Abstract
During development, neural precursors divide to produce new precursors and cells that differentiate as neurons and glia. In Drosophila, apicobasal polarity and orientation of the mitotic spindle play important roles in specifying the progeny of neural precursors for different fates. We examined orientation of zebrafish spinal cord precursors using time-lapse imaging and tested the function of protein kinase C, iota (PrkCi), a member of the Par complex of proteins necessary for apicobasal polarity in the nervous system. We found that nearly all precursors divide within the plane of the neuroepithelium of wild-type embryos even when they must produce cells that have different fates. In the absence of PrkCi function, neural precursor divisions become oblique during late embryogenesis and excess oligodendrocytes form concomitant with loss of dividing cells. We conclude that PrkCi function and planar divisions are necessary for asymmetric, self-renewing division of spinal cord precursors.
Keywords: neural precursors, zebrafish, oligodendrocytes, fate specification
INTRODUCTION
In the developing central nervous system of vertebrates, regulation of cell division influences the balance of neural precursors, neurons, and glia. During early stages of neural development, precursors undergo symmetric, proliferative divisions to expand the precursor population. Later, precursors exhibit asymmetric, self-renewing divisions to produce one precursor and one differentiated cell or symmetric, differentiative divisions to produce two progeny fated to exit the cell cycle. Therefore, mechanisms that regulate the number and type of symmetric versus asymmetric divisions influence brain size and cell composition.
In Drosophila, asymmetric division of embryonic neuroblasts to produce progeny having different fates is regulated by cell polarity and orientation of cell cleavage. An evolutionarily conserved complex of proteins consisting of Par3/Bazooka, Par6 and atypical protein kinase C (aPKC) is localized to the apical membrane of epithelial neuroectodermal cells and thereby to the apical membrane of neuroblasts as they delaminate from the ectoderm (Schober et al., 1999; Wodarz et al., 1999, 2000; Petronczki and Knoblich, 2001). Par complex proteins act through the tumor suppressor Lethal giant larvae to exclude other proteins, including the cell fate determinants Prospero and Numb, from apical membrane, limiting their localization to basal membrane (Betschinger et al., 2003; Plant et al., 2003; Yamanaka et al., 2003). As neuroblasts delaminate from the epithelium, Inscuteable and a cassette of heterotrimeric G protein signaling factors are recruited to the apical membrane and orient the mitotic spindle perpendicular to the plane of the epithelium, resulting in cleavage that is orthogonal to the axis of apicobasal polarity (Kraut et al., 1996; Schaefer et al., 2000; Yu et al., 2000). Consequently, Prospero and Numb are segregated to the basal progeny cell, which becomes a ganglion mother cell (GMC) fated to divide once to produce two neurons. The apical progeny cell, lacking Propero and Numb, remains as a neuroblast (Wodarz and Huttner, 2003).
Regulation of the plane in which neuroepithelial precursors divide by apicobasal polarity cues in the central nervous system (CNS) of vertebrate embryos could provide an effective means for regulating symmetric versus asymmetric divisions. Consistent with this possibility, some investigations have described analyses of fixed tissue and live cell imaging that revealed divisions that were oriented both within and perpendicular to the plane of the neuroepithelium (Chenn and McConnell, 1995; Haydar et al., 2003; Sanada and Tsai, 2005). Additionally, Par complex proteins are localized to the apical membrane at the ventricular surface of neuroepithelial cells (Manabe et al., 2002; Geldmacher-Voss et al., 2003; Afonso and Henrique, 2006; Imai et al., 2006; von Trotha et al., 2006; Kovac et al., 2007), similar to localization of homologous proteins in the Drosophila neuroepithelium. Several other studies, however, indicate that most precursor divisions are planar during both proliferative and neurogenic phases of neural development (Geldmacher-Voss et al., 2003; Lyons et al., 2003; Kosodo et al., 2004; Morin et al., 2007; Noctor et al., 2008). Therefore, the relationship between cell division pattern and cell fate in the vertebrate CNS remains unclear.
In the spinal cord, the ventral pMN precursor domain, defined by expression of the transcription factor-encoding gene Olig2, gives rise first to motor neurons and later to oligodendrocyte progenitor cells (OPCs), which migrate throughout the spinal cord, divide and differentiate as myelinating oligodendrocytes (Lu et al., 2000; Takebayashi et al., 2000; Zhou et al., 2000; Park et al., 2004; Masahira et al., 2006). Our analysis of the clonal progeny of olig2+ neural plate cells in zebrafish suggested the existence of asymmetric divisions that give rise to motor neurons, OPCs and some ventral interneurons (Park et al., 2004). Near the end of embryogenesis remaining olig2+ precursors adopt radial glial characteristics and are maintained as slowly dividing OPC precursors into adulthood (Park et al., 2007). We found that Notch signaling is required continuously during development to maintain olig2+ precursors and regulate the numbers of precursors specified for motor neuron and oligodendrocyte fates (Park and Appel, 2003; Kim et al., 2008), but we are still uncertain about the exact mechanisms that maintain and specify olig2+ precursors and the potential role of cell polarity and cell division orientation.
In this study, we investigated the role of apical cell polarity in spinal cord precursor maintenance and specification using the heart and soul (has) mutation, which disrupts function of PrkCi (Horne-Badovinac et al., 2001). Time-lapse imaging revealed that in has mutant embryos neuroepithelial cells gradually switch from planar to oblique divisions. Concomitant with this switch in cell division pattern is a loss of apical character, loss of neuroepithelial precursors and formation of excess OPCs. We conclude that planar cell division, directed by apically localized PrkCi, is required for maintenance of neuroepithelial precursors.
RESULTS
PrkCi Is Required for Maintenance of Apical Polarity and Adherens Junctions in the Spinal Cord Neuroepithelium
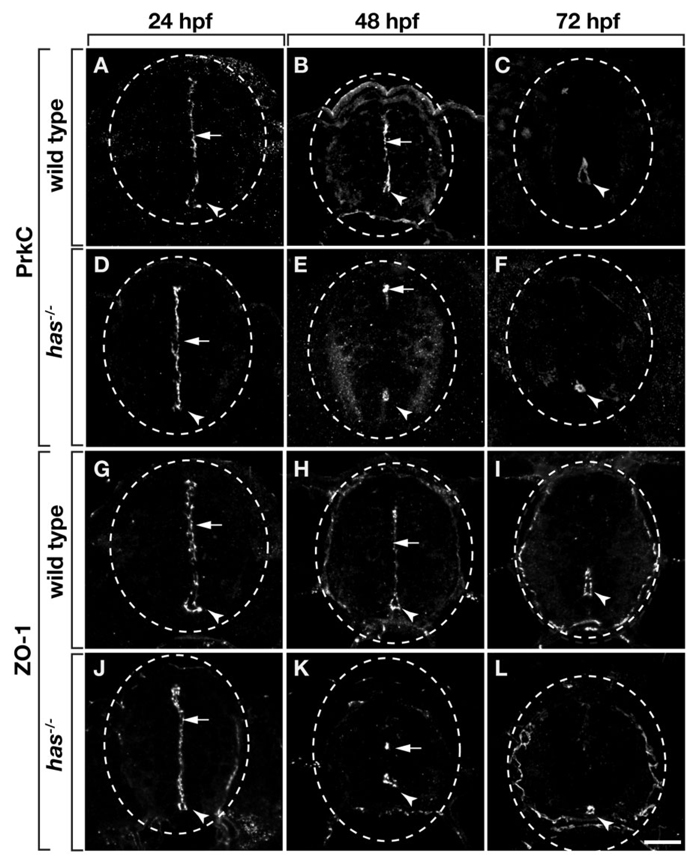
We initiated an analysis of zebrafish spinal cord neuroepithelial polarity by labeling transverse sections with an antibody that recognizes a carboxyl terminal epitope common to PrkCi and Protein kinase C, zeta (PrkCz; Horne-Badovinac et al., 2001; Cui et al., 2007). At 24 hours postfertilization (hpf) and continuing through 48 hpf PrkCi/z proteins were localized to apical cell membranes contacting the spinal cord medial septum and central canal (Fig. 1A,B). By 72 hpf, when most spinal cord cell divisions have ceased (Park et al., 2007), PrkCi/z proteins were diminished at the medium septum, but retained around the central canal (Fig. 1C). Zonula Occludins-1 (ZO-1) antibody, which recognizes a protein associated with apical neuroepithelial adherens junctions (Aaku-Saraste et al., 1996; Manabe et al., 2002; Hurd et al., 2003), revealed a similar pattern of localization (Fig. 1G–I).
Fig. 1.
Zebrafish spinal cord cells have apical polarity, which requires protein kinase C, iota (PrkCi) function. All panels show transverse sections through trunk spinal cord, dorsal up. Dashed circle marks the perimeter of the spinal cord. Arrowheads and arrows indicate central canal and medial septum, respectively. A–F: Sections labeled with anti-PrkCi/z antibody. A,B: PrkC is localized to the medial septum and central canal of wild-type embryos at 24 and 48 hours postfertilization (hpf). C: At 72 hpf, PrkC is absent from the medial septum but remains around the central canal of wild-type larvae. D: At 24 hpf, PrkC localization is normal in has−/− embryos. E: However, by 48 hpf very little PrkC is evident at the medial septum and, although PrkC remains around the central canal, the central canal is reduced in size (E) or entirely absent (not shown). F: Sections of 72 hpf has−/− larvae similarly reveal PrkC localization around an abnormally small and discontinuous central canal. The apical protein ZO-1 has a similar localization pattern to PrkC in wild-type and has−/− embryos and larvae (G–L). Scale bar = 20 µM.
In the Drosophila CNS, apical localization of Par/aPKC complexes is dependent on aPKC function (Wodarz et al., 2000). Consistent with this, targeted mutation of Prkci in mice results in loss of neuroepithelial adherens junctions within the neocortex (Imai et al., 2006). To investigate whether the apical polarity of zebrafish neuroepithelial cells similarly requires PrkCi function, we examined embryos homozygous for the m567 allele of heart and soul (has), which express a truncated, inactive PrkCi protein (Horne-Badovinac et al., 2001). We first assessed the amount and localization of PrkC proteins. Because the hasm567 allele eliminates the antibody epitope from PrkCi, any labeling evident in mutant embryos represents PrkCz and maternally expressed PrkCi (Horne-Badovinac et al., 2001). At 24 hpf, PrkC localization in has−/− embryos was indistinguishable from wild-type embryos (Fig. 1D). However, by 48 hpf, has−/− embryos had diminished levels of PrkC. Particularly, anti-PrkC labeling was nearly absent from the medial septum and revealed a smaller central canal (Fig. 1E). Additionally, PrkC was absent in many sections (data not shown), raising the possibility that the central canal is discontinuous along the length of the spinal cord in mutant embryos. At 72 hpf, diminished levels of PrkC were similarly localized to a smaller, apparently discontinuous central canal (Fig. 1F).
ZO-1 immunocytochemistry revealed a localization pattern identical to that of PrkC in has−/− embryos. At 24 hpf, ZO-1 appeared normal in mutant embryos (Fig. 1J), indicating that in the absence of zygotic PrkCi, PrkCz is sufficient to localize adherens junction proteins. By contrast, at 48 and 72 hpf ZO-1 was mostly absent from the medial septum and outlined a small, discontinuous central canal (Fig. 1K,L). Taken together, these data indicate that apical polarity of spinal neuroepithelial cells is initially normal in the absence of zygotically encoded PrkCi function but that apical polarity, adherens junctions, and central canal integrity gradually degrade.
PrkCi Is Required to Maintain Planar Divisions of Spinal Cord Precursors
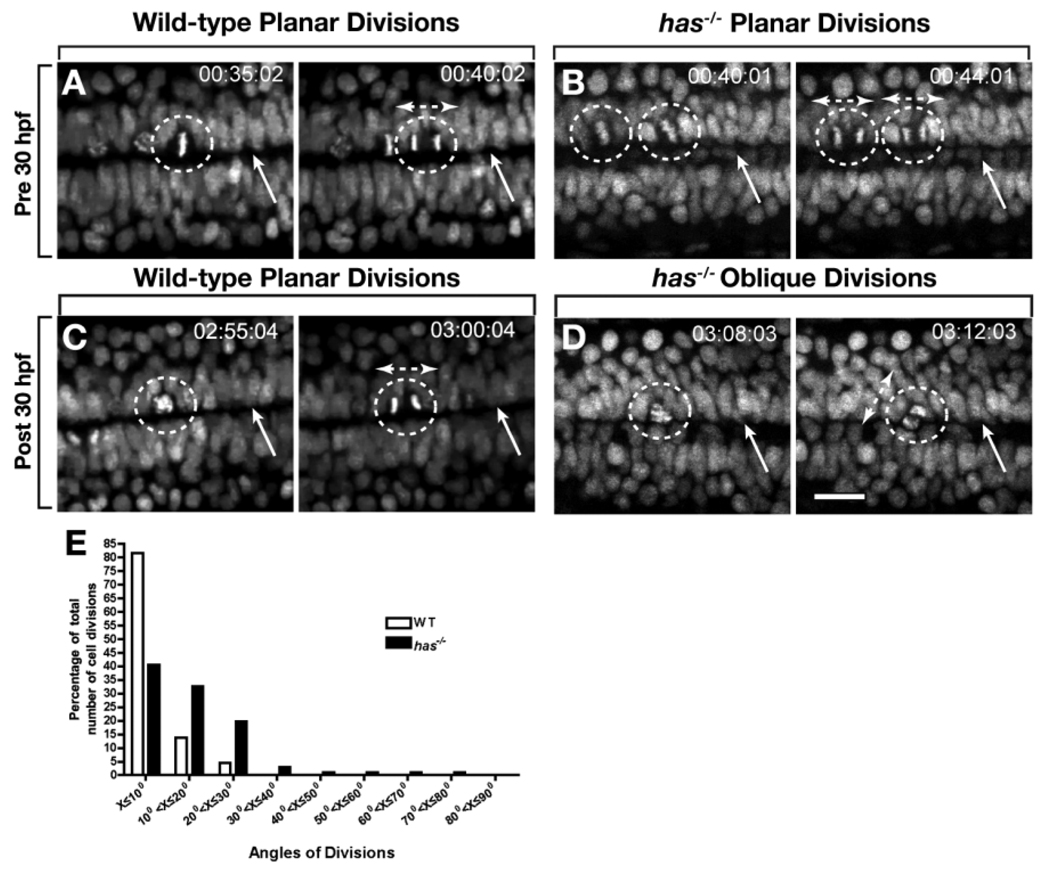
In zebrafish, cell divisions within the medial neural plate are oriented in the mediolateral plane of the neuroepithelium (Concha and Adams, 1998; Geldmacher-Voss et al., 2003). As the neural plate condenses to form the neural keel and then neural rod, cell divisions remain perpendicular to the anteroposterior axis and become orthogonal to the plane of the neuroepithelium (Geldmacher-Voss et al., 2003). Upon formation of the neural tube, cell divisions rotate 90° so that they occur within the plane of the neuroepithelium (Geldmacher-Voss et al., 2003; Lyons et al., 2003). As a prelude to our investigation of neural cell polarity and fate, we performed our own analysis of cell division orientation, but focused on a later period of development than previous studies. To mark dividing cells we used the transgenic reporter Tg(h2afv:gfp), which expresses enhanced green fluorescent protein fused to a histone protein (Pauls et al., 2001). We imaged embryos from a dorsal view, focusing on cells that line the central canal starting at 27 hpf and continuing until 33 hpf. This corresponds to the period during which most spinal cord neurogenesis is completed and precedes OPC formation (Park et al., 2002; Park and Appel, 2003). Consistent with previous reports of cell division patterns in both zebrafish and chick neural tube (Geldmacher-Voss et al., 2003; Lyons et al., 2003; Morin et al., 2007), most divisions of cells bordering the central canal were within 15° of the plane of the neuroepithelium, and therefore parallel or near parallel to the central canal (Fig. 2A,C,E and Supp. Movie S1, which is available online).
Fig. 2.
Protein kinase C, iota (PrkCi) maintains planar divisions of cells that divide along the central canal. A–D: Frames captured from time-lapse movies, from a dorsal view, of wild-type and has−/− embryos carrying the Tg(h2afv:egfp) transgene. Numbers in upper right corners indicate time elapsed from beginning of imaging at 27 hours postfertilization (hpf). Dashed circles outline dividing cells, arrows point to the central canal and bi-directional arrows indicate orientation of the mitotic spindle and angle of division. A,B: In both wild-type and has−/− embryos, divisions that occur before 30 hpf are planar. The central canal is less distinct in has−/− embryos than in wild-type. C,D: Divisions after 30 hpf remain planar in wild-type embryos (C) but become oblique in has−/− embryos (D). The central canal is indistinct and appears to have been replaced by cells. E,F: Quantification of angles of division in wild-type and has−/− embryos (E). In wild-type most division planes are within 15° of the plane of the epithelium, indicated by the central canal. has−/− embryos have numerous divisions greater than 15°. Scale bar = 24 µM.
Time-lapse imaging revealed the orientation of cell divisions changes with time in has mutant embryos. Before 30 hpf, cells bordering the central canal divided similarly to those in wild-type (Fig. 2B). Beginning at approximately 30 hpf, however, more than half of the divisions were greater than 15° from the plane of the neuroepithelium (Fig. 2D,E and Supp. Movie S2). At the same time, the central canal became less distinct, with the space occupied by spinal cord cells (Fig. 2D). Therefore, loss of PrkCi function results in disruption of planar division and a breakdown of the neuroepithelium.
Loss of PrkCi Function Causes Formation of Excess OPCs Without Affecting Motor Neuron Formation
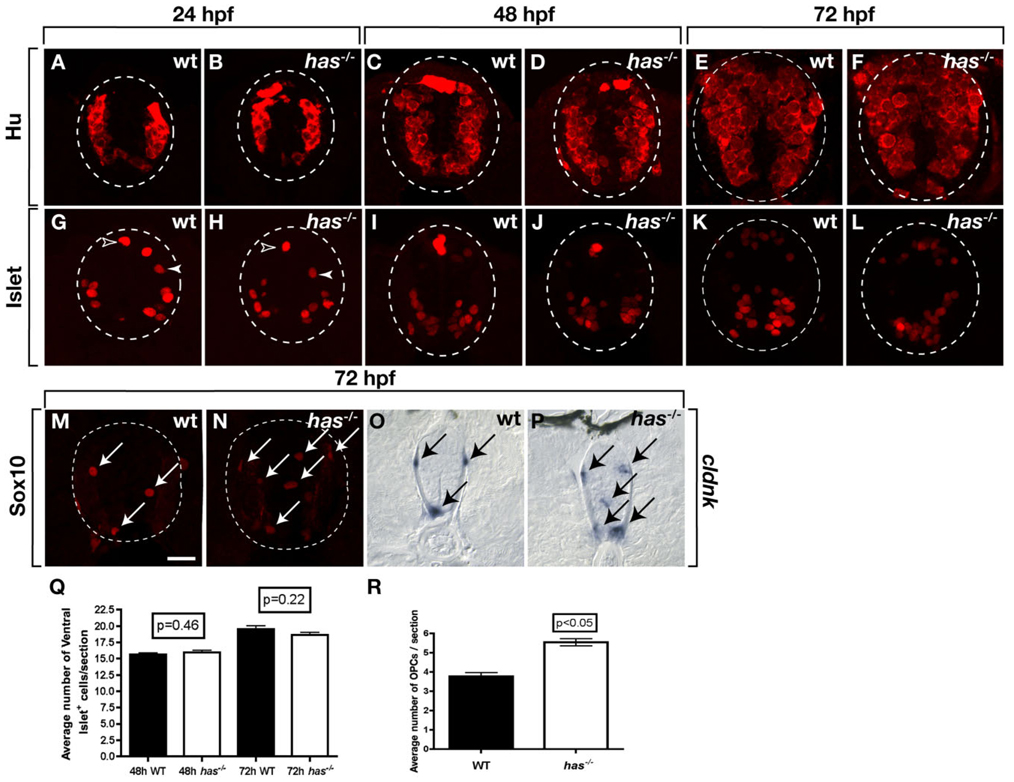
The above data reveal that PrkCi is required for maintenance of apical polarity and planar divisions of spinal cord precursors. Because cell polarity and division pattern often influence cell fate, we assessed formation of neurons and glia using molecular markers. We first labeled transverse sections of wild-type and has−/− spinal cords with anti-Hu antibody, which marks all newly formed neurons (Marusich et al., 1994). No apparent differences in the distribution of Hu+ neurons was evident between wild-type and mutant embryos at 24, 48, and 72 hpf (Fig. 3A–F). To quantify the number of neurons, we labeled sections of 72 hpf wild-type and mutant larvae with anti-Hu antibody and a nuclear stain and determined the proportion of spinal cord cells that had neuronal identity. This revealed little difference in the number of neurons formed by larval stage (data not shown). We also examined Isl+ motor neurons, Rohon-Beard sensory neurons and interneurons, which are mostly formed by approximately 48 hpf. No obvious differences in the number and distribution of Isl+ neurons was apparent between wild-type and mutant through 72 hpf (Fig. 3G–L) and quantification of Isl+ motor neurons revealed no statistically significant differences at 48 and 72 hpf (Fig. 3Q). Therefore, formation of spinal cord neurons is not apparently affected by absence of PrkCi function, consistent with our observation that PrkCz localization, apical polarity and cell division pattern appear normal during neurogenic stages.
Fig. 3.
Loss of protein kinase C, iota (PrkCi) function produces excess oligodendrocyte progenitor cells (OPCs). All images are of transverse sections through trunk spinal cord, dorsal up. Outlined circle marks the perimeter of the spinal cord. A–F: The number and distribution of neurons, marked by anti-Hu labeling, are similar in wild-type and has−/− embryos at 24, 48, and 72 hours postfertilization (hpf). G–L: Isl+ motor neurons (brackets), interneurons (solid arrowheads), and Rohon-Beard sensory neurons (open arrowheads) are similar in wild-type and has−/− embryos and larvae through 72 hpf. M,N: Anti-Sox10 labeling reveals excess OPCs in 72 hpf has−/− larva (N) compared with wildtype (M). O,P: cldnk RNA expression marking differentiating oligodendrocytes. O: In wild-type, cldnk+ cells (arrows) are at the pial surface in dorsal and ventral spinal cord. P: has−/− larvae have excess cldnk+ cells and some occupy ectopic positions in medial spinal cord. Q: Quantification of spinal cord Isl+ motor neurons in the spinal cord between wild-type and has−/− embryos at 48 and 72 hpf. R: Quantification of spinal cord Sox10+ OPCs. Error bars represent SEM. Statistical significance was determined using Student’s t-test. Scale bar = 20 µM.
To investigate formation of OPCs, we labeled sections with anti-Sox10 antibody, which serves as a specific marker of OPCs and differentiating oligodendrocytes (Kuhlbrodt et al., 1998; Park et al., 2005). At 48 hpf, soon after specification of spinal cord OPCs begins, the number and distribution of OPCs was indistinguishable between wild-type and has mutant embryos (data not shown). However, by 72 hpf, both the number and distribution of OPCs was altered in has−/− spinal cords relative to wild-type (Fig. 3M,N). In particular, mutant larvae had 1.4-fold more OPCs than wildtype (Fig. 3R) and some OPCs were abnormally positioned within medial regions of the spinal cord (Fig. 3N). We also performed in situ RNA hybridization to detect expression of cldnk, which specifically marks differentiating oligodendrocytes (N. Takada and B. Appel, unpublished data). This similarly revealed an excess of oligodendrocytes in has−/− spinal cords relative to wild-type (Fig. 3O,P). Additionally, cldnk+ cells sometimes occupied medial spinal cord, where they are normally not found. These data indicate that PrkCi function limits both the formation and differentiation of oligodendrocyte lineage cells.
PrkCi Function and Apical Polarity Are Required to Maintain Spinal Cord Precursors
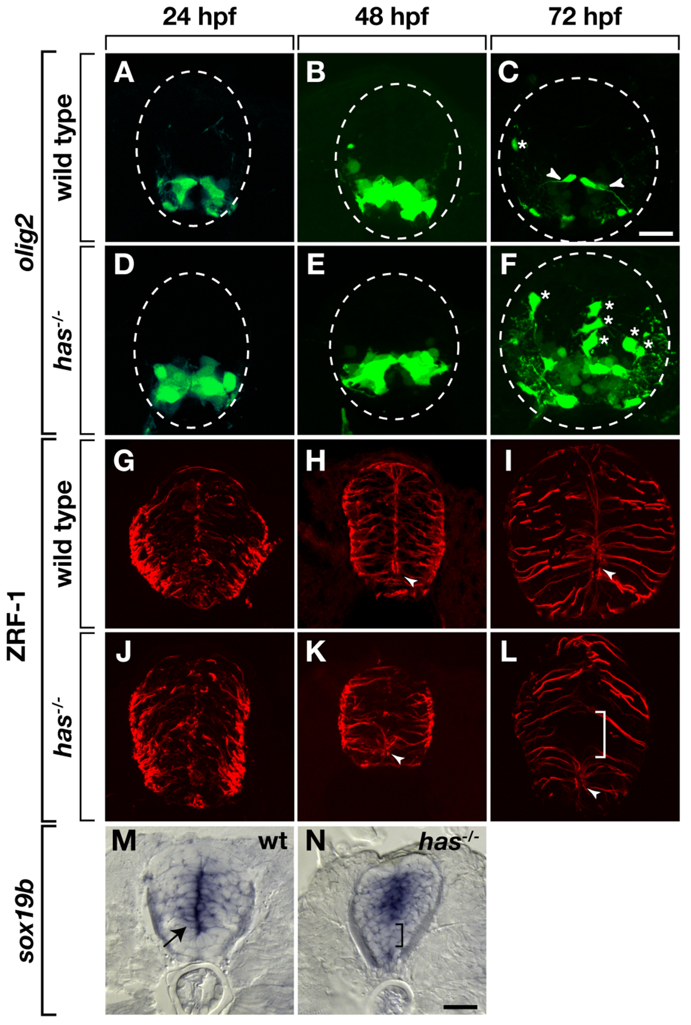
In zebrafish, slowly-dividing olig2+ spinal cord precursors that have radial glial characteristics and continuously give rise to new OPCs are maintained through late embryonic stage into adulthood (Park et al., 2007). One possible explanation for the excess OPC phenotype of has mutant larvae, therefore, is that, in the absence of PrkCi function, olig2+ cells lose precursor characteristics and develop as oligodendrocytes. Consistent with this, whereas olig2+ radial fibers are evident by 72 hpf in transverse sections of wild-type Tg(olig2:egfp) larvae (Fig. 4A–C), similar processes were rare in has−/−; Tg(olig2:egfp) larvae (Fig. 4D–F). We next labeled sections with anti–Zrf-1 antibody, which marks spinal cord radial glia (Trevarrow et al., 1990). Through 48 hpf, Zrf-1+ fibers were similar in wild-type and has mutant embryos (Fig. 4G,H,J,K). However, by 72 hpf, whereas GFAP+ radial glia were distributed uniformly throughout the spinal cord of wild-type larvae (Fig. 4I), radial glia were consistently absent from the spinal cord just dorsal to remnants of the central canal (Fig. 4L). We next used in situ RNA hybridization to detect expression of sox19b, which marks cells of the medial spinal cord that are likely dividing precursors. Similar to Zrf-1, sox19b expression appeared normal through 48 hpf in has mutant embryos (data not shown). By 72 hpf, sox19b expression marks cells near the central canal and medial septum of wild-type larvae (Fig. 4M). By contrast, sox19b expression was frequently absent from ventral spinal cord near the central canal in has mutant larvae but was maintained in more dorsal and ventral cells (Fig. 4N). Taken together, these data indicate that loss of PrkCi function results in loss of spinal cord precursors near the central canal during late embryogenesis.
Fig. 4.
Protein kinase C, iota (PrkCi) is required to maintain ventral spinal cord cells with precursor characteristics. All images are of transverse sections through trunk spinal cord, dorsal up. Outlined circle marks the perimeter of the spinal cord. A–F: Enhanced green fluorescent protein (EGFP) expression driven by the Tg(olig2:egfp) promoter. A,B: In wild-type embryos, EGFP marks motor neurons and pMN precursors through 48 hours postfertilization (hpf). C: By 72 hpf, EGFP+ fibers (arrowheads), marking precursors that persist into larval stage, become evident. D,E: EGFP expression appears normal in has−/− embryos at 24 and 48 hpf. F: At 72 hpf, few EGFP+ radial fibers are evident. Asterisks mark dorsally migrated OPCs. G–L: Zrf-1 immunocytochemistry to label radial glial fibers. G–I: In wild-type embryos and larvae, radial fibers are distributed uniformly through the spinal cord. In ventral spinal cord, the apical membrane of some radial glia surround the central canal (arrowheads). J–L: In has mutants, radial fibers initially appear normal, but by 72 hpf, a gap appears in ventral spinal cord (bracket) and the central canal is reduced or absent. M,N: sox19b RNA expression. M: At 72 hpf in wild-type, sox19b expression marks cells near the central canal (arrow). N: At 72 hpf sox19b is not expressed at its normal position in has−/− larva (bracket). Instead, ventral and dorsal spinal cord cells express sox19b. Scale bar = 20 µM.
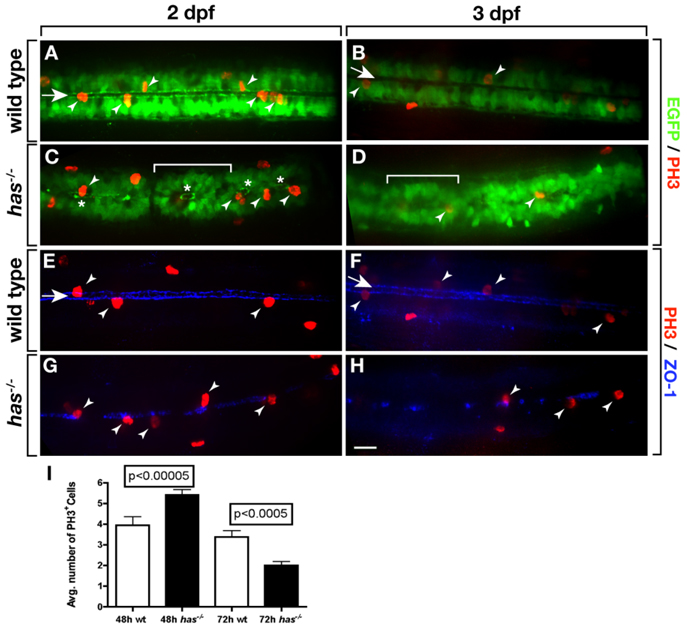
To further investigate the relationship between apical polarity and spinal cord precursor division, we labeled embryos and larvae carrying the Tg(olig2:egfp) transgene with antibody specific to Phospho-Histone H3 (anti-PH3), which labels cells in M phase. At 2 dpf, numerous olig2+ PH3+ cells were evident along the central canal of wild-type embryos (Fig. 5A). By 3 dpf, the number of PH3+ cells was substantially decreased (Fig. 5B), consistent with our previous data showing a significant decline in the number of dividing cells in the spinal cord between 2 and 3 dpf (Park et al., 2007). In has mutant embryos, the central canal, outlined by olig2+ cells, was discontinuous with remaining pockets of central canal surrounded by rosettes of olig2+ cells (Fig. 5C,D). Notably, PH3+ cells were evident, but appeared only within the rosettes. As in wild-type, 3 dpf has mutant larvae had fewer PH3+ cells than at 2 dpf. We also use ZO-1 labeling to examine the distribution of dividing cells. As expected, PH3+ cells in wild-type had apical polarity and were associated with the central canal (Fig. 5E,F). At 2 dpf, has mutant embryos had patches of apical membrane, consistent with the discontinuous nature of the central canal, and PH3+ cells were nearly always associated with ZO-1 labeling (Fig. 5G). We noticed that the density of PH3+ cells seemed to be greater in mutant embryos than in wild-type. In fact, whereas wild-type embryos had 3.9 PH3+ cells within a 288-µm length of trunk spinal cord, mutant embryos had 5.4 PH3+ cells over the same distance (Fig. 5I). At 3 dpf, the gaps in ZO-1 labeling of mutant larvae were larger and more frequent than at 2 dpf (Fig. 5H) and mutant larvae had fewer M phase cells (Fig. 5I). Therefore, has−/− mutant embryos have a transient increase and then deficit of dividing spinal cord precursors.
Fig. 5.
Loss of PrkCi function causes a transient excess and then deficit of dividing spinal cord precursors. A–D: Images, from dorsal view and focused on the trunk spinal cord, of wild-type and has−/− embryos and larvae carrying the Tg(olig2:egfp) reporter and labeled with anti-PH3 antibody to mark M phase cells. In wild-type most mitotic cells (arrowheads) are adjacent to the central canal (arrow). A,B: More cells divide at 2 days postfertilization (dpf; A) than at 3 dpf (B). C,D: In has−/− embryos, many EGFP+ cells form rosettes (brackets) surrounding discontinuous portions of central canal (asterisks). PH3+ are usually only found within rosettes. E–H: Images, from dorsal view and focused on the trunk spinal cord, of wild-type and has−/− embryos and larvae labeled with anti-PH3 antibody (red) and anti–ZO-1 antibody (blue) to mark apical membrane. E,F: In wild-type, ZO-1 localization outlines a continuous central canal closely associated with PH3+ mitotic cells. G,H: In has−/− embryos and larvae, ZO-1 labeling is discontinuous but mitotic cells are nearly always associated with remaining apical membrane. I: Quantification of PH3+ cells at 2 and 3 dpf over a 288 µm length of trunk spinal cord. Error bars represent SEM. Statistical significance was determined using Student’s t-test.
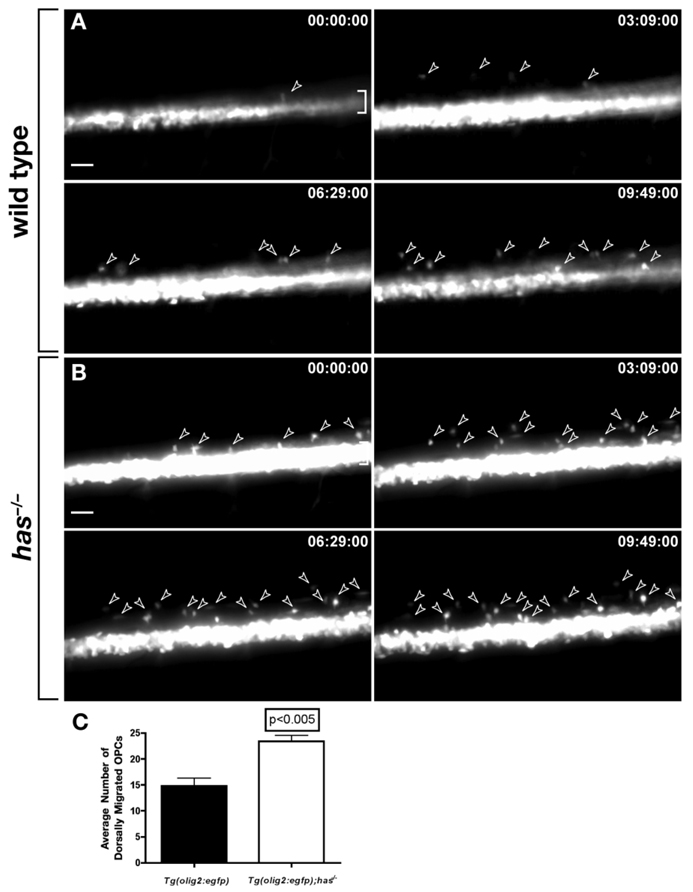
Our data raise the possibility that excess OPCs arise in has−/− mutant embryos due to misallocation of spinal cord precursors to an oligodendrocyte fate. To test this, we performed time-lapse imaging of wild-type and has mutant embryos carrying the Tg(olig2:egfp) transgene. Over a 10-hr period, approximately 1.5-fold more OPCs migrated from ventral spinal cord into dorsal spinal cord in has mutant embryos relative to wild-type (Fig. 6A–Cand Supp. Movie S3 and Supp. Movie S4). Migrating OPCs did not divide more frequently in has mutant embryos than in wild-type. Therefore, excess OPCs arise in has mutant embryos from specification of excess pMN precursors for oligodendrocyte fate.
Fig. 6.
Loss of protein kinase C, iota (PrkCi) function causes formation of excess oligodendrocyte progenitor cells (OPCs) from pMN precursors. Panels show frames captured from time-lapse videos of wild-type and has−/− embryos carrying the Tg(olig2:egfp) reporter. Dorsal is up and time elapsed from beginning of imaging at 50 hours postfertilization (hpf) is indicated in upper right corners of panels. Brackets mark pMN precursor domain in ventral spinal cord. A: Wild-type embryo. Arrowheads mark OPCs migrating dorsally from the ventral pMN precursor domain. B: has−/− embryo. More OPCs (arrowheads) emerge from the pMN domain than in wild-type over a similar period of time. C: Quantification of dorsally migrating OPCs. Data were obtained from time-lapse movies of 6 wild-type and 5 has−/− embryos. Error bars represent SEM. Statistical significance was determined using Student’s t-test.
DISCUSSION
During development, neural precursors divide both to produce new precursors and to give rise to differentiating neurons and glial cells. In vertebrates, most precursors transform into differentiated cell types by early postnatal stages but, at least in some regions of the CNS, others gain characteristics of stem cells (Gotz and Huttner, 2005; Merkle and Alvarez-Buylla, 2006). Therefore, the mechanisms that determine whether precursors undergo self-renewing or differentiative divisions play key roles in neural development and homeostasis.
Extensive investigations of the development of the Drosophila nervous system have uncovered mechanisms for regulation of cell division and cell fate that provide a compelling model for vertebrate neural development (Doe, 2008). The nervous system of the fly embryo is produced from a sheet of ventral neuroectoderm. Divisions of neuroectodermal cells occur within the plane of the epithelium and the resulting progeny cells thereby remain within it. By virtue of a lateral inhibition mechanism mediated by Notch signaling, a select number of neuroectodermal cells are specified as neuroblasts, which begin to delaminate from the epithelium in the basal direction (Egger et al., 2008). Par complex proteins, which are localized to the apical membrane of neuroectodermal cells, remain with apical membrane of the neuroblasts as they delaminate (Yu et al., 2006). The apical localization of Par complex proteins serves to both restrict proteins that specify GMC fate to basal membrane and, by organizing a complex of proteins including Inscuteable, Pins and heterotrimeric G proteins, to direct a 90° rotation of the mitotic spindle (Yu et al., 2006; Egger et al., 2008; Wu et al., 2008). Subsequent division of the neuroblast occurs perpendicular to the plane of the epithelium resulting in distribution of GMC fate determinants to the basal progeny cell but not to the apical cell, which remains as a neuroblast. In embryos, the GMCs then generally divide once to produce a pair of differentiated neurons.
In the neural tube of vertebrate embryos, the mitotic apparatus of dividing neuroepithelial cells is apically positioned so that divisions occur close to the ventricle (Hollyday, 2001). As cells exit the mitotic cycle, they lose contact with the apical surface and migrate basally where they differentiate as neurons or glia. Following the Drosophila example, divisions within the plane of the epithelium might then produce progeny that remain as neuroepithelial precursors whereas those that are perpendicular could be asymmetric, producing one apically positioned precursor cell and one basally positioned differentiated cell. Consistent with this, imaging of living cells in slices of ferret brain revealed both planar and perpendicular divisions as well as some intermediate divisions (Chenn and McConnell, 1995). Imaging of slices from embryonic mouse cortex revealed similar mitotic patterns accompanied by rapid migration of basal progeny produced by perpendicular divisions (Haydar et al., 2003). However, several other studies now indicate that the majority of precursor divisions in the neural tube and retina of vertebrate embryos are planar (Das et al., 2003; Geldmacher-Voss et al., 2003; Lyons et al., 2003; Kosodo et al., 2004; Morin et al., 2007; Konno et al., 2008; Noctor et al., 2008). Our time-lapse imaging of zebrafish spinal cord precursors revealed a complete absence of perpendicular divisions, even during a period when many neurons and OPCs are formed. Therefore, the simple model in which asymmetric neural cell fates result from perpendicular divisions seemingly can not account for the many neurons and glia that arise during development (Huttner and Brand, 1997). Careful examination of neural precursor divisions revealed that some result in unequal segregation of apical membrane to progeny cells, which correlated with asymmetric fate (Kosodo et al., 2004). Whether slight differences in the amount of apical membrane produces asymmetric fates by means of asymmetric distribution of fate determinants and whether these differences can fully account for the decision to remain as a precursor or differentiate remain unknown.
Maintenance of adherens junctions is associated with maintenance of some stem cell populations (Song et al., 2002; Fuchs et al., 2004) and could provide a mechanism by which asymmetric segregation of apical membrane to progeny cells influences fate. Vertebrate embryos localize Par complex proteins to apical adherens junctions of the neuroepithelium (Manabe et al., 2002; Geldmacher-Voss et al., 2003; Afonso and Henrique, 2006; Imai et al., 2006; von Trotha et al., 2006; Kovac et al., 2007) and these are lost from neuroepithelial cells as they differentiate. Therefore, Par complex and adherens junction associated proteins might be necessary for maintaining proliferative neuroepithelial precursors. This idea has been tested by examining mice deficient for Prkci function in Nestin+ neural precursors (Imai et al., 2006). Neuroepithelial character and adherens junctions were lost from mutant mice but, surprisingly, the total number of dividing cells and neurons were normal at embryonic day 15.5 suggesting that, at least in this instance, Prkci is not necessary for neurogenesis.
Other studies have tested the functions of vertebrate proteins homologous to fly proteins that orient the mitotic spindle. Reducing function of the G protein regulator LGN, homologous to Drosophila Pins, in the chick spinal cord randomized the normally planar division patterns and frequently displaced progenitors from the ventricular zone to the mantle but did not overtly affect neurogenesis (Morin et al., 2007). Loss of LGN function in mice similarly shifted planar divisions to oblique and displaced progenitors from the ventricular zone without affecting neuron production (Konno et al., 2008). By contrast, another study showed a higher frequency of nonplanar divisions at slightly later stages of neurogenesis and that blocking Gβλ signaling and function of the Pins homolog AGS3 increases the number of planar divisions that produce two postmitotic neurons (Sanada and Tsai, 2005).
Our own investigation now reveals that loss of PrkCi function and adherens junctions in zebrafish results in the loss of substantial numbers of spinal cord precursors during late embryogenesis, with a concomitant increase in OPCs. In wild-type embryos, ZO-1 labeling reveals adherens junctions at the medial septum and surrounding the central canal of the spinal cord during neurogenic stages. ZO-1 is gradually lost from the medial septum as neurogenesis subsides at late embryonic stage but persists around the central canal, an active site of cell division, into juvenile stage (Park et al., 2007). In has mutant embryos, ZO-1 localization initially appeared normal, probably due to PrkCz function. Accordingly, neural precursor divisions resembled wild-type, remaining within the plane of the neuroepithelium, and neurons were formed in apparently normal number. However, ZO-1 disappeared prematurely from the medial septum and only marked a reduced, apparently discontinuous central canal at late embryonic and early larval stages. At approximately the same time, divisions of many precursors at the level of the central canal shifted from planar to oblique and the central canal appeared to be invaded by cells. These observations indicate that PrkCi maintains the neuroepithelial character of the spinal cord, consistent with the proposed role of PrkCi in the mouse neocortex (Imai et al., 2006). In has mutant embryos, spinal cord cells formed rosettes around remnants of central canal and these contained mitotic cells. After a transient increase in the number of mitotic cells at a late embryonic stage, the number of dividing cells fell below normal and radial glia, revealed by an olig2 transgenic reporter and Zrf-1 labeling, were lost from the region of the central canal by early larval stage. Additionally, by larval stage mutants had a slight but statistically significant increase in the number of OPCs formed from ventral spinal cord olig2+ cells. We conclude that PrkCi is necessary to maintain spinal cord precursors and that in its absence dividing cells exit the cell cycle and differentiate as glia.
In summary, our data indicate that planar divisions directed by PrkCi function are required for both symmetric proliferative and asymmetric self-renewing divisions by spinal cord precursors. The precursor loss and excess formation of OPCs that accompanies the shift to oblique divisions in has mutant embryos suggests a shift to symmetric differentiative divisions at the expense of asymmetric self-renewing divisions. One possible explanation was offered by Konno et al. (2008) who proposed that planar divisions can be asymmetric by segregating both apical and basal processes to one daughter, which remains as a precursor, whereas daughters that only receive apical membrane exit the cell cycle and differentiate. The same pattern holds for symmetric proliferative divisions, suggesting that fate asymmetry must arise through other signaling mechanisms, such as differential regulation of Notch signaling activity (Buchman and Tsai, 2008; Kageyama et al., 2008).
EXPERIMENTAL PROCEDURES
Fish Husbandry
Embryos were produced by pair-wise mating and kept at 28.5°C in egg water or embryo medium. Embryos were staged to hours postfertilization (hpf) or days postfertilization (dpf) according to established zebrafish guidelines (Kimmel et al., 1995). Embryos that were to be used for live imaging, immunocytochemistry, or in situ hybridization were treated in 0.003% phenylthiourea (PTU) in egg water to block pigmentation. The experiments conducted in this study used the following strains of zebrafish: AB, hasm567 (Stainier et al., 1996), Tg(olig2:egfp) (Shin et al., 2003), and Tg(h2afv:gfp) (Pauls et al., 2001).
Immunocytochemistry
Embryos and larvae were fixed in 4% antibody fix (4% paraformaldehyde, 8% sucrose, 1× phosphate buffered saline [PBS]) overnight at 4°C. After fixing, the embryos were embedded in 1.5% agar/ 5% sucrose blocks and placed in 30% sucrose/PBS solution to equilibrate overnight. The blocks were then frozen over 2-methylbutane chilled by liquid nitrogen. We collected 10- to 12-µm sections on superfrost microscope slides using a cryostat microtome. The sections were rehydrated in 1× PBS for 30 min. and then blocked in 2% bovine serum albumin/sheep serum in 1× PBS for 30 min. before incubating with primary antibody overnight at 4°C. For fluorescent detection of antibody labeling, we used Alexa Fluor 488, Alexa Fluor 568, Alexa Fluor 647 goat anti-mouse or goat anti-rabbit conjugates (1:500, Molecular Probes). The primary anti-bodies used included rabbit anti-PkC (#sc-216, 1:200, Santa Cruz Biotechnology, Inc.), mouse anti-Islet (clone # 39.4D5, 1:1,000, Developmental Studies Hybridoma Bank [DSHB], Iowa City, IA), rabbit anti–phospho-Histone-H3 (# 06570, 1:1000, Upstate Biotechnology, Charlottesville, VA), rabbit anti-Sox10 (1:1,000; Park et al., 2005), mouse anti–ZO-1 (#33-9100, 1:200, Invitrogen), and mouse anti–ZRF-1 (1:500, University of Oregon Monoclonal Antibody Facility).
In Situ Hybridization
Embryos and larvae were fixed in 4% paraformaldehyde overnight at 4°C and stored in methanol at −20°C. After linearizing plasmids with the appropriate restriction enzymes, antisense cRNA was transcribed using Roche digoxigenin-labeling reagents and T3, T7, or SP6 RNA polymerases (New England Biolabs). cldnk (Sprague et al., 2001) was identified in a microarray screen for oligodendrocytes-specific genes and will be described elsewhere. sox19b was originally named sox31 (Girard et al., 2001). After processing embryos for in situ RNA hybridization embryos were embedded in agar and sectioned as described above. Sections were rehydrated in 1× PBS for 30 min. then covered with 75% glycerol. Images were obtained using a Retiga EXI camera attached to a Zeiss Axiovert 200 microscope equipped with Openlab software.
In Vivo Time-lapse Imaging
Embryos were raised in egg water containing PTU and at the appropriate stages manually dechorionated using watchmaker forceps. Embryos were anesthetized using 3-aminobenzoic acid ethyl ester (Tricaine) and mounted laterally or dorsally in 35 mm glass bottom Petri dishes containing 0.8% low-melting temperature agarose. Confocal time-lapse movies were obtain by using a ×40 oil immersion objective mounted on a Zeiss Axiovert 200 microscope equipped with a PerkinElmer spinning disk confocal system. Z-stack images were obtained every 3–5 min and compiled into a Quicktime movie using Volocity software (Improvision). Widefield time-lapse movies were obtained using a ×40 objective on a Zeiss Axiovert 200 microscope equipped with a Retiga EXI camera. Z-stacked images were obtained every 10–12 min. Embryos were maintained at 28.5°C using a heated stage chamber during imaging.
Angle of Division Measurements
PTU-treated Tg(h2afv:gfp) and Tg-(h2afv:gfp) has−/− embryos were mounted dorsally at 27 hpf in low melting point agarose. Z-stack images were obtained at the level of the central canal every 3–5 min. These images were then compiled into a Quick-time time-lapse movie using Volocity. Movies were analyzed using Openlab software. Going frame by frame, we tracked dividing cells at the central canal into telophase and then drew a line parallel to the separated chromatids. With the central canal as a reference, we measured the angle between the line and the central canal, using a program called Screen Protractor (Iconico.com).
Supplementary Material
ACKNOWLEDGMENTS
The anti-Isl antibody, developed by T. M. Jessell, and the anti-ZRF1 antibody, developed by B. Trevarrow, were obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the National Institute of Child Health and Development and maintained by The University of Iowa, Department of Biological Sciences, Iowa City, IA 52242. Confocal microscopy was performed using equipment obtained with funds provided by the Vanderbilt Academic Venture Capital Fund. B.A. received a NIH grant and R.K.R. received a Cellular Biochemical and Molecular Sciences Training Grant.
Grant sponsor: NIH; Grant number: NS046668; Grant number: GM008554.
REFERENCES
- Aaku-Saraste E, Hellwig A, Huttner WB. Loss of occludin and functional tight junctions, but not ZO-1, during neural tube closure—remodeling of the neuroepithelium prior to neurogenesis. Dev Biol. 1996;180:664–679. doi: 10.1006/dbio.1996.0336. [DOI] [PubMed] [Google Scholar]
- Afonso C, Henrique D. PAR3 acts as a molecular organizer to define the apical domain of chick neuroepithelial cells. J Cell Sci. 2006;119:4293–4304. doi: 10.1242/jcs.03170. [DOI] [PubMed] [Google Scholar]
- Betschinger J, Mechtler K, Knoblich JA. The Par complex directs asymmetric cell division by phosphorylating the cytoskeletal protein Lgl. Nature. 2003;422:326–330. doi: 10.1038/nature01486. [DOI] [PubMed] [Google Scholar]
- Buchman JJ, Tsai LH. Putting a notch in our understanding of nuclear migration. Cell. 2008;134:912–914. doi: 10.1016/j.cell.2008.09.007. [DOI] [PubMed] [Google Scholar]
- Chenn A, McConnell SK. Cleavage orientation and the asymmetric inheritance of Notch1 immunoreactivity in mammalian neurogenesis. Cell. 1995;82:631–641. doi: 10.1016/0092-8674(95)90035-7. [DOI] [PubMed] [Google Scholar]
- Concha ML, Adams RJ. Oriented cell divisions and cellular morphogenesis in the zebrafish gastrula and neurula: a time-lapse analysis. Development. 1998;125:983–994. doi: 10.1242/dev.125.6.983. [DOI] [PubMed] [Google Scholar]
- Cui S, Otten C, Rohr S, Abdelilah-Seyfried S, Link BA. Analysis of aPK-Clambda and aPrkCzeta reveals multiple and redundant functions during vertebrate retinogenesis. Mol Cell Neurosci. 2007;34:431–444. doi: 10.1016/j.mcn.2006.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das T, Payer B, Cayouette M, Harris WA. In vivo time-lapse imaging of cell divisions during neurogenesis in the developing zebrafish retina. Neuron. 2003;37:597–609. doi: 10.1016/s0896-6273(03)00066-7. [DOI] [PubMed] [Google Scholar]
- Doe CQ. Neural stem cells: balancing self-renewal with differentiation. Development. 2008;135:1575–1587. doi: 10.1242/dev.014977. [DOI] [PubMed] [Google Scholar]
- Egger B, Chell JM, Brand AH. Insights into neural stem cell biology from flies. Philos Trans R Soc Lond B Biol Sci. 2008;363:39–56. doi: 10.1098/rstb.2006.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs E, Tumbar T, Guasch G. Socializing with the neighbors: stem cells and their niche. Cell. 2004;116:769–778. doi: 10.1016/s0092-8674(04)00255-7. [DOI] [PubMed] [Google Scholar]
- Geldmacher-Voss B, Reugels AM, Pauls S, Campos-Ortega JA. A 90-degree rotation of the mitotic spindle changes the orientation of mitoses of zebrafish neuroepithelial cells. Development. 2003;130:3767–3780. doi: 10.1242/dev.00603. [DOI] [PubMed] [Google Scholar]
- Girard F, Crémazy F, Berta P, Renucci A. Expression pattern of the Sox31 gene during zebrafish embryonic development. Mech Dev. 2001;100:71–73. doi: 10.1016/s0925-4773(00)00491-3. [DOI] [PubMed] [Google Scholar]
- Gotz M, Huttner WB. The cell biology of neurogenesis. Nat Rev Mol Cell Biol. 2005;6:777–788. doi: 10.1038/nrm1739. [DOI] [PubMed] [Google Scholar]
- Haydar TF, Ang E, Jr, Rakic P. Mitotic spindle rotation and mode of cell division in the developing telencephalon. Proc Natl Acad Sci U S A. 2003;100:2890–2895. doi: 10.1073/pnas.0437969100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollyday M. Neurogenesis in the vertebrate neural tube. Int J Dev Neurosci. 2001;19:161–173. doi: 10.1016/s0736-5748(00)00093-9. [DOI] [PubMed] [Google Scholar]
- Horne-Badovinac S, Lin D, Waldron S, Schwarz M, Mbamalu G, Pawson T, Jan Y, Stainier DY, Abdelilah-Seyfried S. Positional cloning of heart and soul reveals multiple roles for PKC lambda in zebrafish organogenesis. Curr Biol. 2001;11:1492–1502. doi: 10.1016/s0960-9822(01)00458-4. [DOI] [PubMed] [Google Scholar]
- Hurd TW, Gao L, Roh MH, Macara IG, Margolis B. Direct interaction of two polarity complexes implicated in epithelial tight junction assembly. Nat Cell Biol. 2003;5:137–142. doi: 10.1038/ncb923. [DOI] [PubMed] [Google Scholar]
- Huttner WB, Brand M. Asymmetric division and polarity of neuroepithelial cells. Curr Opin Neurobiol. 1997;7:29–39. doi: 10.1016/s0959-4388(97)80117-1. [DOI] [PubMed] [Google Scholar]
- Imai F, Hirai S, Akimoto K, Koyama H, Miyata T, Ogawa M, Noguchi S, Sasaoka T, Noda T, Ohno S. Inactivation of aPKClambda results in the loss of adherens junctions in neuroepithelial cells without affecting neurogenesis in mouse neocortex. Development. 2006;133:1735–1744. doi: 10.1242/dev.02330. [DOI] [PubMed] [Google Scholar]
- Kageyama R, Ohtsuka T, Shimojo H, Imayoshi I. Dynamic Notch signaling in neural progenitor cells and a revised view of lateral inhibition. Nat Neurosci. 2008;11:1247–1251. doi: 10.1038/nn.2208. [DOI] [PubMed] [Google Scholar]
- Kim H, Shin J, Kim S, Poling J, Park HC, Appel B. Notch-regulated oligodendrocyte specification from radial glia in the spinal cord of zebrafish embryos. Dev Dyn. 2008;237:2081–2089. doi: 10.1002/dvdy.21620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Konno D, Shioi G, Shitamukai A, Mori A, Kiyonari H, Miyata T, Matsuzaki F. Neuroepithelial progenitors undergo LGN-dependent planar divisions to maintain self-renewability during mammalian neurogenesis. Nat Cell Biol. 2008;10:93–101. doi: 10.1038/ncb1673. [DOI] [PubMed] [Google Scholar]
- Kosodo Y, Röper K, Haubensak W, Marzesco AM, Corbeil D, Huttner WB. Asymmetric distribution of the apical plasma membrane during neurogenic divisions of mammalian neuroepithelial cells. EMBO J. 2004;23:2314–2324. doi: 10.1038/sj.emboj.7600223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovac J, Oster H, Leitges M. Expression of the atypical protein kinase C (aPKC) isoforms iota/lambda and zeta during mouse embryogenesis. Gene Expr Patterns. 2007;7:187–196. doi: 10.1016/j.modgep.2006.07.002. [DOI] [PubMed] [Google Scholar]
- Kraut R, Chia W, Jan LY, Jan YN, Knoblich JA. Role of inscuteable in orienting asymmetric cell divisions in Drosophila. Nature. 1996;383:50–55. doi: 10.1038/383050a0. [DOI] [PubMed] [Google Scholar]
- Kuhlbrodt K, Herbarth B, Sock E, Hermans-Borgmeyer I, Wegner M. Sox10, a novel transcriptional modulator in glial cells. J Neurosci. 1998;18:237–250. doi: 10.1523/JNEUROSCI.18-01-00237.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu QR, Yuk D, Alberta JA, Zhu Z, Pawlitzky I, Chan J, McMahon AP, Stiles CD, Rowitch DH. Sonic hedgehog—regulated oligodendrocyte lineage genes encoding bHLH proteins in the mammalian central nervous system. Neuron. 2000;25:317–329. doi: 10.1016/s0896-6273(00)80897-1. [DOI] [PubMed] [Google Scholar]
- Lyons DA, Guy AT, Clarke JD. Monitoring neural progenitor fate through multiple rounds of division in an intact vertebrate brain. Development. 2003;130:3427–3436. doi: 10.1242/dev.00569. [DOI] [PubMed] [Google Scholar]
- Manabe N, Hirai S, Imai F, Nakanishi H, Takai Y, Ohno S. Association of ASIP/mPAR-3 with adherens junctions of mouse neuroepithelial cells. Dev Dyn. 2002;225:61–69. doi: 10.1002/dvdy.10139. [DOI] [PubMed] [Google Scholar]
- Marusich MF, Furneaux HM, Henion PD, Weston JA. Hu neuronal proteins are expressed in proliferating neurogenic cells. J Neurobiol. 1994;25:143–155. doi: 10.1002/neu.480250206. [DOI] [PubMed] [Google Scholar]
- Masahira N, Takebayashi H, Ono K, Watanabe K, Ding L, Furusho M, Ogawa Y, Nabeshima Y, Alvarez-Buylla A, Shimizu K, Ikenaka K. Olig2-positive progenitors in the embryonic spinal cord give rise not only to motoneurons and oligodendrocytes, but also to a subset of astrocytes and ependymal cells. Dev Biol. 2006;293:358–369. doi: 10.1016/j.ydbio.2006.02.029. [DOI] [PubMed] [Google Scholar]
- Merkle FT, Alvarez-Buylla A. Neural stem cells in mammalian development. Curr Opin Cell Biol. 2006;18:704–709. doi: 10.1016/j.ceb.2006.09.008. [DOI] [PubMed] [Google Scholar]
- Morin X, Jaouen F, Durbec P. Control of planar divisions by the G-protein regulator LGN maintains progenitors in the chick neuroepithelium. Nat Neurosci. 10:1440–1448. doi: 10.1038/nn1984. [DOI] [PubMed] [Google Scholar]
- Noctor SC, Martínez-Cerdeño V, Kriegstein AR. Distinct behaviors of neural stem and progenitor cells underlie cortical neurogenesis. J Comp Neurol. 2008;508:28–44. doi: 10.1002/cne.21669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H-C, Appel B. Delta-Notch signaling regulates oligodendrocyte specification. Development. 2003;130:3747–3755. doi: 10.1242/dev.00576. [DOI] [PubMed] [Google Scholar]
- Park HC, Mehta A, Richardson JS, Appel B. olig2 is required for zebrafish primary motor neuron and oligodendrocyte development. Dev Biol. 2002;248:356–368. doi: 10.1006/dbio.2002.0738. [DOI] [PubMed] [Google Scholar]
- Park HC, Shin J, Appel B. Spatial and temporal regulation of ventral spinal cord precursor specification by Hedgehog signaling. Development. 2004;131:5959–5969. doi: 10.1242/dev.01456. [DOI] [PubMed] [Google Scholar]
- Park HC, Boyce J, Shin J, Appel B. Oligodendrocyte specification in zebrafish requires notch-regulated cyclin-dependent kinase inhibitor function. J Neurosci. 2005;25:6836–6844. doi: 10.1523/JNEUROSCI.0981-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HC, Shin J, Roberts RK, Appel B. An olig2 reporter gene marks oligodendrocyte precursors in the postembryonic spinal cord of zebrafish. Dev Dyn. 2007;236:3402–3407. doi: 10.1002/dvdy.21365. [DOI] [PubMed] [Google Scholar]
- Pauls S, Geldmacher-Voss B, Campos-Ortega JA. A zebrafish histone variant H2A.F/Z and a transgenic H2A.F/Z:GFP fusion protein for in vivo studies of embryonic development. Dev Genes Evol. 2001;211:603–610. doi: 10.1007/s00427-001-0196-x. [DOI] [PubMed] [Google Scholar]
- Petronczki M, Knoblich JA. Dm-PAR-6 directs epithelial polarity and asymmetric cell division of neuroblasts in Drosophila. Nat Cell Biol. 2001;3:43–49. doi: 10.1038/35050550. [DOI] [PubMed] [Google Scholar]
- Plant PJ, Fawcett JP, Lin DC, Holdorf AD, Binns K, Kulkarni S, Pawson T. A polarity complex of mPar-6 and atypical PKC binds, phosphorylates and regulates mammalian Lgl. Nat Cell Biol. 2003;5:301–308. doi: 10.1038/ncb948. [DOI] [PubMed] [Google Scholar]
- Sanada K, Tsai LH. G protein beta-gamma subunits and AGS3 control spindle orientation and asymmetric cell fate of cerebral cortical progenitors. Cell. 2005;122:119–131. doi: 10.1016/j.cell.2005.05.009. [DOI] [PubMed] [Google Scholar]
- Schaefer M, Shevchenko A, Shevchenko A, Knoblich JA. A protein complex containing Inscuteable and the Galpha-binding protein Pins orients asymmetric cell divisions in Drosophila. Curr Biol. 2000;10:353–362. doi: 10.1016/s0960-9822(00)00401-2. [DOI] [PubMed] [Google Scholar]
- Schober M, Schaefer M, Knoblich JA. Bazooka recruits Inscuteable to orient asymmetric cell divisions in Drosophila neuroblasts. Nature. 1999;402:548–551. doi: 10.1038/990135. [DOI] [PubMed] [Google Scholar]
- Shin J, Park HC, Topczewska JM, Mawdsley DJ, Appel B. Neural cell fate analysis in zebrafish using olig2 BAC transgenics. Methods Cell Sci. 2003;25:7–14. doi: 10.1023/B:MICS.0000006847.09037.3a. [DOI] [PubMed] [Google Scholar]
- Song X, Zhu CH, Doan C, Xie T. Germline stem cells anchored by adherens junctions in the Drosophila ovary niches. Science. 2002;296:1855–1857. doi: 10.1126/science.1069871. [DOI] [PubMed] [Google Scholar]
- Sprague J, Doerry E, Douglas S, Westerfield M. The Zebrafish Information Network (ZFIN): a resource for genetic, genomic and developmental research. Nucleic Acids Res. 2001;29:87–90. doi: 10.1093/nar/29.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stainier DY, Fouquet B, Chen JN, Warren KS, Weinstein BM, Meiler SE, Mohideen MA, Neuhauss SC, Solnica-Krezel L, Schier AF, Zwartkruis F, Stemple DL, Malicki J, Driever W, Fishman MC. Mutations affecting the formation and function of the cardiovascular system in the zebrafish embryo. Development. 1996;123:285–292. doi: 10.1242/dev.123.1.285. [DOI] [PubMed] [Google Scholar]
- Takebayashi H, Yoshida S, Sugimori M, Kosako H, Kominami R, Nakafuku M, Nabeshima Y. Dynamic expression of basic helix-loop-helix Olig family members: implication of Olig2 in neuron and oligodendrocyte differentiation and identification of a new member, Olig3. Mech Dev. 2000;99:143–148. doi: 10.1016/s0925-4773(00)00466-4. [DOI] [PubMed] [Google Scholar]
- Trevarrow B, Marks DL, Kimmel CB. Organization of hindbrain segments in the zebrafish embryo. Neuron. 1990;4:669–679. doi: 10.1016/0896-6273(90)90194-k. [DOI] [PubMed] [Google Scholar]
- von Trotha JW, Campos-Ortega JA, Reugels AM. Apical localization of ASIP/PAR-3:EGFP in zebrafish neuroepithelial cells involves the oligomerization domain CR1, the PDZ domains, and the C-terminal portion of the protein. Dev Dyn. 2006;235:967–977. doi: 10.1002/dvdy.20715. [DOI] [PubMed] [Google Scholar]
- Wodarz A, Huttner WB. Asymmetric cell division during neurogenesis in Drosophila and vertebrates. Mech Dev. 2003;120:1297–1309. doi: 10.1016/j.mod.2003.06.003. [DOI] [PubMed] [Google Scholar]
- Wodarz A, Ramrath A, Kuchinke U, Knust E. Bazooka provides an apical cue for Inscuteable localization in Drosophila neuroblasts. Nature. 1999;402:544–547. doi: 10.1038/990128. [DOI] [PubMed] [Google Scholar]
- Wodarz A, Ramrath A, Grimm A, Knust E. Drosophila atypical protein kinase C associates with Bazooka and controls polarity of epithelia and neuroblasts. J Cell Biol. 2000;150:1361–1374. doi: 10.1083/jcb.150.6.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu PS, Egger B, Brand AH. Asymmetric stem cell division: lessons from Drosophila. Semin Cell Dev Biol. 2008;19:283–293. doi: 10.1016/j.semcdb.2008.01.007. [DOI] [PubMed] [Google Scholar]
- Yamanaka T, Horikoshi Y, Sugiyama Y, Ishiyama C, Suzuki A, Hirose T, Iwamatsu A, Shinohara A, Ohno S. Mammalian Lgl forms a protein complex with PAR-6 and aPKC independently of PAR-3 to regulate epithelial cell polarity. Curr Biol. 2003;13:734–743. doi: 10.1016/s0960-9822(03)00244-6. [DOI] [PubMed] [Google Scholar]
- Yu F, Morin X, Cai Y, Yang X, Chia W. Analysis of partner of inscuteable, a novel player of Drosophila asymmetric divisions, reveals two distinct steps in inscuteable apical localization. Cell. 2000;100:399–409. doi: 10.1016/s0092-8674(00)80676-5. [DOI] [PubMed] [Google Scholar]
- Yu F, Kuo CT, Jan YN. Drosophila neuroblast asymmetric cell division: recent advances and implications for stem cell biology. Neuron. 2006;51:13–20. doi: 10.1016/j.neuron.2006.06.016. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Wang S, Anderson DJ. Identification of a novel family of oligodendrocyte lineage-specific basic helix-loop-helix transcription factors. Neuron. 2000;25:331–343. doi: 10.1016/s0896-6273(00)80898-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.