Abstract
The phenylalanine transfer RNA of baker's yeast (tRNAPhe) contains a base Y of unknown molecular structure next to the anticodon triplet. Since the base Y fluoresces at room temperature (λmax = 431 nm), its emission properties offer a unique tool for studying conformational and binding properties of tRNAPhe. The results obtained by these experiments include the following: (1) The quantum yield of fluorescence of Y in tRNAPhe (φF) is 0.07 ± 0.01 at high Mg2+ concentrations (>10-2 M) and about half that at 10-3 M or less, indicating a [Mg2+]-dependent conformational change of the anticodon loop. (2) The fluorescence of Y isolated from tRNAPhe (Y+) is red-shifted by 15 nm compared to Y in tRNAPhe which suggests a stacked (more hydrophobic) environment for Y in the intact anticodon loop. φF of Y+ is 0.035. (3) The solvent isotope effect φF(D2O)/φF(H2O) is 1.5 for tRNAPhe and 1.9 for Y+ i.e., Y in tRNA is still hydrated. (4) The temperature dependence of φF in a polar glass shows that quenching occurs only at temperatures at which the glass has sufficiently low viscosity to permit solvent shell relaxation in the excited state. The low-temperature (80°K) fluorescence is blue shifted (λmax = 409 nm) and the phosphorescence has a decay time of 1.5 seconds, a threshold at 392 nm and a spectral shape like that of guanine. (5) In the presence of 10-2 M Mg2+ penta-uridylate, which contains the codon triplet, a small blue shift and a decrease in φF are observed. This shift can be used to establish the formation of a binary complex between the codon and the anticodon with an association constant of 4 × 102 M-1, approximately. A similar complex is formed with poly-uridylate but not with poly-cytidylate. In the absence of Mg2+ the binary complex is not formed.
Full text
PDF

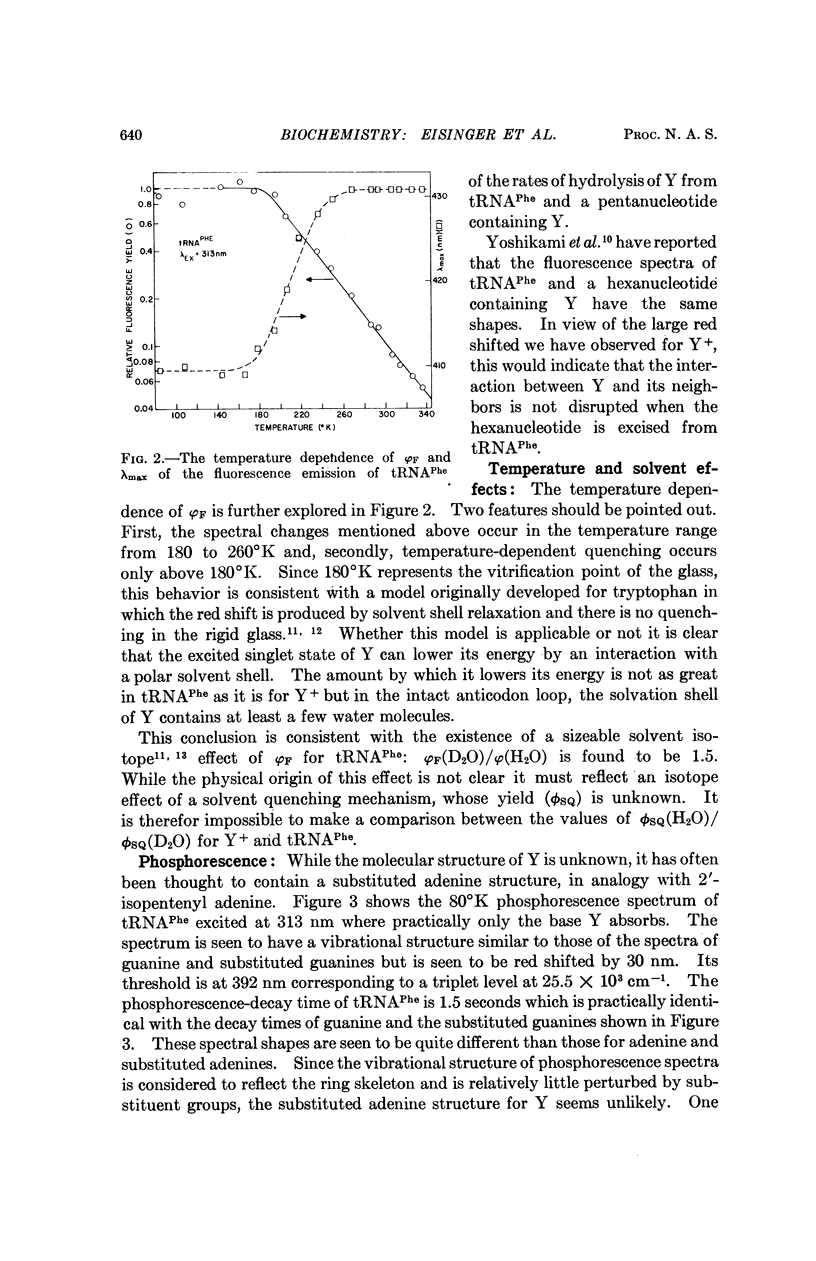
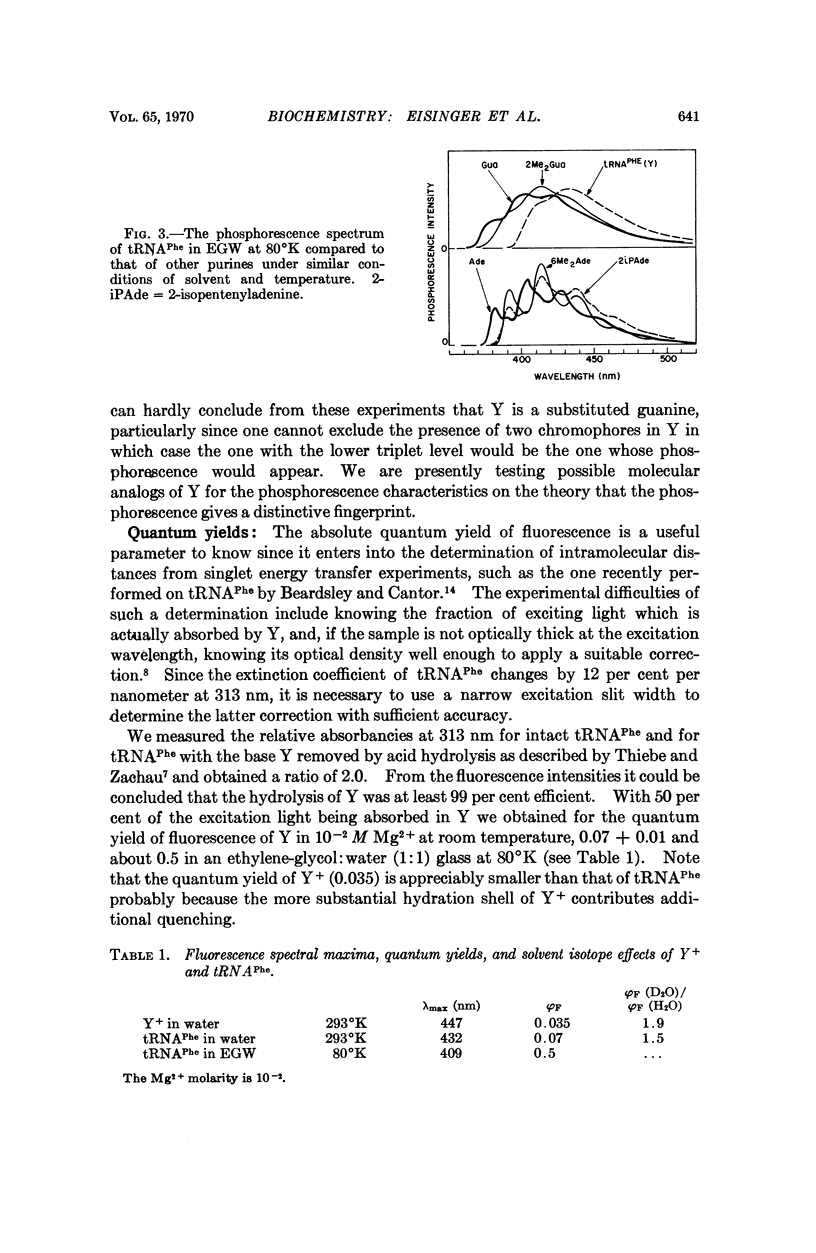
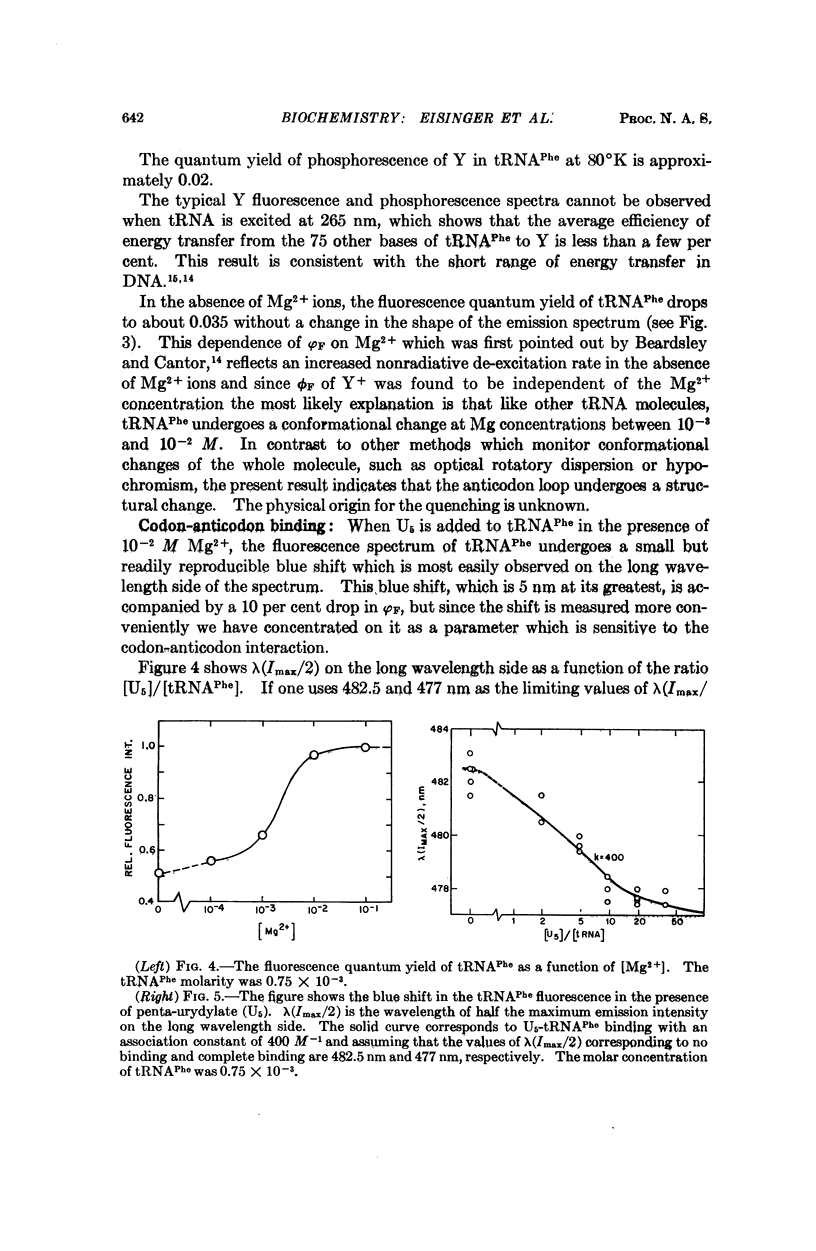


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beardsley K., Cantor C. R. Studies of transfer RNA tertiary structure by singlet-singlet energy transfer. Proc Natl Acad Sci U S A. 1970 Jan;65(1):39–46. doi: 10.1073/pnas.65.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudock B. S., Katz G., Taylor E. K., Holley R. W. Primary structure of wheat germ phenylalanine transfer RNA. Proc Natl Acad Sci U S A. 1969 Mar;62(3):941–945. doi: 10.1073/pnas.62.3.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisinger J. A variable temperature, U.V. luminescence spectrograph for small samples. Photochem Photobiol. 1969 Mar;9(3):247–258. doi: 10.1111/j.1751-1097.1969.tb07289.x. [DOI] [PubMed] [Google Scholar]
- Eisinger J., Navon G. Fluorescence quenching and isotope effect of tryptophan. J Chem Phys. 1969 Mar 1;50(5):2069–2077. doi: 10.1063/1.1671335. [DOI] [PubMed] [Google Scholar]
- Fuller W., Hodgson A. Conformation of the anticodon loop intRNA. Nature. 1967 Aug 19;215(5103):817–821. doi: 10.1038/215817a0. [DOI] [PubMed] [Google Scholar]
- Gillam I., Millward S., Blew D., von Tigerstrom M., Wimmer E., Tener G. M. The separation of soluble ribonucleic acids on benzoylated diethylaminoethylcellulose. Biochemistry. 1967 Oct;6(10):3043–3056. doi: 10.1021/bi00862a011. [DOI] [PubMed] [Google Scholar]
- Guéron M., Eisinger J., Shulman R. G. Excited states of nucleotides and singlet energy transfer in polynucleotides. J Chem Phys. 1967 Nov 15;47(10):4077–4091. doi: 10.1063/1.1701580. [DOI] [PubMed] [Google Scholar]
- Guéron M., Shulman R. G. Energy transfer in polynucleotides. Annu Rev Biochem. 1968;37:571–596. doi: 10.1146/annurev.bi.37.070168.003035. [DOI] [PubMed] [Google Scholar]
- Rajbhandary U. L., Chang S. H., Stuart A., Faulkner R. D., Hoskinson R. M., Khorana H. G. Studies on polynucleotides, lxviii the primary structure of yeast phenylalanine transfer RNA. Proc Natl Acad Sci U S A. 1967 Mar;57(3):751–758. doi: 10.1073/pnas.57.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiebe R., Zachau H. G. A specific modification next to the anticodon of phenylalanine transfer ribonucleic acid. Eur J Biochem. 1968 Sep 24;5(4):546–555. doi: 10.1111/j.1432-1033.1968.tb00404.x. [DOI] [PubMed] [Google Scholar]
- Wimmer E., Maxwell I. H., Tener G. M. A simple method for isolating highly purified yeast phenylalanine transfer ribonucleic acid. Biochemistry. 1968 Jul;7(7):2623–2628. doi: 10.1021/bi00847a026. [DOI] [PubMed] [Google Scholar]
- Yoshikami D., Katz G., Keller E. B., Dudock B. S. A fluorescence assay for phenylalanine transfer RNA. Biochim Biophys Acta. 1968 Oct 29;166(3):714–717. doi: 10.1016/0005-2787(68)90382-1. [DOI] [PubMed] [Google Scholar]



