Abstract
Oxidative DNA damage is important in aging and a variety of diseases. Significant advances have been made in our understanding of the chemistry of radical mediated DNA damage. These studies have been carried out on DNA in the absence of proteins. However, in cells DNA is typically bound by proteins such as in chromatin and transiently by proteins that regulate biochemical processes. How and whether protein binding affects DNA radical reactivity is not well understood. The effect of the DNA binding protein, Hbb on the reactivity of 5-(2'-deoxyuridinyl)methyl radical (1) and 5-(2'-deoxycytidinyl)methyl radical (2) was studied. Hbb bends DNA and disrupts base stacking at the sites of kinking. The reactivity of 1 and 2 are signficantly affected when they are generated at the kinking site in the presence of Hbb. The increased conformational mobility of the radicals results in significantly higher yields of DNA interstrand cross-links. These studies provide the first specific data on how protein binding affects the reactivity of a DNA radical and bring us closer to understanding oxidative DNA damage in cells.
Radical mediated DNA damage is a ubiquitous process that is effected by a variety of methods (e.g. γ-radiolysis, UV photolysis) and reactive species (e.g. hydroxyl radical).1 The consequences of DNA damage can be deleterious, as evidenced by its role in aging and diseases such as cancer and Alzheimer's.2–6 On the other hand, the cytotoxic effects of a number of anticancer agents are attributable to their ability to oxidatively damage DNA. The chemistry of radical mediated DNA damage is complicated by the heterogeneity of the biopolymer, as well as the formation of a variety of radical intermediates, particularly by diffusible species such as hydroxyl radical. Significant advances have been made in our understanding of oxidative DNA damage by independently generating radical intermediates in the biopolymer.7–9 These studies have been carried out in the absence of proteins, which is not how DNA typically exists in cells. In eukaryotic cells, nuclear DNA is wrapped around the octameric complex of histone proteins.10 DNA interacts with other proteins, such as those that regulate transcription, more transiently. Protein binding affects DNA damage by altering its accessibility to oxidizing agents and the efficiency of charge migration through the duplex.11,12 However, its effects on the reactivity of DNA radicals is unknown. Using our ability to independently generate nucleotide radicals we have determined that a protein has a profound effect on DNA radical reactivity.
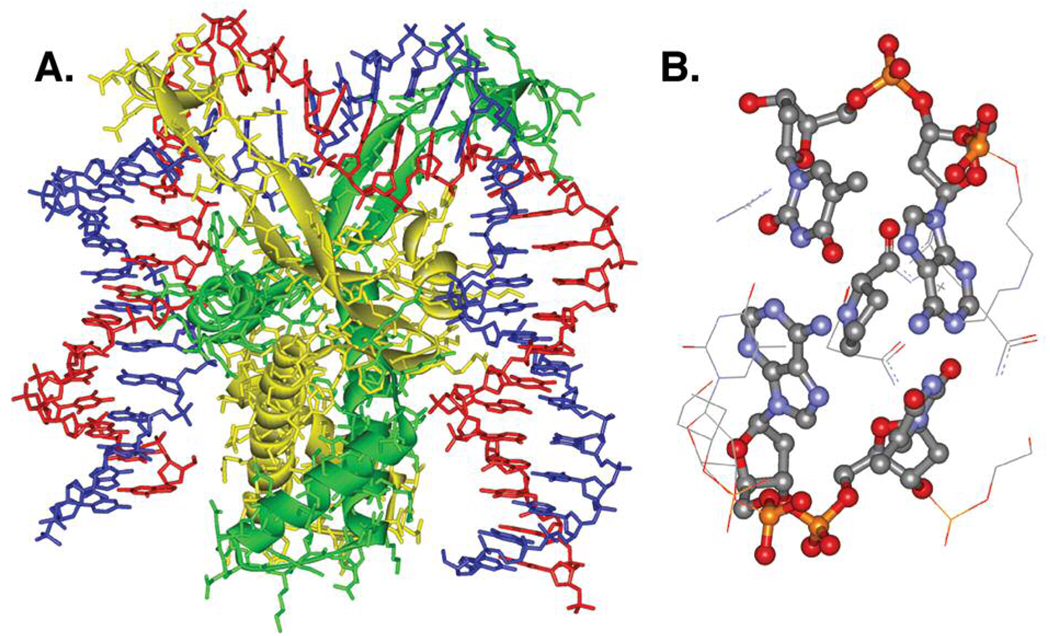
Hbb from Borrelia burgdorferi, the species responsible for lyme disease in humans, is a member of the bacterial integration host factor family of DNA-bending proteins whose functions include chromosomal compaction.13,14 Hbb binds sequence specifically to DNA as a homodimer. X-ray crystallography reveals that each monomeric unit sharply kinks (~80°) the nucleic acid by inserting a proline residue between two base pairs, consequently disrupting base stacking at the site of bending (Figure 1). We rationalized that the disruption in base stacking could significantly alter the reactivity of nucleotide radicals formed at these positions.
Figure 1.
X-Ray crystal structure of Hbb2-DNA complex. A) Homodimeric complex bending DNA. DNA strands: blue, red; Hbb monomers: yellow, green. B) Base stacking disruption at the site of proline intercalation. Structural data from PDB ID: 2NP2.
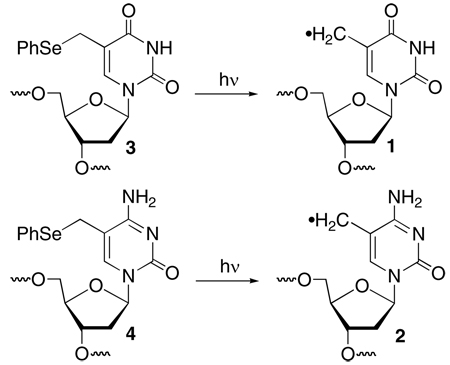
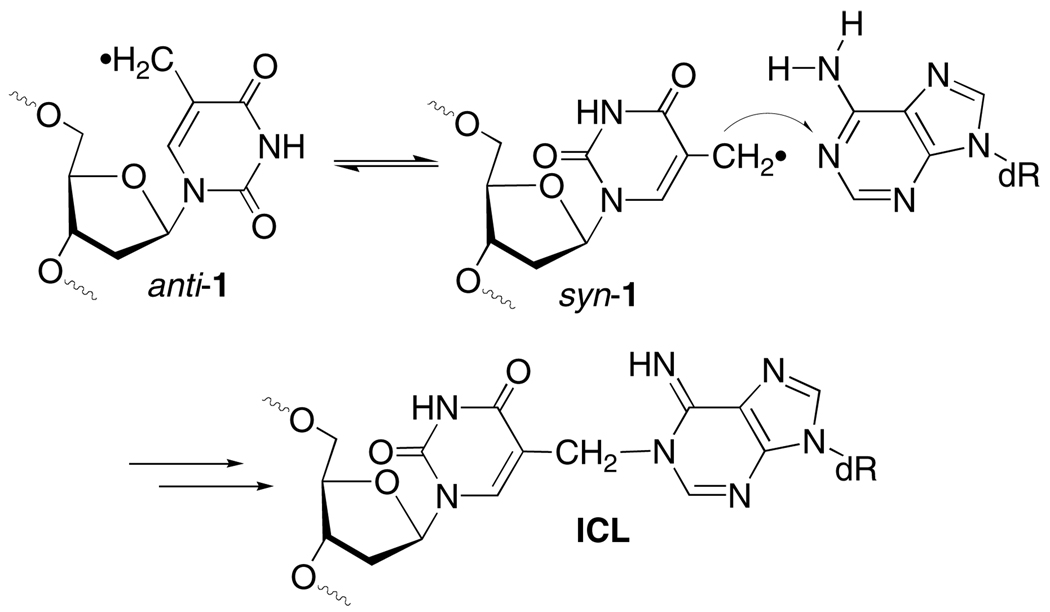
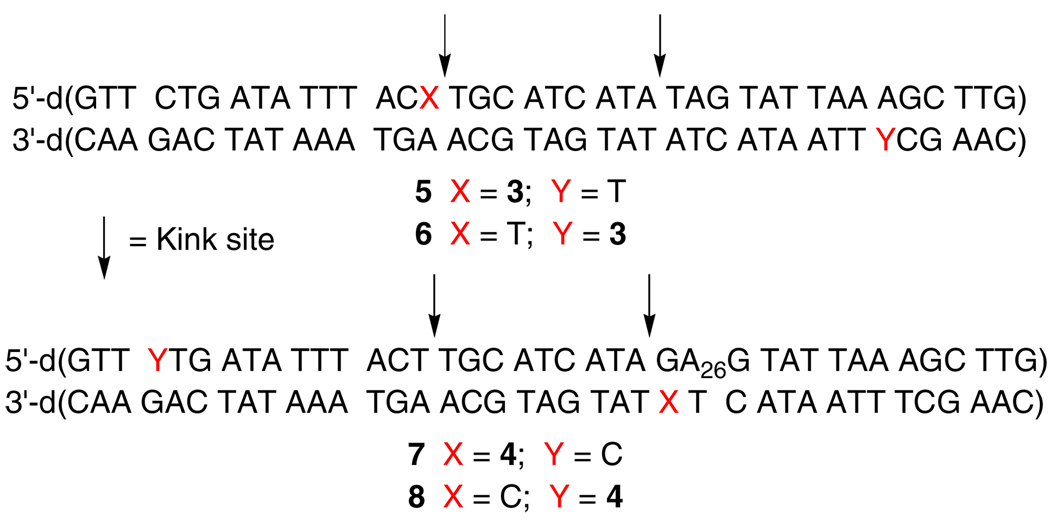
This hypothesis was tested by examining Hbb's effect on interstrand cross-link (ICL) formation from 5-(2'-deoxyuridinyl)methyl radical (1) and 5-(2'-deoxycytidinyl)methyl radical (2). ICLs are a biologically important family of DNA lesions.15,16 The former radical produces ICLs with its opposing dA and by necessity requires adoption of its syn-isomer by rotating about its glycosidic bond in a rate determining step (Scheme 1).17,18 Generation of 1 at a Hbb kink site was expected to increase the ICL yield due to greater conformational freedom arising from the disruption in base stacking. Duplex 5 was prepared based upon the sequence employed in previous Hbb studies.13 The photolabile radical precursor was incorporated in place of a thymidine present at the kink site in the original sequence. Electrophoretic mobility shift assay indicated that 5 (20 nM) was completely bound in the presence of 600 nM Hbb.19 Subsequently, 1 was generated in 5'-32P-5 by photolysis at 350 nm in the absence and presence of Hbb. Denaturing gel electrophoresis analysis revealed that the ICL yield more than doubled when 1 was generated in DNA bound by Hbb (Table 1). Hydroxyl radical cleavage analysis of the cross-linked DNA indicated that the cross-link formed exclusively with the opposing dA, as was observed when 1 was produced in other duplexes in the absence of protein.18,19 Hbb had a far more modest effect on ICL formation in 6 where 1 was generated at a nucleotide that is approximately one helical turn from the kink site.
Scheme 1.
Table 1.
The effect of nucleotide position and Hbb on interstrand crosslink formation.
| Duplex | Hbb | ICL Yield (%)a | kRSH/kICL × 10−3 (M−1)a |
|---|---|---|---|
| 5 | − | 20.4 ± 1.0 | 3.3 ± 0.4 |
| 5 | + | 41.7 ± 2.3 | 0.9 ± 0.1 |
| 6 | − | 19.6 ± 0.8 | 4.9 ± 0.5 |
| 6 | + | 24.2 ± 2.5 | 3.4 ± 0.2 |
| 7 | − | < 0.3 | - |
| 7 | + | 10.6 ± 0.5 | 2.2 ± 0.2 |
Data are the average of at least three experiments. Each experiment consists of 3 replicates.
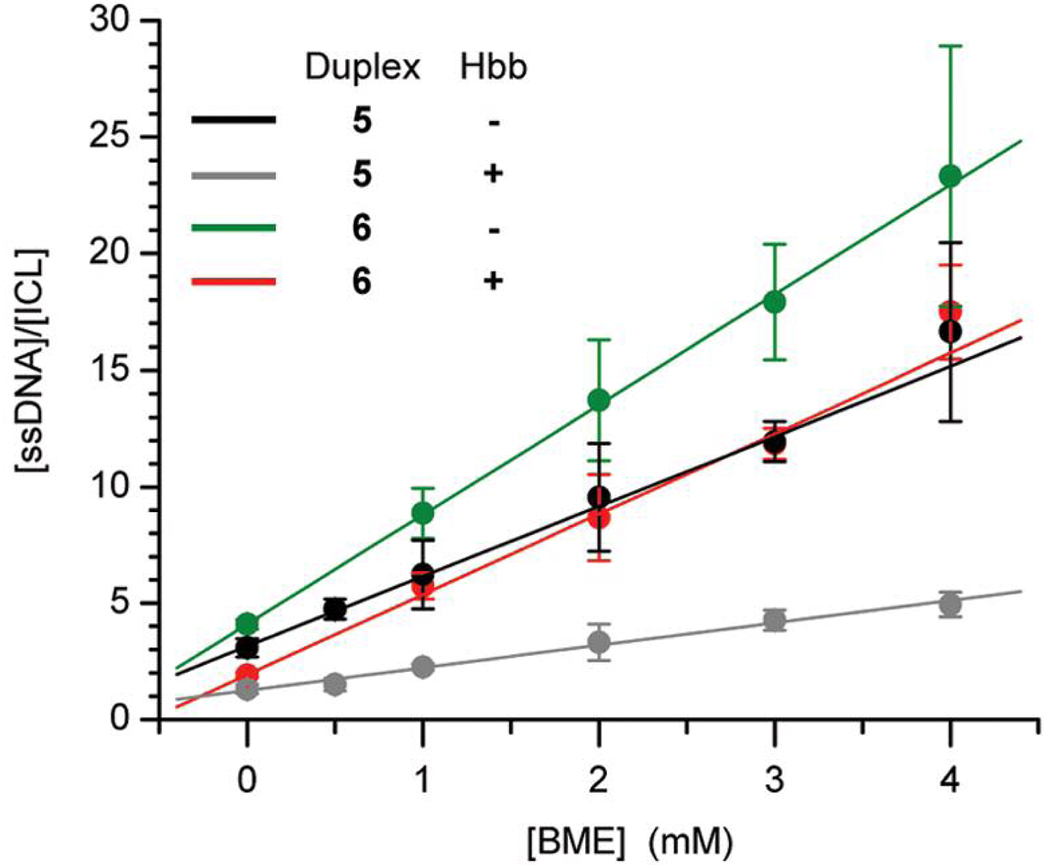
The large increase in ICL yield in the presence of Hbb was also evident from examination of the effect of thiol (RSH, β-mercaptoethanol) on cross-linking by 1 (Table 1, Figure 2). Competition studies (eqn. 1) revealed that the efficiency of thiol trapping relative to cross-linking decreases by ~350% when 1 is generated at the kink site versus ~30% in 6. One cannot rule out that shielding of 1 from the freely diffusible thiol by Hbb contributes to the decrease in the observed ratio of kRSH to kICL. However, this is considered particularly unlikely at the kink site (5) where Hbb binding enhances reaction of a thymidine in unmodified DNA with permanganate.20 We suggest that the majority of the decrease in kRSH/kICL in 5 is due to an increase in the rate constant for cross-link formation when the radical (1) is generated where base stacking is disrupted in the duplex.
| (1) |
Figure 2.
The effect nucleotide position and Hbb on interstrand cross-link formation from 5-(2'-deoxyuridinyl)methyl radical (1) as a function of β-mercaptoethanol (BME) concentration.
The generality of the effect of Hbb on DNA radical reactivity was explored by examining its effect on the reactivity of 5-(2'-deoxycytidinyl)methyl radical (2). Photolysis of 5'-32P-7 or 5'-32P-8 in the absence of Hbb yields <1% ICL. The reluctance of 2 to produce cross-links with dG compared to 5-(2'-deoxyuridinyl)methyl radical (1) with dA is attributed to reduced reactivity of an opposing dG with the radical, and a lower rate constant for glycosidic bond rotation due to stronger base pairing. Support for the latter effect is evident from studies in which 4 is used to form DNA cross-links under nonradical conditions.21 Hbb binding results in <1% cross-linked product when 2 is generated at a position that is remote from the bending site (5'-32P-8). However, irradiation of 5'-32P-7 in which the radical is produced at the kink site in the presence of saturating levels of Hbb produces >10% cross-linked product (Table 1). Hydroxyl radical cleavage shows that the majority of cross-links in 5'-32P-7 involve the guanosine nucleotide and <20% result from reaction with A26.19 We suggest that the greater conformational freedom of 2 in the presence of Hbb enables the radical to react with the more reactive dA adjacent to the opposing dG. The lower ICL yield from 2 than the thymidine analogue (1) is corroborated by the ratio of kRSH/kICL, which is considerably greater than the respective ratio obtained from 1 (Table 1). Assuming that kRSH is approximately the same for reaction with either radical generated in 5'-32P-5 or 5'-32P-7, the thiol dependency suggests that the lower ICL yield from 5-(2'-deoxycytidinyl)methyl radical (2) is partially due to a lower kICL than for 1.
In conclusion, the experiments involving Hbb with 5-(2'-deoxyuridinyl)methyl radical (1) and 5-(2'-deoxycytidinyl)methyl radical (2) reveal how protein binding significantly alters the reactivity of DNA radicals. Although DNA-protein cross-links were not formed in either of the above systems, it is possible that other DNA radical-protein complexes may yield such products. Overall, these studies bring us one step closer to understanding the chemistry of oxidative DNA damage in cells.
Supplementary Material
Scheme 2.
Acknowledgment
We are grateful for support from the National Institute of General Medical Sciences (GM-054996 to MMG, GM-066011 to PAR). We thank Mr. Kent Mouw for helpful advice.
Footnotes
Supporting Information. Experimental procedures, ESI-MS of oligonucleotides, and sample autoradiograms. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.von Sonntag C. Free-Radical-Induced DNA Damage and Its Repair. Berlin: Springer-Verlag; 2006. [Google Scholar]
- 2.Evans MD, Dizdaroglu M, Cooke MS. Mutat. Res. 2004;567:1–61. doi: 10.1016/j.mrrev.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 3.Cadet J, Douki T, Gasparutto D, Ravanat JL. Mutat. Res. 2003;531:5–23. doi: 10.1016/j.mrfmmm.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 4.Moreira PI, Nunomura A, Nakamura M, Takeda A, Shenk JC, Aliev G, Smith MA, Perry G. Free Radical Biol. Med. 2008;44:1493–1505. doi: 10.1016/j.freeradbiomed.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 5.Lombard DB, Chua KF, Mostoslavsky R, Franco S, Gostissa M, Alt FW. Cell. 2005;120:435–567. doi: 10.1016/j.cell.2005.01.028. [DOI] [PubMed] [Google Scholar]
- 6.DePinho RA. Nature. 2000;408:248–254. doi: 10.1038/35041694. [DOI] [PubMed] [Google Scholar]
- 7.Greenberg MM. Org. Biomol. Chem. 2007;5:18–30. doi: 10.1039/b612729k. [DOI] [PubMed] [Google Scholar]
- 8.Giese B. Annu. Rev. Biochem. 2002;71:51–70. doi: 10.1146/annurev.biochem.71.083101.134037. [DOI] [PubMed] [Google Scholar]
- 9.Greenberg MM. Chem. Res. Toxicol. 1998;11:1235–1248. doi: 10.1021/tx980174i. [DOI] [PubMed] [Google Scholar]
- 10.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 11.Rajski SR, Barton JK. Biochemistry. 2001;40:5556–5564. doi: 10.1021/bi002684t. [DOI] [PubMed] [Google Scholar]
- 12.Stisova V, Goffinont S, Spotheim-Maurizot M, Davidkova M. Radiat. Prot. Dosim. 2006;122:106–109. doi: 10.1093/rpd/ncl443. [DOI] [PubMed] [Google Scholar]
- 13.Mouw KW, Rice PA. Mol. Microbiol. 2007;63:1319–1330. doi: 10.1111/j.1365-2958.2007.05586.x. [DOI] [PubMed] [Google Scholar]
- 14.Swinger KK, Rice PA. Curr. Op. in Struct. Biol. 2004;14:28–35. doi: 10.1016/j.sbi.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 15.Noll DM, Mason TM, Miller PS. Chem. Rev. 2006;106:277–301. doi: 10.1021/cr040478b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schärer OD. Chem. Bio. Chem. 2005;6:27–32. doi: 10.1002/cbic.200400287. [DOI] [PubMed] [Google Scholar]
- 17.Hong IS, Ding H, Greenberg MM. J. Am. Chem. Soc. 2006;128:485–491. doi: 10.1021/ja0563657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hong IS, Greenberg MM. J. Am. Chem. Soc. 2005;127:3692–3693. doi: 10.1021/ja042434q. [DOI] [PubMed] [Google Scholar]
- 19.See Supporting Information.
- 20.Kobryn K, Naigamwalla DZ, Chaconas G. Mol. Microbiol. 2000;37:145–155. doi: 10.1046/j.1365-2958.2000.01981.x. [DOI] [PubMed] [Google Scholar]
- 21.Peng X, Hong IS, Li H, Seidman MM, Greenberg MM. J. Am. Chem. Soc. 2008;130:10299–10306. doi: 10.1021/ja802177u. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.