Abstract
Ebolavirus (EBOV) is a highly virulent pathogen capable of causing a severe hemorrhagic fever with 50–90% lethality. The EBOV glycoprotein (GP) is the only virally expressed protein on the virion surface and is critical for attachment to host cells and catalysis of membrane fusion. Hence, the EBOV GP is a critical component of vaccines as well as a target of neutralizing antibodies and inhibitors of attachment and fusion. The crystal structure of the Zaire ebolavirus GP in its trimeric, prefusion conformation (3 GP1 plus 3 GP2) in complex with a neutralizing antibody fragment, derived from a human survivor of the 1995 Kikwit outbreak, was recently determined. This is the first near-complete structure of any filovirus glycoprotein. The overall molecular architecture of the Zaire ebolavirus GP and its role in viral entry and membrane fusion are discussed in this article.
Keywords: cathepsin, ebola, Ebolavirus, filovirus, fusion protein, glycoprotein, membrane fusion, mucin-like domain, viral attachment, viral entry
Ebolavirus (EBOV) is a negative-stranded, membrane-enveloped filovirus that causes a severe hemorrhagic fever in both humans and non-human primates [1]. Since its discovery in central Africa in 1976, five species of EBOV have been isolated: Zaire (ZEBOV), Sudan, Côte d’Ivoire, Reston (REBOV) and the proposed ‘Bundibugyo’ EBOV [2–4]. Zaire, Sudan and Bundibugyo EBOVs have been associated with large outbreaks in Africa with high human case fatalities (25–90%). Côte d’Ivoire EBOV and REBOV can infect humans but no deaths have been reported to date. Recently, domestic swine in the Phillipines have been discovered to host REBOV [5]. Its entry into the human food supply raises significant concern. The natural reservoir of the EBOV and its host range are not completely well defined, although recent studies suggest that fruit bats are involved in the transmission of the virus [6]. Human infections generally occur through mucosal surfaces, skin abrasions or through contaminated needles, after direct contact with the virus from dead or infected people or wildlife. The onset of EBOV-like symptoms generally occurs within a 4–10-day incubation period. In fatal cases, death occurs 6–9 days after onset, as a result of uncontrolled viral replication, multiple organ failure and symptoms resembling severe septic shock [7]. There are currently no approved antiviral therapeutics, although a live attenuated ZEBOV glycoprotein (GP)-pseudotyped Vesicular stomatitis Indiana virus vaccine has shown promise [8–10] and has been used in the case of a laboratory needle-stick accident in Hamburg, Germany [11]. The frequent re-emergence of EBOV in Africa, including the recent emergence of a new species [2], the potential for introduction of the virus into nonendemic countries, concerns from the potential for misuse of the virus [12–14] and the lack of vaccines or treatments make EBOV a considerable public health concern.
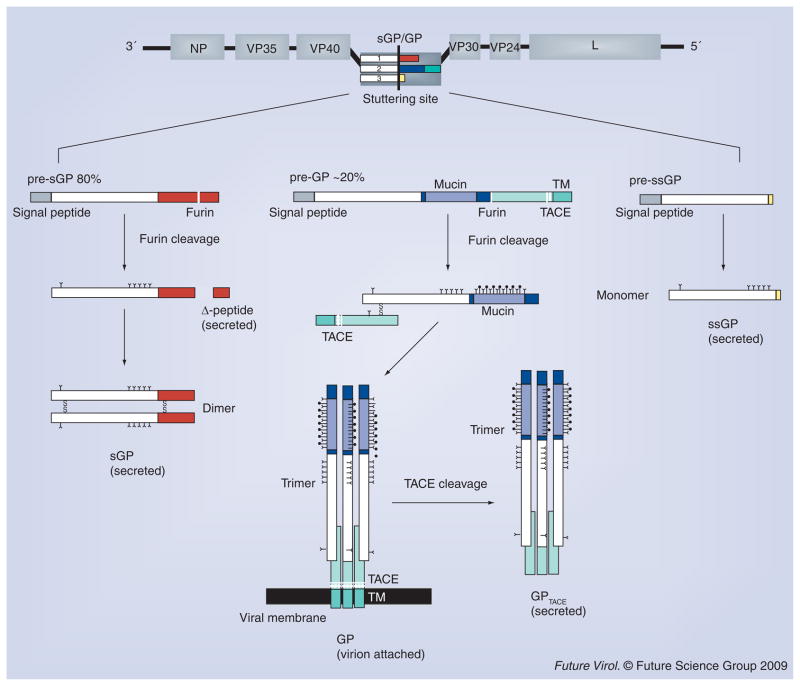
The EBOV genome contains seven genes: NP, VP35, VP40, GP, VP30, VP24 and L. However, more than seven proteins are produced owing to cotranscriptional editing and post-translational processing of the GP gene and gene products (Figure 1) [15–17].
Figure 1. Transcription and processing of Zaire ebolavirus glycoproteins.
The primary open reading frame of ebolavirus GP encodes a sGP (shown as white and red rectangles). Proteolytic cleavage of pre-sGP by furin results in the formation of the mature sGP and a small nonstructural fragment, termed Δ-peptide. Co-transcriptional stuttering of the GP gene results in two additional glycoproteins: GP and ssGP. GP is the virion-attached glycoprotein and proteolytic cleavage of its precursor (pre-GP) by furin results in two subunits, GP1 and GP2, that remain linked by a disulfide bond. The GP1 and GP2 heterodimer trimerizes and forms the viral surface peplomer. TNF-α-converting enzyme can also cleave envelope GP, at a site proximal to the GP2 transmembrane domain, thereby releasing a soluble trimeric GP. ssGP is a small secreted glycoprotein that shares the first 295 amino acids with sGP and GP, but has a different C-terminus (two nonshared residues, as colored in yellow). It has been reported that ssGP forms a monomer in solution. GP: Glycoprotein; sGP: Secreted glycoprotein; ssGP: Small, secreted glycoprotein; TACE: TNF-α-converting enzyme; TM: Transmembrane anchor.
sGP
The primary open reading frame is well conserved among all ebolaviral GP genes and encodes a 364-residue, nonstructural protein termed secreted GP (sGP) [15,16]. Post-translational proteolytic cleavage of pre-sGP by furin yields the mature sGP and a heavily O-glycosylated, small, nonstructural, secreted protein termed Δ-peptide [18]. Monomers of sGP are joined in a parallel orientation by two disulfide bonds between paired Cys53–Cys53 and Cys306–Cys306 residues to form a 110 kDa homodimer that is secreted [17,19–21]. The mature sGP has six predicted N-linked glycosylation sites, but only five sites are used consistently, sGP also contains a rare modification, in which the indole-C2 carbon atom of Trp288 is attached with an α-mannopyranosyl group [22].
It is not yet known whether sGP plays an important role in pathogenesis. Marburgvirus (MARV) does not produce orthologous proteins, yet induces indistinguishable disease in non-human primates and humans. During natural infection with EBOV, however, large amounts of sGP have been detected in the blood of infected patients [15]. Antibodies that are in survivor sera appear to preferentially recognize sGP over GP [23]. Hence, sGP could play a role in the evasion of humoral immune response by absorbing elicited antibodies. sGP has also been suggested to play an anti-inflammatory role by inducing recovery of the endothelial barrier function [24].
GP
The GP gene encodes seven consecutive adenosine nucleotides within a predicted hairpin loop [15,16]. The L polymerase stutters at this site during transcription, resulting in the insertion of an additional adenosine nucleotide in 20% of transcripts. This insertion causes a frameshift of the coding RNA and dictates production of a 676-residue (677 amino acids in REBOV), type I transmembrane GP termed pre-GP [21]. As a result of this frameshift, GP and sGP have an identical 295 amino acid N-terminus, but distinct C-termini. The different C-termini lead to unique patterns of disulfide bonding and different structures. While sGP is a dimer, the final GP is a trimer. The initial EBOV pre-GP is cleaved by furin at a multi-basic motif into two subunits, GP1 and GP2, which remain associated through a disulfide linkage between Cys53 of GP1 and Cys609 of GP2 [25,26]. This heterodimer (GP1 + GP2) assembles into a 450-kDa trimer at the surface of nascent virions. TNF-α-converting enzyme (TACE) can release the virion-attached GP through a cleavage site proximal to the transmembrane anchor [27].
The virion-attached GP is critical in the EBOV life cycle, as it is solely responsible for attachment, fusion and entry of target cells. Moreover, GP is responsible for critical pathogenic differences among viral species. Adenovirally expressed ZEBOV GP causes endothelial cell damage in both human and monkey vessel explants, while REBOV is only cytotoxic to monkey vessels. ZEBOV and REBOV also differ in susceptibility to furin cleavage [26] and amino acid sequence at the TACE cleavage site [27]. Moreover, it has been reported in in vitro and in vivo expression studies in HEK293T cells that overexpression of GP causes cytotoxicity and downregulation of a number of cellular surface proteins, including several involved in cellular adhesion (β1-integrin, α5-integrin and αV-integrin) and immune surveillance (MHC class I) [28–32]. The cytotoxicity caused by GP is dependent on a highly N- and O-linked glycosylated domain, termed the mucin-like domain [31,32]. However, the role of GP cytotoxicity in EBOV pathogenesis remains poorly defined, as constitutive expression of EBOV GP at moderate levels do not cause cell rounding and is not cytotoxic [33].
ssGP
While the addition of an extra adenosine leads to the production of GP, deletion of one or addition of two adenosines during the transcriptional stutter leads to the expression of a third product, a nonstructural, small sGP, termed ssGP [17]. ssGP shares the first 295 N-terminal residues with sGP and GP, but again, differs at the C-terminus, and is thought to be monomeric [16]. It is unclear what the role ssGP plays in EBOV pathogenesis.
We have recently published a crystal structure of the ZEBOV GP in its trimeric, prefusion form in complex with KZ52, a neutralizing antibody identified from a human survivor [34]. The GP molecule reveals a multi-subunit, chalice-shaped trimer, with a thick coating of oligosaccharides on a novel glycan cap and projecting mucin-like domain that, together, probably restrict access to the conserved receptor-binding site (RBS) sequestered inside the chalice bowl. This article will focus on the ZEBOV GPΔmucΔtm structure and its role in viral attachment and membrane fusion.
ZEBOV GPΔmucΔtm prefusion trimer
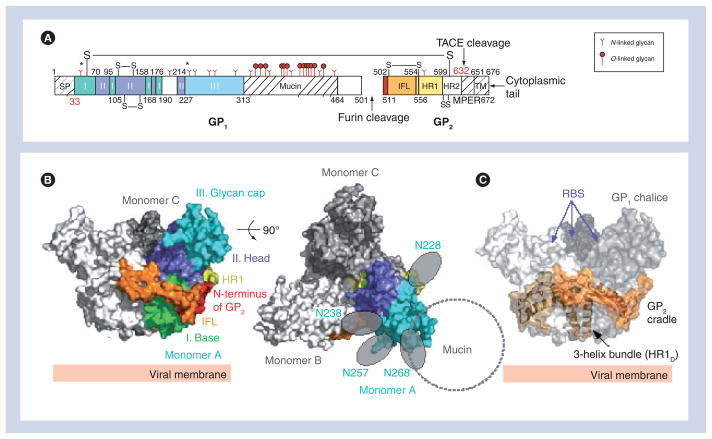
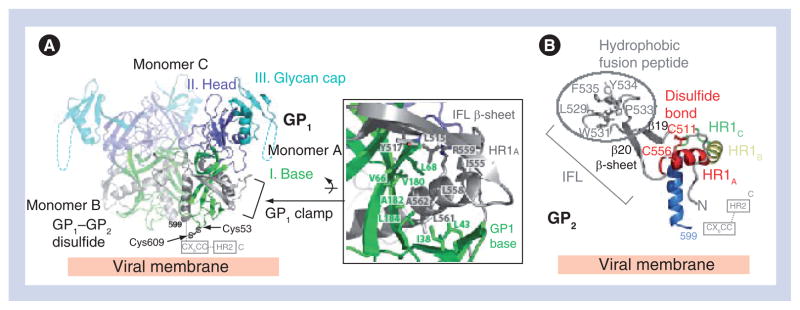
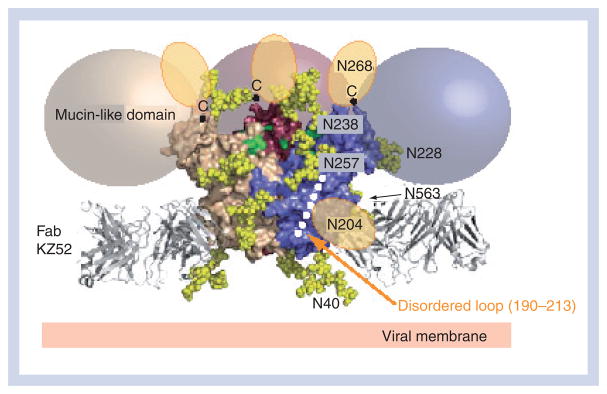
Crystallization of the ZEBOV GP was challenging owing to the intrinsic flexibility and extensive N- and O-linked glycosylation of the oligomeric molecule. In order to promote crystallization, the mucin-like domain and transmembrane domains were excised, two N-linked glycosylation sites at positions 40 and 228 were eliminated and the GP was complexed with an antibody, KZ52, which recognized a conformational GP1/GP2-containing epitope. This nearly complete ZEBOV GP (GPΔmucΔtm; Figure 2A) forms a trimer in solution and is fully capable of mediating virus entry when its transmembrane domain is restored [31,34,35]. The crystal structure illustrates that the prefusion GPΔmucΔtm forms a three-lobed chalice shape with the bowl of the chalice assembled by the three GP1 subunits (Figure 2B). The base of the chalice is formed by three GP2 subunits that cradle and encircle the GP1 trimer (Figure 2C).
Figure 2. Overall structure of Zaire ebolavirus GPΔmucΔtm.
(A) Domain schematic of GP. The disulfide bridges (-S-S-), SP, IFL, HR1, HR2, MPER, TACE cleavage site, TM and cytoplasmic tail are labeled accordingly. White and hash-marked regions designate crystallographically disordered and construct-deleted regions, respectively. (B) Molecular surface of the GP trimer viewed on its side and down its threefold axis. Monomer A is colored according to its subdomains: GP1 base – green; GP1 head – purple; GP1 glycan cap – cyan; GP2 N-terminus – red; GP2 IFL – orange; and GP2 HR1 – yellow. (C) Molecular surface of the Zaire ebolavirus GPΔmucΔtm chalice and cradle. Three lobes of GP1, shown in shades of gray form the GP chalice, while three subunits of GP2 (orange) wrap around the base of the chalice to form the cradle. GP: Glycoprotein; HR: Heptad repeat region; IFL: Internal fusion loop; MPER: Membrane-proximal external region; RBS: Receptor-binding site; SP: Signal peptide; TACE: TNF-α-converting enzyme; TM: Transmembrane anchor.
Adapted from [34].
ZEBOV GP1
The GP1 subunit is responsible for cellular attachment and contains putative receptor-binding regions, as well as a heavily glycosylated region termed the mucin-like domain. GP1 can be separated into three subdomains: base, head and glycan cap. The GP1 base subdomain contains four discontinuous sections (residues 33–69, 95–104, 158–167 and 176–189), and forms a hydrophobic, semicircular surface that interacts with the internal fusion loop and heptad repeat region of GP2 (Figure 3A). The GP1 base forms a clamp that probably stabilizes the GP2 prefusion conformation, thus preventing it from springing prematurely. Also within the GP1 base is Cys53, which forms an intermolecular disulfide bond to Cys609 of the GP2 subunit. Cys53 resides in the β2–β3 loop, proximal to the viral membrane end of the GP1 base subunit. Unexpectedly, electron density was not visible for the side chain of Cys53 and the entire region containing the counterpart GP2 cysteine, suggesting that this region is mobile. The GP1 head is a central subdomain located between the base and glycan cap, composed of residues from four discontinuous sections (residues 70–94, 105–157, 168–175 and 214–226). A significant portion of this subdomain is likely to be exposed to solvent within the bowl of the GPΔmucΔtm chalice, and is thought to contain the residues required for receptor binding. The GP1 glycan cap is composed of a continuous polypeptide chain from residue 227–310 that forms an α/β-dome over the GP1 head subdomain. This subdomain is fully exposed on the outer surface of GPΔmucΔtm and is not involved in any contacts at the trimer interface. Within the GP1 glycan cap, there is a clustering of at least four predicted N-linked oligosaccharides (linked to Asn228, Asn238, Asn257 and Asn268). C-terminal to the glycan cap is the approximately 150-residue GP1 mucin-like domain. This domain is predicted to contain low secondary structural complexity, but is heavily glycosylated with approximately four N- and approximately 13 O-linked oligosaccharides. The crystallization of the ZEBOV GP required the mucin-like domain to be excised in order to improve the chemical and conformational homogeneity of the sample, but at least one protective antibody epitope contained within it has been visualized [36].
Figure 3. Zaire ebolavirus GP1 and GP2.
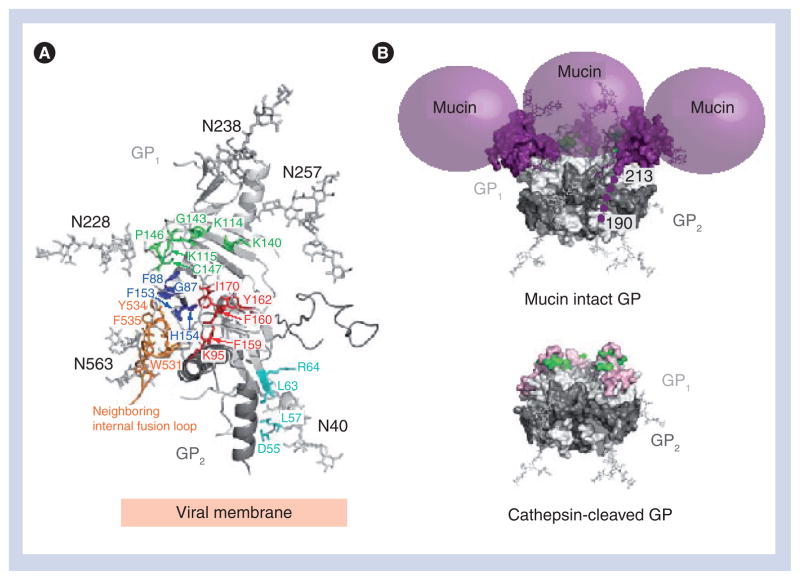
(A) Ribbon diagram of the Zaire ebolavirus GP trimer with each GP1 subdomain colored according to Figure 1B and the three GP2 subunits colored in gray. The GP1 base subdomain forms a hydrophobic, semicircular β-sheet surface that interacts with the hydrophobic face of the GP2 HR1A helix and the β-scaffold of the internal fusion loop (inset box). (B) Ribbon diagram of the prefusion conformation of Zaire ebolavirus GP2. Hydrophobic residues of the GP2 internal fusion loop are displayed on a β-scaffold (gray). The internal fusion loop is stabilized by a disulfide linkage at the base (Cys511–Cys556) and HR1 is separated into four segments (HR1A, HR1B, HR1C and HR1D). Note that the GP1–GP2 disulfide bridge (CX6CC motif) and HR2 region are disordered in the structure; their positions are marked by dashed lines.
GP: Glycoprotein; HR: Heptad repeat region; IFL: Internal fusion loop.
Adapted from [34].
ZEBOV GP2
The GP2 subunit is responsible for fusion of the viral and host cell membranes and contains the hydrophobic internal fusion loop, two heptad repeats (HR1 and HR2), a CX6CC disulfide bond motif, a membrane-proximal external region and a transmembrane anchor [37]. The internal fusion loop, residues 511–556, displays a series of hydrophobic residues (Leu529, Trp531, Ile532, Pro533, Tyr534, Phe535 and Pro537) bounded by an antiparallel, disulfide-linked β-scaffold (Figure 3B). In the prefusion state, the internal fusion loop wraps around the outside of the GP trimer. Its hydrophobic side chains are sequestered from solvent by packing into a neighboring GP1 monomer. Interestingly, although EBOV GP is classified as class I, its internal fusion loop structure is quite different from those of other class I viral GPs. For example, the fusion sequence in influenza virus hemagglutinin (HA) [38] and parainfluenza virus 5 F [39] form helices and reside at the free N-terminus of GP2 released by enzymatic cleavage. Instead, the fusion loop of EBOV more closely resembles the β-sheet structured fusion loops from the class II and III GPs, such as flavivirus E [40–42], vesicular stomatitis virus protein G [43,44] and HSV-1 gB (Figure 4) [45]. Only the first heptad repeat is ordered in the ZEBOV GP crystal structure, and this region can be structurally divided into four segments: HR1A, HR1B, HR1C and HR1D. The first two segments, HR1A and HR1B (residues 554–575), form an α-helix with a approximately 40° kink at Thr565, which delineates HR1A from HR1B. An unusual 3–4–4–3 stutter, rather than the normal 3–4 periodicity of heptad repeats, occurs within the bend between HR1A and HR1B. This stutter has been suggested to act as a conformational switch [46] and has also been observed in parainfluenza virus 5 F [39]. HR1C (residues 576–582) forms an extended coil linker between HR1B and the 16-residue HR1D segment (residues 583–598). HR1D forms an amphipathic helix, the hydrophobic faces of the three helices in the trimer pack together to form the interface of the peplomer.
Figure 4. Class I, II and III prefusion viral glycoprotein fusion peptides and internal fusion loops.
Comparison of prefusion conformation fusion peptide and internal fusion loop structures from class I, II and III GPs. The Zaire ebolavirus GP internal fusion loop more closely resembles those observed in class II and class III GPs than those observed in other trimeric class I prefusion viral GPs. GP: Glycoprotein; HA: Hemagglutinin.
Adapted from [34].
ZEBOV GP glycosylation & vulnerabilities to antibodies
A mixture of complex, oligomannose and hybrid-type glycans are found on the intact, mucin-like domain containing ZEBOV GP1 [47]. The glycans on the main structure of GPΔmucΔtm, outside the mucin-like domain, are likely to be complex-type in nature because the mucin-deleted ZEBOV GP is sensitive to deglycosylation by peptide N-glycosidase F, but not to endoglycosidase H [34]. Molecular modeling of biantennary, complex-type N-linked glycans on the ZEBOV GP structure reveals a thick coating of oligosaccharides on the surface of the GP (Figure 5). This coating extends across the intact GP, and the mucin-like domain contains approximately an additional 17 oligosaccharide chains across approximately 150 residues. The glycocalyx surrounding ZEBOV GPΔmucΔtm probably forms a shield that protects it from humoral immune responses and/or confers stability inside or outside a host. Indeed, most antibodies raised in natural infection are directed towards sGP [23].
Figure 5. Zaire ebolavirus glycoprotein glycosylation.
N-linked biantennary complex-type glycans (Gal2Man3GlcNAc4) are modeled as yellow space-filling spheres onto the Zaire ebolavirus GPΔmucΔtm structure at predicted glycosylation sites: Asn40, Asn204, Asn228, Asn238, Asn257, Asn268 and Asn563. The glycans at Asn204 and Asn268 reside in regions that are poorly ordered and, thus, their tentative locations are shown as orange ovals. The C-terminus of each last ordered residue of GP1, to which each mucin-like domain is linked, is marked with a ‘C’ (top of the chalice). Colored spheres (beige, pink and purple) outline the predicted location of the mucin-like domains. The putative receptor-binding site residues, recessed within the Zaire ebolavirus GP chalice bowl, are colored green.
Adapted from [34].
Even though ZEBOV GP has a thick coating of oligosaccharides, there are regions of vulnerability located at the base of the GP chalice, cathepsin-cleavage site and within the mucin-like domain. Importantly, several monoclonal antibodies have been isolated and characterized to neutralize in vitro or protect in rodent models [48–50]. The epitopes of these anti-EBOV-neutralizing antibodies have been determined by x-ray crystallography, neutralization escape studies and/or by linear peptide dot blots. For a more detailed review, see [51].
The most extensively characterized anti-EBOV-neutralizing antibody is KZ52. This monoclonal antibody was derived from bone marrow RNA donated by a human survivor of the 1995 Kikwit, Zaire (now Democratic Republic of the Congo) outbreak. KZ52 has been found to neutralize ZEBOV in vitro [48] and offers protection from lethal ZEBOV challenge in rodent models [52]. However, passive transfer of the antibody in nonhuman primates was unable to induce sterile immunity [53]. The crystal structure of EBOV GPΔmucΔtm complexed with Fab KZ52 illustrates that the antibody binds to a vulnerable, non-glycosylated site at the base of the GP chalice [34]. Specifically, KZ52 bridges three discontinuous regions of GP1 and GP2: GP1 residues 42–43, GP2 residues 505–514 (N-terminal region released by furin cleavage) and GP2 residues 549–556 (base of the internal fusion loop). The GP2 sequences involved in contacting KZ52 are poorly conserved between the various EBOV species, thus explaining the specificity to the ZEBOV species. While KZ52 requires both GP1 and GP2 for recognition, the crystal structure reveals that KZ52 contacts to GP1 are made through fairly weak van der Waals-type interactions to the main chain nitrogen of Leu43 and side chain carbon of Val42. (Note that at position 42, threonine was mutated to valine to eliminate the N-linked glycosylation site to improve sample homogeneity.) Instead, the majority of interactions are made between KZ52 and GP2. The requirement for GP1, but the relatively few strong contacts to GP1, suggests that the role of GP1 in the KZ52 epitope is primarily to stabilize the prefusion conformation of GP2 for recognition. Indeed, KZ52 does not recognize EBOV GP when the GP1–GP2 disulfide bond has been reduced, nor does it recognize that the postfusion GP2 six-helix bundle. KZ52 was the first antiviral antibody shown to bridge both the receptor-binding and fusion subunits of any viral GP. Recently, however, two broadly neutralizing anti-influenza virus hemagglutinin antibodies were identified that also bridge HA1 and HA2 [54,55], and neutralize by blocking conformational changes associated with fusion.
Another monoclonal antibody termed 133/3.16 recognizes a residue located at the base of the GP chalice. This residue, His549, was identified in neutralization escape experiments and lies near the epitope of KZ52. It is intriguing to speculate whether the base of the GP chalice represents a potential ‘sweet spot’ for neutralization (as reviewed in [51]).
Passive transfer of neutralizing antibodies has offered protection against challenge with a number of acute viruses [56–58]. In EBOV infection, the ability of neutralizing antibodies to serve as passive immunotherapeutics is not well understood. During the ZEBOV outbreak in Kikwit, the convalescent sera from five donors was transfused into eight patients (~11 days postinfection). Of the eight treated patients, only one succumbed to the disease, suggesting that convalescent sera could be useful in treatment of patients recovering from EBOV [59]. During more controlled experiments, with antibodies given closer to the time of exposure, passively transferred convalescent sera or monoclonal antibodies were shown to protect rodents and nonhuman primates against EBOV challenge [50,52,60–62], and may have also improved the survival rates of laboratory workers who were accidentally exposed to EBOV [63,64]. These studies have been cited as evidence for the use of anti-EBOV-neutralizing antibodies as a potential passive immunotherapeutic treatment. However, recent studies cast doubt on the feasibility of neutralizing antibodies to effectively protect humans against EBOV infection [53,65–67]. It is possible that since a single EBOV particle is a lethal dose for a primate, it may be too difficult for any single monoclonal antibody to provide sterile clearance. Perhaps a cocktail of monoclonal antibodies against several unique epitopes might confer improved protection. If so, it will be important to identify multiple, unique antibody epitopes on EBOV GP that could be combined into an antibody cocktail, including, or in addition to, KZ52.
EBOV entry
The EBOV envelope GP directly mediates binding of the virion to the host cell. A number of cellular factors, including DC-SIGN/L-SIGN [68,69], LSECtin [70,71], hMGL [72], β-integrins [73] and Tyro3 family receptors [74], have been implicated as attachment factors, however, none of these proteins individually are necessary and sufficient for viral entry. Hence, a critical cell-surface receptor responsible for EBOV attachment has yet to be identified, despite significant effort. It appears that EBOV enters through a receptor-mediated endocytotic mechanism, but it is still unclear whether clathrin-, caveolae- or cholesterol-dependent processes are used. Recombinant systems using pseudotyped retro-virus particles with ZEBOV GP and live virus studies using chemical inhibitors of clathrin- and caveolae-mediated endocytosis point to the use of caveolae and clathrin in EBOV entry [75,76]. However, cells lacking in caveolae can still be infected with EBOV [77].
Structure reveals a ZEBOV GP site critical for entry
Previous structure-blind studies initially localized receptor-binding regions to residues 54–201 [78–80]. This approximately 150 residue N-terminal region forms the base and head subdomains of GP1 and extends from the bottom to nearly the top of the chalice [81]. Extensive mutational analysis of GP1, performed by several groups, identifies at least 19 residues of GP1 important for infectivity (for these 19 residues, mutation results in secreted, analyzable protein, yet infectivity is abrogated [78–80]). Mapping of these 19 residues onto the ZEBOV GPΔmucΔtm structure allows the assignment of probable functions (Figure 6A). Among these 19, residues Asp55, Leu57, Leu63 and Arg64 reside on the GP1 base subdomain, near the GP1–GP2 disulfide bond and HR1D, and are likely to be important for fusion-mediated conformational changes rather than receptor binding. Residues Phe159, Phe160, Tyr162 and Ile170 are buried in the core of GP1, and probably play a role in structural stability. Residues Gly87, Phe88, Phe153 and His154 pack against the hydrophobic residues from a neighboring internal fusion loop. Mutations to these residues may affect the structural integrity of the fusion loop and GP trimer. The remaining six residues (Lys114, Lys115, Lys140, Gly143, Pro146 and Cys147) reside at or close to the GP1 surface, and cluster together in a approximately 10 × 15 Å site inside the concave bowl of GPΔmucΔtm, which may be the RBS of EBOV. Three lysines in this set of six residues have been recently confirmed as likely cell-contact sites [82]. Inspection of the crystal structure suggests that surrounding residues Phe88, Ile113, Pro116, Asp117, Gly118, Ser119, Glu120, Arg136, Tyr137, Val138, His139, Val141, Ser142, Thr144, Gly145, Arg172 and Gly173 may also be involved in receptor binding. The crystal structure illustrates that this putative RBS is recessed in the bowl of the GPΔmucΔtm chalice and possibly masked by the heavily glycosylated mucin-like domain and GP1 glycan cap. We predict that this site may become exposed or better exposed after entry into the endosome, where cathepsin cleavage occurs.
Figure 6. Zaire ebolavirus glycoprotein site critical for viral entry.
(A) Arrays of GP1 point mutations have identified at least 19 residues critical for viral entry [78–80]. These residues map to four distinct regions (cyan, green, royal blue and red) on the ZEBOV GPΔmucΔtm structure (monomer shown). (B) Zaire ebolavirus GP cathepsin cleavage. A molecular surface representation of the GPΔmucΔtm trimer with N-linked glycans drawn as ball-and-sticks and the mucin-like domain shown as purple spheres. Residues identified by the mutagenesis to be important for viral entry and located on or near the surface of the GPΔmucΔtm structure are colored in green (Lys114, Lys115, Lys140, Gly143, Pro146 and Cys147). Cleavage by cathepsin on a disordered loop (around residue 190) removes the glycan cap and mucin-like domain, exposing additional residues (shown in pink) on the site critical for viral entry [82].
GP: Glycoprotein.
Adapted from [34].
Cathepsin cleavage enhances EBOV binding & infectivity
After transport of the virus to the endosome [76], EBOV GP is processed by cathepsin B (CatB) and/or L (CatL) proteases [35,83,84]. CatB and/or CatL are essential for EBOV infection. Indeed, inhibition of CatL and CatB by specific protease inhibitors or siRNA reduces entry of ZEBOV and ZEBOV GP-pseudotyped viruses alike [35,83,84].
Cathepsin L trims EBOV GP1 from its original (~130 kDa) size to an initial 50-kDa fragment, followed by further cleavage to an approximately 19-kDa species of GP1, which remains linked to GP2 (Figure 6B) [35,83,84]. After digestion by CatL to yield the approximately 19-kDa fragment of GP that is functional in entry, GP may be further digested by CatB in the endosome to peptide-size fragments [83]. Our crystal structure indicated that the probable site of cathepsin cleavage is a loop reaching from residues 189 to 214, which is disordered in the GP structure. The solvent-exposed, mobile and perhaps, flexible nature of this loop render it attractive and accessible to protease cleavage. Indeed, separate and simultaneous biochemical studies confirm this loop (residue 190) to be the site of cathepsin cleavage [82]. Cleavage here would remove the glycan cap and mucin-like regions of GP1, yielding an approximately 19-kDa GP1 core encircled by GP2, with RBS on GP1 more completely exposed. In addition, cathepsin cleavage may expose a set of amino acids (residues 77–82) that resides on a convex surface underneath the glycan cap. The CatB and/or CatL cleavage of GP has been shown to increase binding and infectivity to target cells [35,83,84]. Genetic removal of the mucin-like domain results in cell binding similar to that observed with CatL-treated GP [35]. Taken together, these data suggest that cleavage by CatL facilitates interactions with an entry factor that is blocked by the glycan caps and mucin-like domains prior to cleavage [35]. Interestingly, CatL has also recently been shown to process the GPs of Nipah and Hendra viruses [85–87], as well as the SARS coronavirus [88]. We hypothesize that initial attachment of EBOV to target cells probably occurs via lectins or perhaps also by a low-affinity or low-frequency interactions with the putative RBS (Figure 7A). Higher affinity or facilitated binding of receptor could occur in the endosome, after cathepsin proteolytic cleavage of GP and exposure of the RBS (Figure 7B). A similar example might be HSV-2, as two attachment proteins, gB and gC, form initial low-affinity interactions with cell-surface heparan sulfate proteoglycans followed by specific protein–protein interactions by HSV-2 gD on the cell surface, which lead to entry and fusion (as reviewed in [89]).
Figure 7. Zaire ebolavirus glycoprotein-mediated entryEbolavirus is thought to enter cells through an endocytic mechanism.
(A) Initially, the metastable, prefusion Zaire ebolavirus GP may bind lectins [71] or an unidentified attachment factor at the cell surface (green ovals) via the mucin-like domains (grey spheres) or other sites on GP. (B) Subsequently, ebolavirus is internalized and trafficked to the endosome. Lectins may or may not remain bound to GP, depending on the nature of the individual lectin. In the endosome, host cathepsins cleave GP to remove the glycan cap and mucin-like domain, yielding an approximately 19-kDa GP1 core, disulfide bonded to GP2. (C) The newly exposed surface may allow either tighter binding to a receptor trafficked from the cell surface or binding to an alternate molecule in the endosome. Binding of this molecule, or perhaps further cathepsin cleavage, could then trigger conformational changes in the GP2 fusion subunit. (D) Structural rearrangements in GP2 allow HR1 to form a single 44-residue helix and position the IFL for insertion into the host-endosomal membrane. Upon insertion in the host membrane, the IFL adopts a 310 helix. This is the extended prehairpin intermediate. (E) Based on studies with the influenza virus, more than one trimer of GP2 may be required to complete the membrane fusion process. (F) The HR2 and MPER regions swing from the viral membrane towards the host membrane and HR1. Initial fold-back of the HR2 onto HR1 distorts the virus and host-cell bilayers and brings the two membranes into contact to form a hemifusion stalk. (G) The hemifusion stalk opens up to form the fusion pore and the low energy, postfusion 6HB is formed when three HR2 helices pack into the HR1 trimeric bundle.
6HB: Six helix bundle; CatL/B: Cathepsin L/B; GP: Glycoprotein; HR: Heptad repeat region; IFL: Internal fusion loop; MPER: Membrane-proximal external region; TACE: TNF-α-converting enzyme; TM: Transmembrane domain.
Conformational trigger & membrane fusion
Fusion of the viral and host cell membranes has a very high energetic barrier. To catalyze this fusion reaction, viral fusion proteins undergo a large protein conformational change, using the released free energy to lower the activation barrier. The general steps to class I GP-mediated viral and host cell membrane fusions are fairly well understood from work on the influenza virus HA, parainfluenza virus 5 F and others (see reviews [90–93]). Based on these works and the structure of ZEBOV GPΔmucΔtm, the following mechanism of membrane fusion is proposed (Figure 7).
The first step of viral–host cell membrane fusion requires priming of the envelope GP to a metastable conformation that can be subsequently activated for fusion. For many viruses, such as HIV-1 gp160 and influenza virus HA, this requires the GP to be endoproteolytically cleaved by furin [94,95]. In the case of influenza virus HA, furin cleavage unlocks constraints on the fusion peptide to prime the GP such that it can be subsequently sprung by endosomal low pH [96,97]. In stark contrast, ZEBOV GP does not require furin cleavage for viral infection in a number of cell lines, or in nonhuman primates [98,99]. Based on the crystal structure of EBOV GPΔmucΔtm, the hydrophobic fusion residues are displayed on a loop within a disulfide-bonded, antiparallel β-scaffold and are not conformationally restricted by the lack of a free N-terminus on its GP2 sub-unit. Moreover, the mucin-like domain, which is predicted to have low secondary structural complexity, is N-terminal to the internal fusion loop. We speculate that this flexible domain may provide the additional conformational freedom to the internal fusion loop, in the absence of furin cleavage, during the fusion process.
The next step requires triggering the metastable, prefusion conformation of the GP for fusion. For many viruses, such as influenza and flaviviruses, low pH is the trigger for GP2-like conformational changes necessary for membrane fusion. Here, key histidine residues become protonated in the acidic environment of the endosome [100,101]. These histidines are located in the vicinity of positively charged residues in the pre-fusion conformation but, once protonated, rearrange to instead form electrostatic interactions with negatively charged residues in the postfusion conformation. For other viruses, such as HIV-1, coreceptor-binding triggers conformational rearrangements [102,103]. In other viruses, such as avian leukosis virus-A [104], both receptor binding and low pH are required.
The fusogenic trigger for ZEBOV GP is not well defined. The low pH of the endosome is necessary for GP-mediated cell–cell fusion and infection of ZEBOV GP-pseudotyped Vesicular stomatitis Indiana virus [105–107]. However, low pH dependence is probably not directly involved in triggering, as the ZEBOV GP1–GP2 interface is primarily nonpolar and hydrophobic, lacking the array of histidine and/or acidic residues commonly associated with pH-dependent conformational changes in viral GPs. Furthermore, the prefusion conformation-specific monoclonal antibody KZ52 will bind ZEBOV GPΔmucΔtm at pH 4, suggesting that low pH alone does not trigger GP. Instead, the requirements of low pH are likely to activate the endosomal cathepsins that proteolyze GP. It is unclear whether cathepsin cleavage of ZEBOV GP itself causes some initial conformational changes that trigger the fusogenic conformation [83] or alternatively, if cathepsin cleavage activates GP for triggering by an additional, as yet undiscovered, cellular factor (Figure 7A–C) [84]. Although the identity of the trigger is unknown, from the structure it is clear that the GP1 clamp that stabilizes GP2 HR1 must be released to allow the HR1 and HR2 to rearrange and permit fusion.
Comparison of the prefusion ZEBOV GPΔmucΔtm [34] and the postfusion ZEBOV GP2 fragment structures [46,108] illustrates how the four segments of HR1A–D must unwind from their prefusion ring around GP1, straighten, and assemble into a single 44-residue helical rod (Figure 7D). In the process, the rotational and translational movement of HR1A and HR1B position the internal fusion loops at the top of the trimeric GP2. The hydrophobic residues displayed on the β-scaffold of the internal fusion loop (Leu529, Trp531, Pro533, Tyr534, Phe535 and Pro537) are then able to penetrate the host membrane and adopt a 310 helical character, as described by nuclear magnetic resonance studies of free fusion peptide in sodium dodecyl sulfate micelles or detergent-resistant membrane fractions [109]. It is hypothesized that the conserved Pro537 in the fusion-loop is important to induce bilayer destabilization [110]. The elongated GP2 structure resulting from fusion loop penetration of the target membrane is also called the pre-hairpin intermediate. In HIV-1, the prehairpin intermediate conformation of gp41 has a half-life in the range of minutes, but for other viruses, this conformation may last only seconds [111].
After formation of the prehairpin intermediate, ZEBOV GP2 collapses into a folded-back conformation, where GP2 may flex at its elbow-like CX6CC hinge to allow HR2 to approach the HR1 trimeric bundle (Figure 7F). The collapse of the prehairpin intermediate distorts the viral and host-cell bilayers, draws the two bilayers close to one another to merge into a hemifusion stalk that ultimately opens up into a fusion pore (Figure 7G). Formation of the final postfusion six-helix bundle, in which the internal fusion loop and transmembrane domain are juxtaposed, completes the fusion of the host and virus plasma membranes. A movie of the modeled ZEBOV GP-mediated fusion process can be found at [201].
Future perspective
The EBOV GP is one of the most studied proteins of EBOV owing to its critical role in viral entry and potential as a target for therapeutics. Although the crystal structure of ZEBOV GP has shed light on its many roles, many questions remain unanswered.
What is the identity of the EBOV host receptor? A receptor necessary and sufficient for EBOV entry still eludes researchers. We hypothesize that cathepsin cleavage of ZEBOV GPΔmucΔtm probably leaves behind a convex surface for binding to such a critical entry factor. The fold, location and physiochemical properties of this site should now provide new leads for the search for EBOV receptor(s).
Is the RBS conserved within the filovirus family? Sequence alignments reveal that the EBOV residues involved in entry are well conserved among ZEBOV, Sudan, Côte d’Ivoire and REBOV species. Hence, the ZEBOV GPΔmucΔtm structure should make a representative model that can be used in the design of entry inhibitors and immunotherapeutics against all species of EBOV. However, although previous studies using GP1 fragments suggest that EBOV and MARV compete for binding to the same receptor [81], the sequences of the residues involved in entry are not well conserved between EBOV and MARV. Perhaps MARV binds a different receptor or binds the same receptor in a different way or by using different amino acid residues. Perhaps the receptor binds to the main chain or framework shared between EBOV and MARV. Note that MARV can enter cells independently of CatB [112] and, hence, some structural differences must occur.
What is the trigger for GP2 conformational change? It is unclear whether the precise role of cathepsin cleavage is to prime GP2 for conformational change, or to open up a RBS, or both.
What are the similarities and differences between ssGP, sGP and GP? While the first 295 amino acids of sequence are identical between these three GPs, they each have different C-termini, oligomerization states and preferential reactivity by survivor sera [23,48]. In addition, GP is embedded in the viral surface, while sGP and ssGP are secreted abundantly [1]. It is as yet unclear if the shared sequence adopts a fold that differs between the three GP variants, or if the differential antibody reactivity results from the same fold packaged into alternate quaternary scaffolds. However, for vaccine development, immunogens must be designed to elicit neutralizing antibodies against viral particles rather than secreted and shed proteins.
Executive summary
Zaire ebolavirusGPΔmucΔtm prefusion trimer adopts a bowl-like chalice shape
Three glycoprotein (GP)1 subunits form the bowl of the chalice and three GP2 subunits wrap around the base of the chalice forming a cradle.
GP1 consists of three subdomains: base, head and glycan cap.
Hydrophobic residues from the GP2 internal fusion loop are displayed on a β-scaffold.
Heptad repeat region (HR)1 is separated into four segments: HR1A, HR1B, HR1C and HR1D.
Zaire ebolavirusGP1 acts as a clamp on GP2
Four discontinuous stretches of GP1 fold together to form a hydrophobic β-sheet surface, termed the GP1 base subdomain.
The GP1 base subdomain forms a clamp on the GP2 HR1 prefusion conformation and probably prevents it from prematurely springing into the fusogenic state.
Zaire ebolavirusGPΔmucΔtm is protected by a glycocalyx
Modeling of biantennary, complex-type oligosaccharides on Zaire ebolavirus (ZEBOV) GPΔmucΔtm reveal the surface is cloaked in a thick coating of N-linked glycans.
The extensive glycosylation from the glycan cap and mucin-like domain probably protects the virus from immune surveillance, thus explaining why there are very few neutralizing antibodies elicited during natural infection.
A site critical for viral entry is located inside the bowl of the chalice
The critical ZEBOV GPΔmucΔtm host receptor is unknown.
Structural and mutational studies have identified a patch of lysines (Lys114, Lys115 and Lys140) that resides on the surface of the GPΔmucΔtm. Subsequent mutagenesis experiments reveal these lysines are important for viral attachment.
Three distinct putative receptor-binding sites exist in the GPΔmucΔtm trimer and are buried inside the bowl of the chalice.
Cathepsin cleavage enhances ebolavirus binding & infectivity
Proteolytic cleavage of ZEBOV GP by cathepsin B and/or L is required for viral entry.
Cathepsin B and/or L cleaves ZEBOV GP1 from an approximately 150-kDa subunit to an 18–19-kDa fragment, thus removing the mucin-like domain and glycan cap.
It is suggested that cleavage by cathepsins better exposes the patch of lysines on ZEBOV GP1, and may expose additional residues and surfaces for improved interactions, with an entry factor that is otherwise blocked by the mucin-like domain or glycan cap.
Conformational trigger & membrane fusion
Viral and host cell membrane fusion is mediated by GP2, analogous to other class I viral fusion proteins, such as influenza virus HA2 and HIV-1 gp41.
The trigger to release the GP1 constraints on GP2 is unknown.
Removal of the GP1 constraints allows the GP2 HR1 to form a single 44-residue helix and positions the internal fusion loop at the top of the GP for insertion into the host membrane (prehairpin intermediate).
The extended prehairpin intermediate folds back to distort the viral and host-cell bilayers and facilitates the formation of the hemifusion stalk.
Formation of the low energy six-helix bundle juxtaposes the internal fusion loops and transmembrane domains, facilitating the formation of the fusion pore and final merger of the host and viral membranes.
Acknowledgments
Financial & competing interests disclosure
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
No writing assistance was utilized in the production of this manuscript.
Footnotes
For reprint orders, please contact: reprints@futuremedicine.com
Contributor Information
Jeffrey E Lee, Email: jlee@scripps.edu, Department of Immunology & Microbial Science, The Scripps Research Institute, 10550 North Torrey Pines Road, La Jolla, CA 92037, USA, Tel.: +1 858 784 7976, Fax: +1 858 784 8218.
Erica Ollmann Saphire, Email: erica@scripps.edu, Department of Immunology & Microbial Science and The Skaggs Institute for Chemical Biology, The Scripps Research Institute, 10550 North Torrey Pines Road, La Jolla, CA 92037, USA, Tel.: +1 858 784 8602, Fax: +1 858 784 8218.
Bibliography
Papers of special note have been highlighted as:
▪ of interest
▪▪ of considerable interest
- 1▪.Sanchez A, Geisbert TW, Feldmann H. Filoviridae: Marburg and Ebola viruses. In: Knipe DM, Howley PM, Griffin DE, et al., editors. Fields Virology. Lippincott Williams and Wilkins; Philadelphia, PA, USA: 2007. pp. 1409–1448. Comprehensive review of Filoviruses. [Google Scholar]
- 2.Towner JS, Sealy TK, Khristova ML, et al. Newly discovered ebola virus associated with hemorrhagic fever outbreak in Uganda. PLoS Pathog. 2008;4(11):e1000212. doi: 10.1371/journal.ppat.1000212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3▪.Kuhn JH. History of filoviral disease outbreaks. In: Calisher CH, editor. Filoviruses. Springer Wien; NewYork, Wien, Austria: 2008. Comprehensive review of Filoviruses. [Google Scholar]
- 4.Feldmann H, Geisbert TW, Jahrling PB, et al. Negative sense single stranded RNA viruses. In: Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA, editors. Virus Taxonomy: Classification and Nomenclature of Viruses. Elsevier Academic Press; London, UK: 2005. pp. 645–653. [Google Scholar]
- 5.Barrette RW, Metwally SA, Rowland JM, et al. Discovery of swine as a host for the Reston ebolavirus. Science. 2009;325(5937):204–206. doi: 10.1126/science.1172705. [DOI] [PubMed] [Google Scholar]
- 6.Leroy EM, Kumulungui B, Pourrut X, et al. Fruit bats as reservoirs of Ebola virus. Nature. 2005;438(7068):575–576. doi: 10.1038/438575a. [DOI] [PubMed] [Google Scholar]
- 7.Ksiazek TG, Rollin PE, Williams AJ, et al. Clinical virology of Ebola hemorrhagic fever (EHF): virus, virus antigen, and IgG and IgM antibody findings among EHF patients in Kikwit, Democratic Republic of the Congo, 1995. J Infect Dis. 1999;179(Suppl 1):S177–S187. doi: 10.1086/514321. [DOI] [PubMed] [Google Scholar]
- 8▪.Geisbert TW, DaddariO-Dicaprio KM, Lewis MG, et al. Vesicular stomatitis virus-based ebola vaccine is well-tolerated and protects immunocompromised nonhuman primates. PLoS Pathog. 2008;4(11):e1000225. doi: 10.1371/journal.ppat.1000225. Demonstrates that live-attenuated vesicular stomatitis virus pseudotyped with the Ebolavirus (EBOV) glycoprotein alone is sufficient to protect nonhuman primates from EBOV challenge. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9▪.Geisbert TW, Geisbert JB, Leung A, et al. Single injection vaccine protects nonhuman primates against Marburg virus and three species of Ebola virus. J Virol. 2009;83(14):7296–7304. doi: 10.1128/JVI.00561-09. Demonstrates that live-attenuated vesicular stomatitis virus pseudotyped with the EBOV glycoprotein alone is sufficient to protect nonhuman primates from EBOV challenge. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10▪.Jones SM, Feldmann H, Stroher U, et al. Live attenuated recombinant vaccine protects nonhuman primates against Ebola and Marburg viruses. Nat Med. 2005;11(7):786–790. doi: 10.1038/nm1258. Demonstrates that live-attenuated vesicular stomatitis virus pseudotyped with the EBOV glycoprotein alone is sufficient to protect nonhuman primates from EBOV challenge. [DOI] [PubMed] [Google Scholar]
- 11.Tuffs A. Experimental vaccine may have saved Hamburg scientist from Ebola fever. Br Med J. 2009;338:B1223. doi: 10.1136/bmj.b1223. [DOI] [PubMed] [Google Scholar]
- 12.Geisbert TW, Jahrling PB. Towards a vaccine against Ebola virus. Expert Rev Vaccines. 2003;2(6):777–789. doi: 10.1586/14760584.2.6.777. [DOI] [PubMed] [Google Scholar]
- 13.Rotz LD, Khan AS, Lillibridge SR, Ostroff SM, Hughes JM. Public health assessment of potential biological terrorism agents. Emerg Infect Dis. 2002;8(2):225–230. doi: 10.3201/eid0802.010164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Borio L, Inglesby T, Peters CJ, et al. Hemorrhagic fever viruses as biological weapons: medical and public health management. JAMA. 2002;287(18):2391–2405. doi: 10.1001/jama.287.18.2391. [DOI] [PubMed] [Google Scholar]
- 15.Sanchez A, Trappier SG, Mahy BW, Peters CJ, Nichol ST. The virion glycoproteins of Ebola viruses are encoded in two reading frames and are expressed through transcriptional editing. Proc Natl Acad Sci USA. 1996;93(8):3602–3607. doi: 10.1073/pnas.93.8.3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Volchkov VE, Becker S, Volchkova VA, et al. GP mRNA of Ebola virus is edited by the Ebola virus polymerase and by T7 and vaccinia virus polymerases. Virology. 1995;214(2):421–430. doi: 10.1006/viro.1995.0052. [DOI] [PubMed] [Google Scholar]
- 17.Volchkova VA, Feldmann H, Klenk HD, Volchkov VE. The nonstructural small glycoprotein sGP of Ebola virus is secreted as an antiparallel-orientated homodimer. Virology. 1998;250(2):408–414. doi: 10.1006/viro.1998.9389. [DOI] [PubMed] [Google Scholar]
- 18.Volchkova VA, Klenk HD, Volchkov VE. Δ-peptide is the carboxy-terminal cleavage fragment of the nonstructural small glycoprotein sGP of Ebola virus. Virology. 1999;265(1):164–171. doi: 10.1006/viro.1999.0034. [DOI] [PubMed] [Google Scholar]
- 19.Barrientos LG, Martin AM, Rollin PE, Sanchez A. Disulfide bond assignment of the Ebola virus secreted glycoprotein sGP. Biochem Biophys Res Commun. 2004;323(2):696–702. doi: 10.1016/j.bbrc.2004.08.148. [DOI] [PubMed] [Google Scholar]
- 20.Falzarano D, Krokhin O, Wahl-Jensen V, et al. Structure-function analysis of the soluble glycoprotein, sGP, of Ebola virus. Chembiochem. 2006;7(10):1605–1611. doi: 10.1002/cbic.200600223. [DOI] [PubMed] [Google Scholar]
- 21.Sanchez A, Yang ZY, Xu L, Nabel GJ, Crews T, Peters CJ. Biochemical analysis of the secreted and virion glycoproteins of Ebola virus. J Virol. 1998;72(8):6442–6447. doi: 10.1128/jvi.72.8.6442-6447.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Falzarano D, Krokhin O, Van Domselaar G, et al. Ebola sGP – the first viral glycoprotein shown to be C-mannosylated. Virology. 2007;368(1):83–90. doi: 10.1016/j.virol.2007.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maruyama T, Parren PW, Sanchez A, et al. Recombinant human monoclonal antibodies to Ebola virus. J Infect Dis. 1999;179(Suppl 1):S235–S239. doi: 10.1086/514280. [DOI] [PubMed] [Google Scholar]
- 24.Wahl-Jensen VM, Afanasieva TA, Seebach J, Stroher U, Feldmann H, Schnittler HJ. Effects of Ebola virus glycoproteins on endothelial cell activation and barrier function. J Virol. 2005;79(16):10442–10450. doi: 10.1128/JVI.79.16.10442-10450.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jeffers SA, Sanders DA, Sanchez A. Covalent modifications of the Ebola virus glycoprotein. J Virol. 2002;76(24):12463–12472. doi: 10.1128/JVI.76.24.12463-12472.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Volchkov VE, Feldmann H, Volchkova VA, Klenk HD. Processing of the Ebola virus glycoprotein by the proprotein convertase furin. Proc Natl Acad Sci USA. 1998;95(10):5762–5767. doi: 10.1073/pnas.95.10.5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dolnik O, Volchkova V, Garten W, et al. Ectodomain shedding of the glycoprotein GP of Ebola virus. EMBO J. 2004;23(10):2175–2184. doi: 10.1038/sj.emboj.7600219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang ZY, Duckers HJ, Sullivan NJ, Sanchez A, Nabel EG, Nabel GJ. Identification of the Ebola virus glycoprotein as the main viral determinant of vascular cell cytotoxicity and injury. Nat Med. 2000;6(8):886–889. doi: 10.1038/78645. [DOI] [PubMed] [Google Scholar]
- 29.Chan SY, Ma MC, Goldsmith MA. Differential induction of cellular detachment by envelope glycoproteins of Marburg and Ebola (Zaire) viruses. J Gen Virol. 2000;81(Pt 9):2155–2159. doi: 10.1099/0022-1317-81-9-2155. [DOI] [PubMed] [Google Scholar]
- 30.Francica JR, Matukonis MK, Bates P. Requirements for cell rounding and surface protein down-regulation by Ebola virus glycoprotein. Virology. 2009;383(2):237–247. doi: 10.1016/j.virol.2008.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simmons G, Wool-Lewis RJ, Baribaud F, Netter RC, Bates P. Ebola virus glycoproteins induce global surface protein down-modulation and loss of cell adherence. J Virol. 2002;76(5):2518–2528. doi: 10.1128/jvi.76.5.2518-2528.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sullivan NJ, Peterson M, Yang ZY, et al. Ebola virus glycoprotein toxicity is mediated by a dynamin-dependent protein-trafficking pathway. J Virol. 2005;79(1):547–553. doi: 10.1128/JVI.79.1.547-553.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alazard-Dany N, Volchkova V, Reynard O, et al. Ebola virus glycoprotein GP is not cytotoxic when expressed constitutively at a moderate level. J Gen Virol. 2006;87(Pt 5):1247–1257. doi: 10.1099/vir.0.81361-0. [DOI] [PubMed] [Google Scholar]
- 34▪▪.Lee JE, Fusco MH, Hessell AJ, Oswald WB, Burton DR, Saphire EO. Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature. 2008;454(7201):177–183. doi: 10.1038/nature07082. Describes the first structure of the prefusion, trimeric EBOV glycoprotein in complex with a neutralizing antibody identified from a human survivor of the 1995 Kikwit, Zaire outbreak, and provides a model for EBOV entry and future therapeutic development. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35▪▪.Kaletsky RL, Simmons G, Bates P. Proteolysis of the Ebola virus glycoproteins enhances virus binding and infectivity. J Virol. 2007;81(24):13378–13384. doi: 10.1128/JVI.01170-07. First study to demonstrate that cathepsin-cleaved EBOV glycoprotein exhibits improved attachment to target cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee JE, Kuehne A, Abelson DM, Fusco ML, Hart MK, Saphire EO. Complex of a protective antibody with its Ebola virus GP peptide epitope: unusual features of a V λ × light chain. J Mol Biol. 2008;375(1):202–216. doi: 10.1016/j.jmb.2007.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gallaher WR. Similar structural models of the transmembrane proteins of Ebola and avian sarcoma viruses. Cell. 1996;85(4):477–478. doi: 10.1016/s0092-8674(00)81248-9. [DOI] [PubMed] [Google Scholar]
- 38.Wilson IA, Skehel JJ, Wiley DC. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 Å resolution. Nature. 1981;289(5796):366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- 39.Yin HS, Wen X, Paterson RG, Lamb RA, Jardetzky TS. Structure of the parainfluenza virus 5 F protein in its metastable, prefusion conformation. Nature. 2006;439(7072):38–44. doi: 10.1038/nature04322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rey FA, Heinz FX, Mandl C, Kunz C, Harrison SC. The envelope glycoprotein from tick-borne encephalitis virus at 2 Å resolution. Nature. 1995;375(6529):291–298. doi: 10.1038/375291a0. [DOI] [PubMed] [Google Scholar]
- 41.Modis Y, Ogata S, Clements D, Harrison SC. Structure of the dengue virus envelope protein after membrane fusion. Nature. 2004;427(6972):313–319. doi: 10.1038/nature02165. [DOI] [PubMed] [Google Scholar]
- 42.Nybakken GE, Oliphant T, Johnson S, Burke S, Diamond MS, Fremont DH. Structural basis of West Nile virus neutralization by a therapeutic antibody. Nature. 2005;437(7059):764–769. doi: 10.1038/nature03956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roche S, Bressanelli S, Rey FA, Gaudin Y. Crystal structure of the low-pH form of the vesicular stomatitis virus glycoprotein G. Science. 2006;313(5784):187–191. doi: 10.1126/science.1127683. [DOI] [PubMed] [Google Scholar]
- 44.Roche S, Rey FA, Gaudin Y, Bressanelli S. Structure of the prefusion form of the vesicular stomatitis virus glycoprotein G. Science. 2007;315(5813):843–848. doi: 10.1126/science.1135710. [DOI] [PubMed] [Google Scholar]
- 45.Heldwein EE, Lou H, Bender FC, Cohen GH, Eisenberg RJ, Harrison SC. Crystal structure of glycoprotein B from herpes simplex virus 1. Science. 2006;313(5784):217–220. doi: 10.1126/science.1126548. [DOI] [PubMed] [Google Scholar]
- 46.Weissenhorn W, Carfi A, Lee KH, Skehel JJ, Wiley DC. Crystal structure of the Ebola virus membrane fusion subunit, GP2, from the envelope glycoprotein ectodomain. Mol Cell. 1998;2(5):605–616. doi: 10.1016/s1097-2765(00)80159-8. [DOI] [PubMed] [Google Scholar]
- 47.Ritchie GE. DPhil thesis. Linacre College, University of Oxford; Oxford, UK: 2005. The glycosylation of viral envelope glycoproteins and the effect of glycosidase inhibitors on virus replication and glycoprotein properties. [Google Scholar]
- 48.Maruyama T, Rodriguez LL, Jahrling PB, et al. Ebola virus can be effectively neutralized by antibody produced in natural human infection. J Virol. 1999;73(7):6024–6030. doi: 10.1128/jvi.73.7.6024-6030.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takada A, Feldmann H, Stroeher U, et al. Identification of protective epitopes on ebola virus glycoprotein at the single amino acid level by using recombinant vesicular stomatitis viruses. J Virol. 2003;77(2):1069–1074. doi: 10.1128/JVI.77.2.1069-1074.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wilson JA, Hevey M, Bakken R, et al. Epitopes involved in antibody-mediated protection from Ebola virus. Science. 2000;287(5458):1664–1666. doi: 10.1126/science.287.5458.1664. [DOI] [PubMed] [Google Scholar]
- 51.Lee JE, Saphire EO. Neutralizing Ebolavirus: structural insights into the envelope glycoprotein and antibodies targeted against it. Curr Opin Struct Biol. 2009;19(4):408–417. doi: 10.1016/j.sbi.2009.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Parren PWHI, Geisbert TW, Maruyama T, Jahrling PB, Burton DR. Pre- and post-exposure prophylaxis of Ebola virus infection in an animal model by passive transfer of a neutralizing human antibody. J Virol. 2002;76(12):6408–6412. doi: 10.1128/JVI.76.12.6408-6412.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Oswald WB, Geisbert TW, Davis KJ, et al. Neutralizing antibody fails to impact the course of Ebola virus infection in monkeys. PLoS Pathog. 2007;3(1):e9. doi: 10.1371/journal.ppat.0030009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ekiert DC, Bhabha G, Elsliger MA, et al. Antibody recognition of a highly conserved influenza virus epitope. Science. 2009;324(5924):246–251. doi: 10.1126/science.1171491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sui J, Hwang WC, Perez S, et al. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat Struct Mol Biol. 2009;16(3):265–273. doi: 10.1038/nsmb.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Casadevall A, Scharff MD. Return to the past: the case for antibody-based therapies in infectious diseases. Clin Infect Dis. 1995;21(1):150–161. doi: 10.1093/clinids/21.1.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hangartner L, Zinkernagel RM, Hengartner H. Antiviral antibody responses: the two extremes of a wide spectrum. Nat Rev Immunol. 2006;6(3):231–243. doi: 10.1038/nri1783. [DOI] [PubMed] [Google Scholar]
- 58.Parren PW, Burton DR. The antiviral activity of antibodies in vitro and in vivo. Adv Immunol. 2001;77:195–262. doi: 10.1016/S0065-2776(01)77018-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mupapa K, Massamba M, Kibadi K, et al. Treatment of Ebola hemorrhagic fever with blood transfusions from convalescent patients. J Infect Dis. 1999;179(Suppl 1):S18–S23. doi: 10.1086/514298. [DOI] [PubMed] [Google Scholar]
- 60.Borisevich IV, Mikhailov VV, Krasnianskii VP, et al. Development and study of the properties of immunoglobulin against Ebola fever. Vopr Virusol. 1995;40(6):270–273. [PubMed] [Google Scholar]
- 61.Gupta M, Mahanty S, Bray M, Ahmed R, Rollin PE. Passive transfer of antibodies protects immunocompetent and imunodeficient mice against lethal Ebola virus infection without complete inhibition of viral replication. J Virol. 2001;75(10):4649–4654. doi: 10.1128/JVI.75.10.4649-4654.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mikhailov VV, Borisevich IV, Chernikova NK, Potryvaeva NV, Krasnianskii VP. The evaluation in hamadryas baboons of the possibility for the specific prevention of Ebola fever. Vopr Virusol. 1994;39(2):82–84. [PubMed] [Google Scholar]
- 63.Kudoyarova-Zubavichene NM, Sergeyev NN, Chepurnov AA, Netesov SV. Preparation and use of hyperimmune serum for prophylaxis and therapy of Ebola virus infections. J Infect Dis. 1999;179(Suppl 1):S218–S223. doi: 10.1086/514294. [DOI] [PubMed] [Google Scholar]
- 64.Sadek RF, Khan AS, Stevens G, Peters CJ, Ksiazek TG. Ebola hemorrhagic fever, Democratic Republic of the Congo, 1995: determinants of survival. J Infect Dis. 1999;179(Suppl 1):S24–S27. doi: 10.1086/514311. [DOI] [PubMed] [Google Scholar]
- 65.Jahrling PB, Geisbert J, Swearengen JR, et al. Passive immunization of Ebola virus-infected cynomolgus monkeys with immunoglobulin from hyperimmune horses. Arch Virol Suppl. 1996;11:135–140. doi: 10.1007/978-3-7091-7482-1_12. [DOI] [PubMed] [Google Scholar]
- 66.Jahrling PB, Geisbert JB, Swearengen JR, Larsen T, Geisbert TW. Ebola hemorrhagic fever: evaluation of passive immunotherapy in nonhuman primates. J Infect Dis. 2007;196(Suppl 2):S400–S403. doi: 10.1086/520587. [DOI] [PubMed] [Google Scholar]
- 67.Jahrling PB, Geisbert TW, Geisbert JB, et al. Evaluation of immune globulin and recombinant interferon-α2b for treatment of experimental Ebola virus infections. J Infect Dis. 1999;179(Suppl 1):S224–S234. doi: 10.1086/514310. [DOI] [PubMed] [Google Scholar]
- 68.Alvarez CP, Lasala F, Carrillo J, Muniz O, Corbi AL, Delgado R. C-type lectins DC-SIGN and L-SIGN mediate cellular entry by Ebola virus in cis and in trans. J Virol. 2002;76(13):6841–6844. doi: 10.1128/JVI.76.13.6841-6844.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Simmons G, Reeves JD, Grogan CC, et al. DC-SIGN and DC-SIGNR bind Ebola glycoproteins and enhance infection of macrophages and endothelial cells. Virology. 2003;305(1):115–123. doi: 10.1006/viro.2002.1730. [DOI] [PubMed] [Google Scholar]
- 70.Dominguez-Soto A, Aragoneses-Fenoll L, MartiN-Gayo E, et al. The DC-SIGN-related lectin LSECtin mediates antigen capture and pathogen binding by human myeloid cells. Blood. 2007;109(12):5337–5345. doi: 10.1182/blood-2006-09-048058. [DOI] [PubMed] [Google Scholar]
- 71.Gramberg T, Soilleux E, Fisch T, et al. Interactions of LSECtin and DC-SIGN/DC-SIGNR with viral ligands: differential pH dependence, internalization and virion binding. Virology. 2008;373(1):189–201. doi: 10.1016/j.virol.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Takada A, Fujioka K, Tsuiji M, et al. Human macrophage C-type lectin specific for galactose and N-acetylgalactosamine promotes filovirus entry. J Virol. 2004;78(6):2943–2947. doi: 10.1128/JVI.78.6.2943-2947.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Takada A, Watanabe S, Ito H, Okazaki K, Kida H, Kawaoka Y. Downregulation of beta1 integrins by Ebola virus glycoprotein: implication for virus entry. Virology. 2000;278(1):20–26. doi: 10.1006/viro.2000.0601. [DOI] [PubMed] [Google Scholar]
- 74.Shimojima M, Takada A, Ebihara H, et al. Tyro3 family-mediated cell entry of Ebola and Marburg viruses. J Virol. 2006;80(20):10109–10116. doi: 10.1128/JVI.01157-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Empig CJ, Goldsmith MA. Association of the caveola vesicular system with cellular entry by filoviruses. J Virol. 2002;76(10):5266–5270. doi: 10.1128/JVI.76.10.5266-5270.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sanchez A. Analysis of filovirus entry into Vero E6 cells, using inhibitors of endocytosis, endosomal acidification, structural integrity, and cathepsin (B and L) activity. J Infect Dis. 2007;196(Suppl 2):S251–S258. doi: 10.1086/520597. [DOI] [PubMed] [Google Scholar]
- 77.Simmons G, Rennekamp AJ, Chai N, Vandenberghe LH, Riley JL, Bates P. Folate receptor α and caveolae are not required for Ebola virus glycoprotein-mediated viral infection. J Virol. 2003;77(24):13433–13438. doi: 10.1128/JVI.77.24.13433-13438.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Brindley MA, Hughes L, Ruiz A, et al. Ebola virus glycoprotein 1: identification of residues important for binding and postbinding events. J Virol. 2007;81(14):7702–7709. doi: 10.1128/JVI.02433-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Manicassamy B, Wang J, Jiang H, Rong L. Comprehensive analysis of Ebola virus GP1 in viral entry. J Virol. 2005;79(8):4793–4805. doi: 10.1128/JVI.79.8.4793-4805.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mpanju OM, Towner JS, Dover JE, Nichol ST, Wilson CA. Identification of two amino acid residues on Ebola virus glycoprotein 1 critical for cell entry. Virus Res. 2006;121(2):205–214. doi: 10.1016/j.virusres.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 81.Kuhn JH, Radoshitzky SR, Guth AC, et al. Conserved receptor-binding domains of Lake Victoria Marburgvirus and Zaire Ebolavirus bind a common receptor. J Biol Chem. 2006;281(23):15951–15958. doi: 10.1074/jbc.M601796200. [DOI] [PubMed] [Google Scholar]
- 82▪.Dube D, Brecher MB, Delos SE, et al. The primed ebolavirus glycoprotein (19-kilodalton GP1,2): sequence and residues critical for host cell binding. J Virol. 2009;83(7):2883–2891. doi: 10.1128/JVI.01956-08. Identifies the cathepsin cleavage site on glycoprotein1 and shows that the patch of lysine residues on the surface of glycoprotein1 are involved in viral attachment. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83▪▪.Chandran K, Sullivan NJ, Felbor U, Whelan SP, Cunningham JM. Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science. 2005;308(5728):1643–1645. doi: 10.1126/science.1110656. Describes the original identification of the essential role for cathepsin B and L in EBOV glycoprotein-mediated entry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schornberg K, Matsuyama S, Kabsch K, Delos S, Bouton A, White J. Role of endosomal cathepsins in entry mediated by the Ebola virus glycoprotein. J Virol. 2006;80(8):4174–4178. doi: 10.1128/JVI.80.8.4174-4178.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Diederich S, Thiel L, Maisner A. Role of endocytosis and cathepsin-mediated activation in Nipah virus entry. Virology. 2008;375(2):391–400. doi: 10.1016/j.virol.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pager CT, Craft WW., Jr Patch J, Dutch RE A mature and fusogenic form of the Nipah virus fusion protein requires proteolytic processing by cathepsin L. Virology. 2006;346(2):251–257. doi: 10.1016/j.virol.2006.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pager CT, Dutch RE. Cathepsin L is involved in proteolytic processing of the Hendra virus fusion protein. J Virol. 2005;79(20):12714–12720. doi: 10.1128/JVI.79.20.12714-12720.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Simmons G, Gosalia DN, Rennekamp AJ, Reeves JD, Diamond SL, Bates P. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc Natl Acad Sci USA. 2005;102(33):11876–11881. doi: 10.1073/pnas.0505577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Spear PG. Herpes simplex virus: receptors and ligands for cell entry. Cell Microbiol. 2004;6(5):401–410. doi: 10.1111/j.1462-5822.2004.00389.x. [DOI] [PubMed] [Google Scholar]
- 90▪▪.Harrison SC. Mechanism of membrane fusion by viral envelope proteins. Adv Virus Res. 2005;64:231–261. doi: 10.1016/S0065-3527(05)64007-9. Thorough and informative review on the three classes of viral membrane fusion proteins and their mechanisms of viral entry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91▪▪.Harrison SC. Viral membrane fusion. Nat Struct Mol Biol. 2008;15(7):690–698. doi: 10.1038/nsmb.1456. Thorough and informative review on the three classes of viral membrane fusion proteins and their mechanisms of viral entry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92▪▪.Lamb RA, Jardetzky TS. Structural basis of viral invasion: lessons from paramyxovirus F. Curr Opin Struct Biol. 2007;17(4):427–436. doi: 10.1016/j.sbi.2007.08.016. Thorough and informative review on the three classes of viral membrane fusion proteins and their mechanisms of viral entry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93▪▪.White JM, Delos SE, Brecher M, Schornberg K. Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit Rev Biochem Mol Biol. 2008;43(3):189–219. doi: 10.1080/10409230802058320. Thorough and informative review on the three classes of viral membrane fusion proteins and their mechanisms of viral entry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Stieneke-Grober A, Vey M, Angliker H, et al. Influenza virus hemagglutinin with multibasic cleavage site is activated by furin, a subtilisin-like endoprotease. EMBO J. 1992;11(7):2407–2414. doi: 10.1002/j.1460-2075.1992.tb05305.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hallenberger S, Bosch V, Angliker H, Shaw E, Klenk HD, Garten W. Inhibition of furin-mediated cleavage activation of HIV-1 glycoprotein gp160. Nature. 1992;360(6402):358–361. doi: 10.1038/360358a0. [DOI] [PubMed] [Google Scholar]
- 96.Chen J, Lee KH, Steinhauer DA, Stevens DJ, Skehel JJ, Wiley DC. Structure of the hemagglutinin precursor cleavage site, a determinant of influenza pathogenicity and the origin of the labile conformation. Cell. 1998;95(3):409–417. doi: 10.1016/s0092-8674(00)81771-7. [DOI] [PubMed] [Google Scholar]
- 97.Thoennes S, Li ZN, Lee BJ, et al. Analysis of residues near the fusion peptide in the influenza hemagglutinin structure for roles in triggering membrane fusion. Virology. 2008;370(2):403–414. doi: 10.1016/j.virol.2007.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wool-Lewis RJ, Bates P. Endoproteolytic processing of the Ebola virus envelope glycoprotein: cleavage is not required for function. J Virol. 1999;73(2):1419–1426. doi: 10.1128/jvi.73.2.1419-1426.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Neumann G, Geisbert TW, Ebihara H, et al. Proteolytic processing of the Ebola virus glycoprotein is not critical for Ebola virus replication in nonhuman primates. J Virol. 2007;81(6):2995–2998. doi: 10.1128/JVI.02486-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bullough PA, Hughson FM, Skehel JJ, Wiley DC. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature. 1994;371(6492):37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 101.Kampmann T, Mueller DS, Mark AE, Young PR, Kobe B. The role of histidine residues in low-pH-mediated viral membrane fusion. Structure. 2006;14(10):1481–1487. doi: 10.1016/j.str.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 102.Trkola A, Dragic T, Arthos J, et al. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature. 1996;384(6605):184–187. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- 103.Wu L, Gerard NP, Wyatt R, et al. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature. 1996;384(6605):179–183. doi: 10.1038/384179a0. [DOI] [PubMed] [Google Scholar]
- 104.Smith JG, Mothes W, Blacklow SC, Cunningham JM. The mature avian leukosis virus subgroup A envelope glycoprotein is metastable, and refolding induced by the synergistic effects of receptor binding and low pH is coupled to infection. J Virol. 2004;78(3):1403–1410. doi: 10.1128/JVI.78.3.1403-1410.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chan SY, Speck RF, Ma MC, Goldsmith MA. Distinct mechanisms of entry by envelope glycoproteins of Marburg and Ebola (Zaire) viruses. J Virol. 2000;74(10):4933–4937. doi: 10.1128/jvi.74.10.4933-4937.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Takada A, Robison C, Goto H, et al. A system for functional analysis of Ebola virus glycoprotein. Proc Natl Acad Sci USA. 1997;94(26):14764–14769. doi: 10.1073/pnas.94.26.14764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wool-Lewis RJ, Bates P. Characterization of Ebola virus entry by using pseudotyped viruses: identification of receptor-deficient cell lines. J Virol. 1998;72(4):3155–3160. doi: 10.1128/jvi.72.4.3155-3160.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Malashkevich VN, Schneider BJ, McNally ML, Milhollen MA, Pang JX, Kim PS. Core structure of the envelope glycoprotein GP2 from Ebola virus at 1.9-Å resolution. Proc Natl Acad Sci USA. 1999;96(6):2662–2667. doi: 10.1073/pnas.96.6.2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Freitas MS, Gaspar LP, Lorenzoni M, et al. Structure of the Ebola fusion peptide in a membrane-mimetic environment and the interaction with lipid rafts. J Biol Chem. 2007;282(37):27306–27314. doi: 10.1074/jbc.M611864200. [DOI] [PubMed] [Google Scholar]
- 110.Gomara MJ, Mora P, Mingarro I, Nieva JL. Roles of a conserved proline in the internal fusion peptide of Ebola glycoprotein. FEBS Lett. 2004;569(1–3):261–266. doi: 10.1016/j.febslet.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 111.Chan DC, Fass D, Berger JM, Kim PS. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89(2):263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 112.Chandran K, Li Q, Fabozzi G, Sullivan NJ, Cunningham JM. Cysteine cathepsins are host factors for filovirus infection. Keystone Symposia: Cell Biology of Virus Entry, Replication and Pathogenesis; Victoria, BC, Canada: Keystone Symposia; 2008. p. 29. [Google Scholar]
Website
- 201.Model of EBOV GP-mediated entry. www.nature.com/nature/journal/v454/n7201/index.html.