ablation. Introduction of drug resistance genes into hematopoietic precursor cells can be carried out ex vivo, does not require specific or sustained gene expression, and has the potential to be effective when only a fraction of the cells are transformed, because the transformed cells should exhibit a growth advantage under the selection pressure of drug therapy (Allay et al., 1998). Gene transfer to nondividing cells, including murine and human hematopoietic cells, has been achieved with a variety of vectors. The growth advantage of the transformed cells has been documented in both mice and monkeys by overexpressing wild-type and mutant genes (Budak-Alpdogan et al., 2005).
Thymidylate synthase plays a central role in the de novo biosynthesis of deoxythymidine 5′-monophosphate (dTMP), an essential precursor for DNA synthesis. The enzyme catalyzes reductive methylation of deoxyuridine 5′-monophosphate (dUMP) by (6R,S)-N5,N10-methylene-5,6,7,8-tetrahydrofolate (CH2H4-folate) to produce dTMP and dihydrofolate. Inactivation of TS results in decreased production of the nucleotide precursor dTMP, cessation of DNA synthesis, and subsequent cell death. TS has been a major target for the development of cancer chemotherapeutic agents. It is inhibited by 5-fluorouracil (5-FU) and related fluoropyrimidines. These analogs are commonly employed in the chemotherapy of colon, breast, and head/neck cancers. In fact, 5-FU represents the most extensively used antimetabolite for the treatment of human cancer. 5-FU is metabolized to 5-fluoro-dUMP (FdUMP), which forms an inhibitory, ternary covalent complex with the enzyme. In this complex, the C-6 of FdUMP is located next to a conserved cysteine at position 196, and C-5 of the nucleotide is joined to the methylene carbon of the cosubstrate (Carreras and Santi, 1995).
Although TS is considered to be the chief target of 5-FU-induced cytotoxicity, the production of 5-FU metabolites and their incorporation into RNA (Noordhuis et al., 2004) and deficiencies in dihydropyrimidase in some individuals (van Kuilenburg et al., 2003) have also been associated with 5-FU toxicity. The dosing of fluoropyrimidine inhibitors is frequently limited by bone marrow toxicity (Relling and Dervieux, 2001). Introduction of drug-resistant TS into bone marrow cells could spare patients the severe side effects of these inhibitors, and permit higher dosing and adherence to treatment regimens, which are often curtailed by myelosuppressive toxicity (Gerson, 2004).
Materials and Methods
Genetic selection
For selection of active TS, Escherichia coli χ2913recA cells, lacking TS (ΔthyA572, recA56) and containing the random library, were inoculated (1:100) into 1× YT medium containing thymidine (50 μg/ml), carbenicillin (50 μg/ml), and tetracycline (10 μg/ml), and grown at 37°C until the optical density (OD) at 600 nm attained a value of 0.8 to 1.0. One-milliliter aliquots of the exponentially growing cells were pelleted and resuspended in M9 salts, plated on minimal medium that contained appropriate antibiotics, and incubated at 37°C for 36 hr. Plasmid DNA was isolated from surviving colonies and sequenced. To select for library members that were resistant to killing with 5-fluorodeoxyuridine (5-FUdR), transfectants were plated on the above-described minimal medium that contained increasing amounts of 5-FUdR (0–175 nM) and incubated at 37°C for 48 hr. Colonies formed on 5-FUdR-supplemented agar plates were isolated, and the plasmid from each clone was retransformed into fresh χ2913 E. coli to confirm the drug-resistant phenotype. Each retransformed bacterium was then subjected to the same selection procedure. Plasmid DNA was sequenced from cells that survived 175 nM 5-FUdR.
TS cloning
The TS coding sequence was amplified with primers XhoI-fwd (5′-CTC GAG ATG CCG GTA GCT GGT AGC-3′) and SacII-rev (5′-CCG CGG CTA AAC AGC CAT TTC CAT-3′). The underlined bases indicate XhoI and SacII restriction enzyme sites that were appended to the primers. The polymerase chain reaction (PCR) contained 50 ng of plasmid DNA, 15 pmol of each primer, 250 μmol of each dNTP, 2.5 units of Expand DNA polymerase (Roche Applied Science, Indianapolis, IN), and 1× Expand polymerase buffer with 1.5 mM MgCl2. The reaction was cycled at 94°C for 2 min, followed by 35 cycles of 94°C for 15 sec, 62°C for 30 sec, and 72°C for 1.5 min, and cloned into the pCR 2.1-TOPO vector (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. The plasmids were then subjected to SacII digest, the product was gel-isolated from a 2% low-melt agarose gel, and then digested with XhoI. The gel was isolated again and ligated into a pBMN-GFP vector restricted by an analogous procedure with T4 DNA ligase (Invitrogen) according to the manufacturer's instructions. The product, pBMN-TS-GFP, was transformed into XL10-Gold ultracompetent cells (Stratagene, La Jolla, CA) and carbenicillin-resistant colonies were verified by DNA sequencing.
TS K562 cell line establishment
Twenty micrograms of pBMN-TS-GFP plasmid DNA resuspended in 1 ml of 250 mM CaCl2 (Mallinckrodt/Covidien, Hazelwood, MO) was diluted 1:1 with 1.4 M NaCl, 2.7 M 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (sodium salt), and 80 mM Na2HPO4, pH 6.95. The DNA solution was added drop-wise to Phoenix retrovirus producer cells at 60% confluence in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 1% penicillin–streptomycin, 1% glutamine, and 2.5 μM chloroquine. Twenty-four hours later, the cells were washed with phosphate-buffered saline (PBS) and incubated at 32°C in medium lacking chloroquine for viral production. Forty-eight hours posttransfection, K562 cells (5 × 105 cells/ml) were infected with retroviral supernatant that had been clarified by centrifugation at 1500 rpm for 5 min and passaged through a 0.45-μm pore size filter, to allow for infection at 37°C in the presence of Polybrene (5 μg/ml). Cultures were washed after 24 hr and 5 days later, the K562 cells were resuspended to a final concentration of ∼2 × 107/ml in 20 mM KH2PO4, 150 mM NaCl supplemented with 2% FBS. GFP-positive cells were selected via gating by green fluorescent protein (GFP) fluorescence intensity (with excitation at 488 nm and an emission bandpass filter of 530/30 nm), using a FACSVantage SE cell sorter (BD Biosciences, San Jose, CA).
Results and Discussion
On the basis of the premise that superior drug-resistant TS variants may require a combination of amino acid substitutions that cannot currently be predicted by structure-based computational methods (Encell et al., 1999), we have adopted a directed molecular evolution approach to engineer and identify the most efficacious TS mutants for use in therapeutic gene transfer. In pursuit of this approach, we created two different libraries of human TS that harbor multiple nucleotide substitutions. Our first library, constructed by random nucleotide mutagenesis, consisted of 1.5 million variants of the highly conserved Arg50-loop, a domain in the active site that interacts with the pyrimidine substrate (Landis et al., 2001). We targeted this region for mutagenesis as amino acid substitutions within the Arg50-loop have been recovered from human cells resistant to 5-FUdR and the antifolate Thymitaq (AG337) (Tong et al., 1998). Our second library addressed the possibility that amino acid changes distant from the active site might render TS resistant to 5-FdUMP (Kawate et al., 2002). This library, constructed by whole gene mutagenic PCR, consisted of 2.8 million clones and harbored various nucleotide substitutions throughout the TS cDNA. For each library, we used genetic complementation of TS-deficient E. coli to select for catalytically active mutants, and then screened for mutants that conferred greater 5-FUdR resistance to E. coli than wild-type human TS (Landis et al., 2001; Kawate et al., 2002). We then chose the eight most resistant mutants that, when expressed in E. coli, conferred 50-fold or greater enhancement in resistance to 5-FUdR relative to wild-type human TS.
TS cDNAs encoding both wild-type and the eight E. coli-selected 5-FudR-resistant mutants were individually subcloned into the retroviral vector pBMN-I-GFP for production of amphotrophic retroviruses and used to infect K562 human hematopoietic cells. The TS genes were placed upstream of an internal ribosome entry site (IRES) and were followed by a sequence encoding GFP, permitting proportional expression of TS and GFP (Klefstrom et al., 2002). This approach allowed the use of fluorescence-activated cell sorting (FACS) to obtain stable human hematopoietic cell populations that expressed equivalent amounts of TS. Individual TS-expressing K562 cultures were then exposed to increasing concentrations of 5-FUdR, and drug-induced cytotoxicity was determined in clonogenic survival assays to screen individual mutants for their capacity to confer greater resistance to 5-FUdR than that of wild type. Overexpression of wild-type TS yielded enhanced resistance to 5-FUdR (Fig. 1a), elevating 50% lethal dose (LD50) >5-fold (p < 0.04) (Fig. 1b). This result provides experimental support for the inverse, predictive relationship between the level of TS expression in tumors and response to fluoropyrimidine-based chemotherapy (Salonga et al., 2000), and strengthens the premise that TS is the most critical target of 5-FudR-based chemotherapy. Four of the eight mutants conferred even greater resistance, increasing the LD50 for 5-FUdR an additional 2- to 4-fold over that of the wild-type enzyme (Fig. 1b); all four mutants contained either T51S and/or G52S substitutions in the active site. The mutant that afforded the most protection (T51S, G52S) contained both substitutions; the LD50 for the double mutant was ~4-fold greater than that for wild-type TS (p < 0.04) and ~20-fold greater than that for vector alone (p < 0.016), the latter representing the resistance afforded by endogenous TS. We note that, when purified, the T51S, G52S double mutant protein exhibited 26-fold reduced inhibition (Ki) by FdUMP (Landis et al., 2001). Interestingly, the remaining four mutants did not confer greater-than-wild-type resistance (Fig. 1a). We hypothesize that the discordance in the relative resistance conferred to bacterial and human cells may result from factors unique to the hosts, for example, protein interactions, molecular folding dynamics, and/or chimerization of mutant TS with endogenous wild-type TS in human cells. On the basis of the LD50 values of the mutants (Fig. 1b), we have identified amino acid substitutions that confer resistance to human cells and those that may be neutral, and mapped them on the three-dimensional structure (Phan et al., 2001) of TS (Fig. 2).
FIG. 1.
5-Fluorodeoxyuridine (5-FUdR) resistance conferred to human hematopoietic K562 cells by transduction of either wild-type thymidylate synthase (TS) or each of eight TS mutants. (a) Survival was quantified by clonogenic assay following culture for 3 days in the presence of increasing concentrations of 5-FUdR. Each point represents the mean ± SD of triplicate experiments. (b) The 50% lethal dose (LD50), with 95% confidence limits, was determined by least-squares linear regression analysis of the survival curves. Color coding is the same in (a) and (b).
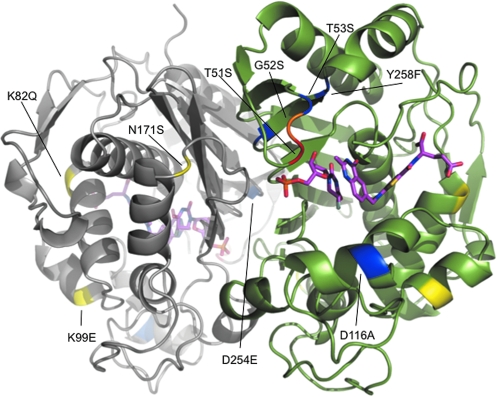
FIG. 2.
Amino acid substitutions in the eight mutants tested for 5-FUdR resistance in human cells are highlighted in the 1.9-Å crystal structure of dimeric human TS complexed with dUMP and the antifolate drug Raltitrexed. The closed conformation is shown. Residues 1–25 are disordered and do not appear in the model. The individual monomers are colored gray or green; for convenience, amino acid substitutions are indicated in only one of the monomers, although all substitutions are present in both. Comparison of LD50 values among mutants (Fig. 1b) permitted the following differentiation of resistance-contributory versus provisionally neutral amino acid substitutions: Substitutions that either alone or together conferred enhanced 5-FUdR resistance (T51S, G52S) are colored red and orange. Substitutions present in mutants that conferred improved 5-FUdR resistance, but appear to be autonomous of this selected property (K82Q, K99E, N171S), are shown in yellow. Substitutions that alone or together did not confer greater 5-FUdR resistance than wild-type TS (D254N, T53S, Y258S, D116A, Y258F) are shown in blue.
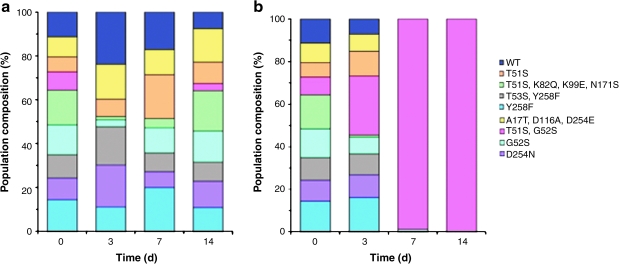
We next used competition experiments to further distinguish the proliferative advantage conferred by the TS mutants to cells growing in the presence of 5-FUdR. Mixed cultures were seeded by FACS to contain equal proportions of cells expressing either wild-type TS or one of the eight TS mutants In the absence of drug, all nine strains were represented at the end of the experiment and no single cell strain was greatly overrepresented in the population (Fig. 3a). In striking contrast, after 7 days in 500 nM 5-FUdR (a concentration similar to that found in blood plasma of patients undergoing continuous, protracted 5-FUdR intravenous therapy; Yamada et al., 2003), cells expressing the T51S, G52S double mutant represented >98% of the population (Fig. 3b). After 14 days of 5-FUdR-mediated selection, only cells containing the T51S, G52S mutant were detected (Fig. 3b). These results are consistent with the survival data obtained for individual cultures (Fig. 1), which also identified the T51S, G52S mutant as most protective. Taken together, these results demonstrate the strong growth advantage that expression of the T51S, G52S mutant confers to human K562 cells in the presence of 5-FUdR. This advantage is greater than that conferred by wild-type TS, single mutant T51S, or single mutant G52S.
FIG. 3.
Equal numbers of GFP expression-normalized wild-type and mutant TS-expressing K562 cells were sorted by FACS and cultured for 14 days (a) under standard cell culture conditions and (b) in medium supplemented with 500 nM 5-FUdR. Genomic DNA extracted from cells after 0, 3, 7, and 14 days of culture was used as template in PCRs employing primers that selectively amplified the retrovirally transduced TS genes. The resulting amplicons were cloned into the pCR 4-TOPO vector and transformed into DH5α Escherichia coli DH5α. The bacteria were plated and incubated overnight, and plasmids from the resultant colonies (n: 150 for each time point) were DNA sequenced with primers flanking the TS gene sequence.
Our results suggest that transfer of the T51S, G52S mutant into hematopoietic precursor cells can be carried out ex vivo and would not require high transfection efficiency or sustained gene expression, as T51S, G52S-transformed cells exhibit a striking growth advantage under the selection pressure of 5-FUdR exposure. Consequently, protection of bone marrow from the toxicity of fluoropyrimidines and other chemotherapeutic drugs could prove to be one of the first successes of cancer gene therapy (Banerjee and Bertino, 2002). We expect that implementation of an analogous directed evolution strategy would be effective for protecting other enzymes/proteins targeted by chemotherapeutic agents, including dihydrofolate reductase, glutathione S-transferase, cytosine deaminase, thymidylate synthase, 8-oxoguanine-DNA glycosylase, topoisomerases I and II, and multiple drug resistance proteins.
Acknowledgments
The authors thank A. Blank for editing the manuscript and A. Blank, D. Deyle, N. Fausto, J. Heddle, C. Heindel, G. M. Martin, R. Monnat, R. Prehn, P. Rabinovitch, J. Salk, and J. Wanagat for insightful comments. The authors are indebted to R. Chung for outstanding technical advice, to E. Adman and P. Murphy for help with computer modeling, and to G. Nolan for providing the pBMN-i-EGFP vector and Phoenix retroviral producer lines. The National Institutes of Health by grants CA78885 and CA102029 (L.A.L.) funded this work, with stipend support for M.W.S. provided by grants AG030314 and GM007266. A Postdoctoral Fellowship from the Natural Sciences and Engineering Research Council of Canada (NSERC), followed by a Canadian Institutes of Health Research (CIHR) Fellowship, and a Terry Fox Foundation Research Fellowship from the National Cancer Institute of Canada, provided support for J.H.B. during the completion of these studies.
Author Disclosure Statement
No competing financial interests exist.
References
- Allay J.A. Persons D.A. Galipeau J. Riberdy J.M. Ashmun R.A. Blakley R.L. Sorrentino B.P. In vivo selection of retrovirally transduced hematopoietic stem cells. Nat. Med. 1998;4:1136–1143. doi: 10.1038/2632. [DOI] [PubMed] [Google Scholar]
- Banerjee D. Bertino J.R. Myeloprotection with drug-resistance genes. Lancet Oncol. 2002;3:154–158. doi: 10.1016/s1470-2045(02)00678-2. [DOI] [PubMed] [Google Scholar]
- Budak-Alpdogan T. Banerjee D. Bertino J.R. Hematopoietic stem cell gene therapy with drug resistance genes: An update. Cancer Gene Ther. 2005;12:849–863. doi: 10.1038/sj.cgt.7700866. [DOI] [PubMed] [Google Scholar]
- Carreras C.W. Santi D.V. The catalytic mechanism and structure of thymidylate synthase. Annu. Rev. Biochem. 1995;64:721–762. doi: 10.1146/annurev.bi.64.070195.003445. [DOI] [PubMed] [Google Scholar]
- Encell L.P. Landis D.M. Loeb L.A. Improving enzymes for cancer gene therapy. Nat. Biotechnol. 1999;17:143–147. doi: 10.1038/6142. [DOI] [PubMed] [Google Scholar]
- Gerson S.L. MGMT: Its role in cancer aetiology and cancer therapeutics. Nat. Rev. Cancer. 2004;4:296–307. doi: 10.1038/nrc1319. [DOI] [PubMed] [Google Scholar]
- Kawate H. Landis D.M. Loeb L.A. Distribution of mutations in human thymidylate synthase yielding resistance to 5-fluorodeoxyuridine. J. Biol. Chem. 2002;277:36304–36311. doi: 10.1074/jbc.M204956200. [DOI] [PubMed] [Google Scholar]
- Klefstrom J. Verschuren E.W. Evan G. c-Myc augments the apoptotic activity of cytosolic death receptor signaling proteins by engaging the mitochondrial apoptotic pathway. J. Biol. Chem. 2002;277:43224–43232. doi: 10.1074/jbc.M206967200. [DOI] [PubMed] [Google Scholar]
- Landis D.M. Heindel C.C. Loeb L.A. Creation and characterization of 5-fluorodeoxyuridine-resistant Arg50 loop mutants of human thymidylate synthase. Cancer Res. 2001;61:666–672. [PubMed] [Google Scholar]
- Noordhuis P. Holwerda U. van der Wilt C.L. van Groeningen C.J. Smid K. Meijer S. Pinedo H.M. Peters G.J. 5-Fluorouracil incorporation into RNA and DNA in relation to thymidylate synthase inhibition of human colorectal cancers. Ann. Oncol. 2004;15:1025–1032. doi: 10.1093/annonc/mdh264. [DOI] [PubMed] [Google Scholar]
- Phan J. Koli S. Minor W. Dunlap R.B. Berger S.H. Lebioda L. Human thymidylate synthase is in the closed conformation when complexed with dUMP and raltitrexed, an antifolate drug. Biochemistry. 2001;40:1897–1902. doi: 10.1021/bi002413i. [DOI] [PubMed] [Google Scholar]
- Relling M.V. Dervieux T. Pharmacogenetics and cancer therapy. Nat. Rev. Cancer. 2001;1:99–108. doi: 10.1038/35101056. [DOI] [PubMed] [Google Scholar]
- Salonga D. Danenberg K.D. Johnson M. Metzger R. Groshen S. Tsaowei D.D. Lenz H.J. Leichman C.G. Leichman L. Diasio R.B. Danenberg P.V. Colorectal tumors responding to 5-fluorouracil have low gene expression levels of dihydropyrimidine dehydrogenase, thymidylate synthase, and thymidine phosphorylase. Clin. Cancer Res. 2000;6:1322–1327. [PubMed] [Google Scholar]
- Tong Y. Liu-Chen X. Ercikan-Abali E.A. Capiaux G.M. Zhao S.C. Banerjee D. Bertino J.R. Isolation and characterization of thymitaq (AG337) and 5-fluoro-2-deoxyuridylate-resistant mutants of human thymidylate synthase from ethyl methanesulfonate-exposed human sarcoma HT1080 cells. J. Biol. Chem. 1998;273:11611–11618. doi: 10.1074/jbc.273.19.11611. [DOI] [PubMed] [Google Scholar]
- van Kuilenburg A.B. Meinsma R. Zonnenberg B.A. Zoetekouw L. Baas F. Matsuda K. Tamaki N. van Gennip A.H. Dihydropyrimidinase deficiency and severe 5-fluorouracil toxicity. Clin. Cancer Res. 2003;9:4363–4367. [PubMed] [Google Scholar]
- Yamada Y. Hamaguchi T. Goto M. Muro K. Matsumura Y. Shimada Y. Shirao K. Nagayama S. Plasma concentrations of 5-fluorouracil and F-β-alanine following oral administration of S-1, a dihydropyrimidine dehydrogenase inhibitory fluoropyrimidine, as compared with protracted venous infusion of 5-fluorouracil. Br. J. Cancer. 2003;89:816–820. doi: 10.1038/sj.bjc.6601224. [DOI] [PMC free article] [PubMed] [Google Scholar]