Abstract
Candida albicans is the most commonly isolated human fungal pathogen and uses a diverse repertoire of morphological transitions to aid colonization and infection. In a recent paper we discuss how one of these transitions, the white-to-opaque switch, is affected both by cell stress and by several other conditions that change the rate of cell growth. Based on our findings, we propose that the master regulator of the white-to-opaque switch, WOR1, acts as a sensitive monitor of both intrinsic and environmental conditions.
Key words: epigenetic, Candida albicans, oxidative stress, genotoxic stress, WOR1, white-opaque switch
In Candida albicans, white-opaque switching is an infrequent stochastic event occurring approximately once every 10,000 generations.1 White and opaque forms are physically distinguishable as white cells are round and form shiny, dome-shaped colonies while opaque cells are elongated and form darker, flatter colonies.2 The switch between white and opaque forms affects many features of C. albicans biology including virulence, biofilm formation and mating.3 The white-opaque switch is controlled by a master transcriptional regulator, WOR1 (White-Opaque Regulator 1). In white cells WOR1 expression is low or absent, but in opaque cells WOR1 expression is high. Positive feedback of Wor1 protein on its own promoter maintains this high level of expression and allows stable inheritance of the opaque state.4–6 A number of exogenous factors influence rates of white-opaque switching including UV irradiation, anaerobiasis and levels of CO2.7–9 In the new work, intrinsic defects in DNA repair, as well as genotoxic and oxidative stress, were shown to increase the frequency of switching from white to opaque.10 Uniting the latter observations was the fact that all of these conditions caused a notable impairment in the rate of growth. Thus, mutants in DNA recombination genes grew slowly, as did wildtype cells when treated with DNA-damaging agents or with hydrogen peroxide. Furthermore, two clinical isolates (P37005 and L26) exhibited slower growth than the laboratory strain of C. albicans, SC5314, and also showed high rates of white-to-opaque switching. The hyper-switching phenotype was directly related to the slow growth rate, as faster growing derivatives of P37005 switched to opaque only at basal levels. Finally, manipulation of WOR1 gene copy number also influenced rates of white-opaque switching; increasing gene number from 1 to 3 increased switching to opaque by nearly two orders of magnitude.10 This result emphasizes the close association between switching frequencies and the expression level of WOR1. Taken together, these findings support the model that increased accumulation of Wor1 occurs in slower growing strains, and this accumulation promotes switching to the opaque form.
These results have direct implications for studies on white-opaque switching. It is possible, for example, that changes in growth rate are responsible for the modulation of white-opaque switching observed in other studies. Alternatively, mechanisms could affect the switch independent of changes in growth rate. Since many factors impinge on cellular growth, the question becomes how to determine if a change in cell generation time is responsible for altered white-opaque switching? One striking observation is that modulation of growth rates did not result in 100% switching to opaque under any of the conditions tested. Instead, slow-growing strains exhibited rates of white-to-opaque switching that were in the 20–70% range (compared to ⊃1% switching in wild type SC5314-derived isolates). This contrasts with experiments in which the transcriptional regulators of the white-opaque switch were targeted; in many cases deletion or overexpression of these factors resulted in an all-or-nothing mode of switching.11–13 Interestingly, CO2 was recently shown to induce >90% switching to opaque in several C. albicans strains, again suggesting that, in this case, increased switching was not simply due to changes in growth rate.
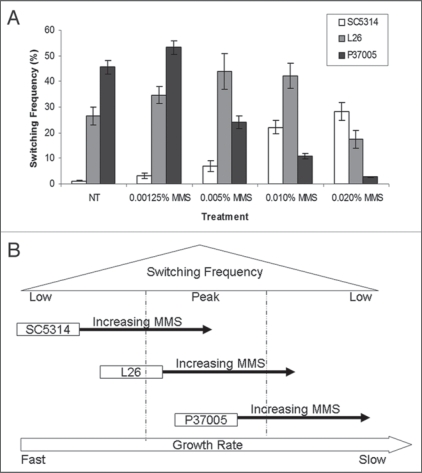
We also note that the relationship between cell generation times and white-opaque switching frequencies is a complex one in C. albicans. For example, high rates of switching to opaque are not observed in extremely slow-growing strains or in strains grown under nutrient limiting conditions.10 A possible explanation is that general RNA and protein synthesis is compromised under these conditions, therefore limiting the potential accumulation of Wor1 protein. There also appears to be a narrow window with respect to growth rates for optimal white-to-opaque switching. Generation times of SC5314-derived strains must be lengthened to enter this window while isolates such as P37005 naturally grow at rates that promote efficient switching. In further support of this model, treatment of strains with the DNA damaging agent MMS (methyl methane sulfonate), was found to have very different effects on switching frequencies. MMS induced high rates of switching to opaque in SC5314 (up to 30%) but had the opposite effect on the already slow-growing isolate, P37005. The latter normally exhibited 46% switching, but the frequency declined in the presence of MMS until, at 0.02% MMS, only 3% of colonies underwent switching (Fig. 1). Interestingly, L26 grew at an intermediate rate between that of SC5314 and P37005, and showed an intermediate response to MMS; low concentrations of MMS increased switching to opaque while higher concentrations inhibited switching. These experiments again suggest that the generation time of SC5314 must be extended to enter the optimal window for white-to-opaque switching, while perturbations to P37005 now result in this strain growing too slowly for efficient switching.
Figure 1.
Effects of MM S on white-opaque switching. (A) Effect of MM S on white-opaque switching in three backgrounds. White phase cells of SC5314, L26 and P37005 were grown on synthetic media containing increasing concentrations of MM S at room temperature. After 7 d colonies were analyzed for the presence of opaque sectors. Error bars represent standard error. (B) Model for how changes in growth rates differentially affects switching frequencies.
In summary, white-opaque switching affords C. albicans a rapid and heritable means of adaptation, and the switch appears acutely sensitive to factors that can alter Wor1 protein levels in the cell. This mechanism ensures that formation of opaques is enhanced in stressful environments, where this form could potentially offer a fitness advantage for the organism. For example, white and opaque cells show marked differences in their interactions with immune cells of the host, and opaque cells may be better suited to avoiding phagocytosis by macrophages.14,15 The white-opaque switch is therefore one component of C. albicans’ repertoire of physical transitions that has evolved to promote survival and adaptation to hostile environments in the mammalian host.
Acknowledgements
We are grateful to Joseph Bliss and Kim Sherwood for reading of the manuscript and helpful discussions. Work in the author’s laboratory was supported by a PATH award from the Burroughs Wellcome Fund (R.J.B.) as well as the NIH (R21AI081560 to R.J.B. and F31DE019752 to K.A.) and a training grant for Graduate Assistance in Areas of National Need (K.A.).
Footnotes
Previously published online: www.landesbioscience.com/journals/cib/article/9487
References
- 1.Rikkerink EH, Magee BB, Magee PT. Opaque-white phenotype transition: a programmed morphological transition in Candida albicans. J Bact. 1988;170:895–899. doi: 10.1128/jb.170.2.895-899.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Slutsky B, Staebell M, Anderson J, Risen L, Pfaller M, Soll DR. “White-opaque transition”: a second high-frequency switching system in Candida albicans. J Bact. 1987;169:189–197. doi: 10.1128/jb.169.1.189-197.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bennett RJ, Johnson AD. Mating in Candida albicans and the search for a sexual cycle. Ann Rev Micro. 2005;59:233–255. doi: 10.1146/annurev.micro.59.030804.121310. [DOI] [PubMed] [Google Scholar]
- 4.Huang G, Wang H, Chou S, Nie X, Chen J, Liu H. Bistable expression of WOR1, a master regulator of white-opaque switching in Candida albicans. Proc Natl Acad Sci USA. 2006;103:12813–12818. doi: 10.1073/pnas.0605270103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Srikantha T, Borneman AR, Daniels KJ, Pujol C, Wu W, Seringhaus MR, et al. TOS9 regulates whiteopaque switching in Candida albicans. Eukaryotic Cell. 2006;5:1674–1687. doi: 10.1128/EC.00252-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zordan RE, Galgoczy DJ, Johnson AD. Epigenetic properties of white-opaque switching in Candida albicans are based on a self-sustaining transcriptional feedback loop. Proc Natl Acad Sci USA. 2006;103:12807–12812. doi: 10.1073/pnas.0605138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang G, Srikantha T, Sahni N, Yi S, Soll DR. CO2 regulates white-to-opaque switching in Candida albicans. Curr Biol. 2009;19:330–334. doi: 10.1016/j.cub.2009.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morrow B, Anderson J, Wilson J, Soll DR. Bidirectional stimulation of the white-opaque transition of Candida albicans by ultraviolet irradiation. J Gen Micro. 1989;135:1201–1208. doi: 10.1099/00221287-135-5-1201. [DOI] [PubMed] [Google Scholar]
- 9.Ramirez-Zavala B, Reuss O, Park YN, Ohlsen K, Morschhauser J. Environmental induction of white-opaque switching in Candida albicans. PLoS Pathog. 2008;4:1000089. doi: 10.1371/journal.ppat.1000089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alby K, Bennett R. Stress-Induced Phenotypic Switching in Candida albicans. Mol Biol Cell. 2009;20:3178–3191. doi: 10.1091/mbc.E09-01-0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zordan RE, Miller MG, Galgoczy DJ, Tuch BB, Johnson AD. Interlocking transcriptional feedback loops control white-opaque switching in Candida albicans. PLoS Biol. 2007;5:256. doi: 10.1371/journal.pbio.0050256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vinces MD, Kumamoto CA. The morphogenetic regulator Czf1p is a DNA-binding protein that regulates white opaque switching in Candida albicans. Microbiology. 2007;153:2877–2884. doi: 10.1099/mic.0.2007/005983-0. [DOI] [PubMed] [Google Scholar]
- 13.Sonneborn A, Tebarth B, Ernst JF. Control of white-opaque phenotypic switching in Candida albicans by the Efg1p morphogenetic regulator. Infect Immun. 1999;67:4655–4660. doi: 10.1128/iai.67.9.4655-4660.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geiger J, Wessels D, Lockhart SR, Soll DR. Release of a potent polymorphonuclear leukocyte chemoattractant is regulated by white-opaque switching in Candida albicans. Infect Imm. 2004;72:667–677. doi: 10.1128/IAI.72.2.667-677.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lohse MB, Johnson AD. Differential phagocytosis of white versus opaque Candida albicans by Drosophila and mouse phagocytes. PLoS ONE. 2008;3:1473. doi: 10.1371/journal.pone.0001473. [DOI] [PMC free article] [PubMed] [Google Scholar]