Abstract
An AccQ•Tag Ultra performance liquid chromatography-electrospray ionization-tandem mass spectrometry (AccQ•Tag -UPLC-ESI-MS/MS) method for fast, reproducible and sensitive amino acid quantitation in biological samples, particularly, the malaria parasite Plasmodium falciparum is presented. The Waters Acquity TQD UPLC/MS system equipped with photodiode array (PDA) detector was used for amino acid separation and detection. The method was developed and validated using amino acid standard mixtures containing acidic, neutral, and basic amino acids. For MS analysis, the optimum cone voltage implemented, based on direct infusion analysis of a few selected AccQ•Tag amino acids with multiple reaction monitoring, varied from 29-39 V, whereas the collision energy varied from 15-35 V. Calibration curves were built using both internal and external standardization. Typically, a linear response for all amino acids was observed at concentrations ranges of 3 × 10−3-25 pmol/μL. For some amino acids, concentration limits of detection were as low as 1.65 fmol. The coefficients of variation for retention times were within the ranges of 0.08-1.08%. The coefficients of variation for amino acid quantitation, determined from triplicate UPLC-MS/MS runs, were below 8% on the average. The developed AccQ•Tag-UPLC-ESI-MS/MS method revealed good technical and biological reproducibility when applied to P. falciparum and human red blood cells samples. This study should provide a valuable insight into the performance of UPLC-ESI-MS/MS for amino acid quantitation using AccQ•Tag derivatization.
Keywords: Amino acid quantitation, derivatization, AccQ•Tag, UPLC, MRM, MS/MS, Plasmodium falciparum, malaria
INTRODUCTION
Amino acids are the key building blocks of proteins and other essential biomolecules and play important roles in the chemical reactions vital to life. Accurate analysis of amino acids is imperative to the life science and food industry.1, 2 Detection and accurate quantitation of amino acids in complex human tissues and body fluids, plants, and food samples is highly desirable, but often represents several challenges.3, 4,5 For example, long sample preparation time, when derivatization methods are used to enhance the sensitivity of amino acid detection, along with unwanted side reactions, and byproducts from derivatizing reagents used to chemically modify amino acids; the presence of interfering compounds, which can hinder separation and detection of amino acids when analyzing complex biological samples; as well as insufficient sensitivity, selectivity and robustness of analysis. Hence, the implementation of improved methodology for amino acid analysis capable of providing high selectivity, sensitivity, and reproducibility of analysis, with minimal sample preparation, reduced analysis time, and which performs well within a broad dynamic range of analyte concentrations would be greatly beneficial for applications requiring amino acid analysis in biological material.
Currently, multiple analytical platforms using diverse modes of separation are used for the analysis of amino acids in various matrices. These techniques include liquid chromatography, capillary electrophoresis, and gas chromatography, combined with various detection systems, such as fluorescence, ultraviolet, electrochemical, and mass spectrometry.6-12 Amino acid analysis has also been performed with microfluidic devices.13
Both direct analysis and chemical derivatization approaches have been implemented for amino acid detection. Liquid chromatography and capillary electrophoresis can be used for direct analysis of amino acids.14-17 Additionally, direct tandem mass spectrometric analysis has been developed.18 Direct analysis provides the advantage of being simple and rapid; however, it is insufficiently sensitive due to the presence of interfering compounds when analyzing complex matrices. This highlights the need for alternative techniques that can deliver adequate sensitivities and that are well suited to the analysis of crude extracts and complex samples.
Pre and post column chemical derivatization has been proposed in an effort to increase the sensitivity of amino acid analysis. This has led to the development of several derivatizing reagents that can react with the amino and/or carboxylic acid group of the amino acid, each exhibiting certain advantages and limitations.19-40 For example, Uutela et al.19 compared the performance of three commonly used amino acid derivatization reagents and showed 2-60 times improvement in limits of detection as a result of amino acid derivatization.19 The main challenge facing many derivatization strategies is sample interferences, particularly when non-specific detectors are used. In a recent study, Guo et al.41 developed a novel differential isotope labeling method that makes use of 12C-/13C-dansyl chloride to derivatize amine and phenol containing metabolites. The authors demonstrated that chemically altering the analyte improves the chromatographic retention of highly polar compounds on the reversed phased stationary phase. Signal enhancements of up to 3 orders of magnitude for dansyl-labeled analytes were reported in this study.
Among the chemical derivatization procedures introduced for amino acid analysis, the AccQ•Tag technology continues to gain widespread acceptance.39, 42, 43 This technique utilizes 6-aminoquinolyl-N-hydroxysuccinimidyl carbamate to transform primary and secondary amines into highly stable fluorescent derivatives.39, 42-44 An attractive feature of this technology is that the derivatization reaction is straightforward and can be completed within 10 minutes. Gardener et al.42 reported the use of the AccQ•Tag derivatization and high performance liquid chromatography (HPLC) for the analysis of amino acid variability in flowering plants.42 The AccQ•Tag technology has also been used in conjunction with ultra performance liquid chromatography (UPLC), offering new opportunities for faster analysis. Boogers et al.43 compared the performance of PicoTag-HPLC and AccQ•Tag-UPLC for the evaluation of amino acids in casein and bovine serum albumin hydrolysates. AccQ•Tag-UPLC improved limits of detection (LOD) up to 3 times while reducing the analysis time 2.5 fold. 43 The AccQ•Tag derivatization was also utilized in the LC-MS approach using an HPLC separation on a C18 column and detection with a 3-D ion trap mass spectrometer.45
To the best of our knowledge, up to now there have been no studies exploring the use of AccQ•Tag derivatization in connection with UPLC coupled to tandem quadrupole mass spectrometric detection for fast, selective and sensitive amino acid quantitation in biological samples. Here we report the development, validation and application of a fast and sensitive AccQ•Tag-UPLC-ESI-MS/MS method for amino acid detection and quantitation. The applicability of this method to the analysis of amino acids in the intraerythrocytic malaria parasite P. falciparum and human erythrocyte (red blood cell, RBC) samples is demonstrated.
EXPERIMENTAL SECTION
Chemicals and Reagents
Amino acid standards, including physiological acidics, neutrals, and basics, were purchased from Sigma (Saint Louis, MS, USA). Isotopically labeled amino acid standards [histidine (ring 2-13C); serine-2,3,3-d3; arginine (guanido-15N2); glycine-d5; L-asparagine (guanido-15N2); citrulline-ureido-13C; L-glutamic acid-2,3,4-d3; threonine (13C, 15N); D-L-alanine-2,3,3,3-d4; proline-2,5,5-d3; ornitine-3,3,4,4,5,5-d6; lysine-3,3,4,4,4,5,5,6,6-d8-01; 4-hydroxyphenyl-2,6-d2-alanine-2-d1-01; methionine-methyl-d3; valine-d8; leucine-d10; phenyl-d5-alanine; tryptophan-2′,4′,5′,6′,7′-d5(indole-d5)-01] were from Cambridge Isotope Laboratories (Andover, MA, USA). AccQ•Tag Ultra eluent concentrates and AccQ•Tag Ultra derivatization kit were purchased from Waters Corporation (Milford, MA, USA). Methanol was from J.T. Baker (Phillipsburg, NJ, USA). Deionized water was from a MilliQ Ultrapure water system (Millipore, Bedford, MA, USA). Ultra high purity argon and nitrogen gas for mass spectrometric analysis were obtained from Speciality Gases (Radnor, PA, USA).
Amino Acid Extraction from Biological Samples
Amino acids were extracted from P. falciparum and RBCs samples with an ice cold 50% (v/v) methanol:water solution, spiked with isotopically labeled internal standards at 4 μg/mL. A description on the protocol implemented for Plasmodium Falciparum culture is provided in the Supporting Information. In order to normalize parasite cell numbers between ring and trophozoite stages, 10 μl of extraction buffer were used per 1 nmol of hemozoin in ring stage, and 3.3 μl of extraction buffer per 1 nmol of hemozoin in trophozoite stage. The reason for this normalization is that trophozoites have approximately 3-5 times as much hemozoin than rings for an equivalent number of parasites.46 Samples were vortexed for 5 min, sonnicated in a water bath for 2 min, and incubated on dry ice for 5 min. Three cycles of water bath sonication and dry ice incubation were completed. Cellular debris was removed by centrifugation at 8000 g for 15 min and 250 μl of the extract were transferred to a clean tube and dried under a stream of N2. The dried extract was dissolved in 50 μl of the 50% (v/v) methanol:water solution.
AccQ•Tag Derivatization
Amino acid derivation with AccQ•Tag reagents was conducted according to the manufacturer’s protocol. Briefly, 10 μL of either a standard amino acid mix solution, or a biological extract were mixed with 70 μL of AccQ•Tag Ultra borate buffer, and 20 μL of AccQ•Tag reagent previously dissolved in 1.0 mL of AccQ•Tag Ultra reagent diluent were added. The reaction was allowed to proceed for 10 min at 55 °C.
UPLC Analysis
Liquid chromatographic analysis was performed on a Waters Acquity UPLC system, equipped with a binary solvent manager, an autosampler, a column heater, a PDA detector, and interfaced to a tandem quadrupole detector. The separation column was a Waters AccQ•Tag Ultra column (2.1 mm i.d. × 100 mm, 1.7 μm particles). The column heater was set at 55 °C and the mobile phase flow rate was maintained at 0.7 mL/min. Eluent A was 10% AccQ•Tag Ultra concentrate solvent A, and eluent B was 100% AccQ•Tag Ultra solvent B. The non-linear separation gradient was 0-0.54 min (99.9% A), 5.74 min (90.0% A), 7.74 min (78.8% A), 8.04-8.64 min (40.4% A), 8.73-10 min (99.9% A). A VanGuard™ Waters column (2.1 mm i.d. × 5 mm, 1.7 μm particles) was used as the guard column. One microliter of sample was injected for analysis. The PDA detector was set at 260 nm, with a sampling rate of 20 points/sec.
Total Heme Determination
Total heme content was determined by decrystallizing heme crystal in aliquots of parasite lysates diluted into 1 ml of 20 mM sodium hydroxide:2% SDS, incubating the suspension at room temperature for 2 h, and then reading the OD at 400 nm (Beckman DU 640 spectrophotometer). The Molar Extinction Coefficient for heme is 100,000 at 400 nm in 20 mM sodium hydroxide:2%SDS.
Direct Infusion ESI-MS/MS
One mg/mL solutions of selected amino acid standards were derivatized with AccQ•Tag Ultra as described above, and further diluted 5 times with deionized water to a final concentration of 20 μg/mL. These derivatized amino acid solutions were individually infused into the ESI ion source of the Waters TQD mass spectrometer at a low flow rate of 20 μL/min or at a combined flow. For the low flow setting (20 μL/min), the desolvation temperature and gas flow rate were 250 °C and 500 L/hour, respectively. For the combined flow setting (200 μL/min), these values were changed to 350 °C and 600 L/hour, respectively.
ESI-MS/MS
MS detection was carried out using a Waters TQD tandem quadrupole detector interfaced with an Acquity UPLC system via an ESI probe. The mass spectrometer was automatically tuned and calibrated using the Waters IntelliStart software. A standard tuning and calibration solution (Waters Corporation, Milford, MA, USA) was directly infused into the mass spectrometer ion source at a flow rate of 20 μL/min. The ESI source was operated in the positive ionization mode at 150 °C, with a desolvation temperature of 450 °C, a 900 L/hour desolvation gas flow rate, and a capillary voltage set at 4.4 kV. The extractor and radiofrequency voltage were fixed to 3.0 and 0.10 V respectively. The cone voltage varied from 29-37 V depending on the amino acid investigated. Argon was used as the collision gas at a flow rate of 0.10 mL/min, and collision energies of 19-35 eV. MS methods, with multiple reaction monitoring (MRM), were designed with five functions covering the 10 min LC separation time window. The dwell time was set to either 0.01 or 0.02 s. Data analysis and quantitation was performed using the Waters MassLynx and QuanLynx software.
Method Validation
Method validation involved the determination of limits of detection, dynamic range, low limits of linearity, high limits of linearity, reproducibility of amino acid identification, and quantification accuracy. Calibration curves were built using serial dilutions of a 0.5 μmol/mL AccQ•Tag derivatized amino acid standard mix solution, spiked with isotopically labeled internal standards (listed in the chemicals and reagents section above) at 4 μg/mL. For serial dilutions a Biomek 2000 Beckman Coulter laboratory automation workstation (Fullerton, CA, USA) was used. The concentration levels of amino acids spanned from 0.25 μmol/mL to 470.68 fmol/mL, whereas the concentration of all internal standards was maintained constant at 2 μg/mL. Three replicate injections were made for each calibration level.
Amino Acid Quantitation in Biological Samples
Amino acid concentrations in Plasmodium falciparum and RBCs were calculated from calibration curves. The technical and biological reproducibility of amino acid detection and quantification were evaluated. Three replicate UPLC-MS/MS analyses were considered for technical reproducibility. Four biological replicates, in the case of ring stage, and five biological replicates, in the case of trophozoite stage, were used to estimate the biological reproducibility.
RESULTS AND DISCUSSION
Method Development
This study aimed at the development of a fast, selective and sensitive AccQ•Tag-UPLC-ESI-MS/MS method using MRM for amino acid measurement in complex biological samples, the malaria parasite P. falciparum and human RBCs in particular. The AccQ•Tag derivatization method uses the N-hydroxysuccinimide-activated heterocyclic carbamate to react with primary and secondary amines, yielding stable fluorescent derivatives. The reaction is performed at a pH value of ~9.0 and at 55 °C. Under these conditions, amino acid derivatization takes place within seconds, but it is allowed to proceed for as long as 10 minutes to guarantee complete conversion and high stability of the formed derivatives. Un-reacted AccQ•Tag reagent hydrolyzes readily within 1 minute, forming 6-aminoquinoline which can be easily separated from the derivatives, along with the non-interfering compounds N-hydroxysuccinimide and carbon dioxide. The chemistry behind the AccQ•Tag derivatization reaction is depicted in Figure S-1. Because the amino acid derivatization reaction with AccQ•Tag is well understood and fully developed,23, 36, 39 yielding a ~100% amino acid conversion, no efforts were made to optimize the reaction conditions.
Although the applicability of the AccQ•Tag derivatization technique using fluorescence detection has already been published, the method presented here has been developed specifically for mass spectrometric detection. Hence, the proposed method offers high sensitivity and specificity of analysis for critical applications requiring selective detection of low-abundance amino acids in complex biological samples. The methodology should offer alternative assays to the biotechnology, pharmaceutical, biomedical, and food industry in need of effective amino acid analysis methods that deliver high-throughput, selectivity and sensitivity.
The main novelty of the study reported here is the implementation of the AccQ•Tag method in an Acquity UPLC system combined with a tandem quadrupole detector, operated in the MRM mode, as well as the demonstration of the applicability of this method for high-throughput sensitive and targeted amino acid measurement in biological material. The major advantage of using MRM, in contrast to full-scan MS detection, is increased sensitivity and selectivity by avoiding interfering background signals generated from substances that might be either part of the sample or the mobile phase used for analyte separation.
A method for amino acid quantitation was initially developed and evaluated using a amino acid standard solution and was applied to amino acid analysis in complex biological samples, such as the P. falciparum ring or trophozoite stages and RBCs. Method development entailed adjusting liquid chromatographic and mass spectrometric conditions (source and analyzer settings), as well as implementing amino acid extraction protocols for P. falciparum and RBCs extracts. When the concentration of AccQ•Tag Ultra concentrate A in the mobile phase A was 10% and the mobile phase flow rate was set at 0.7 mL/min, all amino acids were resolved within less than 10 min (data not shown). Therefore, these chromatographic conditions were maintained for all further experiments.
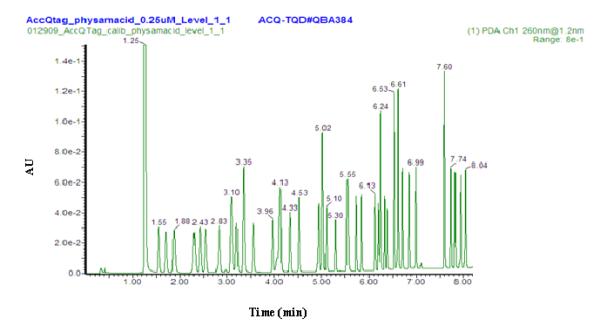
Figure 1 shows a typical chromatogram for the UPLC analysis of a standard mixture of AccQ•Tag derivatized amino acid standards using a PDA detector. All chromatographic separations were performed with a guard column installed at the front of the analytical column. Notice from Figure 1 that amino acid separation was accomplished within 8.5 minutes, and the chromatographic peaks were usually less than 2 seconds wide. The peak observed at ~1.25 min corresponded to the major hydrolysis product (6-aminoquinoline, AMQ) of un-reacted AccQ•Tag reagent. Having an excess of AccQ•Tag reagent in the sample ensures that the reaction goes to completion. Since the main focus of this study was to develop an AccQ•Tag-UPLC-ESI-MS/MS method for rapid, sensitive and accurate targeted amino acid quantification in biological samples, and because the UPLC analysis of AccQ•Tag amino acid derivatives has already been reported in the literature,43 no attempts were made to conduct an in depth optimization of the chromatographic separation of amino acids in terms of efficiency, resolution, and peak capacity.
Figure 1.
Typical chromatogram for UPLC-PDA analysis of AccQ•Tag derivatized physiological amino acid standards. UPLC-PDA system; 2.1 × 100 mm AccQ•Tag column; 0.7 mL/min; 250 pmol/mL standard concentration; 1 μL sample injected.
MS method development included the determination of AccQ•Tag amino acid derivatives’ retention time, their respective intense and specific parent-daughter transitions, the estimation of optimal cone voltage and collision energy, as well as the adjustment of number of MRM functions, time window per function, number of MRM transitions monitored per function, and the dwell time.
In order to create an MRM-MS method for high-throughput targeted amino acid quantification, retention times and parent ions for each particular AccQ•Tag derivatized physiological amino acid had to be determined. To this aim, experiments were first conducted with 0.25 μmol/mL standard amino acid solutions using full scan MS detection. Supplemental Table S-1 shows the predicted parent ions, the experimentally observed retention times and parent ions for AccQ•Tag amino acids, and the isotopic amino acid derivatives.
Optimal cone voltage and collision energy settings for MRM transitions were found for a few selected AccQ•Tag amino acid standards by means of direct infusion into a tandem quadrupole (TQD) mass spectrometer, as described in the experimental section-direct infusion ESI-MS/MS. The purpose for optimizing these values for individual amino acids was to estimate experimental conditions, yielding adequate sensitivity and selectivity of analysis. The amino acids selected for this optimization study were chosen in a way to include at least one amino acid with side chains from each group (i.e. polar, non-polar, acidic, basic, and neutral) in order to have a truly representative group of amino acids with a variety of chemical properties. This is an important aspect to consider when developing a method to be applied to the analysis of biological samples, where compound chemical nature diversity needs to be taken into consideration.
The MRM transitions that were used for targeted amino acid detection corresponded to the intense parent-daughter ion transitions characteristics of each particular amino acid. Since all amino acids were derivatized with AccQ•Tag reagents, and upon MS/MS fragmentation this reagent generates the 171.01 m/z diagnostic ion, this specific ion was selected as the daughter ion in all MRM transitions. Thus, all amino acids shared the same daughter ion, but differed in their precursor ion. Table S-2 displays the cone voltage and collision energy determined for 10 AccQ•Tag derivatized amino acids, including, alanine, arginine, glutamic acid, glycine, 4-hydroxy-L-proline, isoleucine, proline, serine, threonine and valine. Two replicate infusions were performed for every amino acid and the cone voltage and collision energy values that were implemented in the final MS method corresponded to the average of these two measurements. Amino acid infusion into the mass spectrometer was carried out using both low flow and combined flow. In the low flow mode, only the sample (20 μL/min) was infused, whereas in the combined flow mode this same volume of sample was combined with a mobile phase flow rate of 180 μL/min, for a total flow rate of 200 μL/min. The outcome of these two different infusion modes was similar. However, the advantage of using combined flow instead of low flow, for this particular application, is that signal suppression, due to the presence of the non-volatile borate buffer introduced in the sample during amino acid derivatization with the AccQ•Tag reagent, is less problematic as a result of a concomitant dilution of the borate ions with the mobile phase during sample infusion. We noticed that when performing direct infusion experiments, dilution of the AccQ•Tag derivatized sample with an aqueous solution was necessary prior to mass spectrometric analysis to decrease the amount of borate buffer in the sample and obtain reproducible signals. Evidently the dilution approach would become impractical for the analysis of biological material where analyte concentrations are typically low. Additionally, the source required more frequent cleaning to remove interfering material. These observations are not surprising, as it is well-known that the presence of non-volatile inorganic substances in the sample adversely affects analyte ionization efficiency. For example, Nagy, et al.18 observed a 95% loss of signal intensity in electrospray for a solution of 23 standard amino acids containing 130 mmol/L sodium chloride. Thus, when infusing the sample directly into the mass spectrometer without any prior chromatographic separation, care must be taken to maintain the borate buffer at very low concentrations to minimize the formation of salt deposits on the source. Therefore, for direct infusion applications, it would be of interest to investigate the efficacy of alternative volatile buffers for carrying out the AccQ•Tag derivatization reaction.
Optimum cone voltage and collision energy for the evaluated amino acids were on the average close to 30 V and 25 eV, respectively. Therefore, for all remaining amino acids that were not individually optimized, a 30 V cone voltage, and a 25 eV collision energy was applied through all the experiments.
A MS method based on MRM was built for accurate quantification of AccQ•Tag amino acid derivatives. The parameters that were explored for MS method development encompassed: total number of functions, number or MRM transitions per function, time window per function, and dwell time. Based on the results reported in Tables S-1 and S-2, an MRM-MS method consisting of 5 time functions was implemented for the analysis of AccQ•Tag derivatized amino acids by UPLC-ESI-MS/MS. It is generally accepted that as the number of scans within a MS peak increases, a more accurate profile of the peak is obtained, which results in more reliable quantitation. It has been suggested that for quantitation purposes, the number of scans within a peak should be ~15 or higher. Hence, the function time window and dwell time were varied in an effort to generate as many data points per peak as possible. Several MS methods were developed with the dwell time kept at 0.02 s, whereas the number of functions, number of MRM transitions per function, and time window were varied. Preliminary attempts to develop a MS method for AccQ•Tag amino acid measurement resulted in quantification coefficients of variation as high as 60% (data not shown here). This was, primarily, the result of the few scans (<7) obtained within a peak. Nevertheless, when the 10 min long chromatographic run was divided into 5 functions, each with a duration time within the range of 1.4-2.8 min, a reduced number of MRM transitions per function, and a dwell time of either 0.01 or 0.02 s, a marked improvement in quantitation reproducibility was observed. These results will be addressed later in more detail. The two final MS methods that rendered satisfactory amino acid quantification reproducibility are listed in Table 1.
Table 1.
MS methods for analysis of AccQ•Tag derivatized physiological amino acid standards by UPLC-MS/MS using five multiple reaction monitoring functions. Experimental conditions: 2.1 × 100 mm AccQ•Tag colum; 0.25 μmol/mL AccQ•Tag derivatized physiological amino acids, 1 μL sample injection volume; positive ionization mode.
| Function # | MRM Transitions | Time window (min) | Dwell time (s) | |
|---|---|---|---|---|
| Method #1 | Method # 2 | |||
| 1 | 11 | 1-3 | 0.01 | 0.02 |
| 2 | 18 | 2.5-4.5 | 0.01 | 0.02 |
| 3 | 13 | 4.1-6.5 | 0.01 | 0.02 |
| 4 | 17 | 6.1-7.5 | 0.01 | 0.02 |
| 5 | 11 | 7.2-10 | 0.01 | 0.02 |
The coefficients of variation of peak area of AccQ•Tag amino acids derivatives analyzed by UPLC-ESI-MS/MS using the two methods shown in Table 1 were calculated from triplicate UPLC-ESI-MS/MS analysis. Both 0.02 s and 0.01 s dwell times generated low peak areas coefficients of variation, and the majority of these coefficients of variation were below 8% for the 0.01 s dwell time (See Supplemental Table S-3 in the Supporting Information). Nevertheless, lower coefficients of variation for peak areas were generally observed for a 0.01 s dwell time versus a 0.02 s dwell time. Thus 0.01 s was selected for further experiments. It is noteworthy to mention that the reproducibility of amino acid quantitation reported with either CE-MS/MS,47 or LC-MS/MS40 based methods has been on the average below 10%. However, this reproducibility is contingent to amino acid abundance. Hence, an improvement in quantitation reproducibility has been observed with an increase in amino acid concentration as shown in reference. 47, 40 Therefore, in terms of quantitation reproducibility, our method proved comparable to existing methods.
It is noteworthy to mention that ion suppression effects due to the borate buffer used to adjust the pH of the sample prior to derivatization with AccQ•Tag were not observed throughout the experiments, even after 200 consecutive UPLC-MS/MS analyses. The reason for this is that in spite of the borate buffer being a non-friendly mass spectrometric solution as a result of its poor volatility, the volume of sample injected (1-2 μL) into the separation column is negligible in comparison to the mobile phase flow rate (0.7 mL/min) used. The AccQ•Tag amino acid derivatives displayed high stability. For example, when stored at room temperature, the derivatives were stable for at least 2 weeks and when stored at 4 °C the stability was extended to ~1 month (data not shown). However, storage at room temperature is preferred to prevent analyte precipitation at low temperatures.
Method Validation
Method validation addressed the evaluation of coefficients of variation of retention times for AccQ•Tag derivatized amino acids, building of calibration curves, determination of dynamic range, low limits of linearity, high limits of linearity, limits of detection and reproducibility of quantitation.
The MRM transitions and average retention times with their respective standard deviations and coefficients of variation for the studied AccQ•Tag amino acid and isotopic standard derivatives are presented in Table 2. Approximately 70% of the amino acids were detected within the first 3 MRM functions, comprising the 1-6.5 min time window. Due to the narrow peak widths and highly reproducible separations achievable with UPLC, as well as the selectivity provided by MRM, reliable identification and quantitation of isobaric and/or isomeric amino acids, which represents a challenge in the majority of mass spectrometric based methods especially without a precluding chromatographic separation,18 could be accomplished with the method described herein. This method could easily discriminate among hydroxyl-L-proline, isoleucine and leucine. In addition, 1-methyl histidine and 3-methyl histidine could be differentiated. L-Sarcosine, β-alanine L-alanine is yet another example. The results reported in this table were calculated from the data obtained across the entire calibration curve (20 levels, 3 replicate injections each). Notice the highly reproducible separations. The retention time coefficients of variations were as low as 0.08% and not larger than 1.08%.
Table 2.
Reproducibility of retention times across the entire calibration curves for AccQ•Tag derivatized physiological amino acid standards, and isotopically labeled amino acids, analyzed by UPLC-MS/MS. Experimental conditions were the same as in Table 1.
| Amino acid | MRM transition | Function # | Average RT (min) | STD | CV (%) |
|---|---|---|---|---|---|
| Hydroxy-L-proline | 302.11 > 171 | 1 | 1.61 | 0.02 | 1.08 |
| Histidine (ring 2-13C) | 327.21 > 171 | 1 | 1.71 | 0.02 | 0.09 |
| L-Histidine | 326.21 > 171 | 1 | 1.70 | 0.01 | 0.68 |
| 1-Methyl-histidine | 340.21 > 171 | 1 | 2.06 | 0.02 | 0.74 |
| Taurine | 296.11 > 171 | 1 | 2.19 | 0.02 | 0.79 |
| 3-Methyl-histidine | 340.21 > 171 | 1 | 2.30 | 0.01 | 0.5 |
| Serine-2,3,3-d3 | 279.11 > 171 | 1,2 | 2.60 | 0.01 | 0.38 |
| L-Serine | 276.11 > 171 | 1,2 | 2.63 | 0.01 | 0.47 |
| Arginine (guanido-15N2) | 347.36 > 171 | 2 | 3.99 | 0.01 | 0.14 |
| L-Arginine | 345.21 > 171 | 2 | 2.86 | 0.02 | 0.58 |
| Glycine-d5 | 248.25 > 171 | 2 | 2.99 | 0.01 | 0.19 |
| Glycine | 246.08 > 171 | 2 | 3.01 | 0.01 | 0.33 |
| L-Carnosine | 397.21 > 171 | 2 | 2.84 | 0.01 | 0.39 |
| Ethanolamine | 232.09 > 171 | 2 | 3.16 | 0.01 | 0.27 |
| L-Asparagine-15-N2 | 305.27 > 171 | 2 | 3.39 | 0.01 | 0.17 |
| L-Aspartic acid | 304.11 > 171 | 2 | 3.38 | 0.01 | 0.27 |
| L-Sarcosine | 260.17 > 171 | 2 | 3.83 | 0.01 | 0.13 |
| Citrulline-ureido-13C | 347.35 > 171 | 2 | 3.99 | 0.01 | 0.14 |
| L-Glutamic acid-2,4,4-d3 | 321.11 > 171 | 2 | 3.96 | 0.01 | 0.15 |
| L-Glutamic acid | 318.11 > 171 | 2 | 3.97 | 0.01 | 0.19 |
| L-Citrulline | 346.21 > 171 | 2 | 3.99 | 0.01 | 0.15 |
| β-Alanine | 260.17 > 171 | 2,3 | 4.20 | 0.01 | 0.18 |
| Threonine-d5 | 295 > 171 | 3 | 4.43 | 0.01 | 0.13 |
| L-Threonine (13C, 15N) | 295.11 > 171 | 3 | 4.43 | 0.01 | 0.13 |
| L-Threonine | 290.11 > 171 | 3 | 4.43 | 0.01 | 0.13 |
| L-Alanine | 260.1 > 171 | 3 | 4.86 | 0.01 | 0.15 |
| D-L-Alanine-2,3,3,3-d4 | 264.11 > 171 | 3 | 4.86 | 0.01 | 0.12 |
| γ-Amino-n-butyric acid | 274.11 > 171 | 3 | 5.03 | 0.01 | 0.12 |
| Amino adipic acid | 332 > 171 | 3 | 5.24 | 0.00 | 0.08 |
| D-L-β-Aminoisobutyric acid | 274.11 > 171 | 3 | 5.48 | 0.01 | 0.27 |
| Proline-2,5,5-d3 | 289 > 171 | 3 | 5.49 | 0.01 | 0.11 |
| L-Proline | 286.16 > 171 | 3 | 5.51 | 0.03 | 0.58 |
| δ-hydroxylysine | 33.21 > 171 | 3 | 5.23 | 0.01 | 0.23 |
| L-α-Amino-n-butyric acid | 274.11 > 171 | 3 | 6.09 | 0.02 | 0.33 |
| Ornitine-3,3,4,4,5,5-d6 | 479.22 > 171 | 4 | 6.19 | 0.02 | 0.37 |
| L-Ornitine | 303.21 > 171 | 4 | 6.20 | 0.01 | 0.08 |
| L-Lysine | 317.21 > 171 | 4 | 6.60 | 0.01 | 0.19 |
| 4-Hydroxyphenyl-2,6-d2-alanine-2-d1-01 | 355.22 > 171 | 4 | 6.68 | 0.01 | 0.09 |
| Tyrosine | 352.21 > 171 | 4 | 6.69 | 0.01 | 0.01 |
| Methionine-methyl-d3 | 323.13 > 171 | 4 | 6.83 | 0.01 | 0.08 |
| L-Methionine | 320.21 > 171 | 4 | 6.86 | 0.05 | 0.7 |
| Valine-d8 | 296.11 > 171 | 4 | 6.95 | 0.01 | 0.08 |
| L-Valine | 288.21 > 171 | 4 | 6.98 | 0.01 | 0.09 |
| L-Isoleucine | 302.21 > 171 | 5 | 7.73 | 0.02 | 0.19 |
| Leucine-d10 | 312.24 > 171 | 5 | 7.78 | 0.01 | 0.07 |
| L-Leucine | 302.21 > 171 | 5 | 7.82 | 0.03 | 0.40 |
| Phenyl-d5-alanine | 341.21 > 171 | 5 | 7.92 | 0.02 | 0.26 |
| Phenyl-alanine | 336.21 > 171 | 5 | 7.92 | 0.01 | 0.12 |
| Tryptophan-2′,4′,5′,6′,7′-d5(indole-d5)-01 | 380.21 > 171 | 5 | 8.01 | 0.01 | 0.07 |
The dynamic range, high limits of linearity, low limits of linearity, and detection limits for AccQ•Tag amino acid derivatives were obtained from calibration curves that were built using solutions of standard amino acid mix spiked with isotopically labeled amino acids. These calibration solutions were prepared at 20 concentration levels, expanding from 0.25 μmol/mL to 470 fmol/mL, while the concentration of isotopically labeled amino acids was kept constant at 2 μg/mL in all calibration solutions. Three replicate UPLC-MS/MS analysis were performed for each of the concentration levels. The equation used to calculate the detection limits is shown in the Supplemental equation 1 in the Supporting Information.
The limits of detection and linearity of the calibration curves for AccQ•Tag derivatized physiological amino acid standards are included in the supplemental Table S-4 in the Supporting Information. The values reported in this table were obtained using the external standardization method, in which absolute peak areas are correlated to amino acid concentration. The internal standardization method, in which the amino acid peak areas were correlated to the areas of their corresponding isotopes delivered comparable results (data not included).
Most of the amino acids generated calibration curves that were linear within three orders of magnitude. Some displayed dynamic ranges covering up to four orders of magnitude, while a few exhibited linearity over two orders of magnitude. Typically, linearity of quantitation for amino acid concentrations expanding 2 orders of magnitude, using derivatization methods and MRM, has been demonstrated by other authors.17,47,40 Thus, the dynamic range attained by our method was either equal or better by at least 2 orders of magnitude than previously reported methods.
Detection limits were within 1.65-17.95 fmol/ μL. These detection limits are significantly lower than those reported for amino acid quantitation using alternative methods. For example, Nagy et al.18 introduced a novel direct tandem mass spectrometric method for amino acid analysis in neonatal screening that generated 2-42 pmol/μL detection limits.18 Thiele, et al.16 utilized a derivatization free method based on a strong cation exchange liquid chromatography coupled to tandem mass spectrometry for amino acid quantitation in plant tissues. The detection limits for their elegant approach were estimated to be 0.1-3 pmol/μL.16 Peris et al.6 reported an approach for characterizing proteinaceous glues in old paintings. Their strategy, which relied on amino acid quantitation using o-phtalaldehyde derivatization reagents in combination with HPLC and fluorescence detection, yielded detection limits of 600 fmol/μL.6 More recently, Uutela et al.19 compared the performance of several derivatization reagents for amino acid quantitation in rat brain microdialysates. For derivatized amino acids, the detection limits were 7.5-75 fmol.19 Jinmao et al.2 synthesized a new derivatizing reagent (2-(11H-benzo[α]-carbazol-11-yl) ethyl chloroformate and used it for improved amino acid detection by HPLC and fluorescence detection. Their derivatization reagent enabled sensitive detection of amino acids with detection limits that ranged from 1.6 to 14.0 fmol.2 It worthwhile mentioning that these concentration limits of detection are within the same range afforded by the AccQ•Tag-UPLC-ESI-MS/MS method that we present in this manuscript. However, because this approach used HPLC for amino acids analysis, separation times were as long as 60 minutes. In addition, a clean-up step, as a prelude to chromatographic separation, was necessary to remove excess of derivatizing reagent, which might interfere during the analysis. The AccQ•Tag-UPLC-ESI-MS/MS method described here, in contrast, requires separation times no longer than 10 minutes, and the entire analysis, from sample extraction to analyte detection, can be accomplished within 40 minutes. Because interferences owing to excess of derivatizing reagent are not a concern with the AccQ•Tag technology, sample clean-up is not necessary following amino acid derivatization. This can minimize potential problems arising from sample losses due to excessive sample manipulation.
A recent study by Shimbo, et al.40 reported the use of a novel derivatization reagent [3-aminopyridyl-N-hydroxysuccinimidyl carbamate (APDS)] for high-speed analysis of amines and amino acids in biological fluids by LC-ESI-MS/MS. Over 100 compounds with amino groups could be analyzed within 10 minutes. The limits of quantitation attained were between 0.3 and 9.3 pmol/μL. The dynamic range was within 2 orders of magnitude. With the exception of asparagines, RSD values for retention times were within 0.9 and 1.1% for intra-day and inter-day analysis, respectively. For amino acid quantitation, the RSDs were within 4.5 and 8.0%. For a few amino acids, these RSD values were observed to increase to ~ 15.1% as the amino acid concentration decreased to 5-10 pmol/μL.
Attractive approaches for the direct analysis of amino acids by capillary electrophoresis (CE) have been reported. 17,47 For example, Soga, et al.17 implemented a CE-ESI-MS method for the analysis of free amino acids. Their method provided a 100-fold increase in sensitivity compared to CE-UV based methods. For a 3 nL injection volume, limits of detection for basic amino acids were superior to those of acidic amino acids (i.e., 0.1 pmol/μL vs 6.0 pmol/μL). The linearity of the method covered the range from 10 to 500 pmol/μL, with migration time and quantitation RSDs of ~ 1.2% and 6.5%, respectively. In a subsequent study by Soga et al.,47 a CE-ESIMS/MS method was introduced for quantitation of amino acids without the use of derivatization reagents. The minimum detectable levels for 32 amino acids were between 0.1 and 14 pmol/μL. Calibration curves were linear from 10 pmol/μL to 200 pmol/μL. Overall, RSD values for migration time and peak areas were 0.4% and 8.6%, respectively. Quantitation of underivatized D/L-amino acids at the attomole levels has been demonstrated Moini et al.,48, 49 who used 18-crown-6-tetracarboxylic acid in the background electrolyte as a complexation reagent. The use of this complexation reagent significantly increased the sensitivity of detection. The results from this novel method were high separation efficiencies and nanomolar concentration detection limits.
Using the developed AccQ•Tag-UPLC-ESI-MS/MS for amino acid quantitation reported in the present study, limits of detection were in the low femtomole range, with dynamic ranges covering up to 4 orders of magnitude, and comparable RSD values for retention time and amino acid quantitation. An increase in RSDs for certain amino acids quantified at low concentrations was also observed in this study, which is expected because as it is well known at low analyte concentrations, quantitation is compromised.
Application to Amino Acid Quantitation in Biological System: P. falciparum and Human Red Blood Cells (RBCs)
Our research has a strong interest in understanding the mechanisms of infection of red blood cells by the malaria parasite P. falciparum and evaluating the metabolic response of drug treatment on infection. Therefore, the performance of the developed AccQ•Tag-UPLC-MS/MS method was evaluated for amino acid analysis in malaria parasite P. falciparum at two different developmental, ring or trophozoite, stages within RBCs, as well as within human RBCs. Upon invading an erythrocyte, a P. falciparum merozoite quickly transforms into a ring stage parasite which is very transcriptionally active and relatively resistant to malaria drugs for about 24 hours. The next morphologic trophozoite stage has by definition visible heme crystals. This stage is associated with more than 70% of host hemoglobin ingestion and catabolism to amino acids. Most malaria drugs are effective against this stage. DNA synthesis occurs during late trophozoite stage, before nuclear division, which morphologically marks the schizont stage. Completion of schizogony produces 8-32 merozoites, which begin the 48 hour cycle again.
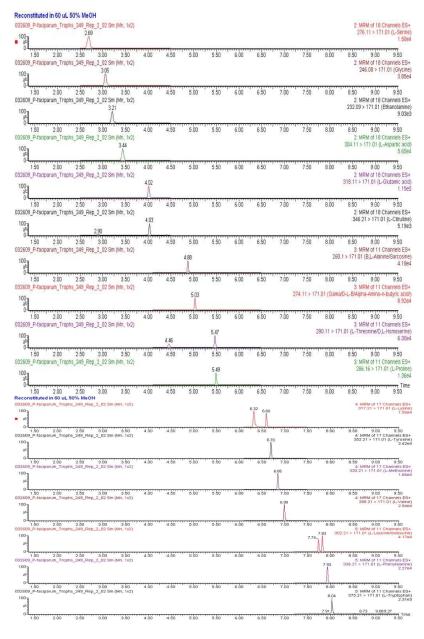
Typical UPLC-ESI-MS/MS chromatograms for the P. falciparum trophozoites, obtained for all five MRM functions within the MS method are introduced in supplemental Figure S-2. A total of 16 amino acids were reliably identified and quantified in the P. falciparum rings or trophozoites, namely serine, glutamic acid, aspartic acid, glycine, ethanolamine, threonine, proline, γ-amino-n-butyric acid, alanine, tyrosine, methionine, valine, tryptophan, phenylalanine, isoleucine, and leucine. Representative MRM transitions for each of these amino acids appear in Figure 2. Analysis of P. falciparum ring stage generated similar UPLC-ESI-MS/MS profiles and ion chromatograms. This separate duplicate data was not included in this manuscript.
Figure 2.
Ion chromatograms of AccQ•Tag derivatized amino acids quantified in P. falciparum, obtained with multiple reaction monitoring. Experimental conditions were the same as in Table 3.
The technical and biological reproducibility of amino acid measurement in the P. falciparum was investigated. The technical reproducibility was calculated based on three consecutive UPLCESI-MS/MS analyses. It was observed that the technical reproducibility of amino acid quantitation was superior for P. falciparum trophozoites than for P. falciparum rings. Whereas the coefficients of variation in the trophozoites were below 5% for the majority of the amino acids, these coefficients of variation increased to ~10% for most of the amino acids in the rings. (See supplemental Table S-5 in Supporting Information) The amino acid concentration in P. falciparum trophozoites was significantly higher than that of the rings. This is the result of the higher metabolic activity of the trophozoites developmental stage compared to the ring stage. Because the amino acid concentration in P. falciparum rings was lower in comparison to the trophozoites, we believe that the reason for the decreased quantitation reproducibility is directly linked to the lower amino acid concentration. Evidently, as the amino acid concentration approaches the limits of detection, the accuracy of quantitation is compromised due to insufficient amount of analyte necessary to produce a reliable signal.
The quantitation reproducibility across biological replicates was calculated from 4 biological replicates in the case of rings, and 5 biological replicates in the case of trophozoites. Note that each biological replicate was analyzed three times by UPLC-MS/MS and the results from these technical replicate analyses were included in the biological replicate reproducibility data. These results are shown in the supporting information (supplemental Figure S-3). Clearly, the reproducibility among biological replicates was lower than for technical replicates. There are two important reasons for the increased standard error observed for biological replicate analysis. First, there is not an accurate way of measuring the number of parasites in the sample, and as a result, we can only normalize all biological replicates based on hemozoin level. Any fluctuation in the number of parasites in the P. falciparum will reflect in the measured amino acid concentration. For instance, an increased number of parasites will result in a higher amino acid concentration and vice versa. Second, the biological reproducibility takes into consideration a greater number of random errors introduced during sample handling and analysis.
The concentration of amino acids in P. falciparum rings and trophozoites determined from biological replicates using absolute peak areas, together with the p-value of these measurements, is summarized in Table 3. Observe from Table 3 that in the P. falciparum rings, L-glutamic acid was the amino acid present at the highest concentrations (~22 pmol/μL), followed by alanine, glycine and aspartic acid, which were present at concentrations between 6-7 pmol/μL. Ethanolamine, tryrosine, tryptophan, proline, isoleucine and methionine were the least abundant amino acids. Phenylalanine, γ-amino-n-butyric acid, threonine, serine, leucine and valine were the second least abundant amino acids. Aspartic acid, glutamic acid and glycine were also the amino acids present at the highest amounts in the P. falciparum trophozoites. Ethanolamine, tryptophan and methionine were the least abundant amino acids in this system. All amino acids that were quantified in the P. falciparum showed a significant change between the two developmental stages, as evidence by the calculated p-values.
Table 3.
Amino acids concentration in P. falciparum rings and trophozoites calculated from biological replicates using the absolute peak areas. Experimental conditions were the same as in Table 1.
| Amino Acid | P. falciparum rings | P. falciparum trophozoites | p- value |
||||
|---|---|---|---|---|---|---|---|
| pmol/μL | STError | Normalized amount (picomol AA per nanomole of hemozoin) |
pmol/μL | STError | Normalized amount (picomol AA per nanomole of hemozoin) |
||
| Serine | 3.9126 | 0.6751 | 7.8252 | 52.0277 | 22.8780 | 34.3499 | 0.032 |
| Glutamic acid | 21.9737 | 2.5666 | 43.9474 | 108.8350 | 37.7964 | 71.8554 | 0.026 |
| Aspartic acid | 6.1297 | 2.5678 | 12.2594 | 129.5683 | 38.5124 | 85.5440 | 0.003 |
| Glycine | 6.6905 | 1.3546 | 13.3810 | 61.9637 | 26.0617 | 40.9099 | 0.031 |
| Ethanolamine | < LOQ | - | - | 1.6462 | 1.3428 | 1.0869 | - |
| Threonine | 2.9726 | 0.4360 | 5.9452 | 35.8404 | 15.5397 | 23.6627 | 0.031 |
| Proline | 0.3824 | 0.3412 | 0.7658 | 16.9832 | 7.5931 | 11.2127 | 0.032 |
| γ-Amino-n- butyric acid |
2.0109 | 0.2745 | 4.0228 | 20.2868 | 6.5311 | 13.3938 | 0.007 |
| Alanine | 7.6203 | 0.7733 | 15.2416 | 48.2945 | 20.5785 | 31.8851 | 0.034 |
| Tyrosine | < LOQ | - | - | 21.2450 | 10.5872 | 14.0264 | - |
| Methionie | 0.9178 | 0.1356 | 1.8366 | 15.5779 | 6.1767 | 10.2849 | 0.016 |
| Valine | 4.5380 | 0.4064 | 9.0760 | 40.0906 | 17.6434 | 26.4687 | 0.032 |
| Tryptophan | 0.3127 | 0.0741 | 0.6254 | 3.3853 | 1.5284 | 2.2351 | 0.038 |
| Phenylalanine | 1.3687 | 0.1482 | 2.7374 | 15.1528 | 6.3066 | 10.0042 | 0.027 |
| Isoleucine | 0.8788 | 0.1922 | 1.7586 | 28.7275 | 13.6388 | 18.9666 | 0.030 |
| Leucine | 4.0228 | 0.3828 | 8.0466 | 46.4224 | 20.8196 | 30.6491 | 0.031 |
Note: LOQ refers to the limits of quantitation.
These p-values were calculated based on the T-test. The number of distribution tails was set to 1, and a two-sample unequal variance of 3 was assumed. Because all estimated p-values were below 0.05, it can be said that a significant difference in amino acid concentration between P. falciparum rings and trophozoites exists. The normalized amino acid amount in P. falciparum in both developmental ring or trophozoite stages was obtained by multiplying the amino acid concentration by the corresponding dilution factors during sample preparation and dividing this number by the average value for hemozoin present in the sample. Notice that hemozoin refers to heme crystals generated from heme from hemoglobin catabolism and it is used as an indirect measurement of the number or parasites in the cell. For the rings stage, this number corresponded to ~1 micromole heme crystal per log 10 parasites, whereas for the trophozoites system this number was ~5-6 micromole heme crystal per log 10 parasites.46
Applicability of this AccQ•Tag UPLC-MS/MS method was also investigated in human RBCs, and the results are displayed in Table 4. Twenty five amino acids were measured in this complex biological system. In normal RBCs, the free amino acids include histidine and arginine and lysine which were not found in the parasites. The coefficients of variation were usually below 10% for technical replicate analysis. Notice again that the biological reproducibility, estimated among 6 biological replicate samples, was higher than the technical reproducibility. Thus, coefficients of variation for technical replicates were within 10% for the majority of the measurements and as low as 1.35%. For technical reproducibility evaluation, three consecutive UPLC-MS/MS runs were considered, whereas for biological reproducibility analysis 6 biological replicates were averaged. In normal RBCs, the free amino acids include histidine and arginine and lysine which were not found in the parasites. To further demonstrate the reproducibility of the method for amino acid quantitation throughout the entire analytical steps-amino acid extraction, derivatization, separation and detection-, results from the internal standard reproducibility data calculated for biological replicates of red blood cells are reported in Table S-6. As can be seen from this table, coefficients of variation for AccQ•Tag derivatized isotopically internal standards, spiked into the read blood cell samples early in the process, were within 3 and 14%.
Table 4.
Technical and biological reproducibility of amino acid concentrations quantified in red blood cells. Experimental conditions were the same as in Table 1. The technical reproducibility was calculated from 3 sequential UPLC-MS/MS analysis. The biological reproducibility was calculated across 6 biological replicates, each analyzed three times by UPLC-MS/MS.
| Amino acid | Technical reproducibility | Biological reproducibility | ||||
|---|---|---|---|---|---|---|
| pmol/μL | CV % | ST Error | pmol/μL | CV % | ST Error |
|
| Histidine | 15.6163 | 9.29 | 0.8373 | 15.0506 | 12.37 | 0.4654 |
| Asparagine | 496.2070 | 1.97 | 5.6581 | 435.5660 | 16.23 | 16.6630 |
| Hydroxy-L-Proline | 12.4053 | 8.58 | 0.6147 | 12.2486 | 7.88 | 0.2275 |
| Taurine | 4.6445 | 9.36 | 0.3075 | 6.3641 | 41.33 | 0.6791 |
| Arginine | 6.3820 | 7.21 | 0.2657 | 8.0786 | 23.12 | 0.4530 |
| Aspartic acid | 35.3750 | 1.35 | 0.2749 | 69.1045 | 50.06 | 8.1540 |
| Serine | 29.0133 | 13.26 | 2.2219 | 28.7161 | 16.49 | 1.1482 |
| Glycine | 36.2560 | 8.82 | 1.8465 | 67.5955 | 44.36 | 7.0675 |
| Citrulline | 4.5825 | 9.77 | 0.3165 | 5.8364 | 23.15 | 0.3900 |
| Glutamic acid | 19.3850 | 3.14 | 0.3512 | 25.7587 | 24.96 | 1.5593 |
| Alanine | 0.4380 | 2.26 | 0.0070 | 0.6801 | 53.65 | 0.1100 |
| Proline | 16.0703 | 9.58 | 0.8888 | 21.8726 | 24.59 | 1.2674 |
| α-amino-n-butyric acid | 0.2523 | 6.94 | 0.0101 | 0.2520 | 25.66 | 0.0179 |
| γ-Amino-n-butyric acid | 0.2015 | 6.67 | 0.0095 | 0.1171 | 60.27 | 0.0166 |
| Tyrosine | 14.8157 | 14.35 | 1.2275 | 16.5172 | 18.27 | 0.7319 |
| Methionine | 2.2285 | 4.09 | 0.0645 | 2.8053 | 28.92 | 0.2342 |
| Lysine | 9.7890 | 2.53 | 0.1750 | 10.4553 | 44.43 | 1.6425 |
| Valine | 5.9700 | 10.47 | 0.4420 | 9.8059 | 42.56 | 1.0435 |
| Tryptophan | 2.0190 | 6.76 | 0.0788 | 2.1402 | 34.73 | 0.1987 |
| Phenylalanine | 1.5917 | 3.94 | 0.0362 | 2.6696 | 55.86 | 0.3986 |
| Leucine | 4.7270 | 4.64 | 0.1550 | 8.9685 | 37.47 | 0.8402 |
| Isoleucine | 6.8557 | 12.92 | 0.5115 | 9.7329 | 33.75 | 0.8481 |
| Threonine | 16.7190 | 1.02 | 0.1210 | 17.3410 | 9.35 | 0.3934 |
| Amino adipic acid | 0.1633 | 10.96 | 0.0103 | 0.2074 | 28.72 | 0.0159 |
| Ornitine | 23.6730 | 3.12 | 0.5220 | 36.8569 | 25.12 | 3.2740 |
CONCLUSIONS
A UPLC-ESI-MS/MS method was developed for the targeted measurement of physiological amino acids in biological samples. The simplicity of the AccQ•Tag technique combined with the speed, resolution and sensitivity characteristics of UPLC, as well as the specificity and sensitivity afforded by mass spectrometry, offers an exciting possibility for sensitive, specific and high-throughput amino acid assays. While traditional amino acid analyses that rely on conventional chromatography or capillary electrophoresis coupled to photometric or fluorescence detection can be laborious, time consuming, and complicated with interfering substances arising from derivatized impurities in the sample. The method reported here can be completed in as little as 40 minutes with all analysis steps included, and exhibits high sensitivity as well as reproducibility of detection and quantitation. For example, limits of detection for the evaluated amino acids were within the range of 1.65-17.95 fmol/μL and the retention time coefficients of variations for amino acid detection were below 1.08%. For amino acid quantification, the coefficients of variation, calculated from three consecutive LC-MS/MS analysis, were typically below 8%. Ion suppression effects, a major concern in ESI-MS, were not manifested, which is an added benefit of this method. When applied to the measurement of amino acids in P. falciparum ring and trophozoite developmental stage, and red blood cells, our method was found to exhibit satisfactory reproducibility compared to previously reported methods.17,47,40 Furthermore, the method could effectively differentiate across two different rings and trophozoite stages of RBCs development in this parasite. Our saponin lysis purification selectively purifies host cytosol from parasite cytosol. The absent amino acids from the parasites were the ones which had the highest limits of detection. Since the malaria parasite relies on hemoglobin as a source of amino acids we can also compare the composition of free amino acids to hemoglobin content at different times. These studies will provide a baseline comparison for the effects of malaria drugs on the parasite and uninfected red blood cells.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by NIH-NIAID grant 2R01AI045774 and NIH NCI grant R01CA120170. An NCCR grant GCRC RR0052 supported the culturing of P. falciparum for the production of malaria parasites.
Footnotes
SUPPORTING INFORMATION AVAILABLE Plasmodium Falciparum Culture Protocol. Figure S-1. Schematic reaction of amino acid derivatization with AccQ•Tag reagent. Figure S-2. Typical chromatograms for AccQ•Tag tag derivatized amino acids detected in P. falciparum trophozoites, obtained with UPLC-MS/MS using MRM. Figure S-3. Biological reproducibility for AccQ•Tag derivatized amino acids detected in P. falciparum rings or trophozoites. Table S-1. Retention times and parent ions observed for AccQ•Tag derivatized physiological amino acid standards and isotopically labeled amino acids, analyzed by UPLC-MS. Table S-2. Optimal cone voltage and collision energy for selected AccQ•Tag derivatized amino acid standards analyzed by ESI-MS/MS, using a triple quadrupole mass spectrometer, and, either, low flow (20 μL/min) or combined flow (200 μL/min). Table S-3. Peak area reproducibility for AccQ•Tag labeled amino acid standards analyzed by UPLC-MS/MS using 001 s and 002 s dwell time. Table S-4. Dynamic range, high limits of linearity, low limits of linearity, and detection limits for AccQ•Tag physiological amino acids standards. Table S-5. Technical reproducibility for AccQ•Tag derivatized amino acids detected and measured in P. falciparum ring and trophozoite developmental stages. Table S-6. Biological reproducibility across peak areas measured for AccQ•Tag derivatized isotopically labeled amino acid internal standards spiked into red blood cells. Equation S-1 used to calculate limits of detection. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- (1).Zhao X, Suo Y. Chromatographia. 2008;67:375–382. [Google Scholar]
- (2).Jinmao Y, Lingjun L, Wenchen Z, Xianen Z, Yourui S, Honglun W, Yulin L. Analytical & Bioanalytical Chemistry. Vol. 387. Springer Science & Business Media B.V.; 2007. pp. 2705–2718. [Google Scholar]
- (3).Meesters RJW, Wolfe RR, Deutz NEP. Journal of Chromatography B. 2009;877:43–49. doi: 10.1016/j.jchromb.2008.11.018. [DOI] [PubMed] [Google Scholar]
- (4).Freeto S, Mason D, Chen J, Scott RH, Narayan SB, Bennett MJ. Ann Clin Biochem. 2007;44:474–481. doi: 10.1258/000456307781646012. [DOI] [PubMed] [Google Scholar]
- (5).Kaspar H, Dettmer K, Gronwald W, Oefner PJ. Anal Bioanal Chem. 2009;393:445–452. doi: 10.1007/s00216-008-2421-1. [DOI] [PubMed] [Google Scholar]
- (6).Peris-Vicente J, Adelantado J. V. Gimeno, Carbó MTD, Castro RM, Reig FB. Talanta. 2006;68:1648–1654. doi: 10.1016/j.talanta.2005.08.050. [DOI] [PubMed] [Google Scholar]
- (7).Poinsot V, Lacroix M, Maury D, Chataigne G, Feurer B, Couderc F. Electrophoresis. 2006;27:176–194. doi: 10.1002/elps.200500512. [DOI] [PubMed] [Google Scholar]
- (8).Herrero M, Ibanez E, Fanali S, Cifuentes A. Electrophoresis. 2007;28:2701–2709. doi: 10.1002/elps.200600599. [DOI] [PubMed] [Google Scholar]
- (9).Lin CC, Liu CY. Electrophoresis. 2004;25:3216–3223. doi: 10.1002/elps.200406037. [DOI] [PubMed] [Google Scholar]
- (10).Husek P. FEBS Lett. 1991;280:354–356. doi: 10.1016/0014-5793(91)80330-6. [DOI] [PubMed] [Google Scholar]
- (11).Simpson JT, Torok DS, Markey SP. Journal of the American Society for Mass Spectrometry. 1995;6:525–528. doi: 10.1016/1044-0305(95)00231-2. [DOI] [PubMed] [Google Scholar]
- (12).Rodier C, Vandenabeele-Trambouze O, Sternberg R, Coscia D, Coll P, Szopa C, Raulin F, Vidal-Madjar C, Cabane M, Israel G, Grenier-Loustalot MF, Dobrijevic M, Despois D. Adv Space Res. 2001;27:195–199. doi: 10.1016/s0273-1177(01)00047-3. [DOI] [PubMed] [Google Scholar]
- (13).Pumera M. Electrophoresis. 2007;28:2113–2124. doi: 10.1002/elps.200600709. [DOI] [PubMed] [Google Scholar]
- (14).Chaimbault P, Petritis K, Elfakir C, Dreux M. J Chromatogr A. 2000;870:245–254. doi: 10.1016/s0021-9673(99)00863-8. [DOI] [PubMed] [Google Scholar]
- (15).Petritis K, Chaimbault P, Elfakir C, Dreux M. J Chromatogr A. 2000;896:253–263. doi: 10.1016/s0021-9673(00)00582-3. [DOI] [PubMed] [Google Scholar]
- (16).Thiele B, Füllner K, Stein N, Oldiges M, Kuhn A, Hofmann D. Analytical and Bioanalytical Chemistry. 2008;391:2663–2672. doi: 10.1007/s00216-008-2167-9. [DOI] [PubMed] [Google Scholar]
- (17).Soga T, Heiger DN. Anal Chem. 2000;72:1236–1241. doi: 10.1021/ac990976y. [DOI] [PubMed] [Google Scholar]
- (18).Nagy K, Takats Z, Pollreisz F, Szabo T, Vekey K. Rapid Commun Mass Spectrom. 2003;17:983–990. doi: 10.1002/rcm.1000. [DOI] [PubMed] [Google Scholar]
- (19).Uutela P, Ketola RA, Piepponen P, Kostiainen R. Analytica Chimica Acta. 2009;633:223–231. doi: 10.1016/j.aca.2008.11.055. [DOI] [PubMed] [Google Scholar]
- (20).Kutlan D, Molnar-Perl I. J Chromatogr A. 2003;987:311–322. doi: 10.1016/s0021-9673(02)01538-8. [DOI] [PubMed] [Google Scholar]
- (21).Molnar-Perl I. J Chromatogr A. 2003;987:291–309. doi: 10.1016/s0021-9673(02)01537-6. [DOI] [PubMed] [Google Scholar]
- (22).Oravec P, Podhradsky D. J Biochem Biophys Methods. 1995;30:145–152. doi: 10.1016/0165-022x(94)00073-m. [DOI] [PubMed] [Google Scholar]
- (23).vanWandelen C, Cohen SA. Journal of Chromatography A. 1997;763:11–22. [Google Scholar]
- (24).Saurina J, HernandezCassou S. Journal of Chromatography A. 1996;740:21–30. [Google Scholar]
- (25).Chang JY, Knecht R, Braun DG. Methods Enzymol. 1983;91:41–48. doi: 10.1016/s0076-6879(83)91009-1. [DOI] [PubMed] [Google Scholar]
- (26).Hill DW, Walters FH, Wilson TD, Stuart JD. Anal Chem. 1979;51:1338–1341. doi: 10.1021/ac50044a055. [DOI] [PubMed] [Google Scholar]
- (27).Farrant M, Zia-Gharib F, Webster RA. J Chromatogr. 1987;417:385–390. doi: 10.1016/0378-4347(87)80133-0. [DOI] [PubMed] [Google Scholar]
- (28).Turnell DC, Cooper JD. J Automat Chem. 1983;5:36–39. doi: 10.1155/S1463924683000115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Fermo I, De Vecchi E, Diomede L, Paroni R. J Chromatogr. 1990;534:23–35. doi: 10.1016/s0378-4347(00)82145-3. [DOI] [PubMed] [Google Scholar]
- (30).O’Hare MM, Tortora O, Gether U, Nielsen HV, Schwartz TW. J Chromatogr. 1987;389:379–388. doi: 10.1016/s0021-9673(01)94449-8. [DOI] [PubMed] [Google Scholar]
- (31).Bartolomeo MP, Maisano F. J Biomol Tech. 2006;17:131–137. [PMC free article] [PubMed] [Google Scholar]
- (32).Liu H, Worthen HG. J Chromatogr. 1992;579:215–224. doi: 10.1016/0378-4347(92)80385-4. [DOI] [PubMed] [Google Scholar]
- (33).Bidlingmeyer BA, Cohen SA, Tarvin TL. J Chromatogr. 1984;336:93–104. doi: 10.1016/s0378-4347(00)85133-6. [DOI] [PubMed] [Google Scholar]
- (34).Stobaugh JF, Repta AJ, Sternson LA, Garren KW. Anal Biochem. 1983;135:495–504. doi: 10.1016/0003-2697(83)90718-2. [DOI] [PubMed] [Google Scholar]
- (35).Haginaka J, Wakai J. J Chromatogr. 1987;396:297–305. doi: 10.1016/s0021-9673(01)94067-1. [DOI] [PubMed] [Google Scholar]
- (36).Cohen SA, De Antonis KM. J Chromatogr A. 1994;661:25–34. doi: 10.1016/0021-9673(93)E0821-B. [DOI] [PubMed] [Google Scholar]
- (37).Betner I, Foldi P. Chromatographia. 1986;22:381–387. [Google Scholar]
- (38).Mcclung G, Frankenberger WT. Journal of Liquid Chromatography. 1988;11:613–646. [Google Scholar]
- (39).Bosch L, Alegria A, Farre R. Journal of Chromatography B-Analytical Technologies in the Biomedical and Life Sciences. 2006;831:176–183. doi: 10.1016/j.jchromb.2005.12.002. [DOI] [PubMed] [Google Scholar]
- (40).Shimbo K, Oonuki T, Yahashi A, Hirayama K, Miyano H. Rapid Commun Mass Spectrom. 2009;23:1483–1492. doi: 10.1002/rcm.4026. [DOI] [PubMed] [Google Scholar]
- (41).Guo K, Li L. Anal Chem. 2009;81:3919–3932. doi: 10.1021/ac900166a. [DOI] [PubMed] [Google Scholar]
- (42).Gardener MC, Gillman MP. Journal of Chemical Ecology. 2001;27:2545–2558. doi: 10.1023/a:1013687701120. [DOI] [PubMed] [Google Scholar]
- (43).Boogers I, Plugge W, Stokkermans YQ, Duchateau AL. J Chromatogr A. 2008;1189:406–409. doi: 10.1016/j.chroma.2007.11.052. [DOI] [PubMed] [Google Scholar]
- (44).Kovacs A, Simon-Sarkadi L, Ganzler K. J Chromatogr A. 1999;836:305–313. doi: 10.1016/s0021-9673(98)00912-1. [DOI] [PubMed] [Google Scholar]
- (45).Callahan Damien L., Kolev Spas D., O’Hair Richard A. J., Salt David E., Baker Alan J. M. New Phytologist. 2007;176:836–848. doi: 10.1111/j.1469-8137.2007.02216.x. [DOI] [PubMed] [Google Scholar]
- (46).Zhang J, Krugliak M, Ginsburg H. Molecular and Biochemical Parasitology. 1999;99:129–141. doi: 10.1016/s0166-6851(99)00008-0. [DOI] [PubMed] [Google Scholar]
- (47).Soga T, Kakazu Y, Robert M, Tomita M, Nishioka T. Electrophoresis. 2004;25:1964–1972. doi: 10.1002/elps.200305791. [DOI] [PubMed] [Google Scholar]
- (48).Moini M, Schultz CL, Mahmood H. Anal Chem. 2003;75:6282–6287. doi: 10.1021/ac034708i. [DOI] [PubMed] [Google Scholar]
- (49).Schultz CL, Moini M. Anal Chem. 2003;75:1508–1513. doi: 10.1021/ac0263925. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.