Abstract
Varicella zoster virus (VZV) becomes latent in ganglionic neurons along the entire neuraxis. Although all predicted VZV open reading frames (ORFs) have been detected by macroarray and microarray analysis in virus-infected cells in culture where virus gene expression is abundant, array technology does not detect all VZV gene transcripts in latently-infected human ganglia, where the abundance of ganglionic RNA is low and VZV gene transcription is highly variable. Using reverse transcription-polymerase chain reaction (RT-PCR) and the GenomeLab Genetic Analysis System (GeXPS), transcripts mapping to all 68 predicted unique VZV ORFs were detected in VZV-infected MeWo cells. Oligonucleotide primers contained both VZV- and cell-specific sequences linked to universal DNA sequences such that PCR amplification products were of predetermined sizes. Amplification products were resolved by capillary gel electrophoresis and detected by fluorescence spectrophotometry. Serial dilutions of total RNA extracted from VZV-infected MeWo cells were analyzed in parallel by GeXPS multiplex RT-PCR and real-time RT-PCR. GeXPS technology detected as few as 20 copies of VZV gene-specific transcripts. Only five multiplex RT-PCR assays were needed to analyze the entire VZV transcriptome. This technology will allow rapid analysis of all VZV genes transcribed during latency in human ganglia.
Keywords: VZV, GeXPS multiplex PCR
1. Introduction
Varicella zoster virus (VZV) is an exclusively human neurotropic alpha-herpesvirus. Primary infection typically causes childhood chickenpox, after which virus becomes latent in ganglionic neurons along the entire neuraxis (Gilden et al., 2003). Decades later, virus can reactivate to produce zoster (shingles), frequently complicated by postherpetic neuralgia (Rogers and Tindall, 1971; Bowsher, 1997). Reactivation can also lead to VZV vasculopathy, myelopathy and retinal necrosis (Nagel and Gilden, 2007).
The 125,000-basepair VZV genome contains 71 identified open reading frames (ORFs) of which 3 are repeated (Davison and Scott, 1986). All predicted VZV ORFs have been detected by macroarray (Cohrs et al., 2003a) and microarray analysis (Kennedy et al., 2005) in virus-infected cells in culture where virus gene expression is abundant. Unfortunately, array technology does not detect all VZV gene transcripts in latently-infected human ganglia, where the abundance of ganglionic RNA is low. In anticipation of studying VZV transcription in latently infected human ganglia, the Beckman Coulter GenomeLab Genetic Analysis System (GeXPS) was adapted to detect transcripts mapping to all predicted VZV ORFs in virus-infected cells in tissue culture and the results were compared with those obtained using quantitative real-time PCR.
2. Materials and methods
2.1. Virus and cells
VZV (Ellen strain) was isolated from a zoster lesion and propagated by co-cultivation in human malignant melanoma MeWo cells (Grose and Brunel, 1978) as described (Cohrs et al., 2002).
2.2. RNA extraction
VZV-infected MeWo cells (2 × 107) were harvested at the peak of virus-induced cytopathic effect (3 days post-infection). Uninfected MeWo cells (2 × 107) were processed in parallel. Uninfected and VZV-infected cells were scraped into phosphate-buffered saline (20 mM phosphate pH 7.4, 150 mM NaCl), pelleted at 1000 × g for 5 min at 4°C, and snap-frozen in liquid nitrogen. Cell pellets were thawed in 9 ml Tri-Reagent (Molecular Research Center, Cincinnati, OH), disrupted by sonication (3 times for 30 sec at 4°C), and total RNA was extracted (Cohrs et al., 1992). Residual DNA was digested with RNase-free DNase (DNase I, amplification grade; Invitrogen, Carlsbad, CA) at room temperature for 15 min. After heat inactivation for 10 min at 65°C in 2 mM EDTA, RNA in 1.5 µl was removed for quantitation (Nanodrop Technologies, Wilmington, DE), and 5 µl was analyzed for RNA integrity by microfiltration on Bioanalyzer chips (Agilent Technologies, Quantum Analytics, Inc., Foster City, CA).
2.3. GeXPS primer design
Five multiplex primer sets were designed to analyze the VZV gene transcription from all 68 predicted unique VZV ORFs (Table 1) using GenomeLab GeXP eXpress Profiler software. Multiplex primer sets were grouped based on the abundance of each VZV gene transcript found earlier (Cohrs et al., 2003a). Multiplex primer sets A and B amplified high-abundance VZV transcripts, and multiplex primer sets C, D and E amplified low-abundance transcripts. Reverse primers consisted of 20 nucleotides complementary to the target gene coupled to a 19-nucleotide universal reverse sequence. Forward primers consisted of 20 nucleotides corresponding to the target gene coupled to an 18 oligonucleotide universal forward sequence. All primers were obtained from Integrated DNA Technologies (Coralville, IA) and supplied at a stock concentration of 100 µM in 96-well microtiter plates. GeXPS primer stocks were diluted 1:100 in nuclease-free water to a final concentration of 1 µM for reverse primer sets AE, and diluted 1:1000 to a final concentration of 0.2 µM for forward primer sets A–E (Table 1).
Table 1.
Oligonucleotide primer sequences and expected PCR product size.
| Multiplex Primer Set |
VZV ORF |
Forward PCR primer (5' - 3')a | Reverse PCR primer (5' - 3')b | PCR Product Size (bp) |
|---|---|---|---|---|
| A | 33 | AGGTGACACTATAGAATAtgctgaagctgacgaagaga | GTACGACTCACTATAGGGAcgcacaatctctggggtaat | 142 |
| A | 18 | AGGTGACACTATAGAATAtttacatttttatcggcggc | GTACGACTCACTATAGGGAcctcgatgcactcttgttca | 152 |
| A | 9 | AGGTGACACTATAGAATAgcccgtttgagacatgaact | GTACGACTCACTATAGGGAcgcttcggtgaatcctctaa | 162 |
| A | 51 | AGGTGACACTATAGAATAacagctgcttgaccgatttt | GTACGACTCACTATAGGGActattggacatgcaagggct | 172 |
| A | 37 | AGGTGACACTATAGAATAtgggttcccttcgtacagac | GTACGACTCACTATAGGGAatcatggttttaaatgccgc | 182 |
| A | 63 | AGGTGACACTATAGAATAtcagaggagagcaccgattc | GTACGACTCACTATAGGGActttcgcggctgtatattcc | 191 |
| A | 58 | AGGTGACACTATAGAATAgcgttgatgcaaatgaaaaa | GTACGACTCACTATAGGGAaggcgttcataaccaagctg | 202 |
| A | 32 | AGGTGACACTATAGAATAcatcatccgtccaaacagtg | GTACGACTCACTATAGGGAaaggtttcgttgcacatgct | 212 |
| A | 62 | AGGTGACACTATAGAATAgacagcccagtggagaaaaa | GTACGACTCACTATAGGGAgaatcatccgaatttgacgg | 222 |
| A | 41 | AGGTGACACTATAGAATAcccagaccttaatgcgagaa | GTACGACTCACTATAGGGAaaaccaatgtgcgaatagcc | 232 |
| A | 42 | AGGTGACACTATAGAATAaccgttgcgaattgtgctat | GTACGACTCACTATAGGGAgtaacgcaacgccctatctc | 237 |
| A | 44 | AGGTGACACTATAGAATAgagtgtggtggtcagacgaa | GTACGACTCACTATAGGGAttccgcatgatttgtgacat | 246 |
| A | 29 | AGGTGACACTATAGAATAgtttataggcatggagggca | GTACGACTCACTATAGGGAggattcgcagctaaatgagg | 256 |
| A | 54 | AGGTGACACTATAGAATAgatcggtggacacatcacaa | GTACGACTCACTATAGGGAccgtaaccgtttcagctctc | 276 |
| A | 38 | AGGTGACACTATAGAATAtgttcttaccactcccccag | GTACGACTCACTATAGGGAcaatgggaattgatgatccc | 286 |
| A | 3 | AGGTGACACTATAGAATAgttggcgtcgtttacacaag | GTACGACTCACTATAGGGAcggtaacagtcgttgggaat | 296 |
| A | 28 | AGGTGACACTATAGAATAaagctttcgcgttgaggtta | GTACGACTCACTATAGGGAtccgacatgcagtcaatttc | 343 |
| B | 1 | AGGTGACACTATAGAATAgttcgggaatctgatagcga | GTACGACTCACTATAGGGAgtggtcttgtttgaacgacg | 137 |
| B | 15 | AGGTGACACTATAGAATAgtccatgatgaaaacctggg | GTACGACTCACTATAGGGAgctgaggttattgcacacga | 147 |
| B | 49 | AGGTGACACTATAGAATAatctgtggattgtgcaaacg | GTACGACTCACTATAGGGAatcggcgtcctctgttacat | 157 |
| B | 61 | AGGTGACACTATAGAATAgaaagggatacccgattggt | GTACGACTCACTATAGGGAatcgtttaacgtctctgccg | 167 |
| B | 24 | AGGTGACACTATAGAATAgcccgttgtgtttttcagtt | GTACGACTCACTATAGGGAacagacacccccgtattttg | 177 |
| B | 68 | AGGTGACACTATAGAATAccgagacttggagctttttg | GTACGACTCACTATAGGGAcggccaactgatcctcttta | 187 |
| B | 53 | AGGTGACACTATAGAATAttactgaaccctcagcctgg | GTACGACTCACTATAGGGAccgaatctcgtaaaggtcca | 197 |
| B | 4 | AGGTGACACTATAGAATAtccggggataaaacaaaaca | GTACGACTCACTATAGGGAacagttcatccgcagactcc | 207 |
| B | 64 | AGGTGACACTATAGAATAcgctatgcgccatatctacc | GTACGACTCACTATAGGGAtaacgataacaccgctctgc | 216 |
| B | 57 | AGGTGACACTATAGAATAcgagaacgtaatgtgtttggaa | GTACGACTCACTATAGGGAacgttgatgagccttgcag | 241 |
| B | 48 | AGGTGACACTATAGAATAtcggaacgtatgaatgtgga | GTACGACTCACTATAGGGAttaacgtggtgtacgtccga | 256 |
| B | 50 | AGGTGACACTATAGAATAccacggttaattcccacaac | GTACGACTCACTATAGGGAacgtagccgtagccttgtga | 270 |
| B | 20 | AGGTGACACTATAGAATAcgtggtattggcttcctgtt | GTACGACTCACTATAGGGAtgcaggataattgtgtggga | 281 |
| B | 67 | AGGTGACACTATAGAATAatacggtggcgttttgtttc | GTACGACTCACTATAGGGAaccaagaatgaaaccatcgg | 291 |
| B | 10 | AGGTGACACTATAGAATAgttcacgtgtgcgtttcatc | GTACGACTCACTATAGGGAtgaggatgtttgaccgcata | 301 |
| B | 2 | AGGTGACACTATAGAATAtgggtatgaatgtggtacgg | GTACGACTCACTATAGGGAtgttagggttgggagagtgg | 305 |
| B | 13 | AGGTGACACTATAGAATAtgggagacttgtcatgttgg | GTACGACTCACTATAGGGActttagcggcgagttctttg | 313 |
| B | 7 | AGGTGACACTATAGAATAacttacgctgcatgttctcg | GTACGACTCACTATAGGGAtcaaacgatgcctgaggttt | 333 |
| C | 25 | AGGTGACACTATAGAATAgtacgaatcggaaaatgcgt | GTACGACTCACTATAGGGAcgggtagaatcagaagaaccc | 142 |
| C | 18 | AGGTGACACTATAGAATAtttacatttttatcggcggc | GTACGACTCACTATAGGGAcctcgatgcactcttgttca | 152 |
| C | 55 | AGGTGACACTATAGAATAacaacccggactttctgttg | GTACGACTCACTATAGGGAtcgcgatttgaatgtgtttc | 157 |
| C | 56 | AGGTGACACTATAGAATAcggttctgacccaacatctc | GTACGACTCACTATAGGGAcgtcaggtctgtttacccgt | 162 |
| C | 43 | AGGTGACACTATAGAATAcccgataacgccttaacaaa | GTACGACTCACTATAGGGAatcaaagaataaaccggggc | 172 |
| C | 14 | AGGTGACACTATAGAATAcatgccttgttacatgcgtc | GTACGACTCACTATAGGGAttcaatatgaagatcggggc | 182 |
| C | 45 | AGGTGACACTATAGAATAatgtttggcgcgtttctatc | GTACGACTCACTATAGGGAgctgcttcttccggtgaata | 192 |
| C | 6 | AGGTGACACTATAGAATAgcgtgatcgcgaatatacct | GTACGACTCACTATAGGGAtgatccactcgcagagacac | 202 |
| C | 32 | AGGTGACACTATAGAATAcatcatccgtccaaacagtg | GTACGACTCACTATAGGGAaaggtttcgttgcacatgct | 212 |
| C | 35 | AGGTGACACTATAGAATAttatgctagcgtcgcatttg | GTACGACTCACTATAGGGAtaaatccaccccacaagctg | 222 |
| C | 41 | AGGTGACACTATAGAATAcccagaccttaatgcgagaa | GTACGACTCACTATAGGGAaaaccaatgtgcgaatagcc | 232 |
| C | 60 | AGGTGACACTATAGAATAtcatcggtatatgaggcgtg | GTACGACTCACTATAGGGAccccgcttttatgttcttca | 242 |
| C | 31 | AGGTGACACTATAGAATAccgtgggattattggttttg | GTACGACTCACTATAGGGAcgacggttcagtgttttgtg | 252 |
| C | 59 | AGGTGACACTATAGAATAtgctgaataccacgcttacg | GTACGACTCACTATAGGGAgaaatggaacacgggacaat | 262 |
| C | 26 | AGGTGACACTATAGAATAtaccgatggagggttaccag | GTACGACTCACTATAGGGAcgcaatctttccaattagcc | 273 |
| C | 5 | AGGTGACACTATAGAATAactcaacgtttttggatggc | GTACGACTCACTATAGGGAgaatggcccaaacatacgac | 282 |
| C | 65 | AGGTGACACTATAGAATAaaaacaccatggagggtgag | GTACGACTCACTATAGGGAgctcccgtcatagcaaataca | 293 |
| C | 13 | AGGTGACACTATAGAATAtgggagacttgtcatgttgg | GTACGACTCACTATAGGGActttagcggcgagttctttg | 313 |
| C | 7 | AGGTGACACTATAGAATAacttacgctgcatgttctcg | GTACGACTCACTATAGGGAtcaaacgatgcctgaggttt | 333 |
| D | 40 | AGGTGACACTATAGAATAaaccctctgtttgcatgtcc | GTACGACTCACTATAGGGAgaaaaggagggattggtggt | 137 |
| D | 52 | AGGTGACACTATAGAATAaacaacggattccaccgtta | GTACGACTCACTATAGGGAaatacgcaaagattggacgg | 147 |
| D | 19 | AGGTGACACTATAGAATAgagccgatatccttcgaaca | GTACGACTCACTATAGGGAgcagcactgcgtatgattgt | 167 |
| D | 21 | AGGTGACACTATAGAATAatgcagatggatatcgcaca | GTACGACTCACTATAGGGAgccaacagtccgatgaagtt | 177 |
| D | 34 | AGGTGACACTATAGAATAcaatatgaacggggacttgg | GTACGACTCACTATAGGGAggaggcaaactgaggaatga | 187 |
| D | 12 | AGGTGACACTATAGAATAtaccggggaatatgtcagga | GTACGACTCACTATAGGGAataacatccacccgtagcca | 197 |
| D | 66 | AGGTGACACTATAGAATAtttggcaaaacggtcttctc | GTACGACTCACTATAGGGAcgggaagagtttggaaaaca | 207 |
| D | 27 | AGGTGACACTATAGAATAatccattttggcgctatcag | GTACGACTCACTATAGGGAtacatcataaaccctccggc | 217 |
| D | 30 | AGGTGACACTATAGAATActgttttccattgtggggag | GTACGACTCACTATAGGGAttctggtcgatccctgtttc | 227 |
| D | 39 | AGGTGACACTATAGAATAcgatcagacctgctaccctt | GTACGACTCACTATAGGGAccccacgacacaggaagtat | 237 |
| D | 17 | AGGTGACACTATAGAATAgttcgctgtcagaccgattt | GTACGACTCACTATAGGGAtgcatatttggaaacggaca | 247 |
| D | 8 | AGGTGACACTATAGAATAgaagatcatcgaaaagccga | GTACGACTCACTATAGGGAttaccgcggatagcataagg | 257 |
| D | 46 | AGGTGACACTATAGAATAaaagaacggaccctcgatct | GTACGACTCACTATAGGGAgcgaataaaatcgggtgcta | 277 |
| D | 22 | AGGTGACACTATAGAATAttgggggttacgtttactgc | GTACGACTCACTATAGGGAggagtcgatgtacggcaaat | 287 |
| D | 23 | AGGTGACACTATAGAATAcgtacgtttaacccgcgtat | GTACGACTCACTATAGGGAagtgcttgactgtgctccct | 298 |
| D | 36 | AGGTGACACTATAGAATAggcgtatggaattggaaaaa | GTACGACTCACTATAGGGAgtccatcaatgccgagattt | 308 |
| E | 50 | AGGTGACACTATAGAATAtcctggctacgttaattgcc | GTACGACTCACTATAGGGAccatctagggtggcgttaaa | 137 |
| E | 65 | AGGTGACACTATAGAATAgcgagcgcaaccaaataata | GTACGACTCACTATAGGGAgcaaaaattcatctgccgtt | 144 |
| E | 54 | AGGTGACACTATAGAATAatgggcgagaacgatacaac | GTACGACTCACTATAGGGAggaacgtacggtttcggtaa | 151 |
| E | 11 | AGGTGACACTATAGAATAgcagcaagtcctctttacgc | GTACGACTCACTATAGGGAtcctgtcttgctctttgggt | 158 |
| E | 7 | AGGTGACACTATAGAATAactcaaaacggcttgctgat | GTACGACTCACTATAGGGAcacttcactttcagcggtca | 165 |
| E | 47 | AGGTGACACTATAGAATAggacaactgcccatttcagt | GTACGACTCACTATAGGGAggggatgtcagatgggagta | 172 |
| E | 61 | AGGTGACACTATAGAATAcaaacctggacctggaaaga | GTACGACTCACTATAGGGAaaagcctgactttttggggt | 179 |
| E | 29 | AGGTGACACTATAGAATAgcccaaattctgcgttgtat | GTACGACTCACTATAGGGAgtggccatctcgatctgttt | 186 |
| E | 5 | AGGTGACACTATAGAATAtttttggatggcctatctgg | GTACGACTCACTATAGGGActgcacgcaccaaaaataag | 193 |
| E | 2 | AGGTGACACTATAGAATActggaggtgcaaggacattt | GTACGACTCACTATAGGGAttaccccgccataaacagag | 200 |
| E | 26 | AGGTGACACTATAGAATAggagacgttttggcgttaga | GTACGACTCACTATAGGGAaattcaaaggcagcgctaaa | 207 |
| E | 64 | AGGTGACACTATAGAATAcggctgatattgatggcata | GTACGACTCACTATAGGGAcgtaggttcttgggactcca | 215 |
| E | 16 | AGGTGACACTATAGAATAttaacacgggatgagacgtg | GTACGACTCACTATAGGGAttcacagattccatgttcgc | 222 |
| E | 3 | AGGTGACACTATAGAATAagtcaacccatccgagtgaa | GTACGACTCACTATAGGGAagcgtgtaaatcatcagccc | 229 |
| E | 13 | AGGTGACACTATAGAATAggtcaaccgattccaaagaa | GTACGACTCACTATAGGGAgctgatcgattccttgctgt | 238 |
| E | 42 | AGGTGACACTATAGAATAttttatatcggccgtcaacc | GTACGACTCACTATAGGGAcgccctatctcactggaaga | 245 |
| E | 62 | AGGTGACACTATAGAATAacccagaacggaagatgttg | GTACGACTCACTATAGGGAgttcatcggtggttttggag | 254 |
| E | 63 | AGGTGACACTATAGAATAacgcgagattcacgaagatt | GTACGACTCACTATAGGGAacattccagtgcgctcctat | 265 |
| Multiplex Set |
Cell Gene (accession number) |
Forward PCR primer (5' - 3')c | Reverse PCR primer (5' - 3')d | PCR Product Size (bp) |
| A, B, C, D, E | GAPdH (NM_002046 ) | AGGTGACACTATAGAATAatcactgccacccagaagac | GTACGACTCACTATAGGGAacctggtgctcagtgtagcc | 339 |
| A, B, C, D, E | Beta-actin (NM_001101 | AGGTGACACTATAGAATAgctatccctgtacgcctctg | GTACGACTCACTATAGGGAgtcaggcagctcgtagctct | 353 |
| A, B, C, D, E | Cyclophilin (BC000689) | AGGTGACACTATAGAATAcctaaagcatacgggtcctg | GTACGACTCACTATAGGGAaggatactgcgagcaaatgg | 357 |
forward universal primer sequence (upper case) and VZV gene-specific primer sequence (lower case) in multiplex primer sets A–E
reverse universal primer sequence (upper case) and VZV gene-specific primer sequence (lower case) in multiplex primer sets A–E
forward universal primer sequence (upper case) and cell gene-specific primer sequence (lower case) in multiplex primer sets A–E
reverse universal primer sequence (upper case) and cell gene-specific primer sequence (lower case) in multiplex primer sets A–E
2.4. GeXPS multiplex RT-PCR
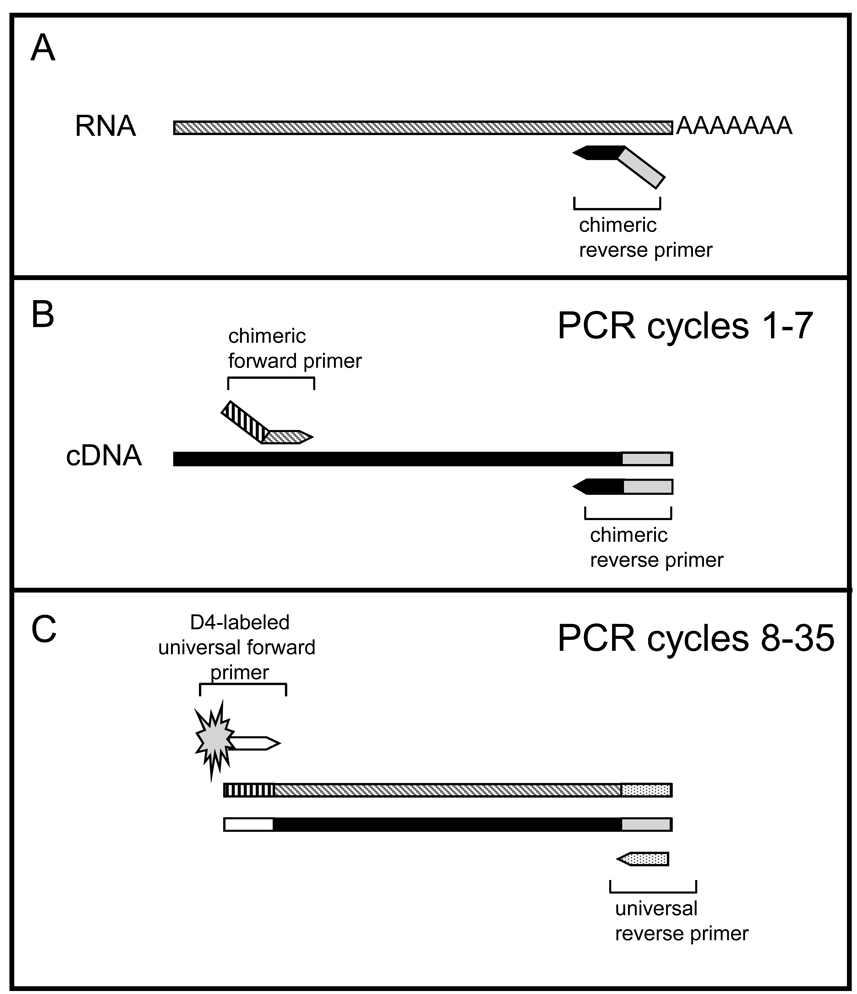
In each GeXPS multiplex RT-PCR experiment, reverse transcription (RT) was followed by PCR (Fig. 1). RT reactions mixtures (20 µl) contained 10–20 ng RNA, 0.1 µM reverse primer set mix, 20 U reverse transcriptase with RNase inhibitor (0.1 U/mL), 250 fM kanamycin resistant (Kanr) RNA and 1x RT Master Mix Buffer containing 10 mM HCl, 50 mM KCl, 2.5 mM MgCl2 10 mM DTT, 1mM each dNTP and 0.05 µM Kanr RT reverse primer (Fig. 1A). RT, RNase inhibitor, Kanr RNA and 1x RT Master Mix Buffer were supplied in kit form (GenomeLab GeXP Start Kit; Beckman Coulter, Fullerton, CA). RT reactions were incubated at 48°C for 1 min, 37°C for 5 min, 42°C for 60 min, and 95°C for 5 min (Fig. 1A).
Fig. 1.
GenomeLab eXpress (GeXPS) multiplex RT-PCR strategy. A. Total RNA extracted from 2 × 107 uninfected or VZV-infected MeWo cells was reverse transcribed using 19–22 chimeric reverse primers each containing 20 nucleotides complementary to the target gene (solid black) coupled with a 19-nucleotide universal reverse sequence (solid gray). B. The cDNA was PCR-amplified in cycles 1–7 using the chimeric primers to synthesize the 19–22 gene-amplification products, each of which contained universal tags at both termini. C. Subsequent PCR amplifications (cycles 7–35) using universal forward and reverse primers to yield D4-fluorescent-labeled amplification products corresponding to each of the 19–22 specific genes tested.
Subsequent PCR was done with each reaction containing 9.3 µl RT reaction, 0.02 µM forward primer set mix, 5 mM MgCl2, 3.5 U Thermo Start Taq DNA polymerase (Thermo Fisher Scientific, Pittsburgh, PA) and 1x PCR Master Mix buffer (GenomeLab GeXP Start Kit; Beckman Coulter) containing 10 mM HCL, 50 mM KCl, 0.3 mMeach dNTP, 0.02 µM Kanr PCR forward primer, 1 µM universal reverse primer, and 1 µM D4-labeled universal forward primer. Amplification conditions consisted of initial denaturation at 95°C for 15 min, followed by 35 two-step cycles of 94°C for 30 sec and 55°C for 30 sec, ending in a single extension cycle of 70°C for 1 min (Fig. 1 B and C).
2.5. GeXP multiplex data analysis
PCR products from multiplex primer sets A and B reactions were diluted 1:100 in water, and 2-µl was added to a 37.75 µl sample loading solution along with 0.25 µl DNA size standard-400 (GenomeLab GeXP Start Kit; Beckman Coulter). For multiplex primer sets C,D, and E reactions, 2-µl of the undiluted PCR product was added to a 37.75 µl sample loading solution along with 0.25 µl DNA size standard-400. The GeXPS system was used to separate PCR products based on size by capillary gel electrophoresis and to measure their dye signal strength in arbitrary units (A.U.) of optical fluorescence, defined as the fluorescent signal minus background.
PCR product sizes were determined using GenomeLab GeXPS software and were compared to the expected PCR product size to identify each transcript. A VZV ORF transcript was considered present in the initial RNA when dye signal of the VZV ORF-specific product was greater than 2,000 A.U. (signal background), and when no corresponding PCR product of the same size was present in RT-PCR reactions with uninfected MeWo cell RNA, VZV-infected MeWo cell RNA without RT, and reactions containing only Kanr RNA.
2.6. Real-time PCR and data analysis
VZV-infected MeWo cell RNA (0.01–20 ng) was added to uninfected MeWo RNA to yield a total of 20 ng RNA in 5 µl. RNA was reverse transcribed in a total reaction volume of 20 µl using oligo-dT primers (Transcriptor First Strand cDNA Synthesis Kit; Roche, Mannheim, Germany). Taqman primer and probes for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and VZV ORFs 21, 29, 62, 63, and 66 (Cohrs and Gilden, 2007) were used to quantitate the abundance of each transcript. These five VZV ORFs were selected for study since they are expressed during latency (Cohrs et al., 1996, 2007). Real-time PCR was performed in a 20-µl volume of 1x qPCR Mastermix (VWR, West Chester, PA) containing 900 nM of each primer, 250 nM probe and 9.3 µl of the cDNA reaction using the 7500-Fast real-time PCR system (Applied Biosystems, Foster City, CA). Amplification conditions consisted of initial denaturation at 95°C for 10 min, followed by 40 two-step cycles of 95°C for 15 sec and 60°C for 1 min. Samples were analyzed in duplicate and VZV DNA (1–106 copies) was used to generate standard graphs (Cohrs and Gilden, 2007).
3. Results
3.1. Detection of VZV transcripts in VZV-infected MeWo cells by GeXPS multiplex RT-PCR
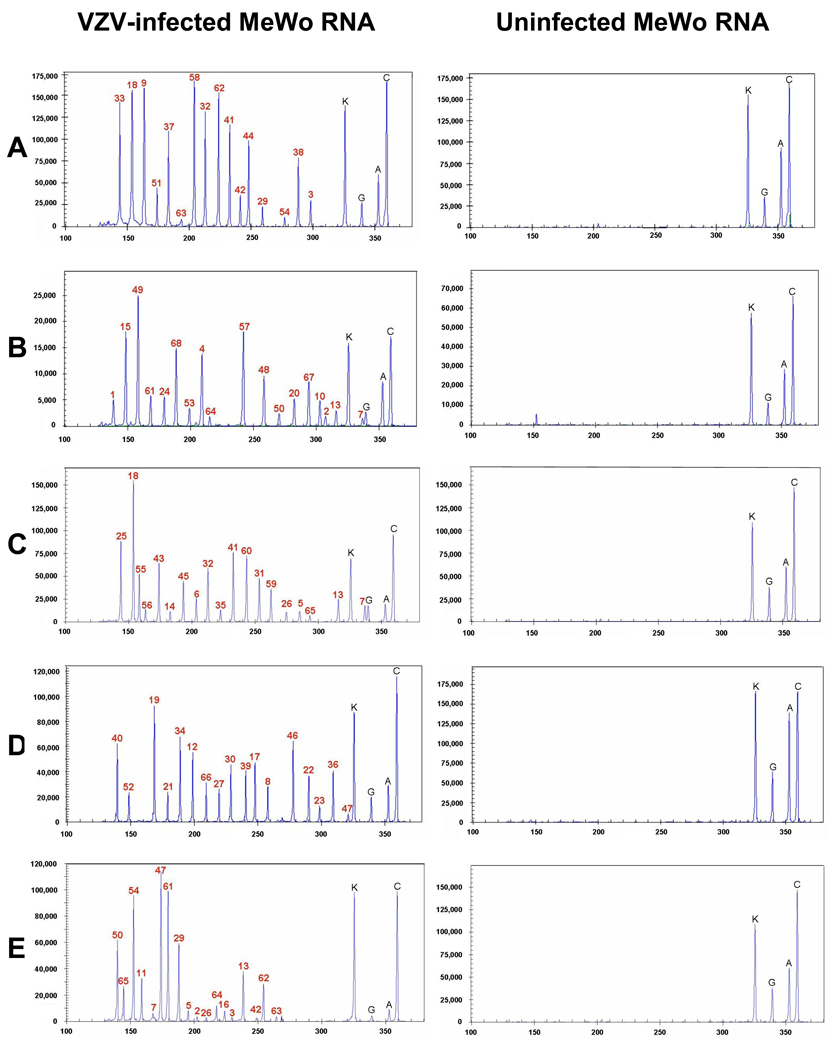
Five independent multiplex PCR reactions using multiplex primers sets A–E detected transcripts from all 68 predicted VZV ORFs in RNA from VZV-infected MeWo cells (Fig. 2, left panels), whereas no PCR products corresponding to any VZV gene transcript were detected in uninfected cells using the multiplex primer sets A-E (Fig. 2, right panels). PCR products corresponding to the internal control Kanr RNA and to cellular GAPDH, β-actin and cyclophilin genes were detected in RNA from uninfected and VZV-infected cells (Fig. 2, all panels). PCR products corresponding to VZV and all control gene transcripts were not detected in DNase treated RNA (data not shown), indicating an absence of contaminating DNA.
Fig. 2.
Electropherogram of D4-labeled PCR products synthesized with multiplex primer sets A–E. D4-labeled PCR products amplified from uninfected MeWo cells (right panels) and VZV-infected MeWo cells (left panels) with multiplex primer sets A–E (left) were separated by capillary electrophoresis and detected by fluorescence spectrophotometry given as dye signal in arbitrary units on the y-axis. Each peak was identified by comparing expected to actual PCR product size (x-axis). Uninfected cells showed peaks corresponding only to the internal control kanamycin-resistance gene (K) and cell control genes GAPDH (G), beta-actin (A) and cyclophilin (C). In VZV-infected MeWo cells, peaks corresponding to amplification products from VZV ORFs incorporated in multiplex primer sets A–E.
3.2. Detection of low-abundance VZV gene transcripts by GeXPS multiplex RT-PCR
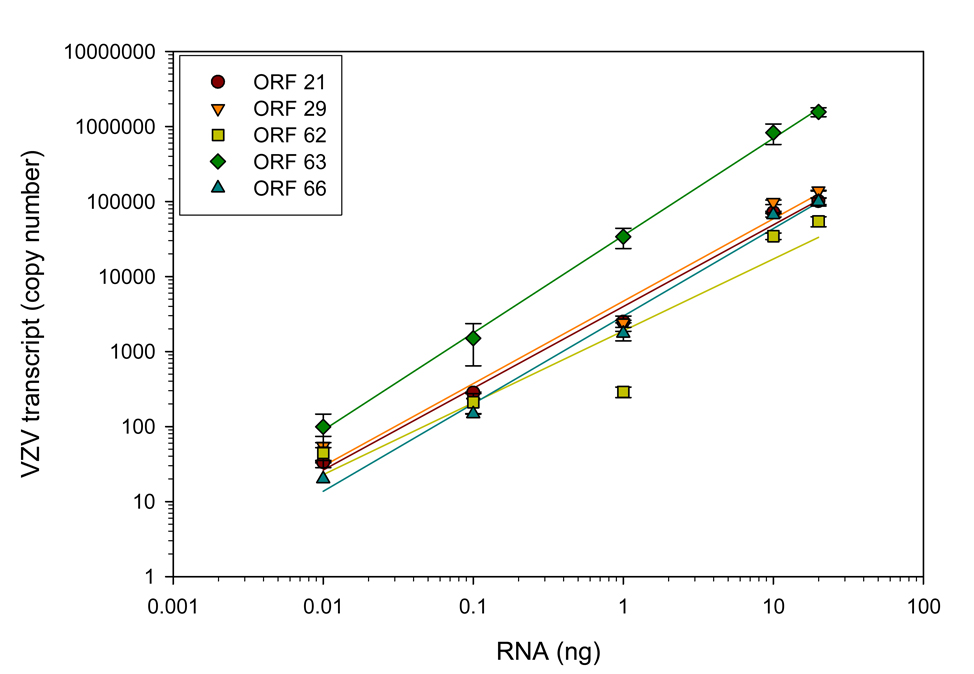
The number of VZV ORF 21, 29, 62, 63 and 66 cDNA copies synthesized from decreasing amounts (20-0.01 ng) of RNA from VZV-infected MeWo cells added to uninfected MeWo cell RNA for a total of 20 ng was quantitated by real-time PCR. Copy number was linear over the range of VZV-infected MeWo cell RNA tested (Fig. 3). In 0.01 ng VZV-infected MeWo cell RNA, there were 33.4 (+/− 0.8) copies of ORF 21, 54.5 (+/− 19.1) copies of ORF 29, 44.5 (+/− 16.3) copies of ORF 62, 99.0 (+/− 46.7) copies of ORF 63 and 20.0 (+/− 0.0) copies of ORF 66 (Fig. 3).
Fig. 3.
Quantitation of VZV transcripts in VZV-infected MeWo cells. Decreasing amounts of RNA (20-0.01 ng) extracted from VZV-infected MeWo cells was added to uninfected MeWo cell RNA to yield a total of 20 ng. cDNA was synthesized and the abundance of VZV ORFs 21, 29, 62, 63 and 66 transcripts was quantitated by real-time PCR. For all five VZV ORFs, there were 20–99 copies of VZV transcripts in 0.01 ng total VZV-infected MeWo RNA.
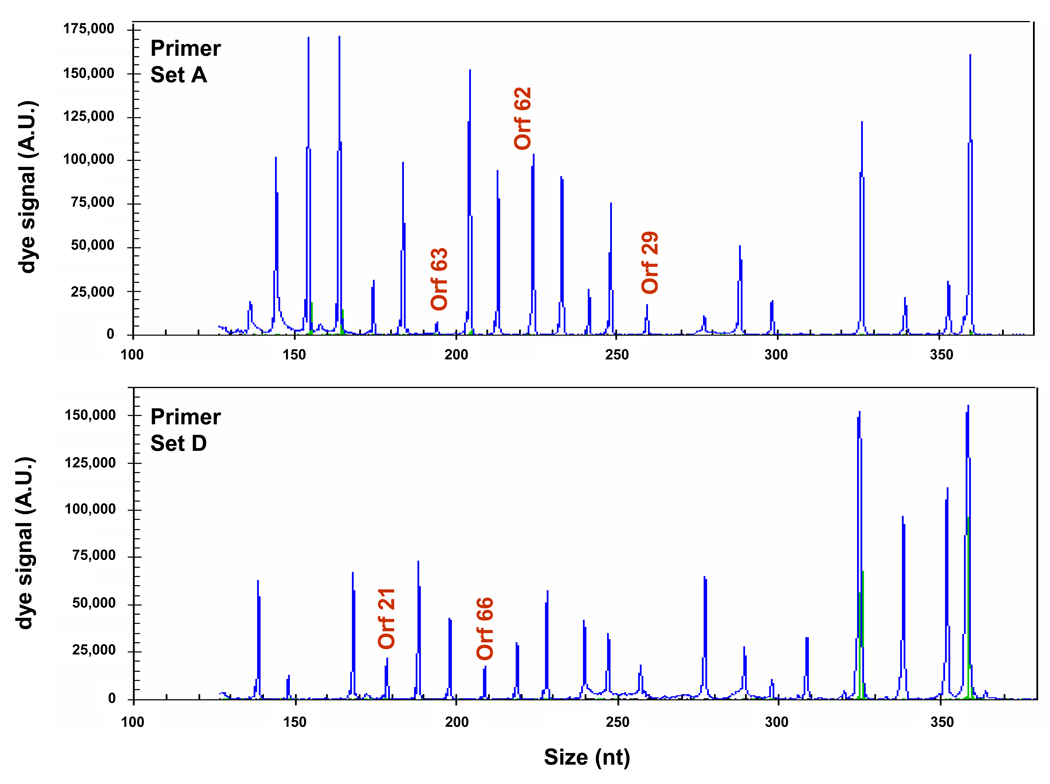
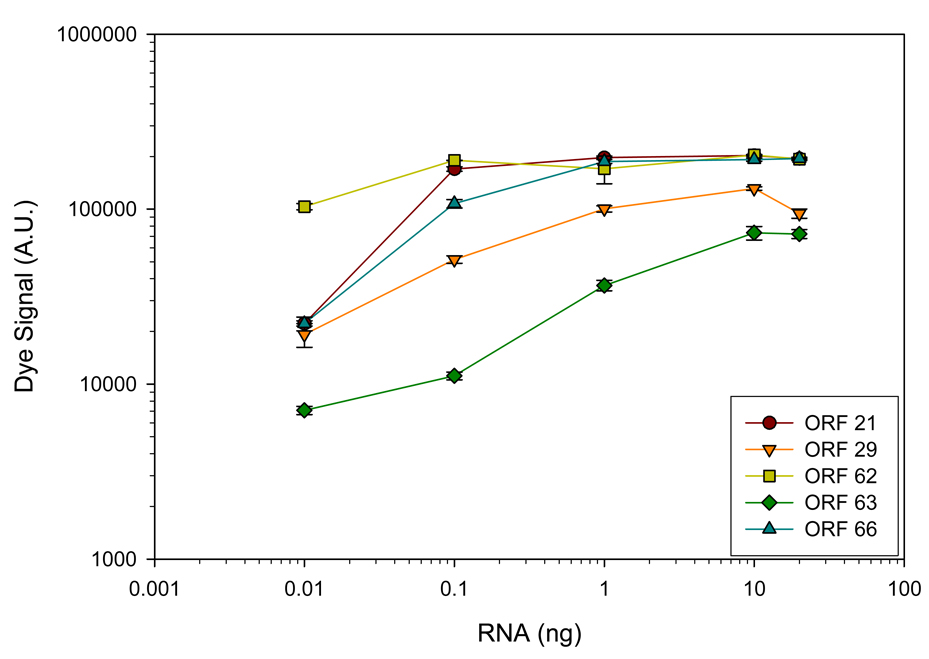
The same decreasing amounts of VZV-infected MeWo cell RNA used in real-time PCR was analyzed by GeXPS multiplex RT-PCR assay using primer sets A and D (Fig. 4, top and bottom panels, respectively). These primer sets were selected to compare realtime PCR quantitation with GeXPS multiplex RT-PCR, since multiplex primer set A contained primers specific for ORFs 63, 62 and 29, and multiplex primer set D contained primers specific for ORFs 21 and 66. At the lowest concentration of VZV-infected MeWo RNA (0.01 ng), PCR products corresponding to ORFs 21, 29, 62, 63, and 66 were detected. A plot of fluorescence intensity of each PCR product as a function of the VZV-infected MeWo RNA revealed a linear relationship between dye signal and RNA concentration when dye signal was <120,000 A.U., plateauing thereafter (Fig. 5).
Fig. 4.
GeXP multiplex technology detection of low-abundance VZV transcripts. PCR products were synthesized using multiplex primer set A (top panel) and D (bottom panel) from 0.01 ng total VZV-infected MeWo cell RNA (shown in Fig. 3) and resolved by capillary gel electrophoresis. Peaks corresponding to VZV ORFs 63, 62, 29, 21 and 66 were detected.
Fig. 5.
GeXP multiplex detection of VZV ORFs in serial dilutions of VZV-infected MeWo cells. Decreasing amounts of RNA (20-0.01 ng) extracted from VZV-infected MeWo cells were added to uninfected MeWo cell RNA to yield a total of 20 ng. PCR products were synthesized using multiplex primer set A and D and resolved by capillary gel electrophoresis. Peaks corresponding to VZV ORFs 63, 62, 29, 21 and 66 at various RNA dilutions were plotted.
4. Discussion
VZV gene transcription has been analyzed by Northern blotting (Ostrove et al., 1985), macroarrays (Cohrs et al., 2003a) and microarrays (Kennedy et al., 2005); however, none of these techniques detects low-abundance transcripts. Herein, a PCR-based assay was developed to identify transcripts corresponding to all predicted VZV ORFs simultaneously. Multiplex RT-PCR detected as few as 20 VZV transcripts in a background of 20 ng total RNA and was specific for each VZV gene transcript, since no amplification product was detected in RNA extracted from uninfected cells.
Over the past decade, characterization of VZV gene transcripts in latently-infected human ganglia has been hindered by: (1) the limited amount of RNA present in human ganglia (~250 µg total RNA or 5 µg mRNA in human trigeminal ganglia); (2) the low abundance of VZV transcripts present in ganglionic neurons (64 to 3,710 transcripts per µg mRNA (Cohrs and Gilden, 2007); and (3) the lack of sensitivity of macroarray and microarray assays (10,000 VZV mRNA copies; data not shown). Consequently, only 12 of the 68 ORFs in the VZV genome have been analyzed for latent VZV gene transcription (Kennedy et al., 1999, 2000; Cohrs et al., 1996, 2003b), and quantitative analysis has been limited to only 5 of these transcripts (Cohrs and Gilden, 2007). Use of our GeXPS multiplex RT-PCR assay system allows detection of low-abundance VZV gene transcripts mapping to all predicted virus ORFs. Additionally, since GeXPS analysis can be performed with small amounts of RNA (100 ng total RNA per reaction), all VZV gene transcripts can be assayed with 500 ng RNA. Thus, it will now be possible to analyze latently-infected human ganglia for the entire VZV transcriptome, a goal previously unattainable. In addition, application of our multiplex RT-PCR technology to animal models of varicella latency (White et al., 2001) will permit analysis not only of virus transcription in latently-infected ganglia, but also after experimentally-induced reactivation.
Supplementary Material
Acknowledgements
This work was supported in part by Public Health Service (PHS) grant NS 32623 from the National Institutes of Health (NIH). Dr. Nagel was supported by PHS grant T32-NS07321 from the NIH. The authors acknowledge Andy Chiang, Dana Griffiths and Irene Newsham at Beckman Coulter, Inc., for their technical assistance. We thank Marina Hoffman for editorial review and Cathy Allen for preparing the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Maria A. Nagel, Email: Maria.Nagel@uchsc.edu.
Don Gilden, Email: Don.Gilden@uchsc.edu.
Ted Shade, Email: Ted.Shade@uchsc.edu.
Bifeng Gao, Email: Bifeng.Gao@uchsc.edu.
Randall J. Cohrs, Email: Randall.Cohrs@uchsc.edu.
References
- Bowsher D. The management of postherpetic neuralgia. Postgrad. Med. J. 1997;73:623–629. doi: 10.1136/pgmj.73.864.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohrs R, Mahalingam R, Dueland AN, Wolf W, Wellish M, Gilden DH. Restricted transcription of varicella-zoster virus in latently infected human trigeminal and thoracic ganglia. J. Infect. Dis. 1992;166 Suppl 1:S24–S29. doi: 10.1093/infdis/166.supplement_1.s24. [DOI] [PubMed] [Google Scholar]
- Cohrs RJ, Barbour M, Gilden DH. Varicella-zoster virus (VZV) transcription during latency in human ganglia: detection of transcripts mapping to genes 21, 29, 62, and 63 in a cDNA library enriched for VZV RNA. J. Virol. 1996;70:2789–2796. doi: 10.1128/jvi.70.5.2789-2796.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohrs RJ, Wischer J, Essman C, Gilden DH. Characterization of varicella-zoster virus gene 21 and 29 proteins in infected cells. J. Virol. 2002;76:7228–7238. doi: 10.1128/JVI.76.14.7228-7238.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohrs RJ, Hurley MP, Gilden DH. Array analysis of viral gene transcription during lytic infection of cells in tissue culture with varicella-zoster virus. J. Virol. 2003a;77:11718–11732. doi: 10.1128/JVI.77.21.11718-11732.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohrs RJ, Gilden DH, Kinchington PR, Grinfeld E, Kennedy PG. Varicella-zoster virus gene 66 transcription and translation in latently infected human ganglia. J. Virol. 2003b;77:6660–6665. doi: 10.1128/JVI.77.12.6660-6665.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohrs RJ, Gilden DH. Prevalence and abundance of latently transcribed varicella-zoster virus genes in human ganglia. J. Virol. 2007;81:2950–2956. doi: 10.1128/JVI.02745-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison AJ, Scott JE. The complete DNA sequence of varicella-zoster virus. J. Gen. Virol. 1986;67:1759–1816. doi: 10.1099/0022-1317-67-9-1759. [DOI] [PubMed] [Google Scholar]
- Gilden DH, Cohrs RJ, Mahalingam R. Clinical and molecular pathogenesis of varicella virus infection. Viral Immunol. 2003;16:243–258. doi: 10.1089/088282403322396073. [DOI] [PubMed] [Google Scholar]
- Grose C, Brunel PA. Varicella-zoster virus: isolation and propagation in human melanoma cells at 36 and 32 degrees C. Infect. Immun. 1978;19:199–203. doi: 10.1128/iai.19.1.199-203.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy PG, Grinfeld E, Gow JW. Latent varicella-zoster virus in human dorsal root ganglia. Virology. 1999;258:451–454. doi: 10.1006/viro.1999.9745. [DOI] [PubMed] [Google Scholar]
- Kennedy PG, Grinfeld E, Bell JE. Varicella-zoster virus gene expression in latently infected and explanted human ganglia. J. Virol. 2000;74:11893–11898. doi: 10.1128/jvi.74.24.11893-11898.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy PG, Grinfield E, Craigon M, Vierlinger K, Roy D, Forster T, Ghazal P. Transcriptional analysis of varicella-zoster virus infection using long oligonucleotide-based microarrays. J. Gen. Virol. 2005;86:2673–2684. doi: 10.1099/vir.0.80946-0. [DOI] [PubMed] [Google Scholar]
- Nagel MA, Gilden DH. The protean neurological manifestations of varicella zoster virus infection. Cleve. Clin. J. Med. 2007;74:489–494. doi: 10.3949/ccjm.74.7.489. [DOI] [PubMed] [Google Scholar]
- Ostrove JM, Reinhold W, Fan CM, Zom S, Hay J, Straus SE. Transcription mapping of the varicella-zoster virus genome. J. Virol. 1985;56:600–606. doi: 10.1128/jvi.56.2.600-606.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers RS, III, Tindall JP. Herpes zoster in the elderly. Postgrad. Med. 1971;50:153–157. doi: 10.1080/00325481.1971.11697705. [DOI] [PubMed] [Google Scholar]
- White TM, Gilden DH, Mahalingam R. An animal model of varicella virus infection. Brain Pathol. 2001;11:475–479. doi: 10.1111/j.1750-3639.2001.tb00416.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.