Abstract
In mature HIV-1 virions, a 26.6 kDa CA protein is assembled into a characteristic cone shaped core (capsid) that encloses the RNA viral genome. The assembled capsid structure is best described by a fullerene cone model that is made up from a hexameric lattice containing a variable number of CA pentamers, thus allowing for closure of tubular or conical structures. In this report, we present a solid-state NMR analysis of the wild type HIV-1 CA protein, prepared as conical and spherical assemblies that are stable and are not affected by magic angle spinning of the samples at frequencies between 10 and 25 kHz. Multidimensional homo- and heteronuclear correlation spectra of CA assemblies of uniformly 13C,15N-labelled CA exhibit narrow lines, indicative of conformational homogeneity of the protein in these assemblies. For the conical assemblies, partial residue-specific resonance assignments were obtained. Analysis of the NMR spectra recorded for the conical and spherical assemblies indicates that the CA protein structure is not significantly different in the different morphologies. The present results demonstrate that the assemblies of CA protein are amenable to detailed structural analysis by solid-state NMR spectroscopy.
Keywords: solid-state NMR, magic angle spinning, MAS, HIV-1, AIDS, CA, capsid protein, resonance assignments, protein assemblies
INTRODUCTION
Human immunodeficiency virus (HIV) is the causative agent of acquired immunodeficiency syndrome (AIDS),1 and at present ca. 33 million people are infected worldwide with at least 25 million people having died of HIV-1 related causes.2 Early stages of HIV infection involve fusion of the viral and the host membranes followed by release of viral particles into the cytoplasm of the host cell.3 Subsequently, the core, which is composed of multiple copies of a 26.6 kDa CA protein, undergoes disassembly in order to allow for reverse transcription of the viral genome and progression through the viral life cycle.4
Despite a plethora of studies, the molecular mechanism of the CA disassembly is poorly understood, partly due to lack of atomic-resolution structures of assembled capsids and their changes during uncoating. The recent discovery of the TRIM5α family of restriction factors, which inhibit retroviral infection in Old World monkeys by causing premature and accelerated capsid disassembly, suggests that capsid disassembly is an exquisitely regulated process.5–7 For humans and New World monkeys, restriction of HIV-1 by TRIM5α is inefficient due to lack of interaction with HIV-1 capsid.8 At the same time, human TRIM5α variants inhibit murine leukemia virus (MLV)8,9 as well as equine infectious anemia virus (EIAV),10,11 and for MLV, accelerated uncoating of the virus capsid has been suggested as a mechanism of restriction.5 However, the exact inhibitory mechanisms for the different TRIM5α variants and retroviruses are still unknown.
While a molecular model of capsid assembled into planar sheets is available from the docking of high-resolution structures of the individual CA protein N- and C-terminal domains12,13 into a 9 Å resolution cryo-EM map,14 as well as the recent high-resolution X-ray structures (1.9 and 2.7 Å resolution) of the hexameric building block of an engineered CA,15 and a cryo-EM structure of a tubular assembly of CA and a high-resolution NMR structure of the CA C-terminal domain (CTD) dimer,16 atomic-resolution structures of full-length CA assemblies have not been yet determined. Their large molecular size and lack of crystalline order have prevented structural characterization by solution NMR or X-ray crystallography. Without such atomic detail structures, processes associated with capsid assembly and disassembly remain poorly understood, and work towards the design of HIV-1 capsid assembly inhibitors is impeded. Similarly, understanding at atomic level resolution the interaction of capsids with TRIM5α factors from rhesus monkeys and humans will be critical for the design of new therapeutic strategies against HIV-1.
Intriguingly, the HIV-1 CA protein assemblies exhibit structural polymorphism.14,17–21 While in vivo the mature virions contain capsids of predominantly conical morphology,22,23 the immature viral particles are composed of spherical assemblies.17 In vitro, depending on the assembly conditions, CA assemblies can adopt conical, spherical, donut-like, or tubular morphologies.14,18–20 Whether this polymorphism is important for the disassembly process and for the recognition of the capsid particles by the restriction factor proteins, remains an unanswered question.
An alternative high-resolution structural methodology that does not require soluble or crystalline preparations and is therefore suited to study CA protein assemblies of different morphologies is Magic Angle Spinning (MAS) NMR spectroscopy. While originally established on microcrystalline protein preparations,24–28 solid-state MAS NMR is becoming a more and more valuable approach for structural studies of biological solids that lack long-range order, such as macromolecular assemblies,29–31 disordered fibrils,32–35 membrane proteins,36–42 and biomaterials.43–45 Recent advances in multidimensional methods for resonance assignments and structural characterization by MAS NMR have made it possible to obtain detailed structural information on uniformly enriched proteins using only 0.1–0.5 μmoles of material.26–28,30,38,46–53
In this report, we present a solid-state NMR study of conical and spherical HIV-1 CA protein assemblies. We discuss the sample preparation protocols for generating conical and spherical assemblies of CA suitable for high-resolution structural analysis by MAS NMR and report partial site-specific resonance assignments by a combination of two- and three-dimensional correlation experiments. Chemical shift analyses indicate that no major differences in the secondary structure of the individual CA proteins in the spherical and conical assemblies exist and that the structural polymorphism most likely is due to the differences in the intermolecular packing. Most importantly, our results illustrate that the CA protein assemblies are amenable to structural analyses by MAS NMR spectroscopy.
RESULTS AND DISCUSSION
Characterization of different assembly morphologies
For the HIV CA protein, prior reports showed that in vitro the protein can assemble into sheets, spheres or cylinders (tubes).14,17–19 Tubular assemblies generally require high salt concentrations (1 M NaCl),20 while conical assemblies can be prepared from lower salt solutions (0.3 M NaCl), and frequently result in mixtures of cones and tubes.22 Spherical assemblies can be generated from concentrated solutions (32 mg/ml) of the R18L CA mutant in the presence of 17.5% PEG-20,000.14 Unfortunately, current conditions used for preparing conical and tubular CA assemblies are not suitable for solid-state NMR, and in the case of the spheres, the mutation might have dictated the assembly mode. We therefore needed to establish protocols for reproducible preparations of CA assemblies in the different morphologies that are compatible with the solid-state NMR requirements. To that end we first carried out extensive screening of possible conditions.
The morphology of the samples was analyzed by TEM, confocal and cryo-SEM microscopy. The best conical assemblies were found when lyophilized WT CA protein was taken up in 17.5% w/v PEG-20,000 to a final concentration of 32 mg/ml, followed by incubation at 37 °C for 1 hour. Under similar conditions, the R18L mutant produces spherical assemblies.14 As illustrated in Figure 1a, varied dimensions and shapes were observed for these assemblies. The TEM images showed ca. 100–200 nm long conical assemblies, and confocal images confirmed CA assembly into 1 μm or smaller diameter particles (Figure 1a). It should be noted that the highest resolution attainable by confocal microscopy is ~0.8 μm, and therefore one cannot derive the exact size and shape of the sub-micron particles. The cryo- SEM images revealed irregularly shaped assemblies ranging in size from 0.5 to 5 μm (Figure 1a). Despite this variation in size and shape, the solid-state NMR spectra exhibit narrow lines, indicating high conformational homogeneity within these assemblies, as described below.
Figure 1.
Assemblies of HIV-1 CA exhibiting conical, spherical, and tubular morphologies. (a), Conical assembly. From left to right: a negatively stained TEM image, confocal image (fluorescence mode), cryo-SEM image, confocal image (fluorescence mode) of the sample after magic angle spinning. (b), Spherical assembly. From left to right: confocal image (fluorescence mode) of the solution containing the assemblies, cryo-SEM image, confocal image (fluorescence mode) of the dried out sample, confocal image (fluorescence mode) of the sample after magic angle spinning. (c), Tubular assembly. From left to right: negatively stained TEM image, confocal image (fluorescence mode), confocal image (fluorescence mode) of the dried out sample, confocal image (fluorescence mode) of the sample after magic angle spinning.
Spherical particles with a narrow size distribution were produced from 16 or 32 mg/ml CA incubated with 3.25% w/v PEG-20,000 on ice. The size of these particles can be controlled by the length of the incubation time. For example, increasing the incubation time from 1 hour to overnight and to one week results in particle sizes of 2, 3, and 3.5 μm. Confocal and cryo-SEM images of such spherical assemblies are illustrated in Figure 1b. It is worth noting that the solutions containing spherical assemblies are not stable over extended periods of time: after several weeks the sample starts turning into an amorphous precipitate, even if kept hydrated. However, drying the solutions that contain the spherical particle under a stream of nitrogen gas, preserved the morphology (Figure 1b), and these dried samples are stable for many weeks. Therefore, these samples are amenable to solid-state NMR experiments.
Tubular assemblies were produced from solutions containing 32 mg/ml CA in 50 mM Tris buffer, pH 8.0, 1 M NaCl, by incubation at 37 °C for 1 hour. TEM and confocal images of these particles are shown in Figure 1c. The requirement for high salt is consistent with the known conditions that produced tubular assemblies used in previous biochemical and cryo-EM studies.18,20 Solid-state NMR studies of such tubular assemblies will be carried out in the future as well.
A common concern with regard to MAS solid-state NMR studies of proteins and protein assemblies relates to the stability of the sample, given the extended periods of extremely fast spinning. We tested the integrity of our samples containing conical, spherical and tubular assemblies after spinning for several days at MAS frequencies between 10 and 24 kHz. A fraction of each sample was taken out of the rotor and subjected to confocal microscopic analysis. As illustrated in Figure 1, no changes were observed in sample morphology by confocal microscopy compared to the appearance prior to the NMR experiments. Therefore, we are confident that during the MAS experiments the CA assemblies remain intact.
DARR spectra of conical CA assemblies
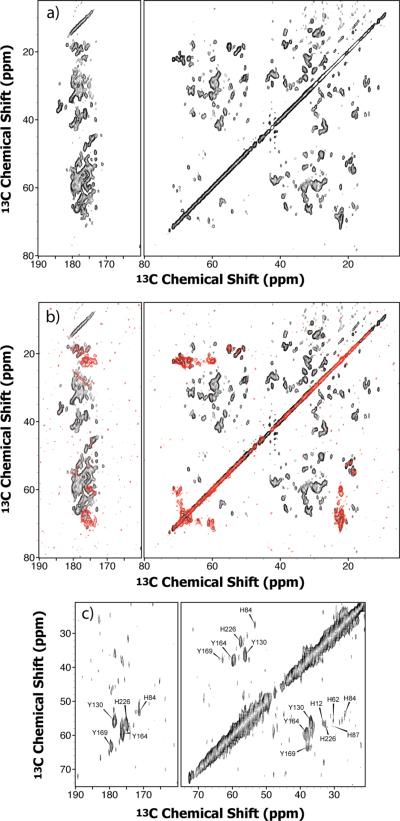
A 2D 13C-13C DARR spectrum of conical assemblies of U-13C,15N CA recorded at 21.1 T is provided in Figure 2. The quality of the data set is remarkably high, allowing extraction of a number of important conclusions, even if complete residue-specific resonance assignments are not available yet. First, the linewidths of the individual cross peaks lie in the range of 0.25–0.5 ppm, indicating that the sample is conformationally homogeneous. Second, despite the size of the protein, numerous well-resolved peaks are present. For example, in the non-overlapped regions of the spectrum a significant number of resonances can be assigned to amino acid type: Ala (12/20), Gly (9/17), Ile (12/15), Pro (9/18), Thr (13/16), Ser (7/9), Val (9/15). Furthermore, the overall number of the cross peaks indicates that most correlations are present in the spectrum. Qualitative analysis of the Cα and Cβ chemical shifts of the Ala and Thr residues additionally reveals that the conformation of the protein is predominantly α-helical, consistent with the prior solution NMR, X-ray diffraction, and cryo-EM studies.12–14 As expected, the spectral resolution and sensitivity are less at lower magnetic field, while the chemical shifts remain the same, as evidenced from the comparison of the spectra acquired at 14.1 T (Figure 3) and 21.1 T (Figure 2). Indeed, in the DARR data sets collected at 14.1 T, several cross peaks in the Ser and Thr Cα-Cβ region are either weak or not observed in the 2D 13C-13C spectra of the uniformly labeled CA assemblies (illustrated in Figure 3a), due to the lower sensitivity at this field.
Figure 2.
DARR spectrum of U-13C,15N HIV-1 CA conical assemblies at 21.1 T. Aliphatic region (right) and carbonyl region (left). The 1D projections in the ω2 and ω1 dimensions corresponding to the resolved cross-peaks for Thr residues are shown and illustrate the remarkable spectral sensitivity and resolution. The spectrum was acquired as (3072 × 216) (complex × real) matrices with 220 scans; the recycle delay was 2 s. The spectral widths were 100 kHz and 50 kHz in the ω2 and ω1 dimensions, respectively, and the DARR mixing time was 50 ms. Cross-peaks corresponding to the different amino acid types appear in distinct, resolved regions of the spectrum and are color coded as follows: Ala, red (Cα-Cβ and Cβ-C' correlations); Gly, orange (Cα-C' correlations); Ile, yellow (Cα-Cδ1, Cβ-Cγ1, Cβ-Cγ2, and Cγ1-Cδ1 correlations); Leu, grey (Cβ-Cγ correlations); Pro, purple (Cα-Cγ correlations); Ser, light green (Cα-Cβ correlations); Thr, blue (Cα-Cβ and Cβ-Cγ correlations); Val, bright green (Cα-Cβ correlations).
Figure 3.
14.1 T DARR spectra of conical assemblies acquired on a set of uniformly and specifically labeled CA. (a) The DARR spectrum of U-13C,15N sample; (b) the superimposed DARR spectra of U-13C,15N sample (black) and U-13C,15N-Ala/U-13C,15N-Thr sample (red), and (c) U-13C,15N-His/U-13C,15N-Tyr sample (expansions of the aliphatic (right) and carbonyl (left) regions illustrate the one-bond correlations for His and Tyr residues). The spectra in (a) were collected as (2000 × 400) (complex × real) matrices with 200 scans, a recycle delay of 2 s, and a spectral width of 50 kHz in both dimensions. The spectra in (b) were obtained as (2000 × 512) (complex × real) matrices with 256 scans, a recycle delay of 1 s, and spectral widths of 50 kHz and 25 kHz in the ω2 and ω1 dimensions, respectively. The spectra in (c) were acquired as (1000 × 128) (complex × real) matrices with 640 scans, and a recycle delay of 1.5 s and spectral widths of 50 kHz and 33.3 kHz in the ω2 and ω1 dimensions, respectively. The DARR mixing time was 10 ms for all data sets.
DARR spectra of spherical CA assemblies
Figure 4 presents the superposition of the 14.1 T DARR spectra of spherical and conical CA assemblies. The spectral resolution is similar or slightly inferior for the spherical particles, and the linewidths lie in the range of 0.4 and 0.8 ppm at this field strength. Interestingly, the sensitivity appeared to be considerably lower than in the conical assemblies, with the signal-to-noise ratio in the one-dimensional CPMAS spectra only ca. 55% that of the conical samples. It may be the case that this noticeable deterioration in spectral quality may be due to dehydration of the spherical assemblies, which is necessary to prevent their conversion to the amorphous precipitate (see above).
Figure 4.
Superposition of DARR spectra recorded on spherical (black) and conical (red) assemblies of U-13C,15N CA at 14.1 T. The spectra of the spherical assemblies were acquired as (1000 × 400) (complex × real) matrices using 80 scans and a recycle delay of 2 s, a spectral width of 50 kHz in both dimensions and the DARR mixing time of 10 ms. The acquisition parameters for the spectra of the conical assemblies are given in the caption to Figure 3.
As can be appreciated from the data presented in Figure 4, no significant differences in 13C chemical shifts can be detected for the spherical and the conical assemblies, suggesting that the secondary and tertiary structure elements in the CA assemblies are the same, irrespective of their macroscopic morphology. At present, it is not possible to carry out any more detailed structural comparison from the DARR spectra alone. Such detailed evaluation will need to await results from three-dimensional heteronuclear correlation spectroscopy.
Resonance assignments of conical CA assemblies
As discussed above, conical assemblies exhibit high-quality spectra permitting the acquisition of an initial set of data. The following three-dimensional homo- and heteronuclear correlation MAS NMR spectra were successfully recorded at 14.1 T: NCACB, NCACX, NCOCX, and CCC (DARR-DARR). Combining these three-dimensional data sets with the DARR spectra at 21.1 T, we have confidently assigned 147 residues to date, about 63% of the entire protein, with an additional 21 residues tentatively assigned.
As illustrated in Figure 2, intra-residue side chain correlations were established for seven amino acid residue types (Ala, Gly, Ile, Pro, Ser, Thr, Val), since these appear in the unique regions of the spectra.24,26,27,50 In addition, due to the unique position of Leu Cβ-Cγ cross peaks in the spectrum, 12 of the expected 18 correlations corresponding to the Leu residues were identified. The two differentially mixed labeled samples, U-13C,15N-Ala/U-13C,15N-Thr and U-13C,15N-His/U-13C,15N-Tyr, were employed for unambiguous identification of the Ala and Thr (Figure 3b), and the His and Tyr (Figure 3c) cross peaks in the DARR spectra. For example, the DARR spectrum of the U-13C,15N-His/U-13C,15N-Tyr sample, allowed the identification of the aliphatic and carbonyl cross peaks corresponding to 4 out of 6 His residues, and 3 out of 4 Tyr residues. In total, the 2D DARR spectra yielded identification for the following amino acids: Ala (12/20), Gly (9/17), Ile (12/15), Pro (9/18), Thr (13/16), Ser (7/9), Val (9/15).
These above spin system assignments were confirmed in NCACX and NCACB 3D experiments, which remove the chemical shift degeneracy in the 13C dimensions for a number of the above amino acid types. As illustrated in Figure 5, many of the 2D 15N planes in the 3D NCACX spectrum exhibit excellent resolution, permitting multiple amino acid residue types to be assigned. In addition to the above listed spin systems, cross peaks corresponding to Cys (2/2), and to Asp (7/7) residues were identified in distinct, well-resolved regions of the spectra.
Figure 5.
Selected 2D planes of the 3D NCACX spectrum of U-13C,15N CA conical assemblies. Spectral resolution in planes a–g and j is high enough to allow assignment of spin systems, while planes h and i exhibit substantial resonance overlap. The spectrum was acquired as (1000 × 40 × 32) (complex × real × real) matrices with 256 scans and a recycle delay of 2 s. The spectral widths were 50 kHz, 4.167 kHz, and 8.333 kHz in the ω3, ω2 and ω1 dimensions, respectively. SPECIFIC-CP (τmix = 6.2 ms) was employed for the first NCA transfer followed by the DARR mixing (τmix = 20 ms) resulting in predominantly one- and two- bond correlations. (k) Sequential backbone connectivity for the sequence stretch A105–L111 in conical assemblies based on 3D NCOCX, NCACX, and NCACB experiments at 14.1 T. The residue names are shown on top of the spectra at their 15N chemical shift plane. Negative cross-peaks resulting from two-bond N-Cβ correlations in the NCACB spectra are displayed in red. The NCOCX spectrum was acquired as (1000 × 36 × 40) (complex × real × real) matrices with 280 scans and a recycle delay of 2 s. The spectral widths were 50 kHz, 4.167 kHz, and 6.250 kHz in the ω3, ω2 and ω1 dimensions, respectively. SPECIFIC-CP (τmix = 6.2 ms) was employed for the first NCO transfer followed by the DARR mixing (τmix = 15 ms) resulting in predominantly one- and two-bond correlations. The NCACB spectrum was acquired as (1000 × 32 × 32) (complex × real × real) matrices with 400 scans and a recycle delay of 2 s. The spectral widths were 50 kHz, 4.167 kHz, and 8.333 kHz in the ω3, ω2 and ω1 dimensions, respectively. SPECIFIC-CP (τmix = 6.2 ms) was employed for the first NCA transfer followed by the DREAM mixing (τmix = 3 ms) to establish the CA-CB correlations.
In total, using a combination of 2D DARR, 3D NCACX, and 3D NCACB experiments, we have identified 100 spin systems for 12 residue types.
Sequence-specific resonance assignments were made possible by the addition of 3D NCOCX spectra to the data analysis, given their excellent resolution shown in the 2D planes, separated by 15N chemical shifts (Figure 5k). Based on the 3D solid-state NMR spectra alone, we have established sequence-specific assignments for 61 residues. A representative backbone walk illustrating the assignments for the sequence stretch A105–L111 is provided in Figure 5k.
A number of residue types (Arg, Asn, Gln, Glu, Lys, Met, Phe, and Trp) present a major challenge with regard to assignment based solely on correlations in the 14.1 T solid-state 2D and 3D spectra alone: their Cα-Cβ cross peaks do not appear in isolated regions and generally exhibit substantial spectral overlap. We therefore compared the 3D NCACB and NCACX solid-state data with the solution NMR chemical shifts of the individual N- and C- terminal54 CA domains. This is a reasonable approach since it has been demonstrated by us and others for a number of proteins that solution and solid-state chemical shifts generally agree well, provided that the secondary structure (backbone torsion angles) for a given amino acid residue is the same in solution and in the solid.24,26–28,51,55,56 Comparison of the firmly assigned solid-state and solution 13C and 15N chemical shifts by residue type (Ala, Asp, Cys, Gly, His, Ile, Leu, Pro, Ser, Thr, Tyr, and Val) revealed excellent agreement within 0.1, 0.4, and 0.3 ppm for NH, Cα , and Cβ, respectively (Table S1 in Supporting information). Based on this comparison we derived tentative sequence-specific assignments for additional residues yielding the following degree of overall assignment: Arg (10/11), Asn (9/11), Gln (13/15), Glu (9/17), Lys (6/11), Met (5/11), Phe (4/4), Trp (5/5). Correlations for the remaining residues were either missing or resided in spectral regions too crowded for unambiguous identification. Based on the comparison between solution and solid-state chemical shifts, an additional 86 residues have been assigned, and tentative intra-residue assignments for 21 more amino acids are available. A summary of all chemical shifts is provided in Tables 1 and S2 (Supporting Information).
Table 1.
13C and 15N Chemical Shift Assignments for the CA Protein in Conical Assemblies.
| Residue | N | Cα | Cβ | C′ | Cγ |
|---|---|---|---|---|---|
| P1 | |||||
| I2 | 121.3 ± 1 | 59.6 ± 0.1 | 37.8 ± 0.1 | 178.7 ± 0.2* | |
| V3 | |||||
| Q4 | 122.4 ± 1 | 54.6 ± 0.2 | 29.6 ± 0.1 | ||
| N5 | 125.1 ± 1 | 53.5 ± 0.3 | 39.3 ± 0.4 | ||
| I6 | |||||
| Q7 | |||||
| G8 | 107.8 ± 1 | 44.9 ± 0.2 | |||
| Q9 | 120.5 ± 1 | 54.3 ± 0.2 | 29.8 ± 0.1 | 180.1 ± 0.1 | |
| M10 | |||||
| V11 | 120.2 ± 1 | 58.9 ± 0.1 | 35.8 ± 0.1 | ||
| H12 | 56.5 ± 0.1 | 33.3 ± 0.2 | 174.9 ± 0.2* | ||
| Q13 | 125.3 ± 1 | 54.0 ± 0.1 | 32.8 ± 0.2 | 176.1 ± 0.3 | |
| A14 | 125.3 ± 1 | 51.7 ± 0.1 | 18.9 ± 0.1 | 178.9 ± 0.2* | |
| I15 | 121.6 ± 1 | 61.5 ± 0.7 | 38.4 ± 0.2 | ||
| S16 | 123.4 ± 1 | 55.7 ± 0.1 | 63.5 ± 0.1 | ||
| P17 | |||||
| R18 | 116.3 ± 1 | 59.2 ± 0.1 | 29.9 ± 0.1 | 179.8 ± 0.1 | |
| T19 | 119.8 ± 1 | 66.3 ± 0.7 | 68.0 ± 0.1 | ||
| L20 | 121.1 ± 1 | 57.8 ± 0.1 | 41.2 ± 0.1 | 179.6 ± 0.2* | |
| N21 | 116.1 ± 1 | 56.1 ± 0.1 | 38.2 ± 0.1 | ||
| A22 | 122.4 ± 1 | 55.1 ± 0.1 | 19.3 ± 0.1 | 176.3 ± 0.4 | |
| W23 | 120.2 ± 1 | 58.4 ± 0.1 | 30.2 ± 0.1 | 178.4 ± 0.8 | |
| V24 | 118.0 ± 1 | 66.2 ± 0.4 | 31.6 ± 0.1 | ||
| K25 | 117.8 ± 1 | 59.4 ± 0.1 | 32.4 ± 0.2 | 179.4 ± 0.2* | |
| V26 | 119.4 ± 1 | 66.1 ± 0.1 | 31.4 ± 0.1 | ||
| V27 | 120.2 ± 1 | 65.7 ± 0.1 | 31.6 ± 0.1 | ||
| E28 | |||||
| E29 | 115.9 ± 1 | 58.1 ± 0.1 | 31.0 ± 0.1 | 183.4 ± 0.3 | |
| K30 | |||||
| A31 | |||||
| F32 | 113.4 ± 1 | 56.9 ± 0.1 | 37.6 ± 0.1 | 176.5 ± 0.2* | |
| S33 | |||||
| P34 | 66.7 ± 0.2* | 32.66 ± 0.2* | 179.5 ± 0.2* | ||
| E35 | 115.9 ± 1 | 59.5 ± 0.2 | 29.4 ± 0.6 | ||
| V36 | 117.0 ± 1 | 65.6 ± 0.1 | 31.9 ± 0.1 | 177.9 ± 0.1 | |
| I37 | 119.2 ± 1 | 67.7 ± 0.1 | 34.9 ± 0.2* | ||
| P38 | |||||
| M39 | 116.2 ± 1 | 59.1 ± 0.4 | 32.3 ± 0.1 | 179.0 ± 0.2 | |
| F40 | 120.7 ± 1 | 61.6 ± 0.1 | 38.5 ± 0.2 | 179.2 ± 0.1 | |
| S41 | |||||
| A42 | 122.7 ± 1 | 54.4 ± 0.7 | 18.2 ± 0.2 | ||
| L43 | 116.3 ± 1 | 56.3 ± 0.1 | 42.4 ± 0.1 | 176.5 ± 0.2* | |
| S44 | 109.6 ± 1 | 57.0 ± 0.1 | 63.6 ± 0.3 | 176.2 ± 0.2 | |
| E45 | 125.3 ± 1 | 58.1 ± 0.3 | 29.1 ± 0.2 | 178.3 ± 0.2 | |
| G46 | 115.6 ± 1 | 46.1 ± 0.1 | 174.6 ± 0.2 | ||
| A47 | 119.7 ± 1 | 53.0 ± 0.5 | 20.5 ± 0.3 | 178.8 ± 0.1 | |
| T48 | 108.6 ± 1 | 59.6 ± 0.1 | 69.1 ± 0.1 | 174.8 ± 0.1 | 22.3 ± 0.2* |
| P49 | |||||
| Q50 | 116.1 ± 1 | 59.8 ± 0.3 | 29.7 ± 0.1 | 179.9 ± 0.2* | |
| D51 | 120.4 ± 1 | 58.0 ± 0.2* | 41.4 ± 0.2 | 178.8 ± 0.1 | |
| L52 | 119.9 ± 1 | 57.8 ± 0.1 | 41.4 ± 0.4 | 179.7 ± 0.2* | |
| N53 | 118.1 ± 1 | 56.3 ± 0.1 | 37.3 ± 0.2 | ||
| T54 | 119.8 ± 1 | 67.0 ± 0.4 | 68.1 ± 0.1 | ||
| M55 | |||||
| L56 | |||||
| N57 | 116.2 ± 1 | 56.0 ± 0.2 | 38.3 ± 0.2 | 176.7 ± 0.1 | |
| T58 | 111.3 ± 1 | 63.9 ± 0.5 | 68.9 ± 0.3 | 176.9 ± 0.1 | |
| V59 | |||||
| G60 | 113.8 ± 1 | 45.4 ± 0.1 | |||
| G61 | 108.9 ± 1 | 45.3 ± 0.2 | |||
| H62 | 56.2 ± 0.1 | 28.5 ± 0.1 | |||
| Q63 | 119.6 ± 1 | 59.8 ± 0.2* | 28.3 ± 0.2* | ||
| A64 | 121.8 ± 1 | 54.6 ± 0.2 | 18.3 ± 0.2 | 181.7 ± 0.2* | |
| A65 | |||||
| M66 | |||||
| Q67 | |||||
| M68 | |||||
| L69 | 122.9 ± 1 | 58.2 ± 0.1 | 41.3 ± 0.1 | 178.8 ± 0.2* | |
| K70 | 119.2 ± 1 | 60.0 ± 0.1 | 32.6 ± 0.3 | ||
| E71 | |||||
| T72 | |||||
| I73 | 122.0 ± 1 | 66.4 ± 0.3 | 37.8 ± 0.1 | ||
| N74 | |||||
| E75 | |||||
| E76 | 120.9 ± 1 | 57.8 ± 0.2* | 26.8 ± 0.5 | ||
| A77 | 124.8 ± 1 | 55.0 ± 0.4 | 17.8 ± 0.2 | 179.0 ± 0.2* | |
| A78 | 119.7 ± 1 | 54.2 ± 0.1 | 18.0 ± 0.3 | 180.1 ± 1.8 | |
| E79 | |||||
| W80 | 121.0 ± 1 | 61.0 ± 0.2* | 27.5 ± 0.1 | ||
| D81 | 119.4 ± 1 | 57.0 ± 0.2* | 39.7 ± 0.1 | ||
| R82 | 120.8 ± 1 | 59.3 ± 0.2* | 29.3 ± 0.2* | 178.7 ± 0.2* | |
| V83 | |||||
| H84 | 117.0 ± 1 | 53.5 ± 0.4 | 26.7 ± 0.2 | 171.3 ± 0.1 | |
| P85 | |||||
| V86 | |||||
| H87 | 125.4 ± 1 | 55.4 ± 0.3 | 30.0 ± 0.3 | ||
| A88 | |||||
| G89 | |||||
| P90 | |||||
| I91 | 122.0 ± 1 | 59.7 ± 0.3 | 37.9 ± 0.5 | ||
| A92 | 131.4 ± 1 | 50.2 ± 0.2* | |||
| P93 | 61.8 ± 0.2 | 32.7 ± 0.9 | 178.8 ± 0.1 | ||
| G94 | 112.2 ± 1 | 45.4 ± 0.1 | 172.0 ± 0.1 | ||
| Q95 | 119.6 ± 1 | 54.1 ± 0.2 | 29.8 ± 0.4 | ||
| M96 | 122.2 ± 1 | 54.8 ± 0.1 | 34.4 ± 0.1 | ||
| R97 | 123.5 ± 1 | 54.8 ± 0.1 | 29.7 ± 0.2* | ||
| E98 | 123.5 ± 1 | 54.8 ± 0.1 | 29.7 ± 0.2* | 179.2 ± 0.2* | |
| P99 | |||||
| R100 | 121.9 ± 1 | 54.6 ± 0.6 | 30.7 ± 0.1 | ||
| G101 | 117.0 ± 1 | 48.8 ± 0.4 | 176.4 ± 0.1 | ||
| S102 | |||||
| D103 | 123.0 ± 1 | 57.1 ± 0.2 | 41.3 ± 0.2 | 180.1 ± 0.1 | |
| I104 | 126.0 ± 1 | 65.5 ± 0.1 | 36.6 ± 0.4 | ||
| A105 | 112.0 ± 1 | 50.2 ± 0.6 | 18.6 ± 0.5 | 178.0 ± 0.1 | |
| G106 | 102.1 ± 1 | 45.7 ± 0.3 | 173.8 ± 0.1 | ||
| T107 | 113.1 ± 1 | 64.0 ± 0.4 | 68.6 ± 0.3 | 177.4 ± 0.1 | 19.8 ± 0.2* |
| T108 | 106.7 ± 1 | 60.8 ± 0.2 | 69.4 ± 0.2 | 173.2 ± 0.2 | |
| S109 | 112.0 ± 1 | 54.3 ± 0.5 | 66.7 ± 0.2 | 174.8 ± 0.1 | |
| T110 | 112.4 ± 1 | 59.9 ± 0.4 | 71.3 ± 0.4 | 176.0 ± 0.3 | 22.5 ± 0.2 |
| L111 | 122.1 ± 1 | 58.0 ± 0.2 | 41.3 ± 0.3 | ||
| Q112 | 115.7 ± 1 | 59.4 ± 0.2 | 27.8 ± 0.2 | ||
| E113 | |||||
| Q114 | 118.0 ± 1 | 59.1 ± 0.4 | 27.7 ± 0.2 | ||
| I115 | 118.1 ± 1 | 65.7 ± 0.1 | 38.3 ± 0.3 | 180.2 ± 0.1 | |
| G116 | 111.4 ± 1 | 47.4 ± 0.2 | 175.4 ± 0.4 | ||
| W117 | 121.7 ± 1 | 61.3 ± 0.8 | 27.7 ± 0.1 | 177.5 ± 0.1 | |
| M118 | 116.2 ± 1 | 59.5 ± 0.4 | 35.2 ± 0.3 | 177.9 ± 0.2 | |
| T119 | 104.4 ± 1 | 60.5 ± 0.1 | 69.8 ± 0.1 | 173.3 ± 0.2* | |
| N120 | 123.4 ± 1 | 58.4 ± 0.2 | 29.0 ± 0.1 | ||
| N121 | |||||
| P122 | |||||
| P123 | |||||
| I124 | 122.9 ± 1 | 59.2 ± 0.1 | 38.0 ± 0.1 | ||
| P125 | 62.6 ± 0.2* | 28.33 ± 0.2* | 179.5 ± 0.2* | ||
| V126 | 115.8 ± 1 | 66.3 ± 0.5 | 30.1 ± 0.1 | 177.2 ± 0.2* | |
| G127 | 106.3 ± 1 | 47.4 ± 0.3 | 175.2 ± 0.2 | ||
| E128 | |||||
| I129 | 121.4 ± 1 | 64.7 ± 0.1 | 38.3 ± 0.3 | 178.9 ± 0.2 | |
| Y130 | 117.0 ± 0.6 | 56.0 ± 0.2 | 37.1 ± 0.3 | 178.5 ± 0.2* | |
| K131 | |||||
| R132 | 117.9 ± 1 | 60.1 ± 0.1 | 29.2 ± 0.1 | ||
| W133 | 122.1 ± 1 | 58.6 ± 0.2* | 29.1 ± 0.1 | ||
| I134 | 119.2 ± 1 | 65.6 ± 0.3 | 38.2 ± 0.1 | 179.0 ± 0.2* | |
| I135 | 121.3 ± 1 | 66.4 ± 0.2* | 37.9 ± 0.3 | ||
| L136 | 122.6 ± 1 | 57.9 ± 0.2* | 41.6 ± 0.1 | 181.1 ± 0.2* | |
| G137 | 108.1 ± 1 | 47.4 ± 0.2 | 175.5 ± 0.2* | ||
| L138 | 122.9 ± 1 | 57.9 ± 0.2* | 42.3 ± 0.1 | 179.5 ± 0.2* | 27.2 ± 0.1 |
| N139 | 117.7 ± 1 | 57.1 ± 0.2* | 38.9 ± 0.2 | 178.6 ± 0.2* | |
| K140 | 119.4 ± 1 | 59.6 ± 0.2* | 32.7 ± 0.1 | 179.1 ± 0.2* | |
| I141 | 120.3 ± 1 | 65.2 ± 0.2 | 38.1 ± 0.2 | 179.6 ± 0.4 | |
| V142 | 118.6 ± 1 | 66.1 ± 0.2* | 31.4 ± 0.1 | ||
| R143 | 117.9 ± 1 | 58.8 ± 0.1 | 29.8 ± 0.1 | ||
| M144 | |||||
| Y145** | 115.1 ± 0.5 | 59.2 ± 0.1 | |||
| S146 | 121.2 ± 1 | 55.8 ± 0.1 | 63.7 ± 0.1 | ||
| P147 | |||||
| T148 | |||||
| S149 | |||||
| I150 | 126.0 ± 1 | 62.1 ± 0.3 | 39.0 ± 0.1 | 178.9 ± 0.3 | |
| L151 | 118.9 ± 1 | 56.6 ± 0.1 | 39.7 ± 0.1 | 177.8 ± 0.2* | |
| D152 | 114.4 ± 1 | 54.6 ± 0.6 | 41.6 ± 0.1 | ||
| I153 | 122.0 ± 1 | 60.9 ± 0.3 | 32.5 ± 0.2 | ||
| R154 | 125.3 ± 1 | 54.2 ± 0.3 | 33.0 ± 0.3 | 175.9 ± 0.1 | |
| Q155 | 127.8 ± 1 | 55.5 ± 0.1 | 26.6 ± 0.6 | 32.9 ± 0.2* | |
| G156 | 116.9 ± 1 | 45.1 ± 0.1 | 173.3 ± 0.2 | ||
| P157 | 130.5 ± 1 | 65.8 ± 0.2* | 32.7 ± 0.2* | 177.4 ± 0.2* | 27.8 ± 0.2* |
| K158 | 114.9 ± 1 | 53.7 ± 0.2 | 32.6 ± 0.2 | 175.8 ± 0.2* | |
| E159 | 125.7 ± 1 | 53.6 ± 0.2 | 31.8 ± 0.6 | ||
| P160 | |||||
| F161 | 128.1 ± 1 | 62.7 ± 0.1 | 39.2 ± 0.2 | 177.2 ± 0.2 | |
| R162 | 115.7 ± 1 | 59.4 ± 0.1 | 29.7 ± 0.2 | 179.3 ± 0.2* | |
| D163 | 118.3 ± 1 | 56.8 ± 0.2* | 39.5 ± 0.1 | ||
| Y164 | 124.6 ± 0.6 | 59.4 ± 0.2 | 38.1 ± 0.1 | 176.1 ± 0.2 | |
| V165 | 120.2 ± 1 | 66.6 ± 0.2 | |||
| D166 | 118.3 ± 1 | 57.5 ± 0.2* | 178.0 ± 0.2 | ||
| R167 | 119.1 ± 1 | 59.3 ± 0.2* | 29.6 ± 0.2 | ||
| F168 | 124.2 ± 1 | 62.1 ± 0.3 | 38.8 ± 0.2 | 178.7 ± 0.2* | |
| Y169 | 117.2 ± 1 | 63.3 ± 0.8 | 38.1 ± 0.3 | 179.3 ± 0.1 | |
| K170 | 120.7 ± 1 | 60.3 ± 0.3 | 32.4 ± 0.2 | ||
| T171 | 117.5 ± 1 | 66.7 ± 0.2 | 68.3 ± 0.1 | 177.0 ± 0.1 | 22.1 ± 0.2* |
| L172 | 124.1 ± 1 | 58.5 ± 0.2 | 41.4 ± 0.2 | 178.3 ± 0.5 | 28.9 ± 0.2* |
| R173 | 116.3 ± 1 | 60.3 ± 0.2* | |||
| A174 | 118.6 ± 1 | 52.0 ± 0.3 | 19.3 ± 0.1 | 176.2 | |
| E175 | |||||
| Q176 | 123.9 ± 1 | 54.3 ± 0.4 | 27.5 ± 0.2 | ||
| A177 | 126.6 ± 1 | 51.4 ± 0.1 | 21.3 ± 0.3 | 176.3 ± 0.3 | |
| S178 | 117.5 ± 1 | 57.6 ± 0.6 | 65.1 ± 0.5 | 175.3 ± 0.2* | |
| Q179 | 121.3 ± 1 | 58.5 ± 0.3 | 28.4 ± 0.1 | ||
| E180 | |||||
| V181 | 121.2 ± 1 | 66.4 ± 0.3 | 31.0 ± 0.1 | ||
| K182 | |||||
| N183 | 119.1 ± 1 | 55.8 ± 0.2 | 37.2 ± 0.1 | ||
| W184 | 122.9 ± 1 | 61.3 ± 0.1 | 27.5 ± 0.7 | ||
| M185 | |||||
| T186 | 112.3 ± 1 | 66.3 ± 0.3 | 68.8 ± 0.4 | 176.5 ± 0.2* | 21.9 ± 0.2* |
| E187 | 118.7 ± 1 | 58.5 ± 0.2* | 30.2 ± 0.2* | ||
| T188 | 111.8 ± 1 | 64.0 ± 0.1 | 68.7 ± 0.2 | 177.2 ± 0.5 | |
| L189 | 121.0 ± 1 | 57.8 ± 0.1 | 41.4 ± 0.2 | ||
| L190 | |||||
| V191 | 115.8 ± 1 | 66.0 ± 0.5 | 31.9 ± 0.2 | ||
| Q192 | 116.7 ± 1 | 59.1 ± 0.7 | 27.7 ± 0.1 | ||
| N193 | 114.8 ± 1 | 52.8 ± 0.2 | 38.2 ± 0.3 | 175.7 ± 0.2* | |
| A194 | 124.6 ± 1 | 52.0 ± 0.2 | 19.0 ± 0.2 | 176.3 ± 0.3 | |
| N195 | 119.2 ± 1 | 51.4 ± 0.2 | 35.1 ± 0.2 | ||
| P196 | 66.9 ± 0.3 | 32.6 ± 0.3 | 179.7 ± 0.3 | 28.3 ± 0.2* | |
| D197 | 115.4 ± 1 | 56.8 ± 0.2 | 40.6 ± 0.3 | ||
| C198 | 116.1 ± 1 | 63.8 ± 0.1 | 28.1 ± 0.3 | 177.2 ± 0.1 | |
| K199 | 121.8 ± 1 | 60.0 ± 0.4 | 32.3 ± 0.5 | 178.6 ± 0.1 | |
| T200 | 113.1 ± 1 | 66.8 ± 0.3 | 68.9 ± 0.2 | 176.8 ± 0.8 | 22.1 ± 0.1 |
| I201 | 122.8 ± 1 | 65.0 ± 0.1 | 38.1 ± 0.1 | 179.4 ± 0.2* | |
| L202 | 118.9 ± 1 | 57.5 ± 0.3 | 41.1 ± 0.3 | ||
| K203 | |||||
| A204 | |||||
| L205 | 118.3 ± 1 | 56.4 ± 0.5 | 42.6 ± 0.1 | 179.3 ± 0.2* | 26.6 ± 0.2* |
| G206 | 104.3 ± 1 | 44.5 ± 0.6 | 172.6 ± 0.1 | ||
| P207 | |||||
| A208 | 121.5 ± 1 | 52.5 ± 0.1 | 20.0 ± 0.2 | 178.1 ± 0.1 | |
| A209 | 122.8 ± 1 | 52.7 ± 0.3 | 20.3 ± 0.2 | 178.7 ± 0.1 | |
| T210 | 113.2 ± 1 | 59.6 ± 0.4 | 71.1 ± 0.3 | 175.7 ± 0.4 | 22.3 ± 0.2* |
| L211 | 122.6 ± 1 | 58.1 ± 0.1 | 41.3 ± 0.1 | 179.7 ± 0.2 | |
| E212 | |||||
| E213 | |||||
| M214 | 120.0 ± 1 | 60.1 ± 0.1 | 32.7 ± 0.1 | ||
| M215 | 116.9 ± 1 | 59.2 ± 0.2 | 32.4 ± 0.1 | ||
| T216 | 117.0 ± 1 | 66.5 ± 0.3 | 68.2 ± 0.2 | 176.9 ± 0.5 | 21.9 ± 0.1 |
| A217 | 122.8 ± 1 | 52.8 ± 0.4 | 177.8 ± 0.1 | ||
| C218 | 110.2 ± 1 | 59.8 ± 0.1 | 28.6 ± 0.3 | 175.6 ± 0.2* | |
| Q219 | 122.4 ± 1 | 59.1 ± 0.8 | 28.3 ± 0.1 | ||
| G220 | 109.9 ± 1 | 45.3 ± 0.3 | |||
| V221 | |||||
| G222 | |||||
| G223 | 108.1 ± 1 | 44.3 ± 0.2 | |||
| P224 | |||||
| G225 | 109.1 ± 1 | 47.2 ± 0.1 | 175.0 ± 0.1 | ||
| H226 | |||||
| K227 | |||||
| A228 | 125.4 ± 1 | 52.8 ± 0.6 | 19.8 ± 0.2 | ||
| R229 | |||||
| V230 | 122.9 ± 1 | 61.0 ± 0.2 | 32.5 ± 0.2 | ||
| L231 |
The chemical shift uncertainties for the nitrogen atoms are determined by the digital resolution of the various 3D data sets acquired at 14.1 T. The chemical shift uncertainties for the carbon atoms are determined by the digital resolution in direct and indirect dimensions of the various homo- and heteronuclear 2D and 3D data sets acquired at 14.1 and 21.1 T.
For these residues, chemical shifts were extracted from a single data set, and the errors reflect average errors across all residues for a given atom type.
For Y145, 13C and 15N chemical shifts exhibit substantial temperature dependences. The chemical shifts reported here were extracted from a 2D NCA spectrum of U-13C,15N-Tyr-labeled CA (conical assembly) acquired at −15 °C.
In order to aim for complete assignments, however, additional data sets at magnetic fields higher than 14.1 T are required and such studies will be performed in the future.
Secondary structure analysis of conical CA assemblies
For the assigned regions of CA, secondary structure analysis was carried out using the Chemical Shift Index (CSI) based on Cα (Figure 6) and C' (Figure S1 of the Supporting Information) chemical shifts,57,58 that uses sequence-corrected random coil values for all amino acids.59 The secondary structural elements derived from Cα CSI for CA in the solid- and solution states were compared (Figure 6). The data clearly indicate that the secondary structures derived from solid state and solution NMR CSI analysis are consistent with the location of helices in the known 3D structure of the individual N- and C-terminal domains.60–63
Figure 6.
Comparison between solid-state and solution secondary chemical shifts for CA conical assemblies. Cα shifts that reside in α-helical regions of the protein exhibit positive secondary shifts, while those located in the β-strands and other secondary structure elements are negative.
CONCLUSION
High-resolution structures of the individual N- and C-terminal domains of HIV-1 CA have been determined by solution NMR61 and X-ray crystallogrphy,62,63 and aided in generating a model of the CA assemblies by cryo-EM.14 However, the cryo-EM studies to date comprise very regular assemblies of CA, most notably in 2D crystalline arrays. Therefore, additional studies are necessary to investigate which and how intermolecular interactions give rise to the observed structural polymorphism of CA assemblies. Without detailed structures of the different capsid morphologies, the mechanisms of viral uncoating and capsid interactions with the restriction factors are not easily understood, impeding the design of new antiretroviral therapies. This lack of knowledge prompted us to embark on a complementary structural methodology, namely multidimensional solid-state MAS NMR spectroscopy.
We have established sample preparation protocols for generating HIV-1 CA assemblies of different morphologies suitable for structural analysis by multidimensional MAS NMR methods. While the large size and high α-helical content of the protein pose considerable challenges for resonance assignments of the U-13C,15N enriched CA, the observed spectral resolution is encouraging and permitted partial resonance assignments with a limited set of 2D and 3D homo- and heteronuclear correlation experiments. Interestingly but perhaps not surprisingly, the chemical shifts did not exhibit noticeable variations depending on sample morphology, indicating that the overall capsid organization is determined by the intermolecular protein interactions rather than by large variations in conformation of individual CA molecules.
In order to alleviate the remaining problems caused by spectral overlap and to complete the resonance assignments, deuteration64 in conjunction with the introduction of paramagnetic T1 relaxation enhancement dopants65–67 will be pursued. These methods should allow us to complete the resonance assignments of the CA protein in differently assembled particle morphologies. Therefore, our present work is an important step towards realizing exciting opportunities in structural analysis of CA assemblies at atomic resolution.
EXPERIMENTS AND METHODS
Materials
All chemicals were purchased from Fisher Scientific or Sigma Aldrich and were used without further purification. 400 mesh copper grids coated with Formval and stabilized with evaporated carbon films were purchased from Electron Microscopy Sciences (Hatfield, PA).
Preparation of HIV-1 CA protein assemblies
The cDNA encoding gag polyprotein, pr55gag was obtained from the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH,68 and the sequence coding for CA was amplified and subcloned into pET21 (EMD chemicals, Inc. San Diego, CA) using NdeI and XhoI sites. U-13C,15N isotopically enriched proteins were expressed in E. coli, Rosetta 2 (DE3), cultured in modified minimal medium using 15NH4Cl and U-13C6-glucose as sole nitrogen and carbon sources, after induction with 0.4 mM IPTG at 23 °C for 16 h. Four additional HIV-1 CA samples were prepared containing isotopic labels in a single amino acid residue type: U-13C,15N-Ala, U-13C,15N-His, U-13C,15N-Thr, and U-13C,15N-Tyr. Proteins were expressed in modified minimal media adding each specific U-13C,15N-amino acid and purified via ionic exchange and gel filtration column chromatography to greater than 99% homogeneity.
For preparation of the CA assemblies, the protein was first dialyzed against 25 mM sodium phosphate buffer, containing 1 mM DTT, and 0.02% NaN3 (pH 5.5). For generating the CA assemblies containing mixed labels, solutions containing U-13C,15N-Ala (or U-13C,15N-Thr) and U-13C,15N-His (or U-13C,15N-Tyr) enriched CA, were mixed 1:1, followed by the assembly step, as described below. Two differentially mixed labeled CA samples were prepared: U-13C,15N-Ala/U-13C,15N-Thr and U-13C,15N-His/U-13C,15N-Tyr.
CA protein assemblies of conical morphology were prepared by the addition of a 17.5% w/v solution of PEG-20,000 to lyophilized protein to a final protein concentration of 32 mg/ml, followed by incubation for 1 hr at 37 °C. Assembled material was recovered as the pellet after centrifugation at 14,000 rpm at 4 °C. Fifteen milligrams of the precipitate were packed into a 3.2 mm Varian rotor and sealed using an upper spacer and a top spinner.
CA assemblies of spherical morphology were prepared by mixing the 32 mg/ml CA solution with 7.5% w/v PEG-20,000 (1:1 volume ratio). The resulting mixture was incubated on ice for 30 min, and then diluted 4-fold. The solution containing the spherical assemblies was dried with N2 gas to remove any excess water. Twelve milligrams of the dried sample were packed into a 3.2 mm Varian rotor and sealed using an upper spacer and a top spinner.
CA assemblies of tubular morphology were prepared by the incubation of the 32 mg/ml CA solution was prepared in 50 mM Tris buffer, pH 8.0, containing 1 M NaCl, at 37 °C for 1 hour.
Transmission electron microscopy (TEM)
The CA assemblies were analyzed using a Zeiss CEM 902 transmission electron microscope operating at 80 kV. Samples were stained with ammonium molybdate (5% w/v), deposited onto 400 mesh, formval/carbon-coated copper grids, and dried for 40 min.
Cryo scanning electron microscopy (cryo-SEM)
Slurries containing conical and spherical assemblies (~1 mm3) were applied to gold carrier plates and frozen in a Leica EM PACT high-pressure freezer. Subsequently, the samples were fractured with a pre-cooled knife and coated with 10 nm of gold. The frozen samples were then transferred to the cryo-stage, warmed to −90 °C for 5–7 min to remove surface water. The images were taken on a Hitachi S-4700 FESEM (Tokyo, Japan) at −125 °C and 1.0 kV with a working distance of approximately 4–5 mm.
Confocal laser scanning microscopy
Confocal images were acquired on a Zeiss LSM 510 NLO laser scanning microscope (Carl Zeiss, Germany) using a Zeiss 40× (NA 1.3) oil immersion objective lens. All data were collected using the 584 nm laser line of a 25 mW Argon laser (LASOS, Germany) with a rhodamine reflector and the transmitted-light channel when appropriate. Protein solutions were stained with the Nile Blue A dye (0.5 g/L) according to the standard protocol, and all images were acquired in fluorescence, transmittance and reflection modes.
NMR spectroscopy
Solid-state NMR spectra were acquired at 14.1 T (600 MHz) on a standard bore Varian InfinityPlus instrument outfitted with a 3.2 mm triple resonance T3 probe; Larmor frequencies are 599.5 MHz for 1H, 150.7 MHz for 13C, and 60.7 MHz for 15N. Dipolar-based two- and three-dimensional DARR, NCA, NCO, NCACB, NCACX, and NCOCX spectra were collected at the MAS frequency of 10.000±0.001 kHz regulated by a Varian MAS controller. The dipolar NCOCX and NCACX experiments utilized the SPECIFIC-CP heteronuclear mixing69 followed by the DARR mixing sequence;70 in the NCACB sequence, homonuclear DREAM mixing71 with a linear ramp was used. The temperature was calibrated at different MAS frequencies using a PbNO3 temperature sensor,72 and the actual temperature at the sample was maintained to within ±0.5 °C throughout the experiments using the Varian temperature controller. The DARR spectra were acquired at temperatures of −5.0 °C, 5.0 °C for spherical and conical assemblies respectively. The heteronuclear 2D and 3D spectra were acquired at 5 °C. 1H, 13C and 15N chemical shifts were referenced with respect to DSS, adamantane and ammonium chloride as external referencing standards.73 The typical pulse lengths in the double resonance mode were 2.6 μs (1H), and 3.1 μs (13C), and in the triple resonance mode 2.6 μs (1H), 5 μs (13C), and 5 μs (15N). 1H- 13C(15N) cross polarization was achieved with a linear amplitude ramp; the 1H radio frequency field was 50 kHz with the center of the ramp Hartmann-Hahn matched to the first spinning sideband. The 1H-13C and 1H-15N contact times were 900 μs and 1.1 ms for conical and spherical assemblies respectively. In the dipolar-based experiments, the DARR mixing time was 10 ms, the SPECIFIC-CP mixing time was 6.2 ms, and the DREAM mixing time was 3.0 ms. TPPM decoupling74 was used, with the decoupling field strengths ranging between 80 and 100 kHz in the different experiments. Recycle delays in all experiments were 2 s. TPPI75 was used for frequency discrimination in the indirect dimensions.
DARR spectra of the conical assemblies were also acquired on a Varian Inova 21.1 T spectrometer outfitted with a 3.2 mm BioMAS HXY probe. The Larmor frequencies were 900.575 MHz and 226.47 MHz for 1H for 13C, respectively. Spectra were collected at a MAS frequency of 14.000±0.001 kHz regulated by a Varian MAS controller, and the temperature was maintained at 15±0.5 °C throughout the experiments using a Varian temperature controller. The pulse lengths in the DARR experiments were 3.7 μs (1H), and 6.3 μs (13C), and 1H-13C cross polarization was performed with a tangent amplitude ramp (80–100%), having the 1H radio frequency field of 68 kHz matched to the second spinning sideband. The 1H-13C contact time was 1 ms. TPPM decoupling (71 kHz) was applied during the evolution and acquisition periods. The 1H field strength during DARR was 14 kHz, and the DARR mixing time was 50 ms. Under these conditions predominantly one-bond correlations are observed in the spectra.
Additional acquisition and processing parameters for data sets collected at 14.1 and 21.1 T are provided in the figure captions and in Table S3 of the Supporting Information.
NMR data processing and analysis
All spectra were processed with NMRpipe76 and analyzed using Sparky.77 For 2D data sets, 90°, 60°, or 30° shifted sine bell apodization followed by a Lorentzian-to-Gaussian transformation was applied in both dimensions. Forward linear prediction to twice the number of the original data points was used in the indirect dimension in some data sets followed by zero filling to twice the total number of points. The 3D data sets were processed by using 30° or 60° shifted sine bells in all dimensions followed by a Lorentzian-to-Gaussian transformation (for resolution enhancement) and by using 90° degree shifted sinebells in all dimensions followed by a Lorentzian-to-Gaussian transformation (for sensitivity enhancement). Detailed information about processing parameters is provided in Table S3 of the Supporting Information.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Institutes of General Medical Sciences (NIH Grant P50GM082251) and is a contribution from the Pittsburgh Center for HIV Protein Interactions. The authors are grateful to Kirk Czymmek and Deborah Powell at the Delaware Biotechnology Institute Bio-Imaging Center at the University of Delaware for their kind assistance with acquiring TEM, cryo-SEM and confocal images. The 21.1 T spectra were acquired at the Environmental Molecular Sciences Laboratory, a national scientific user facility sponsored by the United States Department of Energy's Office of Biological and Environmental Research and located at the Pacific Northwest National Laboratory in Richland, WA. The kind assistance of Andrew Lipton, Jesse Sears, Michael Froehlke, Sarah Burton, David Hoyt and Joseph Ford during our visit to EMSL is gratefully acknowledged.
Footnotes
SUPPORTING INFORMATION Comparison of solid-state and solution chemical shifts for the assigned residues in the conical assemblies of HIV-1 CA. Summary of the resonance assignments for the conical HIV-1 CA assemblies. Acquisition and processing parameters for the solid-state 2D and 3D spectra of conical and spherical HIV-1 CA assemblies. Chemical shift indexes derived from the Cα and C' solid-state shifts. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- (1).Weiss RA. Science. 1993;260:1273. doi: 10.1126/science.8493571. [DOI] [PubMed] [Google Scholar]
- (2).Adamson CS, Salzwedel K, Freed EO. Expert Opin. Ther. Targets. 2009;13:895. doi: 10.1517/14728220903039714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Whittaker GR, Kann M, Helenius A. Annu. Rev. Cell. Dev. Bi. 2000;16:627. doi: 10.1146/annurev.cellbio.16.1.627. [DOI] [PubMed] [Google Scholar]
- (4).Whittaker GR. Adv. Drug Deliver. Rev. 2003;55:733. doi: 10.1016/s0169-409x(03)00051-6. [DOI] [PubMed] [Google Scholar]
- (5).Perron MJ, Stremlau M, Lee M, Javanbakht H, Song B, Sodroski J. J. Virol. 2007;81:2138. doi: 10.1128/JVI.02318-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Stremlau M. Science. 2007;318:1565. doi: 10.1126/science.1152905. [DOI] [PubMed] [Google Scholar]
- (7).Stremlau M, Perron M, Lee M, Li Y, Song B, Javanbakht H, Diaz-Griffero F, Anderson DJ, Sundquist WI, Sodroski J. Proc. Natl. Acad. Sci. USA. 2006;103:5514. doi: 10.1073/pnas.0509996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Lee K, KewalRamani VN. Proc. Natl. Acad. Sci. USA. 2004;101:10496. doi: 10.1073/pnas.0404066101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Yap MW, Nisole S, Lynch C, Stoye JP. Proc. Natl. Acad. Sci. USA. 2004;101:10786. doi: 10.1073/pnas.0402876101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Hatziioannou T, Perez-Caballero D, Yang A, Cowan S, Bieniasz PD. Proc. Natl. Acad. Sci. USA. 2004;101:10774. doi: 10.1073/pnas.0402361101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Keckesova Z, Ylinen LMJ, Towers GJ. Proc. Natl. Acad. Sci. USA. 2004;101:10780. doi: 10.1073/pnas.0402474101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Tang C, Ndassa Y, Summers MF. Nat. Struct. Biol. 2002;9:537. doi: 10.1038/nsb806. [DOI] [PubMed] [Google Scholar]
- (13).Worthylake DK, Wang H, Yoo SH, Sundquist WI, Hill CP. Acta Cryst. D. 1999;55:85. doi: 10.1107/S0907444998007689. [DOI] [PubMed] [Google Scholar]
- (14).Ganser-Pornillos BK, Cheng A, Yeager M. Cell. 2007;131:70. doi: 10.1016/j.cell.2007.08.018. [DOI] [PubMed] [Google Scholar]
- (15).Pornillos O, Ganser-Pornillos BK, Kelly BN, Hua Y, Whitby FG, Stout CD, Sundquist WI, Hill CP, Yeager M. Cell. 2009;137:1282. doi: 10.1016/j.cell.2009.04.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Byeon IJL, Meng X, Jung JW, Zhao GP, Yang RF, Ahn JW, Shi J, Concel J, Aiken C, Zhang PJ, Gronenborn AM. Cell. 2009;139:780. doi: 10.1016/j.cell.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Campbell S, Vogt VM. J. Virol. 1995;69:6487. doi: 10.1128/jvi.69.10.6487-6497.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Gross I, Hohenberg H, Krausslich HG. Eur. J. Biochem. 1997;249:592. doi: 10.1111/j.1432-1033.1997.t01-1-00592.x. [DOI] [PubMed] [Google Scholar]
- (19).Barklis E, McDermott J, Wilkens S, Fuller S, Thompson D. J. Biol. Chem. 1998;273:7177. doi: 10.1074/jbc.273.13.7177. [DOI] [PubMed] [Google Scholar]
- (20).Ehrlich LS, Liu TB, Scarlata S, Chu B, Carter CA. Biophys. J. 2001;81:2992. doi: 10.1016/S0006-3495(01)75725-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Vogt V. In: Retroviruses. Coffin JM, Hughes SH, Varmus HE, editors. Cold Spring Harbor Press; Plainview: 1997. [Google Scholar]
- (22).Ganser BK, Li S, Klishko VY, Finch JT, Sundquist WI. Science. 1999;283:80. doi: 10.1126/science.283.5398.80. [DOI] [PubMed] [Google Scholar]
- (23).Ganser-Pornillos BK, Yeager M, Sundquist WI. Curr. Opin. Struct. Biol. 2008;18:203. doi: 10.1016/j.sbi.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).McDermott A, Polenova T, Bockmann A, Zilm KW, Paulsen EK, Martin RW, Montelione GT. J. Biomol. NMR. 2000;16:209. doi: 10.1023/a:1008391625633. [DOI] [PubMed] [Google Scholar]
- (25).Castellani F, van Rossum B, Diehl A, Schubert M, Rehbein K, Oschkinat H. Nature. 2002;420:98. doi: 10.1038/nature01070. [DOI] [PubMed] [Google Scholar]
- (26).Marulanda D, Tasayco ML, Cataldi M, Arriaran V, Polenova T. J. Phys. Chem. B. 2005;109:18135. doi: 10.1021/jp052774d. [DOI] [PubMed] [Google Scholar]
- (27).Marulanda D, Tasayco ML, McDermott A, Cataldi M, Arriaran V, Polenova T. J. Am. Chem. Soc. 2004;126:16608. doi: 10.1021/ja0464589. [DOI] [PubMed] [Google Scholar]
- (28).Franks WT, Zhou DH, Wylie BJ, Money BG, Graesser DT, Frericks HL, Sahota G, Rienstra CM. J. Am. Chem. Soc. 2005;127:12291. doi: 10.1021/ja044497e. [DOI] [PubMed] [Google Scholar]
- (29).Egawa A, Fujiwara T, Mizoguchi T, Kakitani Y, Koyama Y, Akutsu H. Proc. Natl. Acad. Sci. USA. 2007;104:790. doi: 10.1073/pnas.0605911104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Goldbourt A, Gross BJ, Day LA, McDermott AE. J. Am. Chem. Soc. 2007;129:2338. doi: 10.1021/ja066928u. [DOI] [PubMed] [Google Scholar]
- (31).Paik Y, Yang C, Metaferia B, Tang SB, Bane S, Ravindra R, Shanker N, Alcaraz AA, Johnson SA, Schaefer J, O'Connor RD, Cegelski L, Snyder JP, Kingston DGI. J. Am. Chem. Soc. 2007;129:361. doi: 10.1021/ja0656604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Chimon S, Ishii Y. J. Am. Chem. Soc. 2005;127:13472. doi: 10.1021/ja054039l. [DOI] [PubMed] [Google Scholar]
- (33).Jaroniec CP, MacPhee CE, Bajaj VS, McMahon MT, Dobson CM, Griffin RG. Proc. Natl. Acad. Sci. USA. 2004;101:711. doi: 10.1073/pnas.0304849101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Tycko R. Q. Rev. Biophys. 2006;39:1. doi: 10.1017/S0033583506004173. [DOI] [PubMed] [Google Scholar]
- (35).Wasmer C, Lange A, Van Melckebeke H, Siemer AB, Riek R, Meier BH. Science. 2008;319:1523. doi: 10.1126/science.1151839. [DOI] [PubMed] [Google Scholar]
- (36).Huang L, McDermott AE. Biochim. Biophys. Acta- Bioenergetics. 2008;1777:1098. doi: 10.1016/j.bbabio.2008.01.006. [DOI] [PubMed] [Google Scholar]
- (37).Varga K, Tian L, McDermott AE. Biochim. Biophys. Acta- Prot. Proteom. 2007;1774:1604. doi: 10.1016/j.bbapap.2007.08.029. [DOI] [PubMed] [Google Scholar]
- (38).Frericks HL, Zhou DH, Yap LL, Gennis RB, Rienstra CM. J. Biomol. NMR. 2006;36:55. doi: 10.1007/s10858-006-9070-5. [DOI] [PubMed] [Google Scholar]
- (39).Hong M. Structure. 2006;14:1731. doi: 10.1016/j.str.2006.10.002. [DOI] [PubMed] [Google Scholar]
- (40).Kobayashi M, Matsuki Y, Yumen I, Fujiwara T, Akutsu H. J. Biomol. NMR. 2006;36:279. doi: 10.1007/s10858-006-9094-x. [DOI] [PubMed] [Google Scholar]
- (41).Schneider R, Ader C, Lange A, Giller K, Hornig S, Pongs O, Becker S, Baldus M. 2008:7427. doi: 10.1021/ja800190c. [DOI] [PubMed] [Google Scholar]
- (42).Seidel K, Andronesi OC, Krebs J, Griesinger C, Young HS, Becker S, Baldus M. Biochemistry. 2008;47:4369. doi: 10.1021/bi7024194. [DOI] [PubMed] [Google Scholar]
- (43).Goobes G, Stayton PS, Drobny GP. Prog. NMR Spectr. 2007;50:71. doi: 10.1016/j.pnmrs.2006.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Asakura T, Nakazawa Y. Sen-I Gakkaishi. 2007;63:P261. [Google Scholar]
- (45).Marcotte I, van Beek JD, Meier BH. Macromolecules. 2007;40:1995. [Google Scholar]
- (46).Bockmann A. C. R. Chim. 2006;9:381. [Google Scholar]
- (47).De Paepe G, Bayro MJ, Lewandowski J, Griffin RG. J. Am. Chem. Soc. 2006;128:1776. doi: 10.1021/ja0550430. [DOI] [PubMed] [Google Scholar]
- (48).Ernst M, Detken A, Bockmann A, Meier BH. J. Am. Chem. Soc. 2003;125:15807. doi: 10.1021/ja0369966. [DOI] [PubMed] [Google Scholar]
- (49).Heise H, Hoyer W, Becker S, Andronesi OC, Riedel D, Baldus M. Proc. Natl. Acad. Sci. USA. 2005;102:15871. doi: 10.1073/pnas.0506109102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Igumenova TI, McDermott AE, Zilm KW, Martin RW, Paulson EK, Wand AJ. J. Am. Chem. Soc. 2004;126:6720. doi: 10.1021/ja030547o. [DOI] [PubMed] [Google Scholar]
- (51).Pauli J, Baldus M, van Rossum B, de Groot H, Oschkinat H. Chembiochem. 2001;2:272. doi: 10.1002/1439-7633(20010401)2:4<272::AID-CBIC272>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- (52).Paulson EK, Morcombe CR, Gaponenko V, Dancheck B, Byrd RA, Zilm KW. J. Am. Chem. Soc. 2003;125:15831. doi: 10.1021/ja037315+. [DOI] [PubMed] [Google Scholar]
- (53).Siemer AB, Ritter C, Ernst M, Riek R, Meier BH. Angew. Chem. Int. Ed. 2005;44:2441. doi: 10.1002/anie.200462952. [DOI] [PubMed] [Google Scholar]
- (54).Jung JW, Byeon IJL, Ahn JW, Concel J, Gronenborn AM. Biomol. NMR Assign. 2009 doi: 10.1007/s12104-009-9198-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Yang J, Paramasivan S, Marulanda D, Cataidi M, Tasayco ML, Polenova T. Magn. Reson. Chem. 2007;45:S73. doi: 10.1002/mrc.2092. [DOI] [PubMed] [Google Scholar]
- (56).Sun S, Siglin A, Williams JC, Polenova T. J. Am. Chem. Soc. 2009;131:10113. doi: 10.1021/ja902003u. [DOI] [PubMed] [Google Scholar]
- (57).Wishart DS, Sykes BD. J. Biomol. NMR. 1994;4:171. doi: 10.1007/BF00175245. [DOI] [PubMed] [Google Scholar]
- (58).Wishart DS, Sykes BD, Richards FM. Biochemistry. 1992;31:1647. doi: 10.1021/bi00121a010. [DOI] [PubMed] [Google Scholar]
- (59).Schwarzinger S, Kroon GJA, Foss TR, Chung J, Wright PE, Dyson HJ. J. Am. Chem. Soc. 2001;123:2970. doi: 10.1021/ja003760i. [DOI] [PubMed] [Google Scholar]
- (60).Lanman J, Lam TT, Barnes S, Sakalian M, Emmett MR, M. A, Prevelige PEJ. J. Mol. Biol. 2003;325:759. doi: 10.1016/s0022-2836(02)01245-7. [DOI] [PubMed] [Google Scholar]
- (61).Gitti RK, Lee BM, Walker J, Summers MF, Yoo S, Sundquist WI. Science. 1996;273:231. doi: 10.1126/science.273.5272.231. [DOI] [PubMed] [Google Scholar]
- (62).Gamble TR, Yoo SH, Vajdos FF, vonSchwedler UK, Worthylake DK, Wang H, McCutcheon JP, Sundquist WI, Hill CP. Science. 1997;278:849. doi: 10.1126/science.278.5339.849. [DOI] [PubMed] [Google Scholar]
- (63).Gamble TR, Vajdos FF, Yoo SH, Worthylake DK, Houseweart M, Sundquist WI, Hill CP. Cell. 1996;87:1285. doi: 10.1016/s0092-8674(00)81823-1. [DOI] [PubMed] [Google Scholar]
- (64).Hologne M, Chevelkov V, Reif B. Prog. NMR Spectr. 2006;48:211. [Google Scholar]
- (65).Wickramasinghe NP, Ishii Y. J. Magn. Reson. 2006;181:233. doi: 10.1016/j.jmr.2006.05.008. [DOI] [PubMed] [Google Scholar]
- (66).Wickramasinghe NP, Kotecha M, Samoson A, Past J, Ishii Y. J. Magn. Reson. 2007;184:350. doi: 10.1016/j.jmr.2006.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Linser R, Chevelkov V, Diehl A, Reif B. J. Magn. Reson. 2007;189:209. doi: 10.1016/j.jmr.2007.09.007. [DOI] [PubMed] [Google Scholar]
- (68).Erickson-Viitanen S, Manfredi J, Viitanen P, Tribe DE, Tritch R, Hutchison C. A. r., Loeb DD, Swanstrom R. AIDS Res Hum Retroviruses. 1989;5:577. doi: 10.1089/aid.1989.5.577. [DOI] [PubMed] [Google Scholar]
- (69).Baldus M, Petkova AT, Herzfeld J, Griffin RG. Mol. Phys. 1998;95:1197. [Google Scholar]
- (70).Takegoshi K, Nakamura S, Terao T. Chem. Phys. Lett. 2001;344:631. [Google Scholar]
- (71).Verel R, Baldus M, Ernst M, Meier BH. Chem. Phys. Lett. 1998;287:421. [Google Scholar]
- (72).Neue G, Dybowski C. Solid State NMR. 1997;7:333. doi: 10.1016/s0926-2040(96)01291-x. [DOI] [PubMed] [Google Scholar]
- (73).Morcombe CR, Zilm KW. J. Magn. Reson. 2003;162:479. doi: 10.1016/s1090-7807(03)00082-x. [DOI] [PubMed] [Google Scholar]
- (74).Bennett AE, Rienstra CM, Auger M, Lakshmi KV, Griffin RG. J. Chem. Phys. 1995;103:6951. [Google Scholar]
- (75).Marion D, Wüthrich K. Biochem. Biophys. Res. Comm. 1983;113:967. doi: 10.1016/0006-291x(83)91093-8. [DOI] [PubMed] [Google Scholar]
- (76).Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. J. Biomol. NMR. 1995;6:277. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- (77).Goddard TD, Kneller DG. University of California, San Francisco; San Francisco: [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.