SUMMARY
The major function of farnesoid X receptor (FXR) is to maintain bile acid and lipid homeostasis. Fxr-null mice, in which the levels of hepatic bile acid and lipid have been elevated, develop spontaneous liver tumors. We evaluated differences in hepatic bile acid and triglyceride concentrations, and in generation of oxidative stress between wild-type mice and Fxr-null mice. The hepatic levels of 8-hydroxy-2’-deoxyguanosine (8OHdG), thiobarbituric acid-reactive substance (TBARS) and hydroperoxides, oxidative stress-related genes, and nuclear factor (erythroid-2 like) factor 2 (Nrf2) protein in Fxr-null mice were significantly higher than those in wild-type mice. An increase in the hepatic bile acid concentration in Fxr-null mice fed a cholic acid (CA) diet resulted in an increase in the hepatic levels of hydroperoxides, TBARS and 8OHdG, whereas a decrease in the hepatic concentration in mice fed a diet containing ME3738 (22β-methoxyolean-12-ene-3β, 24(4β)-diol) resulted in a decrease in these oxidative stress marker levels. A good correlation was observed between the hepatic bile acid concentrations and the hepatic oxidative stress marker levels, although there was no significant correlation between the hepatic triglyceride concentrations and oxidative stress. The results show that oxidative stress is spontaneously enhanced in Fxr-null mice, which may be attributable to a continuously high level of hepatic bile acids.
Keywords: FXR, Bile acid, 8OHdG, TBARS, Nrf2, Oxidative stress
INTRODUCTION
Cholestasis is characterized by accumulation of toxic bile acids in the liver and associated with reduced detoxification capacity 1). Imbalanced mitochondrial energy production and hepatic levels of hydrophobic bile acid have been associated with enhancement in generation of reactive oxygen species (ROS) and oxidative damage in hepatocytes 2, 3). It is well known that farnesoid X receptor (FXR) is a member of the nuclear hormone receptor superfamily and plays an essential role in regulating bile acid, lipids and glucose homeostasis 4, 5). Owing to the absence of FXR expression, hepatic bile acid, lipids and glucose accumulate in Fxr-null mice, which results in mild cholestasis, steatosis and type 2 diabetes 4, 5). In addition to controlling the levels of bile acid, lipid and glucose, FXR also helps accelerate normal liver regeneration after 70% hepatectomy, which suggests that FXR also has a role in promoting liver growth after injury 6). Increased bile acids stimulate liver regeneration, in which the function of the bile acid receptor FXR in mice is required. The hepatoprotective role of FXR is essential for maintaining the normal liver physiology and prevention of deleterious effects of bile acids.
Indeed, it was reported that liver tumors spontaneously developed in aged Fxr-null mice 7, 8). These mice showed high levels of hepatic and serum bile acids, increased expression levels of cell cycle-related genes such as cyclin D and E, proinflammatory cytokine interleukin-1β, tumor necrosis factor-α and interleukin-6 (IL-6), and elevated β-catenin and c-myc levels, which are associated with tumorgenesis. One of general reasons for development of endogenous liver tumors is considered to be oxidative stress generation 9). These reports suggest that continuous generation of oxidative stress might occur in Fxr-null mice, and liver tumors would be developed. The relationship between the absence of FXR and generation of oxidative stress, however, has not been investigated yet. Moreover, it is not clear whether increased hepatic bile acid or triglyceride concentration is involved in the generation of oxidative stress.
In the present study, we observed histopathological changes and measured the hepatic levels of oxidative stress markers such as 8-hydroxy-2’-deoxyguanosine (8OHdG), thiobarbituric acid-reactive substance (TBARS) and hydroperoxide. Then we compared the oxidative stress-related genes expression profiles and nuclear factor (erythroid-2 like) factor 2 (Nrf2) protein levels between wild-type and Fxr-null mice. We also tested whether the high level of hepatic bile acids might be involved in generation of hepatic oxidative stress in Fxr-null mice using cholic acid (CA) and an experimental hepatic bile acid lowering compound ME3738 (22β-methoxyolean-12-ene-3β, 24(4β)-diol) that is not a radical scavenger 10).
MATERIALS AND METHODS
Materials
ME3738 was synthesized at Meiji Seika Kaisha, Ltd. (Yokohama, Japan). Highly sensitive enzyme-linked immunosorbent assay (ELISA) kit for mouse 8OHdG was obtained from Japan Institute for the Control of Aging (Shizuoka, Japan). Human Nrf2 polyclonal antibody (c-20) and human histone H1 polyclonal antibody (c-17) were obtained from Santa Cruz Biotechnology, Inc (Santa Cruz, CA). CA, dichlorofluorescein diacetate, dichlorofluorescein, phenylmethylsulfonyl fluoride (PMSF), dithiothreitol, diaminobenzidine, 1,1,3,3-tetraethoxypropane, thiobarbituric acid, 4-Nonylphenylpolyethylene glycol, phosphatase inhibitor cocktail and protease inhibitor cocktail were obtained from Sigma-Aldrich (St. Louis, MO).
Animal treatment and sample collection
The creation of wild-type and Fxr-null mice has been described 4), and these mice were housed under the standard 12 hr light/12 hr dark cycle. Nine-week-old to eleven-week-old male mice were used for all experiments. The mice were fed a standard diet CE-2 (Clea, Tokyo, Japan) and given free access to water for acclimation. As experimental diets, we used the standard diet mixed with 0.15 (w/w)% ME3738, 0.25% CA, or 0.25% CA plus 0.15% ME3738. Eight wild-type mice and 8 Fxr-null mice were fed a control diet. Four Fxr-null mice were fed 0.15% ME3738, 0.25% CA, or 0.25% CA + 0.15% ME3738 diet for 6 days. Wild-type and Fxr-null mice fed a control diet were used in all experiments, and Fxr-null mice fed experimental diets were only used in the measurement of hepatic oxidative stress levels, bile acid, triglyceride and plasma ALT activity. Approximately 0.1 mL of blood was drawn into a heparinized tube and centrifuged at 7,000 × g for 10 min at 4°C. The supernatant in the tube was taken as plasma. After blood collection, the liver was removed quickly, cleaned of blood, and immediately transferred to a container filled with ice-cold saline. The left lateral lobe of the liver was fixed in 10% phosphate-buffered formalin. Approximately 0.5 g of the remaining liver in 4.5 mL of ice-cold 1.15% KCl aqueous solution was homogenized in a Potter-Elvejhem homogenizer under nitrogen gas. The experimental protocol was approved by the Animal Care and Use Committee of Tohoku University. The animals were treated humanely, and animal experiments were performed in accordance with the Guidelines for Animal Experiments of Tohoku University.
Histological examination
After overnight fixation in 10% phosphate-buffered formalin, the livers were embedded in paraffin. The paraffin-embedded livers were cut into consecutive sections (5µm thick) and stained with hematoxylin and eosin (H&E).
Measurement of biochemical parameters
The hepatic 8OHdG concentration was measured with a highly sensitive ELISA kit for 8OHdG (Japan Institute for the Control of Aging, Shizuoka, Japan) according to the manufacturer’s recommended procedure.
The hepatic TBARS concentration was measured by the method of Ohkawa et al. with slight modification 11). The reaction mixture containing 0.05 mL of 0.2% liver homogenate, 0.2 mL of 8.1% sodium dodecyl sulfate (SDS), 0.75 mL of distilled water, and 1.5 mL of 0.8% aqueous solution of thiobarbituric acid was prepared, and heated at 95°C for 60 min. 0. After being cooled with tap water, with the reaction mixture was added with 1.0 mL of distilled water and 5.0 mL of the mixture of n-butanol and pyridine (15:1), and shaken vigorously. After centrifugation of the mixture at 3,000 × g for 10 min, fluorescence of an organic layer (upper layer) was measured at 515 nm excitation and 553 nm emission wavelengths by Fluoroskan Ascent Fluorescence Spectrophotometer (Thermo Fisher Scientific, Inc., Waltham, MA). A standard curve was plotted using 1,1,3,3-tetraethoxypropane.
Hydroperoxides were measured by conversion of nonfluorescent dichlorofluorescein (nonfluorescent DCF) to fluorescent dicholofluorescein (DCFein) 12, 13). Nonfluorescent dichlorofluorescein diacetate (DCF-DA) is hydrolyzed to nonfluorescent DCF by endogenous esterase in liver homogenate. In the presence of intracellular hydroperoxides (hydrogen peroxide and lipid hydroperoxides), nonfluorescent DCF is converted to DCFein. Since DCF-DA itself does not react with hydroperoxides, only hydroperoxides in the liver homogenate are detected in this method. Ten % of the liver homogenate was diluted to 1:200 by adding DCF buffer containing 200 mmol/L sucrose, 100 mmol/L NaCl, 10 mmol/L 3-(N-morpholino)propanesulphonic acid (MOPS), and 2 mmol/L K2PO4 (pH7.4), which was then added with 0.8 mmol/L DFC-DA. After incubation at 37°C for 0, 5, 10 and 20 min, fluorescence was measured at 485 nm excitation and 527 nm emission wavelengths by Fluoroskan Ascent Fluorescence Spectrophotometer (Thermo Fisher Scientific, Inc., Waltham, MA). The amount of DCF-DA converted to DCFein was determined from a standard curve that was constructed using 2,7-dichlorofluorescein solutions added with the liver homogenate.
Plasma ALT activity was measured using a commercially available kit of Transaminase CII-B-test Wako (Wako Pure Chemicals, Osaka, Japan). Hepatic bile acid, cholesterol, and triglyceride concentrations were measured using the following kits, Total Bile Acid Test Wako, Cholesterol E-test Wako, and Triglyceride E-test Wako, respectively (Wako Pure Chemicals, Osaka, Japan).
Plasma IL-6 concentration was measured using a commercially available kit for Mouse IL-6 immunoassay (BioSource International, Inc., Camarillo, CA) according to the manufacturer’s recommended procedure.
Real-time quantitative polymerase chain reaction (PCR)
The expression of mRNA levels for glutathione S-transferase alpha2 (Gsta2), glutathione S-transferase mu3 (Gstm3), heme oxygenase 1 (Hmox1), nicotinamide adenine dinucleotide phosphate (NAD[P]H) quinone oxidoreductase 1 (Nqo1), and metallothionein 1 (Mt1) were determined by quantitative real-time PCR.
The total RNA from mouse liver was isolated with an RNeasy-kit (Qiagen GmbH, Hilden, Germany) according to the manufacturer’s instructions. For cDNA synthesis, Taqman reverse transcription reagents were used (Roche Diagnostics, Indianapolis, IN). Relative quantification of gene expression was performed as described in the manual using glyceraldehyde-3-phosphate dehydrogenase (Gapdh) as an internal control.
The Taqman probes and primers for Gsta2 (assay identification No. Mm00833353_m1, Accession No. NM_010295.1), Gstm3 (assay identification No. Mm00833923_m1, Accession No. NM_010359.1), Hmox1 (assay identification No. Mm00516004_m1, Accession No. NM_010442.1), Nqo1 (assay identification No. Mm00500821_m1, Accession No. NM_008706.3) and Mt1 (assay identification No. Mm00496660_g1, Accession No. NM_013602.2) were assay-on-demand gene expression products (Applied Biosystems, Foster City, CA). Gapdh gene was used as endogenous control (catalog No. 4308313, Accession No. NM_008084.2, Applied Biosystems, Foster City, CA). The gene-specific probes were labeled using reporter dye FAM™, and the Gapdh internal control probe was labeled with a different reporter dye VIC® at the 5' end. A nonfluorescent quencher and a minor groove binder were linked at the 3' end of probe as quenchers. Thermal cycler conditions were as follows: hold for 10 min at 95°C, followed by two-step PCR for 40 cycles at 95°C for 15 s and then at 60°C for 1 min. PCR reactions were carried out using a PCR master mix (Applied Biosystems, Foster City, CA) in a 7900HT Fast real-time PCR System (Applied Biosystems, Foster City, CA). All procedures were repeated in triplicate. Amplification data were analyzed with an Applied Biosystems Sequence Detection Software version 2.2 (Applied Biosystems, Foster City, CA). The efficiency of the target gene amplification and the efficiency of Gapdh amplification were approximately equal, which was proven by examining the absolute value (less than 0.1) of the slope of log input amount versus Δ CT. The ΔΔCT method recommended by the manufacturer was used to compare the relative expression levels.
Extraction of liver nuclei and measurement of Nrf2 and histone H1 proteins
Approximately 0.1g of mouse liver was homogenated by a Potter-Elvejhem homogenizer with 3 mL of ice cold hypotonic buffer A containing 10 mmol/L Hepes-KOH (pH7.4), 10 mmol/L KCl, 0.1 mmol/L ethylenediamine-tetraacetic acid (EDTA), 0.1 mmol/L O'-Bis (2-aminoethyl) ethyleneglycol-N,N,N',N'-tetraacetic acid (EGTA), 1 mmol/L dithiothreitol, 1 mmol/L PMSF, phosphatase inhibitor cocktail, and protease inhibitor cocktail. The homogenate was incubated at 4°C for 10 min and centrifuged at 1,000 × g for 10 min at 4°C. The pellet was suspended in 1.4 mL of ice cold buffer A containing 90 µL of 10% 4-nonylphenyl-polyethylene glycol. The sample was incubated at 4°C for 10 min and centrifuged at 12,000 × g for 30 sec at 4°C. The supernatant was removed, and the residue was added with 0.2 mL of hypertonic buffer B containing 20 mmol/L Hepes-KOH (pH7.4), 400 mmol/L KCl, 1 mmol/L EDTA, 1 mmol/L EGTA, 1 mmol/L dithiothreitol, 1 mmol/L PMSF, phosphatase inhibitor cocktail, and protease inhibitor cocktail. The sample was centrifuged at 12,000 × g for 10 min at 4°C and the supernatant was obtained as nuclei extract.
Liver nuclei extract was loaded onto 10% SDS-polyacrylamide gel (25 µg protein/lane), which was then transferred to polyvinylidene fluoride membrane (Bio-Rad Laboratories, Inc., Hercules, CA) at 15 V for 30 min. The membrane was immunostained with Nrf2 or histone H1 polyclonal antibodies (1:200 dilution) for 2 hr. The membrane was washed five times with 0.01% Tween 20 containing phosphate buffer saline, and incubated with horseradish peroxidase-conjugated goat anti-rabbit IgG (for Nrf2, 1:2000 dilution, Zymed Laboratories, San Francisco, CA) or horseradish peroxidase-conjugated donkey anti-goat IgG (for histone H1, 1:2000 dilution, Santa Cruz Biotechnology, Inc., Santa Cruz, CA) for 1 hr. The membrane was washed five times with 0.01% Tween 20 containing phosphate buffer saline again, and the membrane was stained with ECL Western Blotting Detection Reagents (Amersham Biosciences, Buckinghamshire, UK) and exposed to a film for 30 sec. The film was scanned using Epson GT-8700 scanner, and the band intensities were measured using Scion image software (version beta 4.02, Frederick, MD).
Statistical analysis
All data are shown as mean ± standard deviation (S.D.). Statistically significant differences between wild-type and Fxr-null mice were assessed by Student’s t test. Statistically significant differences among groups in Fxr-null mice were assessed by Analysis of Variance (ANOVA) followed by Tukey’s multiple comparison. Probability values of less than 0.05 were considered to be statistically significant.
RESULTS
Histochemical changes
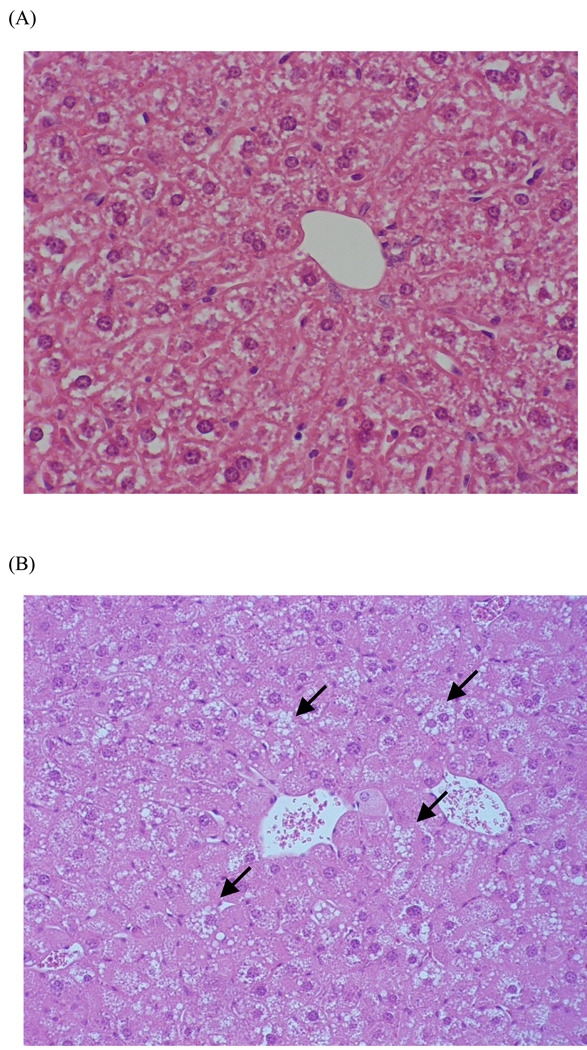
Vacuolation and hypertrophy were observed in hepatocytes of Fxr-null mice (Fig. 1). In contrast, these histochemical changes were not noted in wild-type mice.
Fig. 1.
Histological changes in the livers of wild-type (A) and Fxr-null (B) mice
H&E stained sections of liver tissue show normal morphology in wild-type mice, and vacuolation (Arrows) and hypertrophy in Fxr-null mice (x 50).
Biochemical parameters
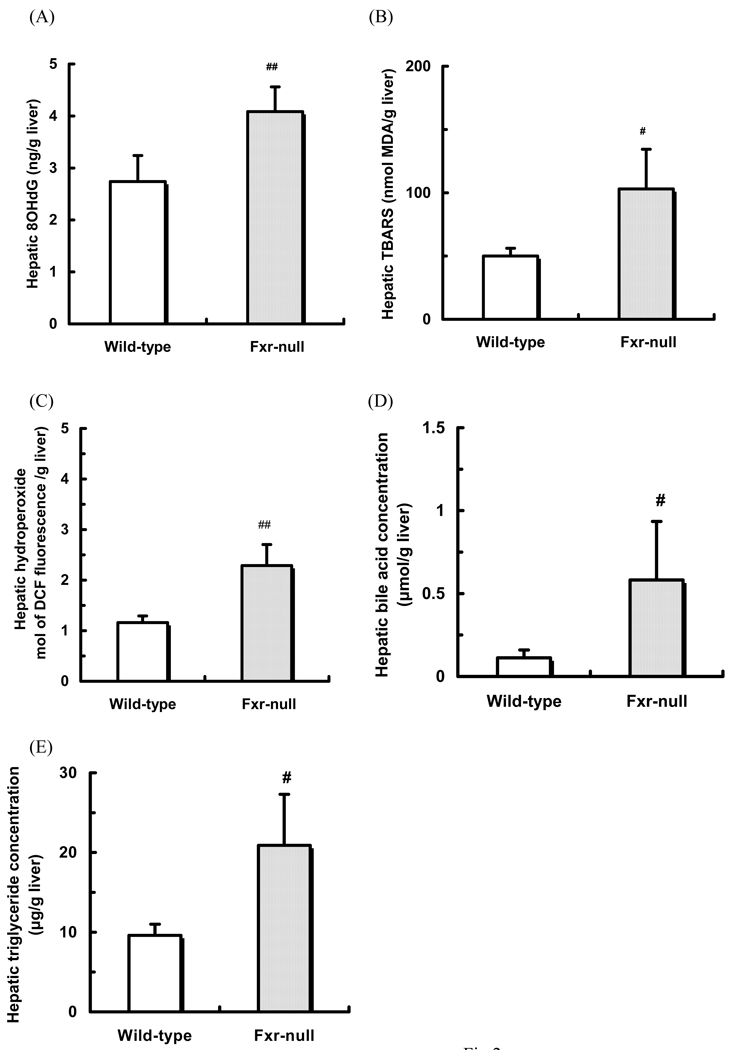
Hepatic 8OHdG concentration was measured by ELISA. The ELISA assay is highly sensitive, and the linearity of hepatic 8OHdG concentration from 0.125 to 10 nmol/L was observed (correlation coefficient (r) = 0.99). The hepatic 8OHdG concentration in Fxr-null mice was significantly higher than that in wild-type mice (Fig. 2A).
Fig. 2.
Hepatic 8OHdG (A), TBARS (B), hydroperoxide (C), bile acid (D) and triglyceride (E) concentrations
Hepatic 8OHdG concentrations were measured by ELISA. Hepatic TBARS, bile acid and triglyceride concentrations were measured by enzyme-colorimetric method. Hepatic hydroperoxide concentrations were measured as fluorescence of dichlorofluorescein.
Data are shown as mean ± S.D. (n=8). #, ##: significantly different from CA group (p<0.05, p<0.01, respectively)
The hepatic TBARS concentration in Fxr-null mice was significantly higher than that in wild-type mice (Fig. 2B). In addition, the hepatic hydroperoxide concentration in Fxr-null mice was also higher than that in wild-type mice (Fig. 2C).
The hepatic bile acid and triglyceride concentrations in Fxr-null mice were approximately 4- and 2-fold higher than those in wild-type mice, respectively, which was consistent with the previous data 4, 14, 15) (Figs. 2D, E). The hepatic cholesterol concentration in Fxr-null mice was 2.8 ± 0.4 mg/g liver, and not significantly higher than that in wild-type mice (2.5 ± 0.6 mg/g liver).
Plasma ALT activity in Fxr-null mice was 108 ± 54 IU/L and higher than that in wild-type mice (10 ± 2 IU/L), which was consistent with the previous reports 14, 15). Furthermore, plasma AST activity in Fxr-null mice was also higher than that in wild-type mice, as noted in the result of ALT activity (Data not shown).
Plasma IL-6 concentration in Fxr-null mice was 9.6 ± 6.7 pg/mL and higher than that in wild-type mice (below limit of quantification (<3.9 pg/mL)).
Real-time quantitative PCR
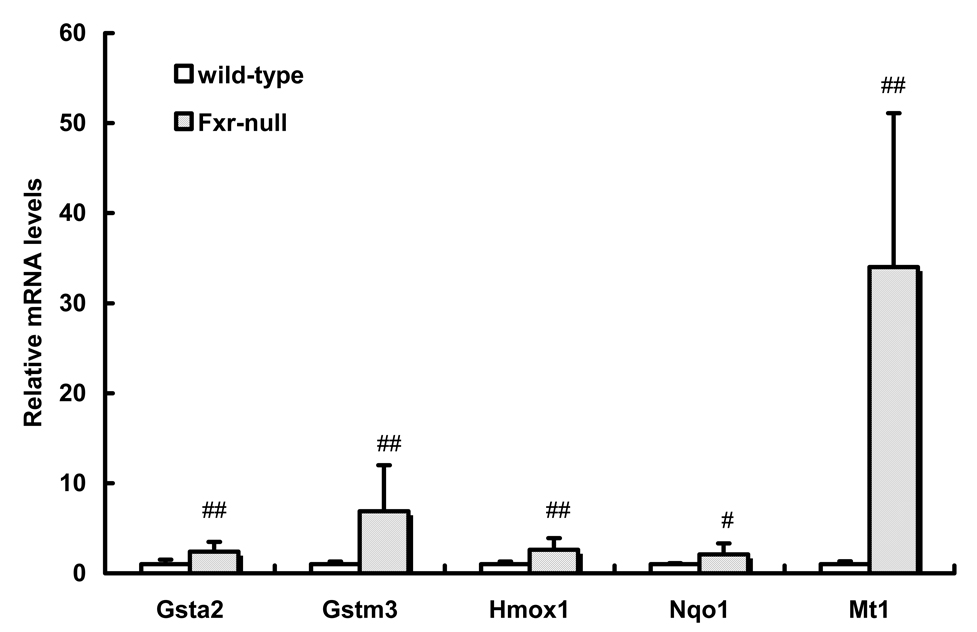
The hepatic oxidative stress marker levels in Fxr-null mice were higher than those in wild-type mice. The levels of hepatic oxidative stress-related gene expression were measured by real-time quantitative PCR. The mRNA levels of Gsta2, Gstm3, Hmox1 and Nqo1 in Fxr-null mice were approximately 2~7-fold higher than those in wild-type mice. Furthermore, the mRNA level of Mt1 was approximately 35-fold higher than that in wild-type mice (Fig. 3).
Fig. 3.
Oxidative stress-related genes expression
Real-time quantitative PCR was carried out using cDNA that was synthesized from the liver of Fxr-null mice and wild-type mice. Data are shown as mean ± S.D. (n=8). #, ##: significantly different from wild-type mice (p<0.05, p<0.01, respectively).
Nrf2 protein level in liver nuclei
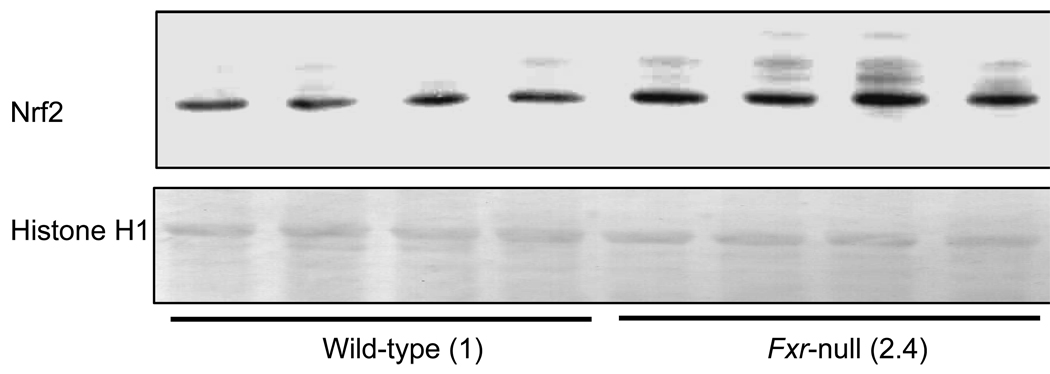
Several kinds of hepatic oxidative stress-related gene expressions in Fxr-null mice were higher than those in wild-type mice and it was reported that these genes were regulated by Nrf2 16–18). We therefore measured the Nrf2 protein level in liver nuclei of wild-type mice and Fxr-null mice. Figure 4 shows a representative result of hepatic Nrf2 protein level in wild-type and Fxr-null mice. Some bands with smaller molecular weights than 66 kDa were detected, and the other bands shown in Fig. 4 were not detected. The histone H1 protein levels were not significantly different between these two types of mice, however the Nrf2 protein level in Fxr-null mice was approximately 2.4-fold higher than that in wild-type mice.
Fig. 4.
Hepatic Nrf2 protein level
Nrf2 protein levels were measured by Western blot analysis. Liver nuclei proteins were subjected to SDS-polyacrylamide gels and electrically transferred to polyvinylidene fluoride membrane for immunostaining with anti-human Nrf2 antibody. Numbers in parenthesis show the expression ratio to wild-type. ##: significantly different from wild-type mice (P<0.01)
Effect of ME3738 in biochemical parameters in Fxr-null mice
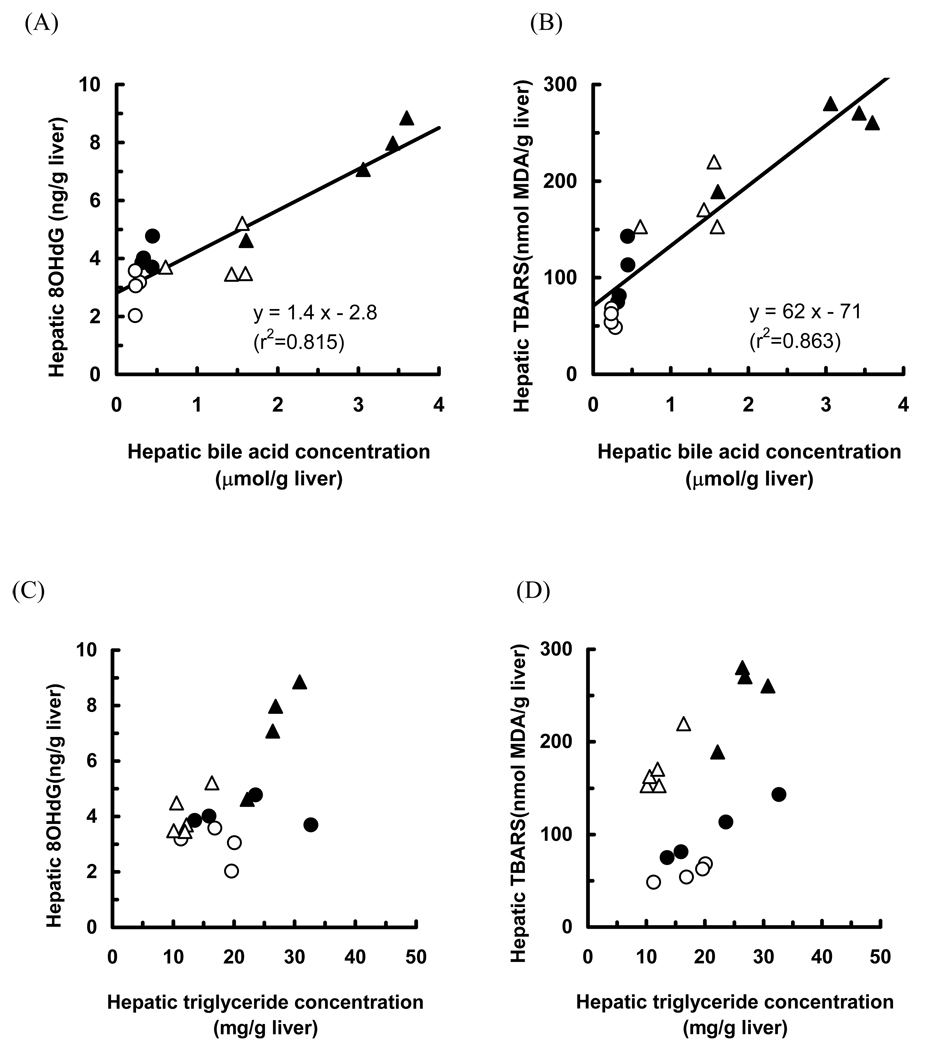
To investigate whether the bile acid level contributes to the generation of oxidative stress, we measured hepatic oxidative stress markers in Fxr-null mice fed a control, and ME3738, CA, or CA plus ME3738 diet. We have reported that feeding of the diet mixed with ME3738 and CA to Fxr-null mice increased biliary bile acid and cholesterol excretion and decreased hepatic bile acid concentrations 10). Feeding with ME3738 decreased the hepatic bile acid, hydroperoxide, TBARS and 8OHdG concentrations and plasma ALT activity (Table 1). Apparent increases in the hepatic bile acid, hydroperoxide, TBARS and 8OHdG concentrations and plasma ALT activity were observed in Fxr-null mice fed CA diet. These concentrations and the activity were lower in the mice fed CA plus ME3738 diet as compared with those in the mice fed CA diet (Table 1). Furthermore, good correlations were observed between the hepatic bile acid concentrations and hepatic 8OHdG or TBARS levels (r2=0.815 and 0.863, respectively). A correlation between the hepatic triglyceride concentrations and hepatic 8OHdG or TBARS levels was, however, coarse (r2=0.341 and 0.206, respectively) when analyzed in all groups of Fxr-null mice fed the control, and ME3738, CA, or CA plus ME3738 diet (Fig. 5).
Table 1.
Hepatic 8OHdG, TBARS, hydroperoxide, bile acid and triglyceride concentrations, and plasma ALT activity in Fxr-null mice
| Liver |
Plasma |
|||||
|---|---|---|---|---|---|---|
| 8OHdG (ng/g liver) |
TBARS (nmol MDA/g liver) |
Hydroperoxide (µmol DCFein/g liver) |
Bile acid (µmol/g liver) |
Triglyceride (mg/g liver) |
ALT (IU/L) |
|
| Control | 4.1 ± 0.5 | 103 ± 31 | 2.29 ± 0.42 | 0.58 ± 0.35 | 20.9 ± 6.4 | 108 ± 54 |
| ME3738 | 2.7 ± 0.5# | 58 ± 9# | 1.60 ± 0.10# | 0.20 ± 0.11# | 17.0 ± 4.1 | 42 ± 23# |
| CA | 6.8 ± 1.5## | 295 ± 108# | 3.73 ± 0.54## | 2.92 ± 0.89## | 19.1 ± 8.8 | 266 ±89## |
| CA+ME3738 | 4.1 ± 0.7** | 172 ± 28* | 2.52 ± 0.58** | 1.35 ± 0.51** | 13.2 ± 4.3 | 158 ± 52* |
Liver homogenate was prepared from Fxr-null mice fed the control, and 0.15% ME3738, 0.25% CA, or CA+ME3738 diet for 6 days. Hepatic 8OHdG concentrations were measured by ELISA. Hepatic TBARS and bile acid concentrations, and plasma ALT activity were measured by enzyme-colorimetric method. Hepatic hydroperoxide concentrations were measured as fluorescence of dichlorofluorescein-diacetate. Data are shown as mean ± S.D. (control; n=8, others; n=4).
significantly different from the control group (p<0.05, p<0.01, respectively),
significantly different from CA group (p<0.05, p<0.01, respectively)
Fig. 5.
Correlations between hepatic bile acid concentrations and 8OHdG (A) or TBARS (B) level, and hepatic triglyceride concentrations and 8OHdG (C) or TBARS (D) level These concentrations and levels in Fxr-null mice fed control diet (closed circle), ME3738 diet (open circle), CA diet (closed triangle) and CA+ME3738 diet (open triangle) for 6 days were plotted.
DISCUSSION
Cholestatic injury is associated with the accumulation of bile acids and activation of proinflammatory cytokines in the liver. Cholestasis causes systemic and intrahepatic retention of potentially toxic bile acids, which results in liver injury and ultimately leads to biliary fibrosis or cirrhosis 19). Elevated bile acid levels are likely to induce toxicity, and therefore bile acid synthesis and enterohepatic circulation are tightly controlled. The present study demonstrated the involvement of FXR in development of liver injury, an early stage of cholestasis, and in generation of hepatic oxidative stress in mouse in vivo.
FXR functions are now well established as a primary bile acid sensor 4). In accordance with this role, many FXR-target genes have been identified to be involved in bile acid and lipid metabolism 20–22). We therefore measured the hepatic bile acid and triglyceride concentrations in both wild-type and Fxr-null mice. As expected, these concentrations in Fxr-null mice were higher than those in wild-type mice (Fig. 2), and vacuolation and hypertrophy were observed in the hepatocytes of only Fxr-null mice (Fig. 1). These results suggested that FXR maintains homeostasis of the hepatic bile acid and lipid levels, and protects against liver injury, an early stage of cholestasis.
ROS induces oxidative damage in various cell constituents such as DNA, protein and lipids. In particular, oxidative DNA damage causes mutations and abnormal expression of genes involved in carcinogenesis 23, 24). 8OHdG, which is produced by oxidation of deoxyguanosine, was reported to be one of the most sensitive markers for oxidative stress 25–27), and an elevation in the 8OHdG level is believed to play an important role in chemically induced carcinogenesis 28). In the present study, the hepatic 8OHdG, TBARS and hydroperoxide levels were significantly higher in Fxr-null mice as compared with wild-type mice (Fig. 2). There was not a large difference in these oxidative stress related parameters between Fxr-null mice and in wild-type mice, because the Fxr-null mouse model does not show drastic pathological changes, but shows light endogenous cholestasis. The hepatic bile acid and triglyceride concentrations in Fxr-null mice were only 4- and 2-fold higher than those in wild-type mice, respectively, which is supported by these results of oxidative stress related parameters.
TBARS is composed primarily of malondialdehyde, an end product of peroxidative decomposition of polyunsaturated fatty acids 29–31). Furthermore, the levels of oxidative stress-related genes such as metallothionein 1 (Mt1), glutathione S-transferase mu3 (Gstm3), alpha2 (Gsta2), NAD(P)H: quinone oxidoreductase 1 (Nqo1), and heme oxygenase 1 (Hmox1) in Fxr-null mice were significantly higher than those in wild-type mice. In particular, the mRNA level of Mt1 in Fxr-null mice was approximately 35-fold higher than that in wild-type mice.
Mt1 is a protective protein against toxicity of heavy metals and ROS, and several lines of evidence indicated that Mt had capacity to scavenge reactive oxygen, particularly hydroxyl radicals 32). Indeed, increased generation of oxidative stress and elevated Mt1 mRNA levels were observed in rats administered with carbon tetrachloride 33). Min et al. reported that an elevation in the Mt level protects against the generation of 8OHdG in rat hepatocytes treated with ferric nitrilotriacetate 34), suggesting that higher Mt1 mRNA expression in Fxr-null mice may be an adaptive response to elevated hepatic oxidative stress levels, especially 8OHdG level. Moreover, it was reported that the Mt1 mRNA levels were inducible by addition of IL-6 33). We measured plasma IL-6 concentrations by ELISA, and compared these concentrations between both types of mice. As a result, the plasma IL-6 concentrations in Fxr-null mice were much higher than those in wild-type mice, suggesting that higher Mt1 mRNA levels in Fxr-null mice may be attributable to higher IL-6 concentration in Fxr-null mice. Glutathione S-transferases catalyze detoxification of electrophilic xenobiotics such as activated chemical carcinogens and chemotherapeutic drugs 35) and were important modulators for the toxicity of such compounds. Hmox1 is a cytoprotective enzyme that plays a critical role in protecting the body against oxidant-induced injury during inflammatory processes 36–38). Hmox1 catalyzed the first and rate-limiting step in the oxidative degradation of heme to CO, biliverdin, and ferrous iron 39–41). Nqo1 is a flavoprotein that catalyzes reduction of quinones and quinone imines, thereby protecting cells against ROS-mediated mutagenicity and carcinogenicity 42). Based on the results of present and earlier studies, oxidative stress is generated spontaneously in Fxr-null mice. Nuclear factor (erythroid-2 like) factor 2 (Nrf2), a basic leucine zipper transcription factor, is known to be a multiorgan protector against various toxic reactive compounds such as chemical carcinogens 43), acetaminophen 44) and hydrophobic bile acid 45). Antioxidant-related genes and phase II drug-metabolising enzymes mentioned above are regulated by Nrf2 16–18). We thus measured the Nrf2 protein levels in liver nuclei of both types of mice. As expected, the Nrf2 protein levels in Fxr-null mice were higher than those in wild-type mice, suggesting that Nrf2 signaling is activated in Fxr-null mice. It supports the results that oxidative stress is spontaneously enhanced in Fxr-null mice.
The bile acid and triglyceride levels, and hepatic oxidative stress markers in Fxr-null mice were higher than those in wild-type mice (Fig.2). In order to determine whether the hepatic bile acid concentration contributes to generation of oxidative stress in Fxr-null mice, we compared the hepatic oxidative stress marker levels in mice fed a control diet, 0.25% CA diet, 0.15% ME3738 diet, or 0.25% CA plus 0.15% ME3738 diet. As a result, an increase in the hepatic concentrations of bile acids in Fxr-null mice fed CA diet led to an increase in the hepatic levels of hydroperoxide, TBARS and 8OHdG. On the other hand, a decrease in hepatic bile acid concentrations in the mice fed ME3738 diet lowered the hepatic oxidative stress marker levels to those in wild-type mice. Furthermore, the hepatic oxidative stress marker levels in the Fxr-null mice fed CA+ME3738 diet were lower than those in CA-fed Fxr-null mice (Table 1). A good correlation between hepatic bile acid concentrations and the level of hepatic 8OHdG or TBARS was observed in Fxr-null mice. The hepatic triglyceride concentrations were not significantly correlated with the hepatic 8OHdG or TBARS level (Fig. 5). These results suggest that chronic high levels of hepatic bile acids may contribute to the generation of oxidative stress in Fxr-null mice. Indeed, as shown in rats intravenously infused with bile acids 46) and in bile-duct ligated rats 47, 48), sustained high level of bile acids was involved in generating oxidative stress.
Some reports described mechanisms for oxidative stress generation by an elevation in the bile acid levels. Krahenbuhl et al. showed that bile acids inhibited the state 3 respiration and reduced the activity of complex I and III of the mitochondrial respiratory chain 1). Moreover, bile acids directly stimulated hydroperoxides generation in hepatic mitochondria, which induced cytochrome c release 13). Mitogen-activated protein (MAP) kinase and c-Jun N-terminal kinase (JNK) signaling pathways appeared to regulate death receptor translocation to the plasma membrane in response to hydrophobic bile acid 49).
Recently, it was reported that aged Fxr-null mice spontaneously developed liver tumors, and the high level of hepatic bile acid may contribute to tumorgenesis 7, 8). One of general reasons for development of endogenous liver tumors is considered to be generation of oxidative stress 9). An increase in hepatic bile acid concentrations through aging in Fxr-null mice is suggested to cause excessive oxidative stress generation, which may develop liver tumors.
It was also reported that absence of FXR disrupted normal glucose homeostasis, and a blood glucose level in Fxr-null mice was higher than that in wild-type mice 5). We, however, could not clarify if the glucose levels contribute to the generation of oxidative stress in Fxr-null mice. Hyperglycemia may lead to increase ROS production in endothelial cells, livers and pancreatic β-cells 50–52), and higher levels of 8OHdG and 4-hydroxynonenal modified proteins are observed in a model of type 2 diabetes mellitus 51). Moreover, obesity is associated with induction of endoplasmic reticulum (ER) stress predominantly in the liver. Under hyperglycemic conditions, the production of glucosamine pathway may initiate ER stress. This stress can promote JNK-dependent serine phosphorylation of insulin receptor substrate 1, which results in suppression of insulin-receptor signaling pathway 53). These reports support that continual generation of oxidative stress is occurred in Fxr-null mice.
In conclusion, the absence of FXR disrupted the hepatic bile acid and lipid levels and spontaneous generation of oxidative stress. The generation of oxidative stress in Fxr-null mice may be attributable to a continuously high level of hepatic bile acids.
Acknowledgements
We would like to express our sincere thanks to Dr. Yoko Shoji, Dr. Shoji Nishiyama, Ms. Mariko Takagi and Dr. Kiyoshi Nagata for skillful technical advice and support. This study was supported by a Grant-in Aid from the Ministry of Education, Science and Culture, Japan and by a Grant-in Aid from the Ministry of Health, Labor and Welfare, Japan.
REFERENCES
- 1.Krahenbuhl S, Talos C, Fischer S, Reichen J. Hepatolgy. 1994;19:471–479. doi: 10.1002/hep.1840190228. [DOI] [PubMed] [Google Scholar]
- 2.Shoda J, Kano M, Asano T, Irimura T, Ueda T, Iwasaki R, Furukawa M, Kamiya J, Nimura Y, Todoroki T, Matsuzaki Y, Tanaka N. Hepatology. 1999;29:1026–1036. doi: 10.1002/hep.510290440. [DOI] [PubMed] [Google Scholar]
- 3.Sokol RJ, Devereaux M, Khandwala RA. J. Lipid Res. 1991;32:1349–1357. [PubMed] [Google Scholar]
- 4.Sinal CJ, Tohkin M, Miyata M, Ward JM, Lambert G, Gonzalez FJ. Cell. 2000;102:731–744. doi: 10.1016/s0092-8674(00)00062-3. [DOI] [PubMed] [Google Scholar]
- 5.Ma K, Saha PK, Chan L, Moore DD. J. Clin. Invest. 2006;116:1102–1109. doi: 10.1172/JCI25604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang W, Ma K, Zhang J, Qatanani M, Cuvillier J, Liu J, Dong B, Huang X, Moore DD. Science. 312:233–236. doi: 10.1126/science.1121435. [DOI] [PubMed] [Google Scholar]
- 7.Kim I, Morimura K, Shah Y, Yang Q, Ward JM, Gonzalez FJ. Carcinogenesis. 2007;28:940–946. doi: 10.1093/carcin/bgl249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang F, Huang X, Yi T, Yen Y, Moore DD, Huang W. Cancer Res. 2007;67:863–867. doi: 10.1158/0008-5472.CAN-06-1078. [DOI] [PubMed] [Google Scholar]
- 9.Maeda S, Kamata H, Luo JL, Leffert H, Karin M. Cell. 2005;121:977–990. doi: 10.1016/j.cell.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 10.Nomoto M, Miyata M, Shimada M, Yoshinari K, Gonzalez FJ, Shibasaki S, Kurosawa T, Shindo Y, Yamazoe Y. Eur. J. Pharmacol. 2007;574:192–200. doi: 10.1016/j.ejphar.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 11.Ohkawa H, Ohishi N, Yagi K. Anal. Biochem. 1979;95:351–358. doi: 10.1016/0003-2697(79)90738-3. [DOI] [PubMed] [Google Scholar]
- 12.Gores GJ, Flarsheim CE, Dawson TL, Nieminen AL, Herman B, Lemasters JJ. Am. J. Physiol. 1989;257:C347–C354. doi: 10.1152/ajpcell.1989.257.2.C347. [DOI] [PubMed] [Google Scholar]
- 13.Sokol RJ, Winklhofer-Roob BM, Devereaux MW, McKim JM., Jr Gastroenterology. 1995;109:1249–1256. doi: 10.1016/0016-5085(95)90585-5. [DOI] [PubMed] [Google Scholar]
- 14.Kok T, Hulzebos CV, Wolters H, Havinga R, Agellon LB, Stellaard F, Shan B, Schwarz M, Kuipers F. J. Biol. Chem. 2003;278:41930–41937. doi: 10.1074/jbc.M306309200. [DOI] [PubMed] [Google Scholar]
- 15.Kitada H, Miyata M, Nakamura T, Tozawa A, Honma W, Shimada M, Nagata K, Sinal CJ, Guo GL, Gonzalez FJ, Yamazoe Y. J. Biol. Chem. 2003;278:17838–17844. doi: 10.1074/jbc.M210634200. [DOI] [PubMed] [Google Scholar]
- 16.Itoh K, Chiba T, Takahashi S, Ishii T, Igarashi K, Katoh Y, Oyake T, Hayashi N, Satoh K, Hatayama I, Yamamoto M, Nabeshima Y. Biochem. Biophys. Res. Commun. 1997;236:313–322. doi: 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- 17.Numazawa S, Yoshida T. J. Toxicol. Sci. 2004;29:81–89. doi: 10.2131/jts.29.81. [DOI] [PubMed] [Google Scholar]
- 18.Copple IM, Goldring CE, Kitteringham NR, Park BK. Toxicology. 2008;246:24–33. doi: 10.1016/j.tox.2007.10.029. [DOI] [PubMed] [Google Scholar]
- 19.Trauner M, Meier PJ, Boyer JL. N. Engl. J. Med. 1998;339:1217–1227. doi: 10.1056/NEJM199810223391707. [DOI] [PubMed] [Google Scholar]
- 20.Song CS, Echchgadda I, Baek BS, Ahn SC, Oh T, Roy AK, Chatterjee B. J. Biol. Chem. 2001;276:42549–42556. doi: 10.1074/jbc.M107557200. [DOI] [PubMed] [Google Scholar]
- 21.Lee H, Zhang Y, Lee FY, Nelson SF, Gonzalez FJ, Edwards PA. J. Lipid Res. 2006;47:201–214. doi: 10.1194/jlr.M500417-JLR200. [DOI] [PubMed] [Google Scholar]
- 22.Hubbert ML, Zhang Y, Lee FY, Edwards PA. Mol. Endocrinol. 2007;21:1359–1369. doi: 10.1210/me.2007-0089. [DOI] [PubMed] [Google Scholar]
- 23.Lau AT, Wang Y, Chiu JF. J. Cell Biochem. 2008;104:657–667. doi: 10.1002/jcb.21655. [DOI] [PubMed] [Google Scholar]
- 24.Nishikawa M. Cancer Lett. 2008;266:53–59. doi: 10.1016/j.canlet.2008.02.031. [DOI] [PubMed] [Google Scholar]
- 25.Kasai H. Mutat. Res. 1997;387:147–163. doi: 10.1016/s1383-5742(97)00035-5. [DOI] [PubMed] [Google Scholar]
- 26.Shibutani S, Takeshita M, Grollman AP. Nature. 1991;349:431–434. doi: 10.1038/349431a0. [DOI] [PubMed] [Google Scholar]
- 27.Yoshiji H, Nakae D, Mizumoto Y, Horiguchi K, Tamura K, Denda A, Tsujii T, Konishi Y. Carcinogenesis. 1992;13:1227–1233. doi: 10.1093/carcin/13.7.1227. [DOI] [PubMed] [Google Scholar]
- 28.Kinoshita A, Wanibuchi H, Imaoka S, Ogawa M, Masuda C, Morimura K, Funae Y, Fukushima S. Carcinogenesis. 2002;23:341–349. doi: 10.1093/carcin/23.2.341. [DOI] [PubMed] [Google Scholar]
- 29.Lee HS, Csallany AS. Lipids. 1987;22:104–107. doi: 10.1007/BF02534861. [DOI] [PubMed] [Google Scholar]
- 30.Esterbauer H, Jurgens G, Quehenberger O, Koller E. J. Lipid Res. 1987;28:495–509. [PubMed] [Google Scholar]
- 31.Esterbauer H, Zollner H. Free Radic. Biol. Med. 1989;7:197–203. doi: 10.1016/0891-5849(89)90015-4. [DOI] [PubMed] [Google Scholar]
- 32.Thirumoorthy N, Manisenthil Kumar KT, Shyam Sundar A, Panayappan L, Chatterjee M. World J. Gastroenterol. 2007;13:993–996. doi: 10.3748/wjg.v13.i7.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Y, Hartley DP, Liu J. Toxicol. Lett. 1998;95:77–85. doi: 10.1016/s0378-4274(98)00009-5. [DOI] [PubMed] [Google Scholar]
- 34.Min KS, Tanaka N, Horie T, Kawano H, Tetsuchikawahara N, Onosaka S. Toxicol. Lett. 2005;158:108–115. doi: 10.1016/j.toxlet.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 35.Dirven HA, Megens L, Oudshoorn MJ, Dingemanse MA, van Ommen B, van Bladeren PJ. Chem. Res. Toxicol. 1995;8:979–986. doi: 10.1021/tx00049a012. [DOI] [PubMed] [Google Scholar]
- 36.Poss KD, Tonegawa S. Proc. Natl. Acad. Sci. U S A. 1997;94:10919–10924. doi: 10.1073/pnas.94.20.10919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yet SF, Perrella MA, Layne MD, Hsieh CM, Maemura K, Kobzik L, Wiesel P, Christou H, Kourembanas S, Lee ME. J. Clin. Invest. 1999;103:R23–R29. doi: 10.1172/JCI6163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wiesel P, Patel AP, DiFonzo N, Marria PB, Sim CU, Pellacani A, Maemura K, LeBlanc BW, Marino K, Doerschuk CM, Yet SF, Lee ME, Perrella MA. Circulation. 2000;102:3015–3022. doi: 10.1161/01.cir.102.24.3015. [DOI] [PubMed] [Google Scholar]
- 39.Abraham NG, Kappas A. Free Radic. Biol. Med. 2005;39:1–25. doi: 10.1016/j.freeradbiomed.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 40.Maines MD, Gibbs PE. Biochem. Biophys. Res. Commun. 2005;338:568–577. doi: 10.1016/j.bbrc.2005.08.121. [DOI] [PubMed] [Google Scholar]
- 41.Ryter SW, Alam J, Choi AM. Physiol. Rev. 2006;86:583–650. doi: 10.1152/physrev.00011.2005. [DOI] [PubMed] [Google Scholar]
- 42.Jaiswal AK. J. Biol. Chem. 1994;269:14502–14508. [PubMed] [Google Scholar]
- 43.Ramos-Gomez M, Kwak MK, Dolan PM, Itoh K, Yamamoto M, Talalay P, Kensler TW. Proc. Natl. Acad. Sci. U S A. 2001;98:3410–3415. doi: 10.1073/pnas.051618798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chan K, Han XD, Kan YW. Proc. Natl. Acad. Sci. U S A. 2001;98:4611–4616. doi: 10.1073/pnas.081082098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tan KP, Yang M, Ito S. Mol. Pharmacol. 2007;72:1380–1390. doi: 10.1124/mol.107.039370. [DOI] [PubMed] [Google Scholar]
- 46.Sokol RJ, McKim JM, Goff MC, Jr, Ruyle SZ, Devereaux MW, Han D, Packer L, Everson G. Gastroenterology. 1998;114:164–174. doi: 10.1016/s0016-5085(98)70644-4. [DOI] [PubMed] [Google Scholar]
- 47.Dueland S, Reichen J, Everson GT, Davis RA. Biochem. J. 1991;280(Pt 2):373–377. doi: 10.1042/bj2800373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang G, Shen H, Rajaraman G, Roberts MS, Gong Y, Jiang P, Burczynski F. Eur. J. Pharmacol. 2007;560:61–68. doi: 10.1016/j.ejphar.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 49.Graf D, Kurz AK, Fischer R, Reinehr R, Haussinger D. Gastroenterology. 2002;122:1411–1427. doi: 10.1053/gast.2002.32976. [DOI] [PubMed] [Google Scholar]
- 50.Ceriello A, dello Russo P, Amstad P, Cerutti P. Diabetes. 1996;45:471–477. doi: 10.2337/diab.45.4.471. [DOI] [PubMed] [Google Scholar]
- 51.Ihara Y, Toyokuni S, Uchida K, Odaka H, Tanaka T, Ikeda H, Hiai H, Seino Y, Yamada Y. Diabetes. 1999;48:927–932. doi: 10.2337/diabetes.48.4.927. [DOI] [PubMed] [Google Scholar]
- 52.Ling PR, Mueller C, Smith RJ, Bistrian BR. Metabolism. 2003;52:868–874. doi: 10.1016/s0026-0495(03)00057-x. [DOI] [PubMed] [Google Scholar]
- 53.Maiese K, Morhan SD, Chong ZZ. Curr. Neurovasc. Res. 2007;4:63–71. doi: 10.2174/156720207779940653. [DOI] [PMC free article] [PubMed] [Google Scholar]