Abstract
c-Fos is a member of the activator protein 1 family that regulates transcription of target genes. c-Fos is transiently induced in specific regions of the brain after a variety of external stimuli including learning and memory formation. Analysis of gene expression in c-Fos-expressing cells of the brain may help identify target genes that play important roles in synaptic strength or neuronal morphology. In the present study, we developed a combined method of laser capture microdissection and 5-bromo-4-chloro-3-indoly-β-D-galactopyranosidase (X-Gal) histology to analyze gene expression in stimulus-induced c-Fos-positive cells. Using transgenic mice carrying a c-fos-lacZ fusion gene, c-Fos-positive cells were easily identified by measuring of β-galactosidase (β-Gal) activity. To establish the fidelity of the reporter transgene, the time course of endogenous c-Fos and the c-fos-lacZ transgene expression in the amygdala induced by LiCl administration was investigated by immunohistochemistry and X-Gal staining. LiCl increased the numbers of c-Fos- and β-Gal-positive cells in the central and basolateral amygdala of the transgenic mice. To ensure that RNA was preserved in X-Gal stained tissue sections, different fixations were examined, with the conclusion that ethanol fixation was best for both RNA preservation and X-Gal staining quality. Finally, in combining X-Gal staining, single-cell LCM and RT-PCR, we confirmed mRNA expression of endogenous c-fos and β-actin genes in LiCl-induced β-Gal-positive cells in the CeA, cortex and hippocampus. Combining LCM and transgenic reporter genes provides a powerful tool with which to investigate tissue- or cell-specific gene expression.
Keywords: c-Fos, X-Gal, c-fos-lacZ, LiCl, amygdala, β-galactosidase, laser capture microdissection
1. Introduction
c-Fos is an immediate early gene product that is a member of the activator protein 1 (AP-1) family of transcription factors. c-Fos is widely expressed in normal tissue cells and neurons during development, differentiation and growth (Herrera and Robertson, 1996). In addition to the basal expression, c-Fos is transiently induced in specific regions of the brain after a variety of external stimuli such as convulsive, noxious, osmotic, somatosensory, stressful stimulation or brain injury (Hoffman et al., 1993; Herrera and Robertson, 1996; Kovacs, 1998). This stimuli-induced c-Fos expression has been used as a molecular marker of the neuronal activation and studied to determine the functional and anatomical maps of neural pathways or populations in the central nervous system (CNS).
Post-synaptic c-Fos induction following certain neural stimuli is known to occur via intracellular second messenger cascades that are triggered from synaptic transmission. Activation of the cyclic adenosine monophosphate (cAMP)-protein kinase A-cAMP response element (CRE) binding protein pathway can induce c-Fos expression (Bravo et al., 1987; Kruijer et al., 1985) via the consensus CRE sites in the c-Fos promoter (Fisch TM, 1989; Sassone-Corsi et al., 1988). In order to form a functional AP-1 transcription factor, c-Fos needs to hetereodimerize with a complementary AP-1 protein, such as Jun members (Rauscher et al., 1998; Chiu et al., 1998; Halazonetis et al., 1988). AP-1 dimers regulate gene transcription by binding to the AP-1 site of target genes’ promoters, although the effect on transcription may depend on the composition of the AP-1 complex (Foletta, 1996; Karin et al., 1997). For example, a heterodimer of c-Fos and c-Jun appears to induce transcription, while a heterodimer of Fra-2 and c-Jun may suppress transcription (e.g., of the Fra-2 gene; Foletta, 1996). Approximately one third of total genes in the human genome contain the consensus AP-1 site in their promoter regions (Zhou et al., 2005), consistent with the involvement of AP-1 transcription factors and their targets in very diverse actions from new cell generation to apoptosis (Foletta, 1996; Karin et al., 1997; Zhou et al., 2005). In the brain, c-Fos induction during learning suggests functional and regional roles of neuronal activation in memory formation. For example, c-Fos expression is increased in the hippocampus during fear conditioning (Huff et al., 2006) and spatial learning (He et al., 2002) and in the amygdala during conditioned taste aversion learning (Lamprecht and Dudai, 1995; Mickley et al., 2004; Wilkins and Bernstein, 2006; Kwon et al., 2008). Inhibition of c-Fos synthesis by microinjection of antisense oligonucleotides into specific brain areas results in an impairment of a variety of memory formation (Lamprecht and Dudai, 1996; Grimm et al., 1997; Guzowski, 2002; He et al., 2002; Yasoshima et al., 2006), indicating that changes in protein synthesis induced by c-Fos may result in long-lasting synaptic changes that are essential for long-term memory formation.
Thus, the analysis of gene expression in c-Fos-expressing cells of the brain may help identify target genes that play important roles in synaptic strength or neuronal morphology. While c-Fos-positive cells can be easily and distinctly identified by e.g. immunohistochemistry, the molecular analysis is complicated by their sparseness. Dissection (i.e. by tissue punch) of regions such as the amygdala that contain hetereogenous cell populations would result in dilution of any intracellular molecular events that are specific to the c-Fos and AP-1 pathways within activated cells. In order to limit the analysis of target genes to activated neurons, it would be desirable to dissect and collect mRNA from c-Fos-positive cells only.
In the present study, we developed a simple method to analyze gene expression in stimulus-induced c-Fos-positive cells. To collect only the cells of interest, we employed the laser capture microdissection (LCM) technique to microdissect out single, c-Fos-positive cells. LCM has been used to collect cell populations for analysis of region- or cell-specific gene expression (Bonner, 1997; Schutze, 1998). The LCM technique has some advantages over dissection with tissue punches. Discrete regions of arbitrary shape can be visualized under a microscope and dissected using laser punches of 7–30 μm diameter. Moreover, sparsely distributed populations of cells or even single cell nuclei can be precisely microdissected from preserved tissue sections using LCM.
To identify c-Fos-positive cells in the brain, we took advantage of c-fos-lacZ transgenic mice carrying a c-fos-lacZ fusion gene. The c-fos-lacZ fusion protein product of the transgene contains 315 N-terminal amino acids from c-Fos and 1050 C-terminal amino acids from lacZ β-galactosidase (β-Gal) enzyme (Schilling et al., 1991). This fusion gene has the same transcriptional regulatory elements including the CRE site within 611 bp of the 5′ untranscribed sequence from the endogenous mouse c-fos gene. Thus, the fusion gene is thought to have the same expression pattern in the transgenic mice as the endogenous c-fos gene (but see discussion for caveats). For example, convulsive kainate treatment induced expression of the c-fos-lacZ fusion gene, which is largely similar with endogenous c-Fos expression, in many brain regions including the amygdala, cortex and hippocampus (Smeyne et al., 1992b and 1993). This line of c-fos-lacZ transgenic mice have also been used to examine regional expression of c-Fos in the developing CNS (Smeyne et al., 1992a), sleep and wake-induced stimulation of c-fos and the fusion gene in the cingulate cortex (Basheer et al., 1997), in the hypothalamic suprachiasmatic nucleus following light stimulation (Crosio et al., 2000) and in motor neurons following sciatic nerve damage (Soares et al., 2001).
Cells which express endogenous c-Fos and lacZ can be readily observed in c-fos-lacZ transgenic mice by measuring of β-Gal activity using 5-bromo-4-chloro-3-indoly-β-D-galactopyranosidase (X-Gal) staining. X-Gal staining is more convenient and requires fewer processing steps compared with immunohistochemistry. Importantly, the buffers for X-Gal staining are more easily kept RNase-free, compared to ex-vivo immunohistochemical reagents. Thus, X-Gal staining may preserve tissue levels of mRNA for subsequent analysis better than immunohistochemical staining. These properties recommended the use of c-fos-lacZ expression to us as a molecular marker of neuronal activation that corresponds with expression of endogenous c-Fos in the transgenic mice.
The integrity of the RNA during the processing of tissue for LCM is critical. RNA is easily degraded during histologic or enzymatic staining. Fixation can preserve RNA, but extraction of useful RNA from fixed tissue for later analysis can be difficult. Previous investigations have shown that fixation with organic solvents such as acetone, methanol and ethanol well preserved mRNA for RT-PCR amplification after LCM (Fend et al., 1999; Goldsworthy et al., 1999; Kohda et al., 2000). More recent studies have suggested that it is possible to analyze gene expression profiles in an immuno-stained cell population in the brain (Lu et al., 2004) and blood-brain barrier (Macdonald et al., 2008) after single-cell LCM using acetone fixation to preserve RNA during immunohistochemical staining. Others also reported gene expression profiles after single-cell LCM of morphological unique cell populations in the testis (Sluka et al., 2002) and inner ear (Pagedar et al., 2006) using acetone or formalin fixation, respectively.
Thus, a successful analysis requires fixation compatible with X-Gal staining, LCM, and RNA preservation. Therefore, in the present study, we examined the effect of different fixations on both X-Gal staining of brain sections and RNA preservation for use in reverse transcription-polymerase chain reaction (RT-PCR).
As a stimulus, we used intraperitoneal (i.p.) injections of lithium chloride (LiCl) to induce c-Fos in the mouse amydgala. Lithium is widely used as a therapeutic drug for bipolar disorder. An acute injection of a high dose of LiCl induces physiological symptoms of malaise in animals such as diarrhea, and it is used as the unconditioned stimulus in conditioned taste aversion learning. The stimulation of chemoreceptive pathways by LiCl causes a high increase in c-Fos expression in specific brain regions such as the central amygdala (CeA) (Yamamoto et al., 1992; Swank, 1999; Spencer and Houpt, 2001; Kwon et al., 2008), the hypothalamic paraventricular nucleus, the parabrachial nucleus and the nucleus of the solitary tract (Yamamoto et al., 1992; Swank and Bernstein, 1994; Houpt et al., 1994; Jahng et al, 2004).
To confirm that the expression of the c-fos-lacZ transgene paralleled the expression of the endogenous c-Fos gene in the amygdala of the transgenic mice, the time course of their expression following LiCl administration was first investigated by immunohistochemistry and X-Gal staining. Co-localization between endogenous c-Fos and transgene expression in the brain was also examined by double labeling with fluorescent immunohistochemistry and X-Gal staining.
Upon determining the time course of c-fos-lacZ induction and the optimal staining and fixation conditions, β-Gal-positive single cells were microdissected out by LCM. Using RT-PCR after X-Gal staining, we confirmed c-Fos mRNA expression in β-Gal-positive single cells of the brain after LiCl treatment. This procedure makes it possible to easily screen expression of a set of genes in a population of c-Fos-specific cells.
2. Materials and methods
2.1 Animals
Transgenic mice (B6C3F1×B6D2) carrying a c-fos-lacZ fusion were generously provided by Dr. James Morgan (St. Jude Children’s Research Hospital). Mice were grouped or individually housed. For RNA preservation studies, adult male Sprague-Dawley rats (Charles River Laboratories, Wilmington, MA) were also used. All animals were housed under a 12-h light – 12-h dark cycle (lights on 07:00) at 25°C with free access to Purina rodent chow and distilled water. All procedures were conducted in the first half of the lights-on period. Anesthesia (halothane, sodium pentobarbital) was used to minimize pain and discomfort, and all experiments were approved by the Florida State University institutional animal care and use committee.
2.2 Detection of transgenic mice
Transgenic males were bred to C57BL/6J females. Offspring were weaned 3–4 weeks after birth. To screen the transgenic mice carrying the c-fos-lacZ fusion gene, a tail biopsy was performed by clipping 0.5 cm from each offspring at the time of weaning. Because the c-fos-lacZ transgene is highly expressed in skin and hair follicles of the tail (Smeyne et al., 1992b), the reporter protein, β-Gal, can be detected in the tail by overnight X-Gal staining at 37°C. After the X-Gal detection, the tails were processed for PCR to confirm the presence of the transgene. Genomic DNA was extracted from each tail and PCR was performed using HotStarTaq Master Mix Kit (Qiagen, Valencia, CA). Only mice that showed positive results from both X-Gal and PCR reactions were used in the present study.
2.3 Reverse transcription-polymerase chain reaction (RT-PCR)
RT-PCR was performed by using the OneStep RT-PCR Kit (Qiagen, Valencia, CA). For each RT-PCR reaction, 2 μl (~10 ng of total RNA for experiment 2) or 8.4 μl (~300 pg of total RNA for experiment 3) out of 40 μl of the total RNA extract from each sample was mixed with 4 μl of 5X buffer, 4 μl of 5X Q-solution, 0.8 μl of 10 mM dNTP mix, 0.4 μl of RNase Inhibitor, 1.6 μl of 10 μM primers (β-actin, sense 5′-TTGTAACCAACTGGGACGATATGG-3′, antisense 5′-GATCTTGATCTTCATGGTGCTAGG-3′; c-Fos, sense 5′GGAGCTGACAGATACGCTCCA-3′, antisense 5′-GCTAATGTTCTTGACCGGCTC-3′) and 0.8 μl of enzyme mix. Some RNase-free H2O was added to make 20 μl of total RT-PCR reaction volume. The RT reaction was performed at 50°C for 30 min first, and then PCR was started with an incubation of 95°C for 15 min. The PCR was performed by 40 cycles of 94°C for 60 s, 55°C for 60 s and 72°C for 60 s. The final incubation of the PCR was performed at 72°C for 5 min. RT-PCR products were visualized with SYBR gold (Invitrogen, Carlsbad, CA) in a 1.6% agarose gel.
2.4 Experiment 1: Time course of transgene expression after LiCl injection
2.4.1 Tissue collection
To compare the expression of the c-fos-lacZ transgene and endogenous c-Fos in the amygdala of transgenic mice after LiCl treatment, mice were injected with LiCl (0.15 M, 40 ml/kg, i.p.) or NaCl (0.15 M). At 1, 3, 6 and 9 h after LiCl or 1 h after NaCl treatment, mice (n=4 at each time point) were anesthetized with sodium pentobarbitol and perfused first with 50 ml of isotonic saline containing 0.5% sodium nitrate and 1000 U heparin, and then with 100 ml phosphate-buffered 4% paraformaldehyde (PFA). Their brains were dissected and cryoprotected in 30% sucrose at 4°C for one day. These time points were selected based on the time course of c-Fos mRNA and protein expression in the rat CeA (Spencer and Houpt, 2001).
2.4.2 Immunohistochemistry
Sections through the amygdalar region were cut at 40 μm on a freezing-sliding microtome and washed in 0.1 M phosphate-buffered saline (PBS) for 10 min twice. Alternate sections were processed for c-Fos immunohistochemistry or X-Gal staining. After PBS washes, sections were washed in 0.2% Triton-1% bovine serum albumin (BSA)-PBS for 30 min, washed in PBS-BSA for 10 min twice, and incubated with a c-Fos antibody (Ab-5, 1:20,000, Oncogene Research, La Jolla, CA) in PBA-BSA for 18 h at room temperature. Sections were then washed in PBS-BSA for 10 min twice, and incubated with a biotinylated goat anti-rabbit antibody (Vector Laboratories, Burlingame, CA) at a dilution of 1:200 in PBA-BSA for 1 h at room temperature. Antibody complexes were amplified using the Elite Vectastain ABC kit (Vector Laboratories, Burlingame, CA), and visualized by a 5-min reaction in 0.05% 3,3-diaminobenzidine tetrahydrochloride. Sections were immediately washed in 0.1 M phosphate buffer twice, mounted on gelatin-coated slides, counterstained with Methyl Green Nuclear Counterstain (Vector Laboratories, Burlingame, CA), and coverslipped with Permount.
2.4.3 X-Gal staining for β-Gal activity
Free-floating brain sections (40 μm) were stained in X-Gal reaction buffer (0.8% NaCl, 8 mM Na2HPO4, 2.7 mM KCl, 1.5 mM KH2PO4, 2 mM MgCl2, 35 mM K4[Fe(CN)6]·3H2O, 35 mM K3[Fe(CN)6], 0.02% Nonidet P-40, 0.01% deoxycholic acid) containing 1 mg/ml of X-Gal to visualize β-Gal activity. In order to increase the reaction rate, nitroblue tetrazolium (1 mg/ml) and phenazine methosulfate (0.02 mg/ml) were added in the X-Gal reaction buffer. The reaction was performed for 20 h at 37°C in darkness. Subsequent to X-Gal staining, sections were washed in 0.1 M phosphate buffer for 10 min twice. Sections were mounted on gelatin-coated slides, stained with Methyl Green Nuclear Counterstain (Vector Laboratories, Burlingame, CA), and coverslipped with Permount.
2.4.4 Quantification and statistical analysis
For the immunohistochemistry and X-Gal staining, cells expressing darkly-positive, nuclear staining were quantified with custom software (MindsEye, T. Houpt; Kwon et al. 2008). Regions were digitally-captured at 40× magnification on a Macintosh computer using an Olympus Provis AX-70 microscope with a Dage-MTI DC-330 CCD camera and Scion LG-3 framegrabber. Cells with dark nuclear staining were automatically detected and counted by the software across each image, based on the relative pixel darkness and circular symmetry of the nuclei relative to surrounding background tissue in the digitized image. To insure a consistent crtieria for the automatic counting, the same threshold parameters were used for all images from all treatments. Counting was restricted to the basolateral amygdala (BLA), CeA, or lateral amygdala (LA) as delineated by a hand-drawn outline. Bilateral cell counts were averaged for 6 sections of the amygdala for each mouse. The individual mean counts for each region were then averaged across mice within experimental groups. Significant effects across treatment groups were detected by one-way ANOVA and Neuman-Keuls post-hoc tests (Kaleidagraph, Synergy Software). All data are presented as the mean ± standard error of the mean.
2.4.5 Co-localization between c-Fos- and β-Gal-positive cells
In order to determine whether the expression of the c-fos-lacZ transgene is co-localized with endogenous c-Fos, brain sections were double-labeled with a c-Fos antibody and X-Gal staining after LiCl treatment. Transgenic mice were injected with LiCl (0.15 M, 40 ml/kg, i.p.). One hour later, their brains were dissected out and immediately frozen with dry ice. The frozen brains were sectioned at 8 μm and mounted on slides in a −20°C cryostat using the CryoJane tape transfer system (Instrumedics, Hackensack, NJ). The frozen sections were briefly defrosted and then fixed with 4% PFA for 5 min at room temperature. Brain sections on slides were washed in 0.1 M PBS for 5 min twice and then stained in X-Gal reaction buffer described above containing 1 mg/ml of X-Gal overnight at 37°C in darkness. After X-Gal staining, sections were washed in 0.1 M PBS for 10 min twice, in 0.2% Triton-1% BSA-PBS for 30 min and in PBS/BSA for 10 min twice. Sections were then incubated with a c-Fos antibody (Ab-5, 1:10,000, Oncogene Research, La Jolla, CA) in PBS-BSA overnight at room temperature. Sections were washed in PBS-BSA for 10 min twice and incubated with a secondary antibody (fluorescein isothiocyanate-conjugated goat anti-rabbit, 1:200, Jackson ImmunoResearch, West Grove, PA) in PBS-BSA for 1.5 h at room temperature in darkness. Sections were washed in PBS for 10 min twice and coverslipped with Vectashield mounting medium (Vector Laboratories, Burlingame, CA) in darkness. Double-labeled sections were examined and photographed under a fluorescence microscope (Nikon AZ 100M, Nikon, Tokyo, Japan).
2.5 Experiment 2: RNA utilization after different fixations
2.5.1 Tissue collection
The time course of RNA degradation was assessed after different fixations. Rats were anesthetized with sodium pentobarbital and decapitated, and then their brains were dissected out. The brains were emmersed in M-1 Embedding Matrix (Shandon, Pittsburgh, PA), immediately frozen with dry ice, and stored at −80°C until sectioning. The frozen brains were cut at 8 μm and mounted on slides in a −20°C cryostat using the CryoJane tape transfer system (Instrumedics, Hackensack, NJ).
2.5.2 RT-PCR after different fixations
Brain sections were fixed with 4% PFA, 70% ethanol, 95% ethanol, acetone, or methanol for 5 min at room temperature. After fixation, brain sections were incubated in 0.1% diethylpyrocarbonate (DEPC)-treated distilled H2O (DEPC-dH2O) for 0, 3 or 20 h at room temperature. Additional brain sections were fixed with 4% PFA for 5 min and then treated with proteinase K (200 μg/ml) at 37°C overnight. A small amount of tissue was scraped from sections after each condition using a sterile micropipette tip. Total RNA was extracted from the tissue scrape with the Total RNA Microprep Kit (Stratagene, La Jolla, CA) according to manufacturer’s protocols. RT-PCR using β-actin primers was performed as described above to confirm the presences of RNA.
The integrity of total RNA extracted from fixed brain sections at each time point was examined in formaldehyde RNA gels. Due to very tiny amount of total RNA (less than 100 ng each sample), however, RNA was hardly visualized in gels. Instead, each 50 mg of frozen brain tissue was collected and fixed with 4% PFA, 70% ethanol, 95% ethanol, acetone, or methanol for 1 h on ice. After fixation, brain tissue was incubated in DEPC-dH2O for 0, 3 or 20 h at room temperature. Additional brain tissue was also fixed with 4% PFA for 1 h and then treated with proteinase K (200 μg/ml) at 37°C overnight. Total RNA was extracted from each tissue sample by using TRIzol (Invitrogen, Carlsbad, CA) according to manufacturer’s protocols. Five micrograms of total RNA per each sample except PFA and PFA-proteinase K samples (less than 0.5 μg) were visualized with SYBR safe (Invitrogen, Carlsbad, CA) in formaldehyde RNA gels.
2.5.3 Quality of X-Gal staining after different fixations
To examine the quality of X-Gal staining after each different fixation, frozen brain sections from LiCl-treated transgenic mice were prepared as described in experiment 3. Brain sections were fixed with 4% PFA, 70% ethanol, 95% ethanol, acetone, or methanol for 5 min at room temperature and then stained overnight in X-Gal reaction buffer containing 1 mg/ml of X-Gal as described above. Subsequent to X-Gal staining, sections were stained with Methyl Green Nuclear Counterstain (Vector Laboratories, Burlingame, CA) and coverslipped with Permount. The quality of X-Gal staining performed after each different fixation was examined under a microscope.
2.5.4 RT-PCR after X-Gal staining
X-Gal solution was also tested to confirm RNA preservation. Frozen rat brain sections were prepared as described above. Brain sections were fixed with 70% ethanol for 5 min at room temperature and washed with DEPC-dH2O for 5 min. Brain sections were then stained in X-Gal reaction buffer containing 1 mg/ml of X-Gal in DEPC-dH2O for 16 h at 37°C. The cortex area of brain sections was scratched once using a pipette tip after X-Gal staining and total RNA was extracted from the scratch with the Total RNA Microprep Kit (Stratagene, La Jolla, CA) according to manufacturer’s protocols. RT-PCR using β-actin primers was performed as described above.
2.6 Experiment 3: Detection of mRNA expression in β-Gal-positive cells after LCM
2.6.1 Tissue collection
In order to confirm mRNA expression and preservation in β-Gal-positive cells, we performed RT-PCR after LCM of single cells. Transgenic mice were injected with LiCl (0.15 M, 40 ml/kg, i.p.). One hour later, they were anesthetized with sodium pentobarbitol, decapitated, and the brains were dissected. The brains were emmersed in M-1 Embedding Matrix (Shandon, Pittsburgh, PA), immediately frozen with dry ice, and stored at −80 °C freezer until used.
2.6.2 X-Gal staining for β-Gal activity
The frozen brains were sectioned at 8 μm and mounted on slides in a −20°C cryostat using the CryoJane tape transfer system (Instrumedics, Hackensack, NJ). The sections were defrosted at room temperature for 30 s and then briefly washed with DEPC-dH2O. The brain sections were fixed in 70% ethanol at room temperature for 5 min and then washed with DEPC-dH2O for 5 min. Brain sections were then stained as above in X-Gal reaction buffer containing 1 mg/ml of X-Gal in DEPC-dH2O. The reaction was performed for 16 h at 37°C in darkness.
2.6.3 LCM of single cells
Subsequent to X-Gal staining, the sections were washed in DEPC-dH2O for 5 min and dehydrated by the following procedure: 70% ethanol for 30 s, 95% ethanol for 30 s, 100% ethanol for 30 s, xylene for 5 min, air dried in a fume hood for 15 min, and then desiccated with Drierite (W.A. Hammond Drierite, Xenia, OH) for 1 h. X-Gal stained cells in the CeA, cortex and hippocampus were microdissected using the PixCell II LCM system (Arcturus, Mountain view, CA). A 7 um diameter laser spot, a 10 ms laser pulse duration, and a 85 mW laser power were used for the microdissection. Cells from cortex and hippocampus were included as positive controls, because those areas have previously been reported to have high levels of c-Fos and lacZ expression (Smeyne et al., 1992b and 1993). Total RNA was extracted from the LCM samples with the Total RNA Microprep Kit (Stratagene, La Jolla, CA) according to the manufacturer’s protocols. RT-PCR using primer pairs for c-Fos and β-actin was performed by using the OneStep RT-PCR Kit (Qiagen, Valencia, CA) as described above. The total RNA extract from each LCM sample of single cells was used in each RT-PCR reaction (15–20 single cells for β-actin and 30–40 single cells for c-Fos).
3. Results
3.1 Experiment 1: Time course of transgene expression after LiCl injection
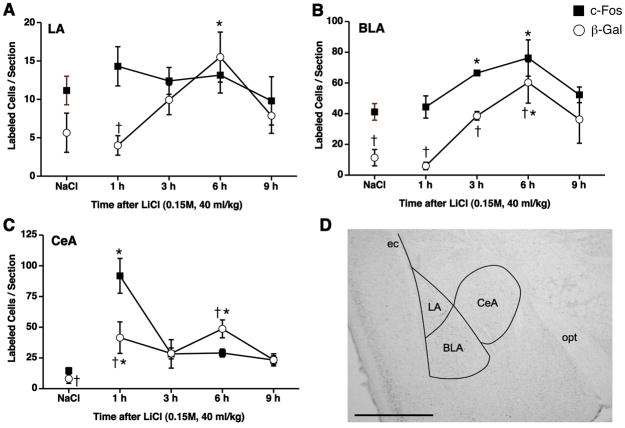
The parallel expression of the c-fos-LacZ transgene and c-Fos in the amygdala of the transgenic mice after LiCl treatment was investigated by X-Gal staining and c-Fos immunohistochemistry at different time points (1, 3, 6 and 9 h). Compared to NaCl, LiCl increased the numbers of c-Fos- and β-Gal-positive cells in the CeA and BLA. Although there were some significant differences among groups in the LA, the absolute numbers of c-Fos- and β-Gal-positive cells in the LA were very low (<20 cells/section) at every time point (Figure 3A).
Figure 3.
Quantification of c-Fos- and β-Gal-positive cells in the LA (A), BLA (B) and CeA (C) 1, 3, 6 and 9 h following LiCl (i.p., 0.15 M, 40 ml/kg) and 1 h following NaCl administrations. LiCl increased c-Fos-positive cells in the CeA at 1 h and BLA at 3 and 6 h. LiCl also increased β-Gal-positive cells in the CeA at 1 and 6 h and BLA and LA at 6 h. β-Gal-positive cells were lower than c-Fos-positive cells in the CeA at 1 h and BLA at 6 h. * p <0.05 vs. NaCl, † p < 0.05 vs. c-Fos. n = 4 per group. A representative coronal section of the mouse brain showing outlines within which c-Fos- and β-Gal-positive cells were quantified (D). BLA, basolateral amygdala; CeA, central amygdala; ec, external capsule; LA, lateral amygdala; opt, optic tract. Scale bar in (D), 1 mm.
In the BLA, the number of c-Fos-positive cells increased slowly in the BLA to 185±29% of NaCl at 6 h (Figure 1 and 3B). The number of c-Fos-positive cells after LiCl was not different from NaCl by 9 h. The number of β-Gal-positive cells was also increased by LiCl compared to NaCl in the BLA at 6 h (Figure 1 and 3B). The increased number of β-Gal-positive cells came back to the level of NaCl in the BLA at 9 h. The number of B-Gal-positive cells was significantly lower than the number of c-Fos-positive cells after NaCl and after LiCl at 1, 3 and 6 h.
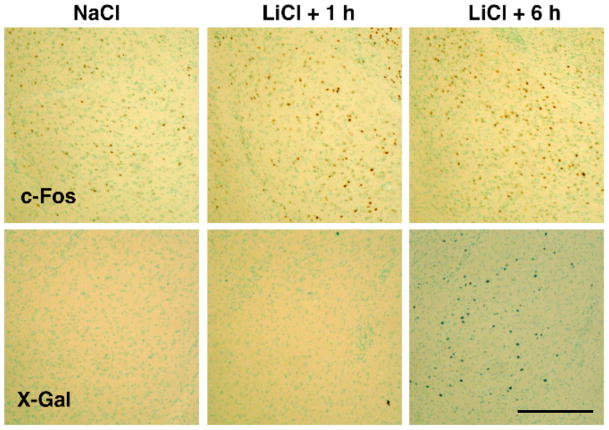
Figure 1.
Photomicrographs of c-Fos immunohistochemistry (top panels) and X-Gal staining (bottom panels) in the BLA 1 and 6 h following LiCl (i.p., 0.15 M, 40 ml/kg) or 1 h following NaCl administration. Scale bar, 300 μm.
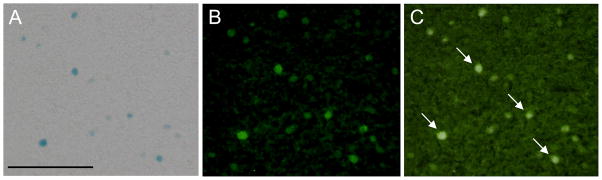
In the CeA, the number of c-Fos-positive cells increased rapidly after LiCl injection to 631±97% of NaCl at 1 h (Figure 2 and 3C). The number of c-Fos-positive cells after LiCl was not different from NaCl by 3 h. The number of β-Gal-positive cells was also increased by LiCl compared to NaCl in the CeA at 1 h and 6 h (Figure 2 and 3C). However, the number of β-Gal-positive cells was significantly lower than the number of c-Fos-positive cells at 1 h after LiCl and higher at 6 h after LiCl. Almost all β-Gal-positive cells were double-labeled with c-Fos-positive cells in the CeA (Figure 4), cortex and hippocampus (not shown) 1 h after LiCl, indicating that detection of transgene expression by X-Gal staining can be used for detection of endogenous c-Fos expression in the brain.
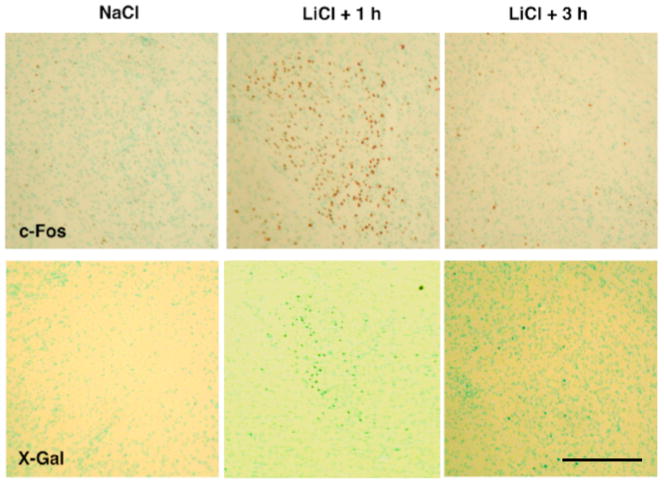
Figure 2.
Photomicrographs of c-Fos immunohistochemistry (top panels) and X-Gal staining (bottom panels) in the CeA 1 and 3 h following LiCl (i.p., 0.15 M, 40 ml/kg) or 1 h following NaCl administration. Scale bar, 300 μm.
Figure 4.
Photomicrographs of β-Gal-positive (blue, A) and fluorescently-labeled c-Fos-positive (green, B) cells and a merged image (C) in the CeA 1 h after a LiCl injection. Arrows in (C) indicate β-Gal and c-Fos double-labeled cells. Scale bar, 100 μm in (A).
The effects of LiCl treatment on X-Gal staining and c-Fos induction at 4 different time points were compared to expression 1 h after a control injection of NaCl. Because the effects of control injections of NaCl were not examined at later time points, it is possible that differences observed at 3 h and 6 h reflect handling and injection stress, and not a specific effect of LiCl. Nonetheless, X-Gal staining and c-Fos induction were generally parallel, suggesting coordinate regulation of c-fos and c-fos-LacZ genes.
Thus, the two genes have a common promoter region, showed a similar expression in the CNS (Schilling et al., 1991; Smeyne et al., 1992b and 1993), and β-Gal was co-expressed with c-Fos. However, the number of β-Gal-positive cells was generally less than that of c-Fos-positive cells in the amygdala at baseline (after NaCl injection) and after induction by LiCl administration.
3.2 Experiment 2: RNA utilization after different fixations
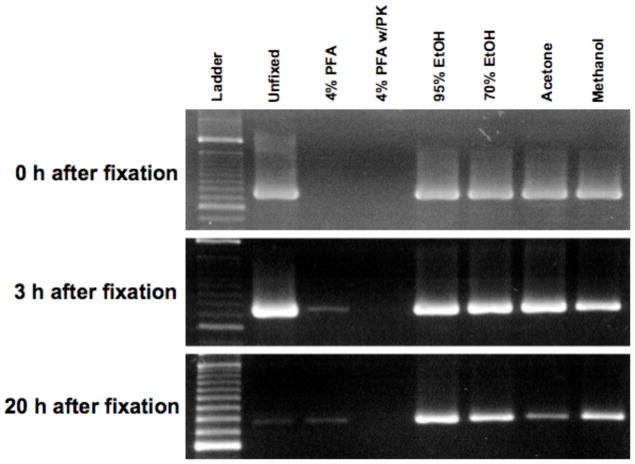
To determine the optimal conditions of fixation for RNA preservation prior to single cell LCM, RNA utilization after different fixations was assessed by RT-PCR with β-actin primers. RT-PCR product was amplified from unfixed control tissue after 0 and 3 h incubation in DEPC-dH2O, but only barely after 20 h incubation (Figure 5), indicating that RNA was almost degraded between 3 and 20 h.
Figure 5.
RT-PCR products of β-actin after different fixations. Brain sections (8 μm) were fixed with DEPC-dH2O (unfixed), 4% PFA, 70% ethanol, 95% ethanol, acetone, or methanol for 5 min at room temperature. Some brain sections fixed with 4% PFA were also treated with proteinase K overnight. After fixations, brain sections were incubated in DEPC-dH2O for 0, 3 and 20 h at room temperature. RT-PCR using β-actin primers was performed with total RNA extracted from each section scratch at the time points. β-actin mRNA was well amplified in the ethanol-, methanol- and acetone-fixed sections at all time points, indicating mRNA preservation sufficient for utilization in RT-PCR. PFA, paraformaldehyde; PK, proteinase K; EtOH, ethanol.
β-actin mRNA was not amplified immediately after a brief 4% PFA fixation, although a faint RT-PCR product was seen after 3 and 20 h incubation in DEPC-dH2O after 4% PFA fixation. It has been previously reported that mRNA can be amplified from PFA-fixed tissue after proteinase K treatment (Davies et al., 1996; Specht et al., 2001). In the current study, however, proteinase K treatment did not rescue mRNA at any time point after 4% PFA fixation.
Robust RT-PCR amplification was seen after ethanol, methanol and acetone fixations even after 0, 3 and 20 h incubation in DEPC-dH2O (Figure 5), indicating good utilization of RNA with these fixatives.
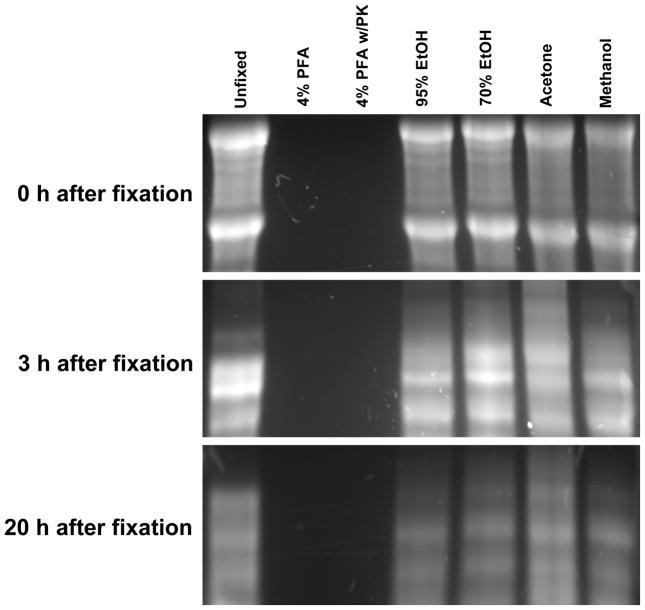
Ethanol-, acetone-, methanol- and un-fixed samples showed intact RNA at 0 h, but showed partially degraded RNA at 3 and 20 h (Figure 6). Because very small amount of total RNA (less than 1 μg RNA form 50 mg brain tissue) was extracted from 4% PFA and 4% PFA-proteinase K treated tissue at all time points, the RNA was scarcely visualized in gels.
Figure 6.
Total RNA isolated from brain tissue after different fixations. Brain tissue (50 mg each) was fixed with DEPC-dH2O (unfixed), 4% PFA, 70% ethanol, 95% ethanol, acetone, or methanol for 1 h on ice. Some brain tissue fixed with 4% PFA was also treated with proteinase K overnight. After fixations, brain tissue was incubated in DEPC-dH2O for 0, 3 and 20 h at room temperature. Total RNA was extracted at the time points and visualized in formaldehyde RNA gels. 5 μg per each sample except PFA and PFA-proteinase K samples (less than 0.5 μg) were loaded. Ethanol-, acetone-, methanol- and un-fixed samples showed intact RNA at 0 h, but showed partially degraded RNA at 3 and 20 h. Total RNA from PFA and PFA-proteinase K samples was scarcely visualized at all time points. PFA, paraformaldehyde; PK, proteinase K; EtOH, ethanol.
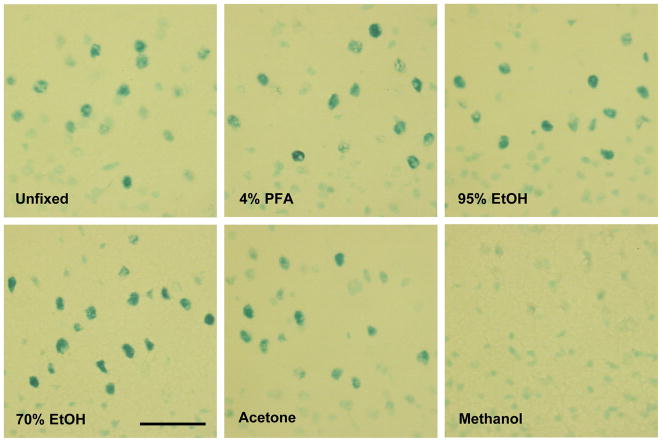
To find the best fixation for X-Gal staining that could be combined with RT-PCR, brain sections from LiCl-treated transgenic mice were processed for X-Gal staining after each fixation. All the fixatives except methanol showed a proper quality of X-Gal staining. However, 70% ethanol allowed the best quality of X-Gal staining among the fixatives (Figure 7). β-actin mRNA was well amplified after X-Gal staining with 70% ethanol fixation (not shown). The result of RT-PCR following X-Gal staining confirmed that mRNA is still usable for RT-PCR amplification after X-Gal staining.
Figure 7.
Photomicrographs of X-Gal staining after different fixations in the cortex 1 h following LiCl (0.15 M, 40 ml/kg, i.p.). PFA, paraformaldehyde; EtOH, ethanol. Scale bar, 50 μm.
3.3 Experiment 3: Detection of mRNA expression in β-Gal-positive cells after LCM
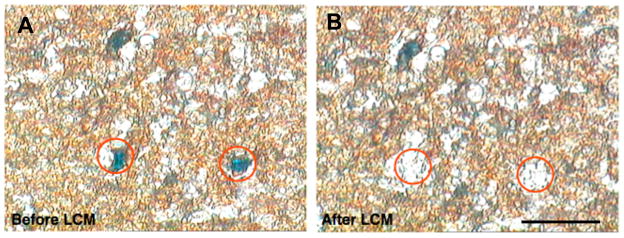
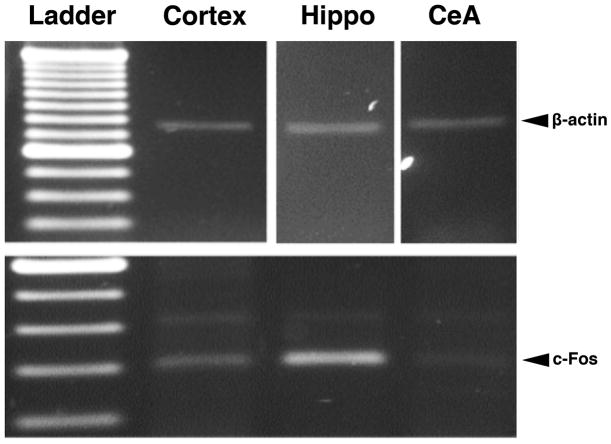
mRNA expression of c-fos and β-actin genes in c-Fos-specific cells was examined after X-Gal staining, single-cell LCM and RT-PCR. Brains of transgenic mice were dissected out 1 h after LiCl injection. Single β-Gal-positive cells in the CeA, cortex and hippocampus were successfully captured by LCM (Figure 8). Using RT-PCR, c-Fos and β-actin mRNA was amplified from total RNA extracted from X-Gal stained cells in the cortex, hippocampus and CeA (Figure 9).
Figure 8.
Laser capture microdissection (LCM) of β-Gal-positive single cells in the CeA. An X-Gal stained and dehydrated 8 μm brain section of a LiCl-treated transgenic mouse is shown before (A) and after (B) microdissection of β-Gal-positive single cells in the CeA. Blue spots in circles indicate β-Gal-positive cells (A). Scale bar, 20 μm.
Figure 9.
RT-PCR products of c-fos (bottom panel) and β-actin (top panels) after LCM of β-Gal-positive single cells. Brain sections (8 μm) from a LiCl-treated transgenic mouse were fixed with 70% ethanol and then stained in X-Gal reaction buffer for 16 h at 37°C. β-Gal-positive single cells were microdissected in the cortex, hippocampus and CeA. The total RNA extract from each LCM sample of single cells (15–20 single cells for β-actin and 30–40 single cells for c-Fos) was used in each RT-PCR reaction. The amplified c-fos (bottom panel) and β-actin (top panels) bands confirmed mRNA expression of c-fos and β-actin genes in LiCl-induced β-Gal-positive cells in the CeA, cortex and hippocampus of the transgenic mice. CeA, central amygdala; Hippo, hippocampus.
4. Discussion
In the present study, we employed a c-fos-lacZ transgenic mouse line to demonstrate RT-PCR expression analysis in microdissected brain cells identified as c-Fos-positive by X-Gal staining. To establish the fidelity of the reporter transgene, the time course of endogenous c-Fos and the c-fos-lacZ transgene expression in the amygdala induced by LiCl administration was investigated by immunohistochemistry and X-Gal staining. Moreover, co-localization between c-Fos- and β-Gal-positive cells in the brain was confirmed by double labeling with fluorescent immunohistochemistry and X-Gal staining. To ensure that RNA was preserved in X-Gal stained tissue sections, different fixations were examined, with the conclusion that ethanol fixation was best for both RNA preservation and X-Gal staining quality. Finally, in combining X-Gal staining, single-cell LCM and RT-PCR, we confirmed mRNA expression of endogenous c-fos and β-actin genes in LiCl-induced β-Gal-positive cells in the CeA, cortex and hippocampus.
Although X-Gal staining may be advantageous for LCM and RT-PCR, the utility of lacZ reporter transgenes is compromised if the fidelity of the reporting is not perfect. As previously reported in mice (Swank, 1999) and rats (Yamamoto et al., 1992; Spencer and Houpt, 2001; Kwon et al., 2008), LiCl increased the numbers of c-Fos-positive cells in the CeA and BLA, and expression of β-Gal was increased in parallel. Moreover, all β-Gal-positive-cells also expressed endogenous c-Fos in the CeA after LiCl stimulation. However, the number of β-Gal-positive cells was much less than that of c-Fos-positive cells, especially in the CeA. Therefore, we assume that only a subpopulation of the c-Fos-positive cells in the amygdala was identified by X-Gal staining after LiCl stimulation. In contrast to the amygdala, in the cortex and hippocampus β-Gal expression closely corresponded to endogenous c-Fos expression after LiCl (present study, data not shown). Similarly, Smeyne et al. (1992b) noted that much less expression of the transgene was observed in the dentate gyrus of c-fos-lacZ transgenic mice after induction of seizure by pentylenetetrazol, although seizure by kainic acid stimulation exhibited same expression patterns between the endogenous c-Fos and c-fos-lacZ transgene. The c-fos-lacZ transgene was thought to have the same expression amount and pattern with the endogenous c-fos gene in the transgenic mice because the c-fos-lacZ transgene has the same transcriptional regulatory elements including the CRE site as the endogenous c-fos gene. Some possible explanations for the discrepancy between the numbers of c-Fos- and β-Gal-positive cells include: 1) an integration effect: the c-fos-lacZ transgene might be incorporated into a less accessible part of the genome, and so respond less vigorously in the amygdala than, e.g., the cortex and hippocampus. 2) altered rates of synthesis or degradation: Due to the shortened promoter or different structure of the transgene, c-fos-lacZ and native c-fos might have different rates of transcription, translation, or degradation. or 3) a methodological difference: X-Gal staining might be less sensitive than immunohistochemistry. Although a small discrepancy in expression occurs in the amygdala, however, the constitutive and stimulated expression of the c-fos-lacZ transgene in the CNS accurately reflects the expression of endogenous c-Fos in transgenic mice (Smeyne et al., 1992a, 1992b and 1993).
We found no large differences in the time course of c-Fos and X-gal activity in the amygdala following LiCl, although there may be differences in the half-life of c-Fos and the β-Gal fusion gene. Specifically, Schilling et al. (1991) reported, in transfected neuroblastoma cells, that the time course for induction of the c-fos-lacZ gene and synthesis of the β-Gal reporter protein was similar to the induction and synthesis of endogenous c-Fos. However, the half-life of the β-Gal protein was longer than endogenous c-Fos. Similarily, in fos-lacZ transgenic mice (Smeyne et al. 1992b), systemic administration of the convulsants kainic acid and pentylenetetrazol rapidly induced expression of both c-Fos and β-Gal within 30 min, but β-Gal levels, although declining rapidly, did not return to baseline for many hours. Thus it appears that the 3.5 kb portion of the c-fos promoter is sufficient to confer rapid induction of the fos-lacZ reporter gene that parallels endogenous c-Fos induction. The β-Gal fusion protein has a longer half-life than the endogenous c-Fos protein, however, presumably because the c-Fos protein contains regulatory regions that confer tighter regulation of protein half-life.
In order to utilize β-Gal-positive cells for RT-PCR, we examined RNA preservation after different fixations for X-Gal staining as measured with RT-PCR of β-actin. The unfixed control showed good mRNA amplification for 3 h, but a poor mRNA amplification at 20 h, indicating that RNA is not usable when kept in DEPC-dH2O at room temperature for 20 h. Consistent with these reports (Goldsworthy et al., 1999; Kohda et al., 2000), we could amplify mRNA with ethanol, methanol and acetone fixations and incubation in DEPC-dH2O at least for 20 h. These organic coagulants fix tissue by precipitating of proteins, which leaves the mRNA available for RT-PCR amplification. In contrast, we were unable to extract proper amount of RNA from 4% PFA-fixed tissue and detected a poor mRNA amplification at all time points. Because PFA preserves tissue by cross-linking of proteins, proteinase K treatment of PFA-fixed tissue has been used to dissociate proteins cross-linked with mRNA prior to RT-PCR (Davies et al., 1996; Specht et al., 2001). However, we were unable to utilize RNA extracted from 4% PFA-fixed tissue followed by proteinase K treatment for RT-PCR amplification. It is possible that proteinase K treatment hardly dissociates the protein cross-linking in PFA-fixed tissue, which may make RNA extraction impossible. Surprisingly, even though RNA degradation partially occurred after 3 and 20 h incubation in DEPC-dH2O after ethanol, methanol and acetone fixations, robust RT-PCR amplifications were performed at all time points after these fixations. Importantly, we also determined that among the different fixatives ethanol fixation was best for ensuring quality X-Gal staining.
Having determined the pattern of reporter gene expression and the optimal fixation and staining conditions for preserving RNA, we microdissected β-Gal-positive cells in the CeA, cortex and hippocampus after LiCl administration and detected mRNA for c-fos and β-actin genes by RT-PCR. Others have analyzed gene expression in immuno-stained single cells using immunohistochemistry, LCM and RT-PCR after acetone fixation (e.g. Lu et al., 2004; Macdonald et al., 2008). The procedure of X-Gal staining, however, is more convenient and simpler than that of immunohistochemistry, and allows for better RNA preservation during processing. Because the bacterial lacZ gene is widely used as a reporter gene in transgenic mice to demonstrate tissue- or cell-specific gene expression, detection of β-Gal along with other reporters such as green fluorescent protein can be used to identify specific subpopulations of cells in which the controlling promoter is active (Wong et al., 2004; Bhattacherjee et al., 2004). Combining LCM and transgenic reporter genes provides a powerful tool with which to investigate tissue- or cell-specific gene expression.
Acknowledgments
This work was supported by National Institute on Deafness and other Communication Disorders grant R01DC03198 (TAH) and a research grant from the B.W. Robinson Memorial Endowment for the Neurosciences (BSK). We thank Dr. Frank Johnson for access to the PixCell II system.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Basheer R, Sherin JE, Saper CB, Morgan JI, McCarley RW, Shiromani PJ. Effects of sleep on wake-induced c-fos expression. J Neurosci. 1997;17:9746–50. doi: 10.1523/JNEUROSCI.17-24-09746.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacherjee V, Mukhopadhyay P, Singh S, Roberts E, Hackmiller R, Greene R, Pisano M. Laser capture microdissection of fluorescently labeled embryonic cranial neural crest cells. Genesis. 2004;39:58–64. doi: 10.1002/gene.20026. [DOI] [PubMed] [Google Scholar]
- Bonner RF, Emmert-Buck M, Cole K, Pohida T, Chuaqui R, Goldstein S, Liotta LA. Laser capture microdissection: molecular analysis of tissue. Science. 1997;278:1481–1483. doi: 10.1126/science.278.5342.1481. [DOI] [PubMed] [Google Scholar]
- Bravo R, Neuberg M, Burckhardt J, Almendral J, Wallich R, Müller R. Involvement of common and cell type-specific pathways in c-fos gene control: stable induction of cAMP in macrophages. Cell. 1987;48:251–60. doi: 10.1016/0092-8674(87)90428-4. [DOI] [PubMed] [Google Scholar]
- Chiu R, Boyle WJ, Meek J, Smeal T, Hunter T, Karin M. The c-Fos protein interacts with c-Jun/AP-1 to stimulate transcription of AP-1 responsive genes. Cell. 1988;54:541–2. doi: 10.1016/0092-8674(88)90076-1. [DOI] [PubMed] [Google Scholar]
- Crosio C, Cermakian N, Allis CD, Sassone-Corsi P. Light induces chromatin modification in cells of the mammalian circadian clock. Nat Neurosci. 2000;3:1241–7. doi: 10.1038/81767. [DOI] [PubMed] [Google Scholar]
- Davies GN, Bevan IS, Lundemose JB, Smith H, Sweet C. Use of proteinase K for RT-PCR of cytokine mRNA in formalin fixed tissue. Clin Mol Pathol. 1996;49:M364–M367. doi: 10.1136/mp.49.6.m364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fend F, Emmert-Buck MR, Chuaqui R, Cole K, Lee J, Liotta LA, Raffeld M. Immuno-LCM: laser capture microdissection of immunostained frozen sections for mRNA analysis. Am J Pathol. 1999;154:61–6. doi: 10.1016/S0002-9440(10)65251-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisch TM, Prywes R, Simon MC, Roeder RG. Multiple sequence elements in the c-fos promoter mediate induction by cAMP. Genes Dev. 1989;3:198–211. doi: 10.1101/gad.3.2.198. [DOI] [PubMed] [Google Scholar]
- Foletta VC. Transcription factor AP-1 and the role of Fra-2. Immunol Cell Bio. 1996;74:121–133. doi: 10.1038/icb.1996.17. [DOI] [PubMed] [Google Scholar]
- Goldsworthy SM, Stockton PS, Trempus CS, Foley JF, Maronpot RR. Effects of fixation on RNA extraction and amplification from laser capture microdissected tissue. Mol Carcinog. 1999;25:86–91. [PubMed] [Google Scholar]
- Grimm R, Schicknick H, Riede I, Gundelfinger ED, Herdegen T, Zuschratter W, Tischmeyer W. Suppression of c-fos induction in rat brain impairs retention of a brightness discrimination reaction. Learn Mem. 1997;3:402–413. doi: 10.1101/lm.3.5.402. [DOI] [PubMed] [Google Scholar]
- Guzowski J. Insights into immediate-early gene function in hippocampal memory consolidation using antisense oligonucleotide and fluorescent imaging approaches. Hippocampus. 2002;12:86–104. doi: 10.1002/hipo.10010. [DOI] [PubMed] [Google Scholar]
- Halazonetis TD, Georgopoulos K, Greenberg ME, Leder P. c-Jun dimerizes with itself and with c-Fos, forming complexes of different DNA binding affinities. Cell. 1988;55:917–24. doi: 10.1016/0092-8674(88)90147-x. [DOI] [PubMed] [Google Scholar]
- He J, Yamada K, Nabeshima T. A role of Fos expression in the CA3 region of the hippocampus in spatial memory formation in rats. Neuropsychopharmacology. 2002;26:259–68. doi: 10.1016/S0893-133X(01)00332-3. [DOI] [PubMed] [Google Scholar]
- Herrera DG, Robertson HA. Activation of c-fos in the brain. Prog Neurobiol. 1996;50:83–107. doi: 10.1016/s0301-0082(96)00021-4. [DOI] [PubMed] [Google Scholar]
- Hoffman GE, Smith MS, Verbalis JG. c-Fos and related immediate early gene products as markers of activity in neuroendocrine systems. Frontiers in Neuroendocrinology. 1993;14:173–213. doi: 10.1006/frne.1993.1006. [DOI] [PubMed] [Google Scholar]
- Houpt TA, Philopena JM, Wessel TC, Joh TH, Smith GP. Increased c-fos expression in nucleus of the solitary tract correlated with conditioned taste aversion to sucrose in rats. Neurosci Lett. 1994;172:1–5. doi: 10.1016/0304-3940(94)90648-3. [DOI] [PubMed] [Google Scholar]
- Huff N. Amygdala regulation of immediate-early gene expression in the hippocampus induced by contextual fear conditioning. J Neurosci. 2006;26:1616–1623. doi: 10.1523/JNEUROSCI.4964-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahng JW, Lee JH, Lee S, Lee JY, Kim GT, Houpt TA, Kim DG. N(omega)-nitro-L-arginine methyl ester attenuates lithium-induced c-Fos, but not conditioned taste aversion, in rats. Neurosci Res. 2004;50:485–92. doi: 10.1016/j.neures.2004.08.016. [DOI] [PubMed] [Google Scholar]
- Karin M, Liu Z-G, Zandi E. AP-1 function and regulation. Curr Opinion Cell Bio. 1997;9:240–246. doi: 10.1016/s0955-0674(97)80068-3. [DOI] [PubMed] [Google Scholar]
- Kohda Y, Murakami H, Moe OW, Star RA. Analysis of segmental renal gene expression by laser capture microdissection. Kidney Int. 2000;57:321–31. doi: 10.1046/j.1523-1755.2000.00824.x. [DOI] [PubMed] [Google Scholar]
- Kovács KJ. c-Fos as a transcription factor: a stressful (re)view from a functional map. Neurochem Int. 1998;33:287–97. doi: 10.1016/s0197-0186(98)00023-0. [DOI] [PubMed] [Google Scholar]
- Kruijer W, Schubert D, Verma IM. Induction of the proto-oncogene fos by nerve growth factor. Proc Natl Acad Sci USA. 1985;82:7330–4. doi: 10.1073/pnas.82.21.7330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon B, Goltz M, Houpt TA. Expression of AP-1 family transcription factors in the amygdala during conditioned taste aversion learning: role for Fra-2. Brain Res. 2008;1207:128–41. doi: 10.1016/j.brainres.2008.01.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamprecht R, Dudai Y. Differential modulation of brain immediate early genes by intraperitoneal LiCl. Neuroreport. 1995;7:289–93. [PubMed] [Google Scholar]
- Lamprecht R, Dudai Y. Transient expression of c-Fos in rat amygdala during training is required for encoding conditioned taste aversion memory. Learn Mem. 1996;3:31–41. doi: 10.1101/lm.3.1.31. [DOI] [PubMed] [Google Scholar]
- Lu L, Neff F, Dun Z, Hemmer B, Oertel WH, Schlegel J, Hartmann A. Gene expression profiles derived from single cells in human postmortem brain. Brain Res Brain Res Protoc. 2004;13:18–25. doi: 10.1016/j.brainresprot.2003.12.003. [DOI] [PubMed] [Google Scholar]
- Macdonald JA, Murugesan N, Pachter JS. Validation of immuno-laser capture microdissection coupled with quantitative RT-PCR to probe blood-brain barrier gene expression in situ. J Neurosci Methods. 2008;174:219–26. doi: 10.1016/j.jneumeth.2008.07.009. [DOI] [PubMed] [Google Scholar]
- Mickley GA, Kenmuir CL, McMullen CA, Yocom AM, Valentine EL, Dengler-Crish CM, Weber B, Wellman JA, Remmers-Roeber DR. Dynamic processing of taste aversion extinction in the brain. Brain Res. 2004;1016:79–89. doi: 10.1016/j.brainres.2004.04.071. [DOI] [PubMed] [Google Scholar]
- Pagedar NA, Wang W, Chen DH, Davis RR, Lopez I, Wright CG, Alagramam KN. Gene expression analysis of distinct populations of cells isolated from mouse and human inner ear FFPE tissue using laser capture microdissection--a technical report based on preliminary findings. Brain Res. 2006;1091:289–99. doi: 10.1016/j.brainres.2006.01.057. [DOI] [PubMed] [Google Scholar]
- Rauscher FJ, Voulalas PJ, Franza BR, Curran T. Fos and Jun bind cooperatively to the AP-1 site: reconstitution in vitro. Genes Dev. 1988;2:1687–99. doi: 10.1101/gad.2.12b.1687. [DOI] [PubMed] [Google Scholar]
- Sassone-Corsi P, Visvader J, Ferland L, Mellon PL, Verma IM. Induction of proto-oncogene fos transcription through the adenylate cyclase pathway: characterization of a cAMP-responsive element. Genes Dev. 1988;2:1529–38. doi: 10.1101/gad.2.12a.1529. [DOI] [PubMed] [Google Scholar]
- Schilling K, Luk D, Morgan JI, Curran T. Regulation of a fos-lacZ fusion gene: a paradigm for quantitative analysis of stimulus-transcription coupling. Proc Natl Acad Sci USA. 1991;88:5665–9. doi: 10.1073/pnas.88.13.5665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutze K, Lahr G. Identification of expressed genes by laser-mediated manipulation of single cells. Nat Biotechnol. 1998;16:737–742. doi: 10.1038/nbt0898-737. [DOI] [PubMed] [Google Scholar]
- Sluka P, O’Donnell L, McLachlan RI, Stanton PG. Application of laser-capture microdissection to analysis of gene expression in the testis. Prog Histochem Cytochem. 2008;42:173–201. doi: 10.1016/j.proghi.2007.10.001. [DOI] [PubMed] [Google Scholar]
- Sluka P. Stage-specific expression of genes associated with rat spermatogenesis: characterization by laser-capture microdissection and real-time polymerase chain reaction. Biol Reprod. 2002;67:820–828. doi: 10.1095/biolreprod.102.004879. [DOI] [PubMed] [Google Scholar]
- Smeyne RJ, Curran T, Morgan JI. Temporal and spatial expression of a fos-lacZ transgene in the developing nervous system. Brain Res Mol Brain Res. 1992a;16:158–62. doi: 10.1016/0169-328x(92)90206-q. [DOI] [PubMed] [Google Scholar]
- Smeyne RJ, Schilling K, Oberdick J, Robertson L, Luk D, Curran T, Morgan JI. A fos-lac Z transgenic mouse that can be used for neuroanatomic mapping. Adv Neurol. 1993;59:285–91. Review. [PubMed] [Google Scholar]
- Smeyne RJ, Schilling K, Robertson L, Luk D, Oberdick J, Curran T, Morgan JI. fos-lacZ transgenic mice: mapping sites of gene induction in the central nervous system. Neuron. 1992b;8:13–23. doi: 10.1016/0896-6273(92)90105-m. [DOI] [PubMed] [Google Scholar]
- Soares HD, Chen SC, Morgan JI. Differential and prolonged expression of Fos-lacZ and Jun-lacZ in neurons, glia, and muscle following sciatic nerve damage. Exp Neurol. 2001;167:1–14. doi: 10.1006/exnr.2000.7558. [DOI] [PubMed] [Google Scholar]
- Specht K, Richter T, Müller U, Walch A, Werner M, Höfler H. Quantitative gene expression analysis in microdissected archival formalin-fixed and paraffin-embedded tumor tissue. Am J Pathol. 2001;158:419–29. doi: 10.1016/S0002-9440(10)63985-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer CM, Houpt TA. Dynamics of c-fos and ICER mRNA expression in rat forebrain following lithium chloride injection. Brain Res Mol Brain Res. 2001;93:113–26. doi: 10.1016/s0169-328x(01)00173-5. [DOI] [PubMed] [Google Scholar]
- Swank MW. Coordinate regulation of Fos and Jun proteins in mouse brain by LiCl. Neuroreport. 1999;10:3685–9. doi: 10.1097/00001756-199911260-00041. [DOI] [PubMed] [Google Scholar]
- Swank MW, Bernstein IL. c-Fos induction in response to a conditioned stimulus after single trial taste aversion learning. Brain Res. 1994;636:202–8. doi: 10.1016/0006-8993(94)91018-9. [DOI] [PubMed] [Google Scholar]
- Sng J, Taniura H, Yoneda Y. Histone modifications in kainate-induced status epilepticus. Eur J Neurosci. 2006;23:1269–1282. doi: 10.1111/j.1460-9568.2006.04641.x. [DOI] [PubMed] [Google Scholar]
- Wong MH, Saam JR, Stappenbeck TS, Rexer CH, Gordon JI. Genetic mosaic analysis based on Cre recombinase and navigated laser capture microdissection. Proc Natl Acad Sci USA. 2000;97:12601–6. doi: 10.1073/pnas.230237997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T. Brain regions responsible for the expression of conditioned taste aversion in rats. Chem Senses. 2006;32:105–109. doi: 10.1093/chemse/bjj045. [DOI] [PubMed] [Google Scholar]
- Yasoshima Y, Sako N, Senba E, Yamamoto T. Acute suppression, but not chronic genetic deficiency, of c-fos gene expression impairs long-term memory in aversive taste learning. Proc Natl Acad Sci USA. 2006;103:7106–11. doi: 10.1073/pnas.0600869103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Zarubin T, Ji Z, Min Z, Zhu W, Downey JS, Lin S, Han J. Frequency and distribution of AP-1 sites in the human genome. DNA Res. 2005;12:139–150. doi: 10.1093/dnares/12.2.139. [DOI] [PubMed] [Google Scholar]