Abstract
Metagenomics is an emerging methodology for the direct genomic analysis of a mixed community of uncultured microorganisms. The current analyses of metagenomics data largely rely on the computational tools originally designed for microbial genomics projects. The challenge of assembling metagenomic sequences arises mainly from the short reads and the high species complexity of the community. Alternatively, individual (short) reads will be searched directly against databases of known genes (or proteins) to identify homologous sequences. The latter approach may have low sensitivity and specificity in identifying homologous sequences, which may further bias the subsequent diversity analysis. In this paper, we present a novel approach to metagenomic data analysis, called Metagenomic ORFome Assembly (MetaORFA). The whole computational framework consists of three steps. Each read from a metagenomics project will first be annotated with putative open reading frames (ORFs) that likely encode proteins. Next, the predicted ORFs are assembled into a collection of peptides using an EULER assembly method. Finally, the assembled peptides (i.e., ORFome) are used for database searching of homologs and subsequent diversity analysis. We applied MetaORFA approach to several metagenomics datasets with low coverage short reads. The results show that MetaORFA can produce long peptides even when the sequence coverage of reads is extremely low. Hence, the ORFome assembly significantly increased the sensitivity of homology searching, and may potentially improve the diversity analysis of the metagenomic data. This improvement is especially useful for the metagenomic projects when the genome assembly does not work because of the low sequence coverage.
Keywords: Metagenomics, ORFome, ORFome assembly, Function annotation
1. INTRODUCTION
Owning to the rapid advancement of the ultra-high throughput DNA sequencing technologies 1, the genomic studies of microorganisms in environmental samples have recently shifted from the focused sequencing of 16sRNA sequences 2 to the shotgun sequencing of the whole DNAs in the sample. This new methodology, now called metagenomics or environmental genomics, has opened a door for biologists to assess the unknown world of the uncultured microorganisms that are believed to be the majority in any environmental sample. The early attempts of this kind can be traced back to a report published in 2002, in which extremely high diversity of uncultured marine viral communities were revealed through genome sequencing 3. However, the most important progress in shotgun metagenomics happened in 2004 4,5,6,7, when two research groups published results from their large-scale environmental sequencing projects. The first project studied the sample from the Sargasso Sea, and revealed ~ 2000 distinct species of microorganisms, including 148 types of bacteria that have never been observed before 8. In the second project, a handful of genomes of bacteria and archaea that had previously resisted attempts to culture them were revealed based on the analysis of the sample from the acid mine drainage 9. Since then, many more metagenomics projects have been conducted, involving broadened applications from ecology and environmental sciences to chemical industry 10 and human health, e.g., the human gut microbiome projects 11,12.
The rapid growth of metagenomic data has posed great challenges to the computational analysis 13,14. Some metagenomics projects applied directly the data analysis pipeline that includes the whole genome assemblers 15,16,17,18 and gene finding programs 19—originally designed for the conventional Whole Genome Shotgun (WGS) sequencing projects—with only some small parameter modifications 8,9,12,20. However, it is unclear how accurate these existing tools for fragment assembly and genome annotation are when applied to metagenomic data. Mavromatis and colleagues have conducted a valuable benchmarking experiment to evaluate the performance of conventional genome assembly and annotation pipeline on simulated metagenomic data 21. In this experiment, sequencing reads were randomly collected from 113 assembled genomes that are mixed at various complexities. Afterwards, the quality of the results from each processing step (i.e., assembly, gene prediction, and phylogenetic binning) was assessed separately by comparison to the corresponding genomes used in the simulation. This experiment delivered an encouraging message that the number of errors made at each step overall is not high, and some errors (e.g., the chimeric contigs) would not be propagated into the subsequent steps (e.g., binning). Nevertheless, we argue that this experiment may not completely reflect the challenge of metagenomic data analysis, especially the difference between metagenomic data and the data from conventional genome sequencing. Conventional genome projects deal with only one or sometimes a few individual genomes from the same species that are isolated prior to sequencing, whereas metagenomics attempts to analyze simultaneously a huge amount of genomes not only from hundreds of different microorganisms, but also from many individuals of each organism. As a result, even the reads from the same species might be quite different from each other since they might be sampled from different individuals’ genomes. Furthermore, those microbial species may exist in the sample at a wide range of abundances. Hence, typically, only a few dominant species can receive good sequence coverage for their genomes, whereas the sequence coverage for the remaining species is low.
More and more metagenomic projects have applied Next-Generation Sequencing (NGS) technologies that produce massive but shorter reads (e.g., ~ 200 bps for 454 pyrosequencing machines)a than those from the Sanger sequencing methods. Therefore, many metagenomic sequencing projects that acquired a merely small number of short sequencing reads often skipped the step of fragment assembly, and directly used the short reads for downstream analysis 3,22,23. For instance, short reads can be used to search against protein database using TBLASTX to identify homologous proteins, in which an arbitrary E-value (e.g., ≤ 1e – 5) was chosen as a cutoff 22. This direct search approach, however, often missed many homologous genes (or proteins) 24, and resulted in a very low false positive rate b but high false negative rate. This drawback may bias the further analysis of species diversity (i.e., how many different species are present in the sample) and functional coverage (i.e., how many functional categories of proteins are present in the sample).
In this paper, we present a novel ORFome assembly approach to assembling metagenomic sequencing reads. Different from the conventional genome analysis pipeline that first assembles sequencing reads into contigs (or scaffolds) and then predicts protein coding regions within the contigs, our method first identifies putative protein coding regions (i.e., open reading frames, or ORFs) within unassembled reads, and then focuses on the assembly of only these sequences (i.e., ORFome). The ORFome assembly approach has several advantages. First, it significantly simplifies the task of fragment assembly that is often complicated by the repetitive sequences present mainly in non-coding regions 25. Meanwhile, we argue that ORFome assembly does not lose much useful information by neglecting the non-coding sequences due to several reasons: (1) the set of proteins (or the ORFome that encodes them) carry the most important information for the downstream analysis; (2) the microbial genomes are often very compact and protein coding regions comprise a major fraction of them; and (3) microbial proteins are mainly encoded by continuous nonsplit open reading frames (ORFs), thus the prediction of coding sequences prior to assembly is relatively straightforward. Second, from ORFome assembly, complete proteins (or long peptides) may be derived, thus higher sensitivity and specificity can be achieved in the step of database searching for homologs 24. Furthermore, most single nucleotide polymorphisms are synonymous mutations that do not change the encoding amino acids so that ORFome assembly does not even feel them. So by working on the peptide sequences (translated from sequencing reads in silico) instead of the raw DNA sequences, the ORFome assembly alleviates the assembly difficulty caused by the differences among individual genomes at polymorphic sites. We used four marine viral metagenomic datasets of short reads, acquired using 454 sequencing technique, to test our ORFome assembly method—no genome assemblies are available for these metagenomic datasets because the reads are extremely short and the sequence coverage is low.
2. METHODS
The computational framework of ORFome assembly consists of three steps (Fig. 1 (e-f)): (1) each read is assessed individually and the putative open reading frames (ORFs) that likely encode proteins are annotated; (2) the annotated ORFs are assembled into a collection of peptides using a modified EULER assembly method 26; and (3) the assembled peptides are used for the database searching of homologs.
Fig. 1.
A schematic comparison of the ORFome assembly approach with the Whole Genome Assembly (WGA) pipeline for the metagenomic sequence analysis. Both approaches attempt to characterize the protein coding genes in the shotgun sequencing reads from the metagenomic analysis of an environmental sample containing a number of different microorganisms (the reads are shown as double-barreled, as currently several NGS techniques are capable of generating such data; however, some early metagnomics projects, including the datasets used in this paper, did not produce double-barreled sequencing reads, and thus the scaffolding step is not feasible) (a). The whole genome assembly (WGS) pipeline (b-d) first assembles the reads into contigs and scaffolds, and then annotates the genes in the assembled sequences. In comparison, ORFome assembly approach (e-g) first applies gene finding in the unassembled reads, and then assembles only those annotated (partial) ORFs into peptides. These peptides may be further connected to form scaffolds if there are mate-pairs available from double-barreled sequencing (g).
A major difference between the ORFome assembly approach and the conventional whole genome assembly is that the former approach conducts gene annotation (at this stage we used all six frame translations; but a dedicated gene finder will be developed in the future to provide better prediction of ORFs) followed by the assembly of identified short peptides, whereas the latter approach conducts gene annotation after assembly of DNA sequences. Conventional fragment assembly algorithms are mostly based on the analysis of overlap graph, in which the reads are represented by vertices and the overlaps between reads are represented by edges 27. The presence of repeats in the genomes often induce many spurious edges in the overlap graph, which is a major challenge in fragment assembly. There are two additional aspects in the metagenomic data that make fragment assembly even more challenging. First, metagenomics projects often apply NGS technique, and produce shorter reads (~ 200 bps) than Sanger sequencing methods (500-1000 bps). As a result, many short repeats (with lengths between 200 bps and 500 bps) may increase the complexity of the overlap graph, and cause many more mis-assemblies 28. Second, unlike the conventional genome shotgun sequencing, which handles a single species, metagenomics sequencing reads are collected from a large amount of different genomes. Hence, we anticipate these reads should be assembled into not one but many sequences that may even share high similarity on multiple regions. Therefore, the straightforward application of conventional fragment assemblers may encounter difficulties. In contrast, the ORFome assembly approach attempts to assemble only the most important portions of the target genomes, i.e., the protein coding regions, which can highly reduce the complexity of the overlap graph and thus improve the assembly quality.
It is worth pointing out the idea of ORFome assembly can be viewed as an extension of the repeat masking approach used in whole genome assembly of large eukaryotic (including human) genomes. To avoid the complication induced by the many interspersed repeat copies present in most eukaryotic genomes, Celera Assembler first masked out putative repeats in the unassembled reads, and then focused on the assembly of the remaining reads from non-repetitive regions 29,30. The resulting overlap graph, which consists of a number of connected components each representing reads from continuous non-repetitive regions, is much simpler and easy to be analyzed. Similarly, the ORFome assembly approach divides the complex overlap graph into a number of components each representing reads from a single gene or several highly similar genes from the same family.
We applied the ORFome assembly approach to several metagenomics datasets from Ocean samples with low coverage and short reads 22. The results show that MetaORFA can produce long peptides even when the sequence coverage of reads is extremely low. Hence, further analysis of assembled peptides significantly increased the sensitivity for subsequent homology searching, and may potentially improve the diversity analysis of the metagenomic data.
2.1. ORFome Assembly Algorithm
We implemented a tool called MetaORFA in C/C++ under linux platforms for the ORFome assembly. MetaORFA consists of two programs. One program takes as input a set of reads and predicts a number of putative ORFs; and the other program (EULER-ORFA) takes as input the set of putative ORFs, and reports a set of peptides corresponding to the assembled ORFs. Prior to be supplied to MetaORFA, the original reads were first processed by MDUST (a tool for autonomous masking from TIGR, which implements the DUST algorithm 31) to mask out low-complexity regions, and then processed by Tandem Repeat Finder (TRF V4.0) 32 to mask out short tandem repeats.
In this preliminary study, we adopted a very simple method for ORF prediction. For each read (and its reverse complement), a region from the beginning (i.e., position 1, 2, or 3, depending on the frame) or a start codon to the end of the read or a stop codon is considered as a potential ORF. Only ORFs with more than a threshold K (default K = 25) codons were reported. These ORFs will then be transformed into peptide sequences, and subsequently assembled using EULER-ORFA algorithm, modified from the original EULER algorithm designed for DNA fragment assembly 26. In this process, we first build a de Bruijn graph using all k-mers (default k = 10) in the putative peptides from previous step, and then apply the equivalent transformations as described in Ref. 26 to resolve short repeats among peptides. Unlike many other genome assemblers that assemble reads into linear contigs, EULER aims at constructing from the reads a repeat graph that represents not only the unique regions but also the repeat structures 33. Although we anticipate there are not many repeats in the coding sequences, the similar parts of homologous proteins from the same family may act like repeats during the ORFome assembly. In addition to peptide assembly, EULER-ORFA can report a compact graph structure, called the protein family graph, to represent the architecture of domain combinations, including domain recurrences and shuffling34 among homologous proteins in the same sample.
Fig. 2 illustrates the EULER-ORFA process using a synthetic example. Assume that two homologous proteins from different microorganisms are encoded in the metagenome. Due to the short read length, it is difficult to reconstruct the complete sequences for both proteins. However, using a de Bruijn graph approach, we can assemble them into a protein family graph by glueing together all tuples longer than K (here K ≥ 2). The common and distinct parts between two (or more) homologous proteins are represented by separate edges, and each protein sequence corresponds to a path in the protein family graph.
Fig. 2.
A synthetic example for the ORFome assembly resulting into a protein family graph. Two homologous proteins are encoded in the metagenome. Due to the short read length, it is difficult to reconstruct the complete sequences of these two proteins. The EULER-ORFA approach assembles them into a protein family graph, in which the common and distinct parts between two proteins are represented by separate edges, and each protein corresponds to a path in the graph.
Notably, the protein family graph is not always a partial order graph 35,36. When domain reorganization happens between multiple homologous proteins in a family, the protein family graph may contain cycles, as demonstrated in the EULER multiple alignment algorithm 34. However, we expect this will rarely be encountered in ORFome assembles because (1) there are far fewer multi-domain proteins in bacteria; and (2) metagenomic sequencing may rarely cover the full lengths of long multi-domain proteins. Therefore, the resulting protein family graphs will likely be partial order graphs, and can be compared with similar protein sequences by a network matching algorithm, to deduce the full length protein sequences in the sample. An alternative strategy is to traverse in the family graph and collect all the paths each corresponding to a potential peptide. However this strategy will result in many peptides—which may slow down the further similarity search—and the potential chimeric peptides may complicate the database search.
We note the further analysis of the ORFome assembly results, as described below, has not fully taken advantages of the protein family graph representation. Rather, we searched the individual assembled peptide sequences corresponding to each edge in the protein family graph after assembly against the target protein sequence database. Nevertheless, our preliminary analysis has already demonstrated that even this simple analysis revealed—in the metagenome sample—more reads with similarity to known proteins.
2.2. Functional Coverage Assessment
The ORFome, i.e., the set of assembled peptides, is ready for further computational analysis with different purposes, e.g., searching against database for homologous sequences, or mapping to biological pathways to study metabolic diversity 37. Here we show that we can improve the functional coverage of metagenomics sequences by using assembled peptides instead of unassembled reads. There are various ways to estimate functional coverage of a sample. In this study we used PANTHER (Protein ANalysis THrough Evolutionary Relationships) protein family classification 38 for such assessment. The comparison of the functional coverage between different ORFomes is then straightforward. We can simply count the number of families (subfamilies) found in assembled ORFome and unassembled reads, and calculate their differences.
In the PANTHER classification system, proteins are classified into families and subfamilies of shared function by experts. Families and subfamilies are presented as Hidden Markov Models (HMMs). We downloaded the PANTHER HMM library Version 6.1 (release date December 17, 2007) from ftp://ftp.pantherdb.org, which contains 5547 protein family HMMs, divided into 24,582 functionally distinct protein subfamily HMMs. We also downloaded the HMM searching tool (pantherScore.pl, version 1.02), which utilized fast BLAST search prior to the more sensitive but time-consuming HMM matching procedure to speed up the process. The query protein sequence will first be blasted against the consensus sequences of each PANTHER HMMs, and then based on the results, some heuristics are applied to determine which HMMs (i.e., protein families or subfamilies) that the query should be compared with using hmmsearch from the hmmer package (http://hmmer.janelia.org).
2.3. Metagenomic Sequences Datasets
We tested our algorithm on four datasets each containing metagenomics sequences of a major oceanic region community (the four regions are Sargasso Sea, Coast of British Columbia, Gulf of Mexico, and Arctic Ocean) (referred to as Ocean Virus datasets) 22. The reads were acquired by 454 sequencing machine, and they are typically very short. All the metagenomic sequences were downloaded from CAMERA website (http://camera.calit2.net/) 39.
3. RESULTS
First we tested the performance of MetaORFA using different length cutoffs of input ORFs. Then we chose the best cutoff and applied the MetaORFA to assemble the four Ocean Virus datasets. The assembly of a dataset took about from several minutes to half an hour for the four datasets we used here (on a linux machine with Intel(R) Core(TM)2 CPU@ 2.40GHz). The unassembled reads and assembled peptides were searched against Integrated Microbial Genomics (IMG) database 40 using BLASTP to identify known homologous proteins in pre-sequenced microbial genomes. To show the improvement of functional coverage after the ORFome assembly, we also searched both sets of sequences against PANTHER families and subfamilies. Below we first report the basic statistics of the assembled peptides as compared to the unassembled reads, and then show the annotation of the ORFs by BLAST search and PANTHER family annotation. Finally we show that MetaORFA can assemble sequences with synonymous mutations, demonstrating the advantage of using ORF assembly over the assembly of DNA sequences.
3.1. Optimization of the length cutoff of input ORFs
The length cutoff of input ORFs for MetaORFA is an important parameter, which influences the quality of ORF assembly as well as the speed of MetaORFA. We tested lengths of 15, 20, 25, 30 and 35 amino acids. We evaluated the assembly quality using two measures: the total number of long peptides (e.g., peptides of at least 60 amino acids), and the length of the longest assembled peptide. Our tests show that MetaORFA performs roughly the same when using cutoff of 20 or 25 in all four datasets. Fig 3 shows the performance of the MetaORFA versus the length cutoff of the input ORFs for the Sargasso Sea dataset. When a high cutoff (e.g., cutoff = 35) is applied, fewer ORFs will be included in assembly; as a result, fewer long peptides will be assembled. On the other hand, using too many short ORFs (e.g., cutoff = 15) will increase the noise for assembly, thus worsening the assembly results. Considering that using more ORFs as the input will slow down MetaORFA, we chose 25 as the length cutoff of predicted ORFs as input for MetaORFA.
Fig. 3.
Comparison of the MetaORFA performance using different length cutoffs of input ORFs as shown in the total number of long assembled peptides (of at least 60aa)(a), and the length of the longest peptide (b).
3.2. Assembled Peptides from the ORFome Assembly
Table 1 shows the statistics of the reads, unassembled putative ORFs and assembled peptides for the four Ocean Viruses datasets. For all four datasets, the ORFome assembly successfully produced long peptides (≥ 60) that are not present in the unassembled reads. However, the number and the length of long peptides are different from one dataset to another. For example, the ORFome assembly produced the largest number (13,547) of long peptides with longest average length (37 aa) in the Arctic Ocean dataset, even though comparable number of sequencing reads were acquired in each of these four datasets. This may indicate either the diversity of the microorganisms in Arctic Ocean sample is lower than the diversity in the other samples, or the microorganism genomes in this sample are more compact than the genomes in the other samples.
Table 1.
Statistics of the ORFs for Ocean Virus datasets
| Sample | Num | Min | Max | Ave | Num60 | |
|---|---|---|---|---|---|---|
| Arctic Ocean | Reads | 688590 | 35 | 370 | 99 | - |
| UA-ORF | 1015432 | 30 | 58 | 33 | 0 | |
| A-Pep | 359023 | 30 | 162 | 37 | 13547 | |
| Sargasso Sea | Reads | 399343 | 36 | 282 | 104 | - |
| UA-ORF | 345411 | 30 | 49 | 33 | 0 | |
| A-Pep | 211922 | 30 | 202 | 34 | 1813 | |
| Coast of British Columbia |
Reads | 16456 | 37 | 254 | 102 | |
| UA-ORF | 426666 | 30 | 61 | 33 | 1 | |
| A-Pep | 300227 | 30 | 196 | 36 | 3165 | |
| Gulf of Mexico | Reads | 771849 | 38 | 246 | 95 | - |
| UA-ORF | 467085 | 30 | 54 | 33 | 0 | |
| A-Pep | 204595 | 30 | 155 | 34 | 2166 |
Num, Min, Max and Ave represent the total number, the minimum, maximum, and average length of the reads (in nucleotides), unassembled ORFs (UA-ORF, in amino acid residues) and assembled peptides (A-Pep, in amino acid residues), respectively. Num60 represents the total number of unassembled ORFs and assembled peptides of length ≥ 60. We note that MetaORFA only reports assembled peptides of at least 30 aa. So in this statistics we use 30 as the minimum length of unassembled ORFs, just for the comparison purpose. Also there are still many short ORFs that can not be assembled or assembled well for these four datasets, causing the low average length of assembled peptides. This also re ects the difficulties of assembling of these four datasets.
We use the longest peptide assembled from the Gulf of Mexico dataset as an example to illustrate the advantages of the ORFome assembly. Fig. 4 shows that 18 putative ORFs detected from different short reads were assembled into the long peptide (155 aa) by the ORFome assembly, which shows strong similarity across the entire peptide with an annotated protein in IMG database. In the Sargasso sea dataset, an even longer peptide was assembled (contig216592), which has 202 amino acid residues. Homology search against IMG database shows this peptide is similar to a major coat protein from Enterobacteria phage alpha3 (IMG ID: 638278159) with E-value = 2e-24.
Fig. 4.
A long peptide with 155 aa (contig196081, highlighted in bold line) assembled from 18 putative ORFs (represented as thin lines below the contig) in the Gulf of Mexico dataset shows strong similarity with proteins in IMG database with known function (a). (b) shows the BLAST alignment between the peptide and the PhoH-like protein from Roseophage SIO1 in IMG database.
3.3. Homology Search of Assembled Peptides
One of the commonly used analysis of metagenomic data is the searching of the unassembled reads against databases of known microbial proteins in an attempt to use the identified homologous proteins to assess the function and species diversity in the sample 41,23. In this type of analysis, a quite high cutoff is often chosen for the BLAST E-values (i.e., less significant) because the query sequences (i.e., reads) are quite short. As a result, there may be many false hits included in the final list of homologous proteins, which can mislead the diversity analysis. Comparing with this straightforward approach, we anticipate the homology search using the assembled peptides from the ORFome assembly can achieve higher sensitivity and result in more hits with higher significance (i.e., lower E-values).
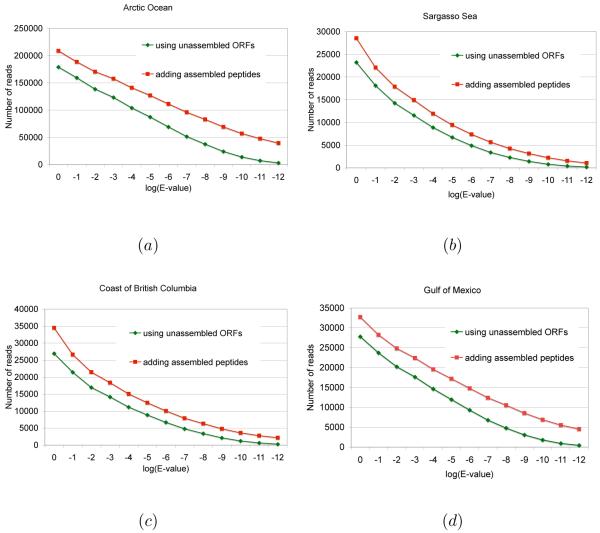
We compared the results of homology searches using assembled peptides with the results using unassembled reads. The four Ocean Virus datasets were tested separately against IMG database. As reported in Ref. 22 c, only few reads hit proteins in the database. We emphasize that the assembled peptides increase the number of significant hits (i.e., E-value ≤ 1e – 5) in all four datasets, from 40.5% in the Sargasso Sea dataset (i.e., 2,728 read hits were added to 6,726 read hits received from the searching using unassembled reads) to 45.3% in the Arctic Ocean dataset (39,658 read hits were added to 87,487 original read hits). Fig. 5 shows the detailed comparison of the added number of read hits when various E-value cutoffs were applied. For all four datasets, a nearly constant number of read hits can be added by using assembled peptides at different similarity significance levels (E-values). In comparison, a majority of read hits from the similarity searching using unassembled reads received high E-values. For instance, there are only 14,127 read hits in the Arctic Ocean dataset with E-values ≤ 1e-10, whereas 43,098 additional read hits (i.e., 305% more!) can be added from the similarity searching using assembled peptides.
Fig. 5.
Detailed comparison of the total number of read hits in IMG database using unassembled and the total number of read hits including those read hits belonging to the assembled peptides at different BLAST E-value cutoffs. The deviation between the two lines indicates the gain of read hits by using assembled peptides from the ORFome assembly.
3.4. Novel Assignments of Functional Categories by Assembled Peptides
We further assessed the performance of the ORFome assembly in improving the function annotation on the Ocean Virus datasets. Table 2 summarizes the statistics of the number of matched families in PANTHER database for all four datasets. Both the number of hits from the searching of unassembled reads as well as the additional number of hits from the searching of assembled peptide are listed. Although the additional numbers of families detected by using assembled peptides are relatively low for all datasets, there are still some new protein families (or novel protein functions) that can be annotated when assembled peptides were used. For example, in the Gulf of Mexico dataset, the assembled peptides hit additional 25 PANTHER protein families, one of which is ATP synthase mitochondrial F1 complex assembly factor 2 (Panther family ID PTHR21013). The results suggest that we may be able to improve the protein function annotation using assembled peptides.
Table 2.
Summary of the family annotation of assembled peptides versus unassembled reads for the four ocean virus datasets
| Sample | Family | Add-on | Example |
|---|---|---|---|
| Arctic Ocean | 598 | 34 | PTHR22748 |
| Sargasso Sea | 270 | 5 | PTHR11527 |
| Coast of British Columbia | 361 | 9 | PTHR10566 |
| Gulf of Mexico | 438 | 25 | PTHR17630 |
The “Family” column lists the total number of protein families that are found from unassembled reads. The “Add-on” column lists the additional PANTHER protein families that are detected by using assembled peptides. The last column gives a few examples of the additional protein families (or functions) that are annotated based the assembled peptides only: PTHR22748, AP endonuclease (E-value = 5.4e-12); PTHR11527 (subfamily SF15), heat shock protein 16 (E-value = 1.5e-07); PTHR10566 (subfamily SF7), ubiquinone biosynthesis protein AARF (E.coli)/ABC (Yeast)-related (E-value = 7.3e-11); PTHR17630 (subfamily SF20), carboxymethylenebutenolidase (Evalue = 4.7e-08) .
3.5. MetaORFA can Assemble Sequences with Synonymous Polymorphism
We further checked if MetaORFA can assemble sequences that have synonymous polymorphic sites. The results show that about one third of the assembled peptides include at least one amino acid that involves synonymous mutation. For example, in the Sargasso Sea dataset, out of 2,558 assembled peptides (here only the peptides that hit IMG sequences with E-value ≤ 1e-5 were included, considering they are more likely to be real proteins as compared to other peptides that have no homologs), total 825 peptides involve synonymous mutations. Fig. 6 shows an example of these proteins with 11 synonymous polymorphic sites. These results demonstrate that ORFome assembly does not feel the mutation at the DNA level that does not change the amino acids (i.e., synonymous mutations), which is one of the MetaORFA’s main features. Once we have the protein sequence assembled (which can be not done otherwise at the DNA level because of the mutations), we can map them back to the DNA sequences and further study their polymorphism.
Fig. 6.
A peptide involving 11 synonymous polymorphic sites (starting from position 30, ending at position 60) assembled from the Sargasso Sea dataset. In the graph, the aligned protein sequences are shown on the top and the corresponding DNA sequences are shown on the bottom; the mutations are highlighted in bold and italic (if there are only two sequences covering the site, one arbitrary codon is highlighted).
4. DISCUSSION
One of the main issues in whole genome assembly is the chimeric contigs that are resulted from mis-assemblies. Tremendous finishing efforts have to be invested in order to identify and correct these errors. This issue is expected to be more serious in metagenomic data analysis because of the higher complexity of metagenomic sample and the short read length. Although it remains unclear whether the mis-assemblies will dramatically influence the conclusion on the principal aims of metagenomics, such as the assessment of species diversity in the sample, many metagenomic projects avoided assembling sequencing reads, and analyzed the original reads directly. The ORFome assembly provides a simple solution to bypass the assembly obstacle, i.e. to conduct a small-scale but accurate assembly of protein coding regions that can improve the sensitivity of homology search. In this study, although we showed the homology searching was improved after the ORFome assembly, we have not systematically evaluated the influence of these improvements on the diversity analysis. Our next step is to apply the ORFome assembly approach to more datasets with various sequence coverage and sample complexities (i.e., the approximate number of species and the range of abundances among these species). Our intention is to estimate the minimal sequencing efforts required to get a good assessment of species diversity for samples with different complexities.
There are several ways to further improve the ORFome assembly algorithm described here. For example, the current method for predicting putative ORFs in sequencing reads can be improved by incorporating additional features of gene coding sequences (e.g., the codon usages) and utilizing sophisticated probabilistic models. This indicates that there is still room for the further improvement of the ORFome assembly method by selecting more appropriate parameters. Finally, as we mentioned in the METHODS section, the advantages of the ORFome assembly have not been fully taken in the downstream data analysis in this study. The EULER-ORFA method used here can assemble putative ORFs into a protein family graph, in addition to the peptides represented by edges in the graph. Therefore, we can adopt a network matching approach as used in Ref. 36 to achieve a more sensible database searching (which, however, may be slower than BLAST based database searching). The network matching of a family graph against a sequence database can also help to define the path that corresponds to the most likely peptide among all the possible ones (including the chimeric ones).
Finally we point out that the basic method we adopted for ORF prediction may generate some spurious peptides, and some of the assembled ORFs may be not real proteins. Those spurious peptides may not cause serious problems in applications such as the homology search based annotations as used in this paper. However, we should not neglect their impact on other types of applications, such as comparison of the number of protein clusters (families) among different metagenomic datasets. In the latter case, we may need to rely on non-homology based approaches to filter out the spurious peptides.
5. CONCLUSION
We present a novel ORFome assembly approach to metagenomics data analysis. The application of this method on four metagenomics datasets achieved promising results. Even with low coverage short reads from these datasets, our method has assembled many long peptides, which can hit annotated proteins by similarity searching that are not detectable otherwise. The ORFome assembly provides a useful tool to retrieve rich information from metagenomic sequencing reads, and it shows potential to facilitate an accurate assessment of the species and functional diversity in metagenomics.
6. ACKNOWLEDGEMENT
This research was supported by the Indiana METACyt Initiative of Indiana University (funded in part through a major grant from the Lilly Endowment, Inc), and NIH grant 1R01HG004908-01. The authors thank the University Information Technology Services team in Indiana University for their help with high-performance computing (for BLAST search).
Footnotes
454 sequencing machines can now produce longer reads
For example, the MEGAN analysis based on the direct BLAST search method has achieved a 0 false positive rate 23!
We note that a direct comparison is not feasible since different databases were used for homology searching in these two studies.
References
- 1.Mardis E. Anticipating the 1,000 dollar genome. Genome Biol. 2006;7:112. doi: 10.1186/gb-2006-7-7-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lane D, Pace B, Olsen G, et al. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc Natl Acad Sci USA. 1985;82:6955–6959. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Breitbart M, Salamon P, Andresen B, et al. Genomic analysis of uncultured marine viral communities. Proc Natl Acad Sci USA. 2002;99:14250–14255. doi: 10.1073/pnas.202488399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Galperin M. Metagenomics: from acid mine to shining sea. Environ Microbiol. 2004;6:543–545. doi: 10.1111/j.1462-2920.2004.00652.x. [DOI] [PubMed] [Google Scholar]
- 5.Eyers L, George I, Schuler L, et al. Environmental genomics: exploring the unmined richness of microbes to degrade xenobiotics. Appl Microbiol Biotechnol. 2004;66:123–130. doi: 10.1007/s00253-004-1703-6. [DOI] [PubMed] [Google Scholar]
- 6.Streit W, Schmitz R. Metagenomics–the key to the uncultured microbes. Curr Opin Microbiol. 2004;7:492–498. doi: 10.1016/j.mib.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 7.Riesenfeld C, Schloss P, Handelsman J. Metagenomics: genomic analysis of microbial communities. Annu Rev Genet. 2004;38:525–552. doi: 10.1146/annurev.genet.38.072902.091216. [DOI] [PubMed] [Google Scholar]
- 8.Venter J, Remington K, Heidelberg J, et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- 9.Tyson G, Chapman J, Hugenholtz P, et al. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature. 2004;428:37–43. doi: 10.1038/nature02340. [DOI] [PubMed] [Google Scholar]
- 10.Lorenz P, Eck J. Metagenomics and industrial applications. Nat Rev Microbiol. 2005;3:510–516. doi: 10.1038/nrmicro1161. [DOI] [PubMed] [Google Scholar]
- 11.Turnbaugh P, Ley R, Mahowald M, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 12.Gill S, Pop M, Deboy R, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen K, Pachter L. Bioinformatics for whole-genome shotgun sequencing of microbial communities. PLoS Comput Biol. 2005;1:106–112. doi: 10.1371/journal.pcbi.0010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Foerstner K, von Mering C, Bork P. Comparative analysis of environmental sequences: potential and challenges. Philos Trans R Soc Lond B Biol Sci. 2006;361:519–523. doi: 10.1098/rstb.2005.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Batzoglou S, Jaffe D, Stanley K, et al. ARACHNE: a whole-genome shotgun assembler. Genome Res. 2002;12:177–189. doi: 10.1101/gr.208902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jaffe D, Butler J, Gnerre S, et al. Whole-genome sequence assembly for mammalian genomes: Arachne 2. Genome Res. 2003;13:91–96. doi: 10.1101/gr.828403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huson D, Reinert K, Kravitz S, et al. Design of a compartmentalized shotgun assembler for the human genome. Bioinformatics. 2001;17(Suppl 1):S132–139. doi: 10.1093/bioinformatics/17.suppl_1.s132. [DOI] [PubMed] [Google Scholar]
- 18.Aparicio S, Chapman J, Stupka E, et al. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science. 2002;297:1301–1310. doi: 10.1126/science.1072104. [DOI] [PubMed] [Google Scholar]
- 19.Azad R, Borodovsky M. Probabilistic methods of identifying genes in prokaryotic genomes: connections to the HMM theory. Brief Bioinformatics. 2004;5:118–130. doi: 10.1093/bib/5.2.118. [DOI] [PubMed] [Google Scholar]
- 20.Yooseph S, Sutton G, Rusch D, et al. The Sorcerer II Global Ocean Sampling expedition: expanding the universe of protein families. PLoS Biol. 2007;5:e16. doi: 10.1371/journal.pbio.0050016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mavromatis K, Ivanova N, Barry K, et al. Use of simulated data sets to evaluate the fidelity of metagenomic processing methods. Nat Methods. 2007;4:495–500. doi: 10.1038/nmeth1043. [DOI] [PubMed] [Google Scholar]
- 22.Angly F, Felts B, Breitbart M, et al. The marine viromes of four oceanic regions. PLoS Biol. 2006;4:e368. doi: 10.1371/journal.pbio.0040368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huson D, Auch A, Qi J, et al. MEGAN analysis of metagenomic data. Genome Res. 2007;17:377–386. doi: 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wommack K, Bhavsar J, Ravel J. Metagenomics: read length matters. Appl Environ Microbiol. 2008;74:1453–1463. doi: 10.1128/AEM.02181-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tang H. Genome assembly, rearrangement, and repeats. Chem Rev. 2007;107:3391–3406. doi: 10.1021/cr0683008. [DOI] [PubMed] [Google Scholar]
- 26.Pevzner P, Tang H, Waterman M. An Eulerian path approach to DNA fragment assembly. Proc Natl Acad Sci USA. 2001;98:9748–9753. doi: 10.1073/pnas.171285098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Myers E. Toward simplifying and accurately formulating fragment assembly. J Comput Biol. 1995;2:275–290. doi: 10.1089/cmb.1995.2.275. [DOI] [PubMed] [Google Scholar]
- 28.Chaisson M, Pevzner P, Tang H. Fragment assembly with short reads. Bioinformatics. 2004;20:2067–2074. doi: 10.1093/bioinformatics/bth205. [DOI] [PubMed] [Google Scholar]
- 29.Myers E, Sutton G, Delcher A, et al. A whole-genome assembly of Drosophila. Science. 2000;287:2196–2204. doi: 10.1126/science.287.5461.2196. [DOI] [PubMed] [Google Scholar]
- 30.Venter J, Adams M, Myers E, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 31.Morgulis A, Gertz E, Scher A, et al. A fast and symmetric DUST implementation to mask low-complexity DNA sequences. J Comput Biol. 2006;13:1028–1040. doi: 10.1089/cmb.2006.13.1028. [DOI] [PubMed] [Google Scholar]
- 32.Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pevzner PA, Tang H, Tesler G. De novo repeat classification and fragment assembly. Genome Res. 2004;14(9):1786–1796. doi: 10.1101/gr.2395204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raphael B, Zhi D, Tang H, et al. A novel method for multiple alignment of sequences with repeated and shuffled elements. Genome Res. 2004;14:2336–2346. doi: 10.1101/gr.2657504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee C, Grasso C, Sharlow MF. Multiple sequence alignment using partial order graphs. Bioinformatics. 2002;18(3):452–64. doi: 10.1093/bioinformatics/18.3.452. [DOI] [PubMed] [Google Scholar]
- 36.Ye Y, Jaroszewski L, Li W, et al. A segment alignment approach to protein comparison. Bioinformatics. 2003;19:742–749. doi: 10.1093/bioinformatics/btg073. [DOI] [PubMed] [Google Scholar]
- 37.Rodriguez-Brito B, Rohwer F, Edwards R. An application of statistics to comparative metagenomics. BMC Bioinformatics. 2006;7:162. doi: 10.1186/1471-2105-7-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thomas P, Campbell M, Kejariwal A, et al. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003;13:2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seshadri R, Kravitz S, Smarr L, et al. CAMERA: a community resource for metagenomics. PLoS Biol. 2007;5:e75. doi: 10.1371/journal.pbio.0050075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Markowitz V, Szeto E, Palaniappan K, et al. The integrated microbial genomes (IMG) system in 2007: data content and analysis tool extensions. Nucleic Acids Res. 2007 doi: 10.1093/nar/gkm846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Edwards R, Rohwer F. Viral metagenomics. Nat Rev Microbiol. 2005;3:504–510. doi: 10.1038/nrmicro1163. [DOI] [PubMed] [Google Scholar]