Abstract
Cold temperature, dietary restriction, reduced insulin/insulin-like growth factor signaling, and mutations in mitochondrial genes have all been shown to extend the lifespan of Caenorhabditis elegans (Kenyon et al., Nature 366:461–464, 1993; Klass, Mech Ageing Dev 6:413–429, 1977; Lakowski and Hekimi, Science 272:1010–1013, 1996). Additionally, all of them extend the lifespan of mice (Bluher et al., Science 299:572–574, 2003; Conti et al., Science 314:825–828, 2006; Holzenberger et al., Nature 421:182–187, 2003; Liu et al., Genes Dev 19:2424–2434, 2005; Weindruch and Walford, Science 215:1415–1418, 1982). The mechanism by which these treatments extend lifespan is currently unknown, but our study uses an epistatic approach to show that these four manipulations are mainly additive in terms of lifespan. Classical interpretation of this data suggests that these manipulations are independent of each other. However, using a Gompertz mortality rate analysis, the maximum mortality rate doubling time can be achieved through the use of only dietary restriction and cold temperature, suggesting that the mechanisms by which cold temperature and caloric restriction extend lifespan are the only independent mechanisms.
Keywords: Caenorhabditis elegans, Aging, Lifespan, Gompertz analysis
Introduction
Although the number of manipulations that increase the lifespan of Caenorhabditis elegans has continued to grow, the exact mechanism by which these manipulations extend lifespan is still unknown. Of further interest, the exact cause of senescence, in this article defined as the increase in mortality rate seen with increasing age, is also unknown. If a single cause of senescence, such as free radicals generated by mitochondria, exists, then all these life-extending manipulations may have one mechanism in common. To determine if there is a single pathway by which senescence is retarded, we have examined four separate manipulations that have been shown to increase lifespan in C. elegans: dietary restriction (DR), cold-/hypothermic-induced longevity (CHIL), decreased insulin signaling, and mutations influencing mitochondrial respiration (Ball et al. 1947; Conti et al. 2006; Fernandes et al. 1976; Houthoofd et al. 2002; Liu et al. 2005; Mair et al. 2003).
Dietary restriction has been shown to extend lifespan by up to 50% in mammals (Davis et al. 1983; Yu et al. 1985; Weindruch and Walford 1982). In several different worm models of dietary restriction, evidence has revealed that DAF-16, a forkhead box O (FOXO) transcription factor that is required for life-extension via the insulin pathway, is not required for the life-extending effects of dietary restriction (Houthoofd et al. 2003; Lakowski and Hekimi 1998). This observation suggests, contrary to the overwhelmingly compelling hypothesis, that the insulin-like pathway does not mediate the effects of dietary restriction. Using epistatic analysis, studies in both C. elegans (Houthoofd et al. 2003) and mice (Bartke et al. 2001) indicated that mutations in the insulin-like pathway and dietary restriction produced completely additive effects on maximum lifespan. However, these studies did not account for the fact that caloric availability produces a U-shaped function, the optimum of which could be influenced by the presence of insulin-like signaling. Addressing this question in more detail, Partridge and coworkers (Clancy et al. 2002) used epistatic analysis combined with a dose–response curve of caloric availability and found that caloric restriction and the insulin-like pathway were, in fact, not additive at optimum levels. They concluded that caloric restriction actually does increase maximum lifespan by reducing insulin-like signaling. Thus, at present, there is no consensus regarding this key question.
CHIL has been used to extend lifespan for many decades (Klass 1977; Liu and Walford 1970). While CHIL is often dismissed as a natural result of the Arrhenius equation, relating temperature to reaction rate, several sources of data point to the contrary. Although metabolic rate tends to scale with temperature in exothermic animals (Tribe and Bowler 1968), metabolic rate clearly does not determine lifespan (Yen et al. 2004). Work in flies, worms, and mice has shown that metabolism and longevity are not always correlated (Arking et al. 1988; Dillin et al. 2002; Holloszy and Smith 1986; Houthoofd et al. 2002; Hulbert et al. 2004). The effects of cold temperature on an organism are more complicated than usually appreciated. In mussels and worms, cold temperature has been associated with an increase in total protein and neurosecretory activity (Rao 1962). Zebrafish, in response to cold temperature, increase specific antioxidant enzymes such as superoxide dismutase (SOD)-1 and SOD-2 (Malek et al. 2004), and carp also show a specific gene expression profile in response to cold temperature that is reminiscent of those changes seen in calorically restricted mice (Gracey et al. 2004; Han and Hickey 2005; Lee et al. 1999, 2000). C. elegans also displays a specific physiological change in response to cold temperatures (Madi et al. 2003; Paul et al. 2000), and it has even been noted that cold temperature is a type of mild stress, which suggests a possible hormetic effect. Wong et al. have shown that clk-1 mutants are insensitive to temperature changes during embryogenesis, which demonstrates that developmental adjustments to changing temperatures is an active process requiring genes such as clk-1 (Wong et al. 1995).
The first single gene mutation that increased lifespan in C. elegans was named age-1 (Klass 1983). This mutation was later found to have a defective form of PI-3-kinase, a protein found in the insulin-signaling pathway (Morris et al. 1996). The daf-2 mutation also increases maximum lifespan, and it was subsequently discovered that this gene codes for an insulin/insulin-like growth factor (IGF) receptor (Kenyon et al. 1993; Kimura et al. 1997). The primary transcription factor that is responsible for the activation of the antisenescence program was discovered to be DAF-16, a member of the forkhead family of transcription factors. Deletion of daf-16 prevents the daf-2 or age-1 mutations from conferring their usual antisenescence phenotype (Kenyon et al. 1993; Tissenbaum and Ruvkun 1998). Defective insulin/IGF signaling is also associated with an increased lifespan in several other species. In Drosophila, chico flies that have reduced insulin signaling and flies that have had their insulin-like peptide-producing median neurosecretory cells ablated experience an increase in lifespan (Broughton et al. 2005; Clancy et al. 2001). Overexpression of dFOXO, a homolog of DAF-16, has also been shown to increase lifespan in flies (Giannakou et al. 2004; Hwangbo et al. 2004). Fat-specific insulin receptor knockout (KO) mice have an increased lifespan (Bluher et al. 2003), as do both growth hormone receptor-deficient and IGF receptor KO mice (Coschigano et al. 2003; Holzenberger et al. 2003).
Another category of life extending manipulation is disruption of the mitochondrial proteins involved in the electron transport chain (Feng et al. 2001). Excluding complex II, RNAi of any complex of the electron transport chain increases the lifespan of worms (Curran and Ruvkun 2007; Dillin et al. 2002). The complete mechanism by which mitochondrial mutations extend lifespan has not been elucidated, but evidence has suggested that it does not require daf-16 and is partially dependent on aak-2 (Dillin et al. 2002). A specific mutant, clk-1, has been shown to extend lifespan (Wong et al. 1995), possibly through its preferential usage of complex II instead of complex I (Kayser et al. 2004). This mutant will be used in our subsequent studies. The mechanism by which clk-1 extends lifespan is controversial. Studies have both shown that clk-1 does and does not extend lifespan when the worms undergo dietary restriction (Braeckman et al. 2000; Lakowski and Hekimi 1998). This difference may be explained by the difference in application of caloric restriction. Lakoswki and Hekimi utilized the eat-2 mutant while Braeckman et al. used axenic media. What has been established is that clk-1 has an additive effect when crossed with daf-2 worms (Lakowski and Hekimi 1996). Mitochondrial mutations in mice have also been shown to extend lifespan. One mammalian study on clk-1 has shown a 15% increase in lifespan (Liu et al. 2005). In addition, mice lacking p66shc live 30% longer than wild type (Migliaccio et al. 1999). Recent work has shown that p66shc is associated with mitochondrial membranes and has been shown to regulate mitochondrial transmembrane potential (Orsini et al. 2004).
In the field of aging research, epistatic analysis using survival curves and average lifespan has become a standard method of determining the independence of genes and pathways (Bartke et al. 2001; Bonkowski et al. 2006; Braeckman et al. 2000; Clancy et al. 2002; Kenyon et al. 1993; Lakowski and Hekimi 1998; Tissenbaum and Ruvkun 1998). As such, this study uses a similar epistatic design. In addition to analyzing survival curves and average lifespan, these studies will also analyze the mortality rate as modeled by the Gompertz equation.
The Gompertz equation is an empirically derived formula that relates the arithmetic progression of time to the geometric progression of mortality rate and is written as:
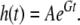 |
A is the initial mortality rate, and G is the Gompertz variable, which quantifies the age-dependent acceleration of mortality rate. G is inversely related to the mortality rate doubling time (MRDT) where MRDT = ln2/G. Human studies have shown that the MRDT is stable even under conditions where extrinsic mortality rates are highly variable (Finch 1990; Finch et al. 1990). In Drosophila, Spencer and Promislow compared seven different wild-caught strains and found that although they all had different average lifespans, their Gompertz variable was not significantly different (Spencer and Promislow 2005).
Material and methods
Worm strains used
The following strains were used in the experiments:
| Description | Mutation | Strain (allele) |
| Wild type | None | N2 |
| clk-1 mutant | clk-1 | CB4876(e2519) |
| clk-1 mutant | clk-1 | MQ130 (qm30) |
| Insulin/IGF receptor mutation | daf-2 | CB1370(e1370) |
| Double mutant | clk-1;daf-2 | mq513 (e1370; e2519) |
Data from both strains of clk-1 mutants were pooled together, as there was no statistical difference between the lifespans and survival curves and did not make a difference in the analysis.
Lifespan assays
Lifespan studies were performed as in Adachi and Ishii (2000). Eggs were collected from gravid nematodes by standard hypochlorite treatment and grown on standard nematode growth media (NGM) plates at 16°C to prevent dauer formation in daf-2 worms until L4-adult stage. The worms were then transferred to media that is supplemented with 5-fluorodeoxyuridine to inhibit growth of progeny and transferred to fresh media every week or month for monoxenic or axenic cultures, respectively. Worms were scored at least every 3 days to check for dead worms. Worms that were not moving or did not respond to gentle prodding from a platinum wire were scored as dead. Any worms that died of internal hatching or crawled off the plate were censored on the date that they were last observed. All worms were maintained at 25°C for the control temperature or 16°C for the cold temperature.
Animals
C. elegans were maintained on NGM plates seeded with OP50 strain of Escherichia coli or axenic media supplemented with cholesterol. OP50 is a uracil auxotrophic strain that has limited growth ability to facilitate the counting of the worms on the plate. Axenic medium is a liquid medium devoid of any bacteria and is made of 3% w/v soy peptone, 3% w/v yeast extract, 0.5 mg/ml hemoglobin, and 5 μg/ml cholesterol. This medium was supplemented with 50 µg/ml ampicillin and 25 µg/ml tetracycline to prevent bacterial growth.
Gompertz analysis
Maximum likelihood estimates (MLE) for G and A variables were obtained with WinModest and reconfirmed with a separate script written for the freely available R statistical programming environment (Yen et al. 2008). Hypothesis testing for the MLE values to test for significant changes in Gompertz variables utilized a Student’s t test on the values obtained by at least three independent replications (average number of animals per cohort = 101). MLE are asymptotically (meaning, as the number of samples tend to infinity) unbiased, efficient, and normally distributed (Eliason 1993).
Results and discussion
Average lifespan analysis
As can be seen in Tables 1 and 2 and Fig. 1, cold temperature significantly increased average lifespan in all but one group. Ad lib fed, daf-2 worms did not experience a significant increase with cold temperature, but there was a trend (p = 0.067). Dietary restriction significantly increased the average lifespan for all but three groups. All three groups had the daf-2 mutation (daf-2 or clk-1;daf-2 worms). Two were at 16°C while the third daf-2 at 25°C showed a slight trend toward significant (p = 0.09). In our hands, the clk-1 mutation did not significantly increase lifespan for most conditions. Two notable conditions where addition of the clk-1 mutation significantly increased lifespan were both at 25°C and under dietary restriction. When compared to wild-type worms at 25°C under dietary restriction, the addition of the clk-1 mutation significantly increased the average lifespan by 29.6%. The double mutant clk-1;daf-2 worms maintained at 25°C under dietary restriction also had a significant increase of 20.0% in average lifespan compared to daf-2 worms at 25°C under dietary restriction. The addition of the insulin/IGF signaling mutation, daf-2, significantly increased the average lifespan in all but three conditions. Decreasing insulin signaling did not significantly increase the lifespan of any group at 16°C under dietary restriction whether it was wild-type worms or clk-1. The third group that did not have a significant change with decreased insulin signaling was wild-type, ad lib fed worms at 16°C, although there was a trend (p = 0.06).
Table 1.
Average lifespan of each treatment
| Temperature (°C) | Diet | Mutation | Average lifespan (days) ± SE | Number of independent replications |
|---|---|---|---|---|
| 25 | Ad lib | None | 15.9 ± 1.7 | 3 |
| 16 | Ad lib | None | 32.6 ± 0.7 | 3 |
| 25 | DR | None | 24.6 ± 0.7 | 3 |
| 16 | DR | None | 45.8 ± 2.1 | 3 |
| 25 | Ad lib | clk-1 | 17.1 ± 0.2 | 4 |
| 16 | Ad lib | clk-1 | 34.6 ± 2.4 | 4 |
| 25 | DR | clk-1 | 31.9 ± 0.5 | 4 |
| 16 | DR | clk-1 | 66.5 ± 10.1 | 4 |
| 25 | Ad lib | daf-2 | 32.1 ± 4.5 | 4 |
| 16 | Ad lib | daf-2 | 45.3 ± 5.1 | 4 |
| 25 | DR | daf-2 | 42.2 ± 1.9 | 3 |
| 16 | DR | daf-2 | 54.4 ± 5.0 | 3 |
| 25 | Ad lib | clk-1;daf-2 | 35.0 ± 3.3 | 3 |
| 16 | Ad lib | clk-1;daf-2 | 52.3 ± 6.1 | 3 |
| 25 | DR | clk-1;daf-2 | 50.7 ± 3.3 | 3 |
| 16 | DR | clk-1;daf-2 | 63.5 ± 4.6 | 3 |
A table indicating significant comparisons can be found in Table 2
DR dietary restriction, SE standard error
Table 2.
Comparison of average lifespan
| Compared to | Days change | Percent change |
|---|---|---|
| Does cold temperature change average lifespan? | ||
| 25°C, WT, ad liba | 16.70 | 104.71 |
| 25°C, WT, DRa | 21.19 | 86.19 |
| 25°C, clk-1, ad liba | 17.49 | 102.29 |
| 25°C, clk-1, DRa | 34.66 | 108.83 |
| 25°C, daf-2, ad lib | 13.27 | 41.39 |
| 25°C, daf-2, DRa | 12.13 | 28.71 |
| 25°C, clk-1;daf-2, ad liba | 17.28 | 49.33 |
| 25°C, clk-1;daf-2, DRa | 12.75 | 25.15 |
| Does daf-2 change average lifespan? | ||
| 25°C, WT, ad liba | 16.12 | 101.11 |
| 16°C, WT, ad lib | 12.70 | 38.90 |
| 25°C, WT, DRa | 17.66 | 71.86 |
| 16°C, WT, DR | 8.61 | 18.81 |
| 25°C, clk-1, ad liba | 17.94 | 104.88 |
| 16°C, clk-1, ad liba | 17.73 | 51.24 |
| 25°C, clk-1, DRa | 18.86 | 59.20 |
| 16°C, clk-1, DR | −3.06 | −4.60 |
| Does clk-1 change average lifespan? | ||
| 25°C, WT, ad lib | 1.16 | 7.25 |
| 16°C, WT, ad lib | 1.95 | 5.98 |
| 25°C, WT, DRa | 7.27 | 29.58 |
| 16°C, WT, DR | 20.75 | 45.34 |
| 25°C, daf-2, ad lib | 2.97 | 9.26 |
| 16°C, daf-2, ad lib | 6.98 | 15.40 |
| 25°C, daf-2, DRa | 8.46 | 20.04 |
| 16°C, daf-2, DR | 9.09 | 16.71 |
| Does DR change average lifespan? | ||
| 25°C, WT, ad liba | 8.64 | 54.16 |
| 16°C, WT, ad liba | 13.13 | 40.21 |
| 25°C, daf-2, ad lib | 10.18 | 31.74 |
| 16°C, daf-2, ad lib | 9.04 | 19.93 |
| 25°C, clk-1, ad liba | 14.75 | 86.26 |
| 16°C, clk-1, ad liba | 31.92 | 92.28 |
| 25°C, clk-1;daf-2, ad liba | 15.67 | 44.73 |
| 16°C, clk-1;daf-2, ad lib | 11.14 | 21.29 |
WT wild type, DR dietary restriction
aSignificant difference
Fig. 1.
Kaplan–Meier graphs of one experimental set: a wild-type (WT) worms, bclk-1 worms, cdaf-2 worms, and dclk-1;daf-2 worms
Gompertz analysis
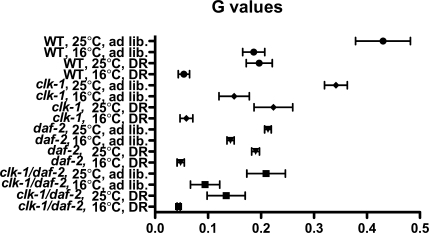
As seen in Tables 3 and 4 and Fig. 2, cold temperature significantly decreased the Gompertz variable for all but two groups. Both clk-1;daf-2 groups did not show a significant decrease in G, but both had a trend toward significance (p < 0.07). Dietary restriction also significantly decreased G in all but three groups. daf-2 worms at 25°C and clk-1;daf-2 worms at 25°C or 16°C did not have a significant decrease in G. The clk-1 mutation did not show a significant decrease in G for any condition. Decreased insulin signaling only decreased G for ad lib fed, wild-type, and clk-1 worms maintained at 25°C. As seen in Fig. 3, the minimal Gompertz value achieved occurred with DR and cold temperature. Further addition of various treatments, although extending average lifespan, had no significant effect on G.
Table 3.
Estimated parameters of the Gompertz mortality rate equation
| Temperature (°C) | Diet | Mutation | G ± SE | ln(A) ± ln(SE) | MRDT (days) |
|---|---|---|---|---|---|
| 25 | Ad lib | None | 0.43 ± 0.06 | −7.67 ± −8.16 | 1.61 |
| 16 | Ad lib | None | 0.19 ± 0.03 | −8.05 ± −8.66 | 3.72 |
| 25 | DR | None | 0.20 ± 0.03 | −6.80 ± −7.76 | 3.52 |
| 16 | DR | None | 0.05 ± 0.01 | −5.61 ± −6.28 | 12.74 |
| 25 | Ad lib | clk-1 | 0.34 ± 0.02 | −7.32 ± −8.45 | 2.03 |
| 16 | Ad lib | clk-1 | 0.15 ± 0.03 | −6.95 ± −7.73 | 4.64 |
| 25 | DR | clk-1 | 0.22 ± 0.04 | −7.44 ± −7.36 | 3.10 |
| 16 | DR | clk-1 | 0.06 ± 0.01 | −6.37 ± −6.84 | 11.74 |
| 25 | Ad lib | daf-2 | 0.20 ± 0.02 | −7.85 ± −8.57 | 3.45 |
| 16 | Ad lib | daf-2 | 0.14 ± 0.01 | −8.01 ± −8.43 | 5.06 |
| 25 | DR | daf-2 | 0.19 ± 0.01 | −10.20 ± −12.27 | 3.65 |
| 16 | DR | daf-2 | 0.05 ± 0.01 | −5.98 ± −7.46 | 14.34 |
| 25 | Ad lib | clk-1;daf-2 | 0.21 ± 0.04 | −7.75 ± −7.78 | 3.31 |
| 16 | Ad lib | clk-1;daf-2 | 0.09 ± 0.03 | −6.72 ± −7.21 | 7.35 |
| 25 | DR | clk-1;daf-2 | 0.13 ± 0.04 | −7.84 ± −8.01 | 5.16 |
| 16 | DR | clk-1;daf-2 | 0.04 ± 0.00 | −6.30 ± −8.22 | 15.59 |
MRDT is equal to ln(2)/G. A table indicating significant comparisons can be found in Table 4
G quantifies the age-dependent acceleration of mortality rate, A initial mortality rate variable, MRDT mortality rate doubling time, SE standard error
Table 4.
Comparison of age-dependent mortality variable (G)
| Compared to | Absolute difference | Percent change |
|---|---|---|
| Does cold temperature change G? | ||
| 25°C, WT, ad liba | 0.24 | −56.73 |
| 25°C, WT, DRa | 0.14 | −72.34 |
| 25°C, clk-1, ad liba | 0.19 | −56.28 |
| 25°C, clk-1, DRa | 0.16 | −73.57 |
| 25°C, daf-2, ad liba | 0.06 | −31.80 |
| 25°C, daf-2, DRa | 0.14 | −74.53 |
| 25°C, clk-1;daf-2, ad lib | 0.12 | −55.00 |
| 25°C, clk-1;daf-2, DR | 0.09 | −66.89 |
| Does daf-2 change G? | ||
| 25°C, WT, ad liba | 0.23 | −53.35 |
| 16°C, WT, ad lib | 0.05 | −26.48 |
| 25°C, WT, DR | 0.01 | −3.53 |
| 16°C, WT, DR | 0.01 | −11.16 |
| 25°C, clk-1, ad liba | 0.13 | −38.59 |
| 16°C, clk-1, ad lib | 0.05 | −36.79 |
| 25°C, clk-1, DR | 0.09 | −39.88 |
| 16°C, clk-1, DR | 0.01 | −24.69 |
| Does clk-1 change G? | ||
| 25°C, WT, ad lib | 0.09 | −20.66 |
| 16°C, WT, ad lib | 0.04 | −19.84 |
| 25°C, WT, DR | −0.03 | 13.56 |
| 16°C, WT, DR | 0.00 | 8.52 |
| 25°C, daf-2, ad lib | −0.01 | 4.45 |
| 16°C, daf-2, ad lib | 0.04 | −31.09 |
| 25°C, daf-2, DR | 0.06 | −29.23 |
| 16°C, daf-2, DR | 0.00 | −8.02 |
| Does DR change G? | ||
| 25°C, WT, ad liba | 0.23 | −54.30 |
| 16°C, WT, ad liba | 0.13 | −70.79 |
| 25°C, daf-2, ad lib | 0.01 | −5.49 |
| 16°C, daf-2, ad liba | 0.09 | −64.70 |
| 25°C, clk-1, ad liba | 0.12 | −34.59 |
| 16°C, clk-1, ad liba | 0.09 | −60.45 |
| 25°C, clk-1;daf-2, ad lib | 0.08 | −35.96 |
| 16°C, clk-1;daf-2, ad lib | 0.05 | −52.88 |
WT wild type, DR dietary restriction
aSignificant difference
Fig. 2.
Linearized mortality rate curves: a wild-type (WT) worms, bclk-1 worms, cdaf-2 worms, and dclk-1;daf-2 worms
Fig. 3.
Gompertz variable graph with standard error bars. The minimum Gompertz variable, which inversely relates to mortality rate doubling time, is achieved with dietary restriction and cold temperature. WT wild-type worms, DR dietary restriction
Few changes in the initial mortality rate (A) were detected. The lack of a significant finding is not surprising, as it has been shown that A is highly variable (Promislow et al. 1999). CHIL increased A for both daf-2 and clk1/daf-2 worms on DR, and dietary restricted daf-2 worms had a decreased A compared to dietary restricted N2 worms. Lastly, a significant increase in A was observed in ad lib daf-2 worms at 16°C that were subjected to DR.
Epistatic analysis can be performed using several different methods (Gems et al. 2002). When specifically considering average lifespan analysis, it is unknown how independent mechanisms will affect lifespan. Does one add the absolute number of days gained for each single treatment (i.e., each treatment increases lifespan + days), add the percent increase for each treatment (i.e., each treatment increases lifespan + percent of wild type), or multiply the percent increase for each treatment (i.e., each treatment increases lifespan × percent on top of the previous treatment)? Empirically, it seems that multiplying the percent increase allows for the closest approximation to the actual data in this study.
Based on the epistatic analysis of average lifespan, several interactions between treatments are observed. Temperature has an antagonistic effect on insulin/IGF signaling. At 16°C, the effect of the decreased insulin/IGF signaling on lifespan is decreased by 50%. This interaction between temperature and the daf pathway has been noted previously by Golden and Riddle (1984). Dietary restrictions by axenic media and insulin/IGF signaling also seem to share overlapping mechanisms. Under control conditions, DR or decreased insulin/IGF signaling increases lifespan 50% and 100%, respectively. When combined, the effects of either DR or the daf-2 mutation are ameliorated and the average lifespan of the combination are less than would be predicted. The effects of the clk-1 mutation on lifespan were less predictable, and the lack of replication of clk-1 effects on lifespan under standard conditions may be due to the fact that our control worms lived an average of 15.9 days compared to those of Lakowski and Hekimi that lived an average of 9.2 days (Lakowski and Hekimi 1996). While no effect on lifespan was observed under control conditions, there was a significant effect of clk-1 under DR conditions at 25°C either on wild-type background or combined with the daf-2 mutation, indicating that there is an interaction between the two pathways.
Analysis of the Gompertz variable does not corroborate the interaction between temperature and the daf-2 mutation, as the change in Gompertz variable with cold treatment was indistinguishable between daf-2 mutants and other groups. This suggests that the interaction between daf-2 and temperature seen with average lifespan analysis may be due to changes in initial mortality rate. Although there is no interaction between daf-2 and temperature on the Gompertz variable, we were able to corroborate the interaction between DR and the daf-2 mutation. In addition, there was a surprising and significant interaction between DR and temperature. The addition of cold temperature to worms under DR causes a significant decrease in G (72% decrease) when compared to ad lib fed worms that are placed in cold temperatures (50% decrease). This synergistic effect may indicate an interaction between the two treatments. The clk-1 mutation did not have any significant effect on G, and this suggests that clk-1 does not affect senescence, which may also apply to other mitochondrial mutations.
As with any epistatic analysis, there are caveats to the interpretation of the data (Gems et al. 2002). In particular, nonnull mutations can give the false impression that pathways are independent when they are not. In this study, the clk-1 mutations tested are functional null mutations (e2519 is a missense mutation and qm30 is a deletion mutation) that both completely suppress the synthesis of ubiquinone (UQ9; Ewbank et al. 1997; Miyadera et al. 2001). While the daf-2 mutation used is not a null mutation (null mutations prevent the worms from developing into an adult), the worms are phenotypically similar to null mutations (i.e., they form 100% dauers) at our control temperature of 25°C. This, nonetheless, may explain why we find an additive effect on lifespan of insulin/IGF signaling in combination with other treatments. In fact null mutants of PI3K, a downstream kinase of the insulin/IGF signaling pathway, have extreme longevity beyond that of daf-2 mutants (Ayyadevara et al. 2008). The optimization of DR protocol used in this study, for technical reasons, was not possible with this method of DR. Although it is easy to get an additive effect with suboptimal epistatic analysis, our study highlights the importance of alternative analysis such as the Gompertz analysis. The finding that only DR and CHIL are the only independent pathways is actually strengthened, given that not all our parameters were optimized.
Contrary to some of the previous studies performed in other species (Bartke et al. 2001; Houthoofd et al. 2003; Panowski et al. 2007), our data clearly find that the insulin/IGF pathway and DR have significant interactions and may ultimately use the same pathways to extend lifespan. This has also been demonstrated in flies (Clancy et al. 2002). The minimum Gompertz variable value occurs with DR and cold temperature, suggesting that the other manipulations utilize a common pathway to reduce age-related mortality rate, and we find that even DR and CHIL have a synergistic relationship that may indicate an interaction between the two.
Although we have not directly identified the pathways involved in senescence in this study, there are many possible connections between DR and insulin/IGF signaling (reviewed in Narasimhan et al. 2009). On a biochemical level, free radicals are implicated in senescence and in mouse studies, DR decreases free-radical production from the mitochondria (Gredilla et al. 2001). In worms, insulin/IGF mutants have an increase in free-radical resistance (Gredilla et al. 2001; Honda and Honda 1999). Although free radicals have been correlated with senescence, in both mice and worms, there is increasing evidence that they may not be the causative agent (Doonan et al. 2008; Ran et al. 2007; Van Raamsdonk and Hekimi 2009; Van Remmen et al. 2003; Yen et al. 2009). The target of rapamycin pathway, which may also link DR and insulin/IGF signaling, regulates autophagy, and this is important for both DR and insulin/IGF mediated longevity (Hansen et al. 2008; Kaeberlein et al. 2005; Kapahi et al. 2004; Melendez et al. 2003).
Our results suggest that there may be a final common pathway by which all life-extending manipulations decrease senescence, and concentration on the mechanisms by which DR and CHIL extend lifespan may explain the cause of senescence. Because all of these treatments also extend the lifespan of mice, it may indicate that there is a common cause of senescence for all organisms.
Acknowledgements
Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the NIH National Center for Research Resources (NCRR).
References
- Adachi H, Ishii N. Effects of tocotrienols on life span and protein carbonylation in Caenorhabditis elegans. J Gerontol A Biol Sci Med Sci. 2000;55:B280–B285. doi: 10.1093/gerona/55.6.b280. [DOI] [PubMed] [Google Scholar]
- Arking R, Buck S, Wells RA, et al. Metabolic rates in genetically based long lived strains of Drosophila. Exp Gerontol. 1988;23:59–76. doi: 10.1016/0531-5565(88)90020-4. [DOI] [PubMed] [Google Scholar]
- Ayyadevara S, Alla R, Thaden JJ, et al. Remarkable longevity and stress resistance of nematode PI3K-null mutants. Aging Cell. 2008;7:13–22. doi: 10.1111/j.1474-9726.2007.00348.x. [DOI] [PubMed] [Google Scholar]
- Ball ZB, Barnes RH, Visscher MB. The effects of dietary caloric restriction on maturity and senescence, with particular reference to fertility and longevity. Am J Physiol. 1947;150:511–519. doi: 10.1152/ajplegacy.1947.150.3.511. [DOI] [PubMed] [Google Scholar]
- Bartke A, Wright JC, Mattison JA, et al. Longevity: extending the lifespan of long-lived mice. Nature. 2001;414:412. doi: 10.1038/35106646. [DOI] [PubMed] [Google Scholar]
- Bluher M, Kahn BB, Kahn CR. Extended longevity in mice lacking the insulin receptor in adipose tissue. Science. 2003;299:572–574. doi: 10.1126/science.1078223. [DOI] [PubMed] [Google Scholar]
- Bonkowski MS, Rocha JS, Masternak MM, et al. Targeted disruption of growth hormone receptor interferes with the beneficial actions of calorie restriction. Proc Natl Acad Sci USA. 2006;103:7901–7905. doi: 10.1073/pnas.0600161103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braeckman BP, Houthoofd K, Vanfleteren JR. Patterns of metabolic activity during aging of the wild type and longevity mutants of Caenorhabditis elegans. Age (Omaha) 2000;23:55–73. doi: 10.1007/s11357-000-0007-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broughton SJ, Piper MDW, Ikeya T, et al. Longer lifespan, altered metabolism, and stress resistance in Drosophila from ablation of cells making insulin-like ligands. Proc Natl Acad Sci USA. 2005;102:3105–3110. doi: 10.1073/pnas.0405775102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy DJ, Gems D, Harshman LG, et al. Extension of life-span by loss of CHICO, a Drosophila insulin receptor substrate protein. Science. 2001;292:104–106. doi: 10.1126/science.1057991. [DOI] [PubMed] [Google Scholar]
- Clancy DJ, Gems D, Hafen E, et al. Dietary restriction in long-lived dwarf flies. Science. 2002;296:319. doi: 10.1126/science.1069366. [DOI] [PubMed] [Google Scholar]
- Conti B, Sanchez-Alavez M, Winsky-Sommerer R, et al. Transgenic mice with a reduced core body temperature have an increased life span. Science. 2006;314:825–828. doi: 10.1126/science.1132191. [DOI] [PubMed] [Google Scholar]
- Coschigano KT, Holland AN, Riders ME, et al. Deletion, but not antagonism, of the mouse growth hormone receptor results in severely decreased body weights, insulin, and insulin-like growth factor I levels and increased life span. Endocrinology. 2003;144:3799–3810. doi: 10.1210/en.2003-0374. [DOI] [PubMed] [Google Scholar]
- Curran SP, Ruvkun G. Lifespan regulation by evolutionarily conserved genes essential for viability. PLoS Genet. 2007;3:e56. doi: 10.1371/journal.pgen.0030056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis TA, Bales CW, Beauchene RE. Differential effects of dietary caloric and protein restriction in the aging rat. Exp Gerontol. 1983;18:427–435. doi: 10.1016/0531-5565(83)90021-9. [DOI] [PubMed] [Google Scholar]
- Dillin A, Hsu A-L, Arantes-Oliveira N, et al. Rates of behavior and aging specified by mitochondrial function during development. Science. 2002;298:2398–2401. doi: 10.1126/science.1077780. [DOI] [PubMed] [Google Scholar]
- Doonan R, McElwee JJ, Matthijssens F, et al. Against the oxidative damage theory of aging: superoxide dismutases protect against oxidative stress but have little or no effect on life span in Caenorhabditis elegans. Genes Dev. 2008;22:3236–3241. doi: 10.1101/gad.504808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliason SR. Maximum likelihood estimation logic and practice. Newbury Park: Sage; 1993. [Google Scholar]
- Ewbank JJ, Barnes TM, Lakowski B, et al. Structural and functional conservation of the Caenorhabditis elegans timing gene clk-1. Science. 1997;275:980–983. doi: 10.1126/science.275.5302.980. [DOI] [PubMed] [Google Scholar]
- Feng J, Bussiere F, Hekimi S. Mitochondrial electron transport is a key determinant of life span in Caenorhabditis elegans. Dev Cell. 2001;1:633–644. doi: 10.1016/S1534-5807(01)00071-5. [DOI] [PubMed] [Google Scholar]
- Fernandes G, Yunis EJ, Good RA. Influence of diet on survival of mice. Proc Natl Acad Sci USA. 1976;73:1279–1283. doi: 10.1073/pnas.73.4.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch CE. Longevity, senescence, and the genome. Chicago: The University of Chicago Press; 1990. [Google Scholar]
- Finch CE, Pike MC, Witten M. Slow mortality rate accelerations during aging in some animals approximate that of humans. Science. 1990;249:902–905. doi: 10.1126/science.2392680. [DOI] [PubMed] [Google Scholar]
- Gems D, Pletcher S, Partridge L. Interpreting interactions between treatments that slow aging. Aging Cell. 2002;1:1–9. doi: 10.1046/j.1474-9728.2002.00003.x. [DOI] [PubMed] [Google Scholar]
- Giannakou ME, Goss M, Junger MA, et al. Long-lived Drosophila with overexpressed dFOXO in adult fat body. Science. 2004;305:361. doi: 10.1126/science.1098219. [DOI] [PubMed] [Google Scholar]
- Golden JW, Riddle DL. A pheromone-induced developmental switch in Caenorhabditis elegans: temperature-sensitive mutants reveal a wild-type temperature-dependent process. Proc Natl Acad Sci USA. 1984;81:819–823. doi: 10.1073/pnas.81.3.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gracey AY, Fraser EJ, Li W, et al. Coping with cold: an integrative, multitissue analysis of the transcriptome of a poikilothermic vertebrate. Proc Natl Acad Sci USA. 2004;101:16970–16975. doi: 10.1073/pnas.0403627101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gredilla R, Sanz A, Lopez-Torres M, et al. Caloric restriction decreases mitochondrial free radical generation at complex I and lowers oxidative damage to mitochondrial DNA in the rat heart. FASEB J. 2001;15:1589–1591. doi: 10.1096/fj.00-0764fje. [DOI] [PubMed] [Google Scholar]
- Han E-S, Hickey M. Microarray evaluation of dietary restriction. J Nutr. 2005;135:1343–1346. doi: 10.1093/jn/135.6.1343. [DOI] [PubMed] [Google Scholar]
- Hansen M, Chandra A, Mitic LL, et al. A role for autophagy in the extension of lifespan by dietary restriction in C. elegans. PLoS Genet. 2008;4:e24. doi: 10.1371/journal.pgen.0040024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloszy JO, Smith EK. Longevity of cold-exposed rats: a reevaluation of the “rate-of-living theory”. J Appl Physiol. 1986;61:1656–1660. doi: 10.1152/jappl.1986.61.5.1656. [DOI] [PubMed] [Google Scholar]
- Holzenberger M, Dupont J, Ducos B, et al. IGF-1 receptor regulates lifespan and resistance to oxidative stress in mice. Nature. 2003;421:182–187. doi: 10.1038/nature01298. [DOI] [PubMed] [Google Scholar]
- Honda Y, Honda S. The daf-2 gene network for longevity regulates oxidative stress resistance and Mn-superoxide dismutase gene expression in Caenorhabditis elegans. FASEB J. 1999;13:1385–1393. [PubMed] [Google Scholar]
- Houthoofd K, Braeckman BP, Lenaerts I, et al. Axenic growth up-regulates mass-specific metabolic rate, stress resistance, and extends life span in Caenorhabditis elegans. Exp Gerontol. 2002;37:1371–1378. doi: 10.1016/S0531-5565(02)00173-0. [DOI] [PubMed] [Google Scholar]
- Houthoofd K, Braeckman B, Johnson T, et al. Life extension via dietary restriction is independent of the Ins/IGF-1 signalling pathway in Caenorhabditis elegans. Exp Gerontol. 2003;38:947–954. doi: 10.1016/S0531-5565(03)00161-X. [DOI] [PubMed] [Google Scholar]
- Hulbert AJ, Clancy DJ, Mair W, et al. Metabolic rate is not reduced by dietary-restriction or by lowered insulin/IGF-1 signalling and is not correlated with individual lifespan in Drosophila melanogaster. Exp Gerontol. 2004;39:1137–1143. doi: 10.1016/j.exger.2004.04.006. [DOI] [PubMed] [Google Scholar]
- Hwangbo DS, Gersham B, Tu M-P, et al. Drosophila dFOXO controls lifespan and regulates insulin signalling in brain and fat body. Nature. 2004;429:562–566. doi: 10.1038/nature02549. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M, Powers RW, III, Steffen KK, et al. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science. 2005;310:1193–1196. doi: 10.1126/science.1115535. [DOI] [PubMed] [Google Scholar]
- Kapahi P, Zid BM, Harper T, et al. Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway. Curr Biol. 2004;14:885–890. doi: 10.1016/j.cub.2004.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayser E-B, Sedensky MM, Morgan PG, et al. Mitochondrial oxidative phosphorylation is defective in the long-lived mutant clk-1. J Biol Chem. 2004;279:54479–54486. doi: 10.1074/jbc.M403066200. [DOI] [PubMed] [Google Scholar]
- Kenyon C, Chang J, Gensch E, et al. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- Kimura KD, Tissenbaum HA, Liu Y, et al. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science. 1997;277:942–946. doi: 10.1126/science.277.5328.942. [DOI] [PubMed] [Google Scholar]
- Klass MR. Aging in the nematode Caenorhabditis elegans: major biological and environmental factors influencing life span. Mech Ageing Dev. 1977;6:413–429. doi: 10.1016/0047-6374(77)90043-4. [DOI] [PubMed] [Google Scholar]
- Klass MR. A method for the isolation of longevity mutants in the nematode Caenorhabditis elegans and initial results. Mech Ageing Dev. 1983;22:279–286. doi: 10.1016/0047-6374(83)90082-9. [DOI] [PubMed] [Google Scholar]
- Lakowski B, Hekimi S. Determination of life-span in Caenorhabditis elegans by four clock genes. Science. 1996;272:1010–1013. doi: 10.1126/science.272.5264.1010. [DOI] [PubMed] [Google Scholar]
- Lakowski B, Hekimi S. The genetics of caloric restriction in Caenorhabditis elegans. Proc Natl Acad Sci USA. 1998;95:13091–13096. doi: 10.1073/pnas.95.22.13091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C-K, Klopp RG, Weindruch R, et al. Gene expression profile of aging and its retardation by caloric restriction. Science. 1999;285:1390–1393. doi: 10.1126/science.285.5432.1390. [DOI] [PubMed] [Google Scholar]
- Lee C-K, Weindruch R, Prolla TA. Gene-expression profile of the ageing brain in mice. Nat Genet. 2000;25:294–297. doi: 10.1038/77046. [DOI] [PubMed] [Google Scholar]
- Liu RK, Walford RL. Observations on the lifespans of several species of annual fishes and of the world’s smallest fishes. Exp Gerontol. 1970;5:241–246. doi: 10.1016/0531-5565(70)90044-6. [DOI] [PubMed] [Google Scholar]
- Liu X, Jiang N, Hughes B, et al. Evolutionary conservation of the clk-1-dependent mechanism of longevity: loss of mclk1 increases cellular fitness and lifespan in mice. Genes Dev. 2005;19:2424–2434. doi: 10.1101/gad.1352905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madi A, Mikkat S, Ringel B, et al. Mass spectrometric proteome analysis for profiling temperature-dependent changes of protein expression in wild-type Caenorhabditis elegans. Proteomics. 2003;3:1526–1534. doi: 10.1002/pmic.200300490. [DOI] [PubMed] [Google Scholar]
- Mair W, Goymer P, Pletcher SD, et al. Demography of dietary restriction and death in Drosophila. Science. 2003;301:1731–1733. doi: 10.1126/science.1086016. [DOI] [PubMed] [Google Scholar]
- Malek RL, Sajadi H, Abraham J, et al. The effects of temperature reduction on gene expression and oxidative stress in skeletal muscle from adult zebrafish. Comp Biochem Physiol C Toxicol Pharmacol. 2004;138:363. doi: 10.1016/j.cca.2004.08.014. [DOI] [PubMed] [Google Scholar]
- Melendez A, Talloczy Z, Seaman M, et al. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science. 2003;301:1387–1391. doi: 10.1126/science.1087782. [DOI] [PubMed] [Google Scholar]
- Migliaccio E, Giorgio M, Mele S, et al. The p66shc adaptor protein controls oxidative stress response and life span in mammals. Nature. 1999;402:309–313. doi: 10.1038/46311. [DOI] [PubMed] [Google Scholar]
- Miyadera H, Amino H, Hiraishi A, et al. Altered quinone biosynthesis in the long-lived clk-1 mutants of Caenorhabditis elegans. J Biol Chem. 2001;276:7713–7716. doi: 10.1074/jbc.C000889200. [DOI] [PubMed] [Google Scholar]
- Morris JZ, Tissenbaum HA, Ruvkun G. A phosphatidylinositol-3-OH kinase family member regulating longevity and diapause in Caenorhabditis elegans. Nature. 1996;382:536. doi: 10.1038/382536a0. [DOI] [PubMed] [Google Scholar]
- Narasimhan SD, Yen K, Tissenbaum HA (2009) Converging pathways in Lifespan Regulation. Curr Biol (in press) doi:10.1016/j.cub.2009.06.013 [DOI] [PMC free article] [PubMed]
- Orsini F, Migliaccio E, Moroni M, et al. The life span determinant p66Shc localizes to mitochondria where it associates with mitochondrial heat shock protein 70 and regulates trans-membrane potential. J Biol Chem. 2004;279:25689–25695. doi: 10.1074/jbc.M401844200. [DOI] [PubMed] [Google Scholar]
- Panowski SH, Wolff S, Aguilaniu H, et al. PHA-4/Foxa mediates diet-restriction-induced longevity of C. elegans. Nature. 2007;447:550–555. doi: 10.1038/nature05837. [DOI] [PubMed] [Google Scholar]
- Paul RJ, Gohla J, Foll R, et al. Metabolic adaptations to environmental changes in Caenorhabditis elegans. Comp Biochem Physiol B Biochem Mol Biol. 2000;127:469–479. doi: 10.1016/S0305-0491(00)00284-4. [DOI] [PubMed] [Google Scholar]
- Promislow DEL, Tatar M, Pletcher SD, et al. Below-threshold mortality: implications for studies in evolution, ecology and demography. J Evol Biol. 1999;12:314–328. doi: 10.1046/j.1420-9101.1999.00037.x. [DOI] [Google Scholar]
- Ran Q, Liang H, Ikeno Y, et al. Reduction in glutathione peroxidase 4 increases life span through increased sensitivity to apoptosis. J Gerontol A Biol Sci Med Sci. 2007;62:932–942. doi: 10.1093/gerona/62.9.932. [DOI] [PubMed] [Google Scholar]
- Rao KP. Physiology of acclimation to low temperature in poikilotherms. Science. 1962;137:682–683. doi: 10.1126/science.137.3531.682. [DOI] [PubMed] [Google Scholar]
- Spencer CC, Promislow DEL. Age-specific changes in epistatic effects on mortality rate in Drosophila melanogaster. J Hered. 2005;96:513–521. doi: 10.1093/jhered/esi071. [DOI] [PubMed] [Google Scholar]
- Tissenbaum HA, Ruvkun G. An insulin-like signaling pathway affects both longevity and reproduction in Caenorhabditis elegans. Genetics. 1998;148:703–718. doi: 10.1093/genetics/148.2.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tribe MA, Bowler K. Temperature dependence of “standard metabolic rate” in a poikilotherm. Comp Biochem Physiol. 1968;25:427–436. doi: 10.1016/0010-406X(68)90351-4. [DOI] [PubMed] [Google Scholar]
- Raamsdonk JM, Hekimi S. Deletion of the mitochondrial superoxide dismutase sod-2 extends lifespan in Caenorhabditis elegans. PLoS Genet. 2009;5:e1000361. doi: 10.1371/journal.pgen.1000361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remmen H, Ikeno Y, Hamilton M, et al. Life-long reduction in MnSOD activity results in increased DNA damage and higher incidence of cancer but does not accelerate aging. Physiol Genomics. 2003;16:29–37. doi: 10.1152/physiolgenomics.00122.2003. [DOI] [PubMed] [Google Scholar]
- Weindruch R, Walford RL. Dietary restriction in mice beginning at 1 year of age: effect on life-span and spontaneous cancer incidence. Science. 1982;215:1415–1418. doi: 10.1126/science.7063854. [DOI] [PubMed] [Google Scholar]
- Wong A, Boutis P, Hekimi S. Mutations in the clk-1 gene of Caenorhabditis elegans affect developmental and behavioral timing. Genetics. 1995;139:1247–1259. doi: 10.1093/genetics/139.3.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen K, Mastitis JW, Mobbs CV. Lifespan is not determined by metabolic rate: evidence from fishes and C. elegans. Exp Gerontol. 2004;39:947–949. doi: 10.1016/j.exger.2004.03.016. [DOI] [PubMed] [Google Scholar]
- Yen K, Steinsaltz D, Mobbs CV. Validated analysis of mortality rates demonstrates distinct genetic mechanisms that influence lifespan. Exp Gerontol. 2008;43:1044–1051. doi: 10.1016/j.exger.2008.09.006. [DOI] [PubMed] [Google Scholar]
- Yen K, Patel HB, Lublin AL, et al. SOD isoforms play no role in lifespan in ad lib or dietary restricted conditions, but mutational inactivation of SOD-1 reduces life extension by cold. Mech Ageing Dev. 2009;130:173–178. doi: 10.1016/j.mad.2008.11.003. [DOI] [PubMed] [Google Scholar]
- Yu BP, Masoro EJ, McMahan CA. Nutritional influences on aging of Fischer 344 rats: I. Physical, metabolic, and longevity characteristics. J Gerontol. 1985;40:657–670. doi: 10.1093/geronj/40.6.657. [DOI] [PubMed] [Google Scholar]